Abstract
Ebola (EBOV) and Zika viruses (ZIKV) are responsible for recent global health threats. As no preventive vaccines or antiviral drugs against these two re-emerging pathogens are available, we evaluated whether the molecular tweezer CLR01 may inhibit EBOV and ZIKV infection. This small molecule has previously been shown to inactivate HIV-1 and herpes viruses through a selective interaction with lipid-raft-rich regions in the viral envelope, which results in membrane disruption and loss of infectivity. We found that CLR01 indeed blocked infection of EBOV and ZIKV in a dose-dependent manner. The tweezer inhibited infection of epidemic ZIKV strains in cells derived from the anogenital tract and the central nervous system, and remained antivirally active in the presence of semen, saliva, urine and cerebrospinal fluid. Our findings show that CLR01 is a broad-spectrum inhibitor of enveloped viruses with prospects as a preventative microbicide or antiviral agent.
Keywords: Zika virus, Ebola virus, Broadly active antiviral agents, Virus inactivation, Lipid rafts
Highlights
-
•
The molecular tweezer CLR01 inhibits EBOV and ZIKV infection.
-
•
ZIKV infection of cells derived from the anogenital tract and the central nervous system is blocked.
-
•
Anti-ZIKV activity is retained in semen, urine, saliva, and CSF.
-
•
CLR01 represents a broad-spectrum inhibitor of enveloped viruses with prospects as a microbicide or antiviral agent.
1. Introduction
1.1. Zika virus
Zika virus (ZIKV) was first isolated in 1947 from a febrile rhesus macaque in the Zika Forest of Uganda (Dick et al., 1952). Since then, sporadic ZIKV infections occurred in Africa and Asia (Haddow et al., 2012, Hayes, 2009). Until 2015, ZIKV infection usually has been associated with mild symptoms and thus, the virus had not been considered a threatening pathogen. However, since the recent outbreaks it is evident that ZIKV causes drastic birth defects, most prominently microcephaly (Mlakar et al., 2016, Rasmussen et al., 2016), and is associated with neurological disorders, such as Guillain-Barré syndrome (Cao-Lormeau et al., 2016, Krauer et al., 2017). Since 2007, 84 countries and territories reported ongoing ZIKV transmission (http://www.who.int/emergencies/zika-virus/en/). In Brazil alone, 1.3 million persons had been infected (Shuaib et al., 2016), prompting the WHO to declare ZIKV a public health emergency of international concern. Currently, there are no specific treatments or vaccines against ZIKV making development of effective preventive measures an urgent public health need (Barrows et al., 2016, Paixão et al., 2016). ZIKV is transmitted mainly via mosquito bites but cases of sexual transmission also have been reported (D'Ortenzio et al., 2016, Moreira et al., 2017, Petersen et al., 2016). Like other members of the Flaviviridae family (e.g. Dengue virus), Zika virions are surrounded by a host-membrane-derived lipid bilayer containing envelope glycoproteins responsible for cell entry (Sirohi et al., 2016).
1.2. Ebola virus
Ebola virus (EBOV) was discovered in 1976 near the Ebola River in the former Zaire. Since then, outbreaks sporadically occurred in Africa, with the most severe in 2014, reaching epidemic proportions and a death toll of more than 11,000 people (Centers for Disease Control and Prevention (CDC), 2016). EBOV belongs to the family of Filoviridae, forms enveloped filamentous infectious particles, and causes hemorrhagic fever, a rare and deadly disease with a high fatality rate. In addition to being a global health concern, the virus also is considered a potential biological threat agent. EBOV likely is transmitted from infected bats to humans where it may spread through personal contact, contaminated objects, or sexual intercourse (Mate et al., 2015, Pandey et al., 2014). Noteworthy, even after recovery, EBOV is found in some body fluids, including semen, up to several months (Deen et al., 2017). As for now, no licensed vaccine or medicine is available to prevent or manage future EBOV outbreaks (Sharma and Ketki Jangid, 2017).
1.3. The molecular tweezer CLR01
Molecular tweezers are novel drug candidates for the treatment of amyloidosis and related conditions. A lead compound, CLR01, has been used in many in vitro and in vivo studies to date (Attar et al., 2014, Schrader et al., 2016). Binding of CLR01 to lysine residues in polypeptides disrupts noncovalent molecular interactions that are important for the abnormal self-assembly that leads to the formation of toxic oligomers and amyloid aggregates (Schrader et al., 2016, Sinha et al., 2011). In fact, CLR01 prevents assembly and promotes disaggregation of amyloid fibrils that are associated with neurodegenerative diseases (Attar et al., 2012, Ferreira et al., 2014, Prabhudesai et al., 2012, Richter et al., 2017). Therapeutic effects of CLR01 have been demonstrated in animal models of Parkinson's disease (Lulla, 2016, Prabhudesai et al., 2012, Richter et al., 2017), Alzheimer's disease (Attar et al., 2012, Malik et al., 2017), Familial Amyloidotic Polyneuropathy (FAP) (Ferreira et al., 2014), and desmin-related cardiomyopathy (Xu et al., 2017) and was found to be safe in mice at doses 250-times higher than those showing therapeutic effects (Attar et al., 2014). We have recently established that CLR01 also disrupted the formation of amyloid fibrils in semen (Lump et al., 2015), which enhance HIV-1 infection (Münch et al., 2007, Usmani et al., 2014). Surprisingly, CLR01 also inhibited HIV-1 infection through a direct virus-inactivating mechanism: the tweezer preferentially bound to raft-rich regions in the viral membrane resulting in the disruption of the lipid bilayer and loss of infectivity. In line with the membrane targeting activity, CLR01 is inactive against non-enveloped viruses such as human adenovirus but destroyed other enveloped viruses, including hepatitis-C virus, human cytomegalovirus, and herpes simplex virus, demonstrating that CLR01 represents a broadly active antiviral compound with prospects for microbicide or drug development (Lump et al., 2015). Thus, we evaluated here whether CLR01 might also block infection of re-emerging enveloped viruses. We show that CLR01 indeed inhibits infection of replication-competent EBOV and ZIKV as well as Marburg, Rabies, and SARS Corona virus (SARS-CoV) pseudoparticles. CLR01 lost antiviral activity in the presence of serum, but remained active in the presence of semen, urine, saliva or cerebrospinal fluid. Thus, this broadly active compound represents a promising lead for further development as a microbicide to protect from the sexual acquisition of ZIKV or EBOV, or as a topically applied drug for the therapy of viral infections of the skin, mucosa or the respiratory tract.
2. Materials and methods
2.1. Reagents
CLR01 and CLR03 were synthesized as described previously (Fokkens et al., 2005, Talbiersky et al., 2008) and 7.4 mM stock solutions were prepared in PBS. ZIKV envelope protein had been ordered from Fitzgerald Industries International, Acton, USA.
2.2. Cells and virus stocks
Vero E6 cells were used for propagation and infection with ZIKV as described (Müller et al., 2017). TCID50 values (Tissue Culture Infectious Dose 50; TCID50/ml) were determined according to Reed and Muench, multiplied with a factor of 0.7 to obtain PFU/ml (Plaque Forming Units) and, by normalizing to the number of cells, used to calculate the respective MOI (Multiplicity of Infection). SW480 (human epithelial colon carcinoma) cells were kindly provided by Ninel Azoitei (Center for Internal Medicine I, University of Ulm, Ulm, Germany). HFF (human foreskin fibroblasts), human glioblastoma (A172) or neuroglioma (H4) cells were kindly provided by Jens von Einem and Karin Danzer (Ulm University Medical Center, Ulm, Germany). Hepatic Huh-7 cells were kindly provided by S. Pöhlmann (Göttingen, Germany). The reporter cell line TZM-bl was obtained through the NIH ARRRP. Virus stocks of R5-tropic HIV-1 NL4-3 92TH014 were generated by transient transfection of 293T cells as described (Münch et al., 2007).
Methods describing the effect of CLR01 on pseudotyped lentiviral particles (2.3.), Ebola virus infection (2.4.), the detection of ZIKV infection by a colorimetric MTT assay (2.5.) or by cell-based ZIKV immunodetection assay (2.6.), flow cytometry (2.7.) and confocal microscopy (2.8.) as well as the RNA release assay (2.9.) and the antiviral activity of CLR01 in body fluids (2.10) can be found in the supplement.
3. Results
3.1. The antiviral activity of CLR01 is independent of viral glycoproteins
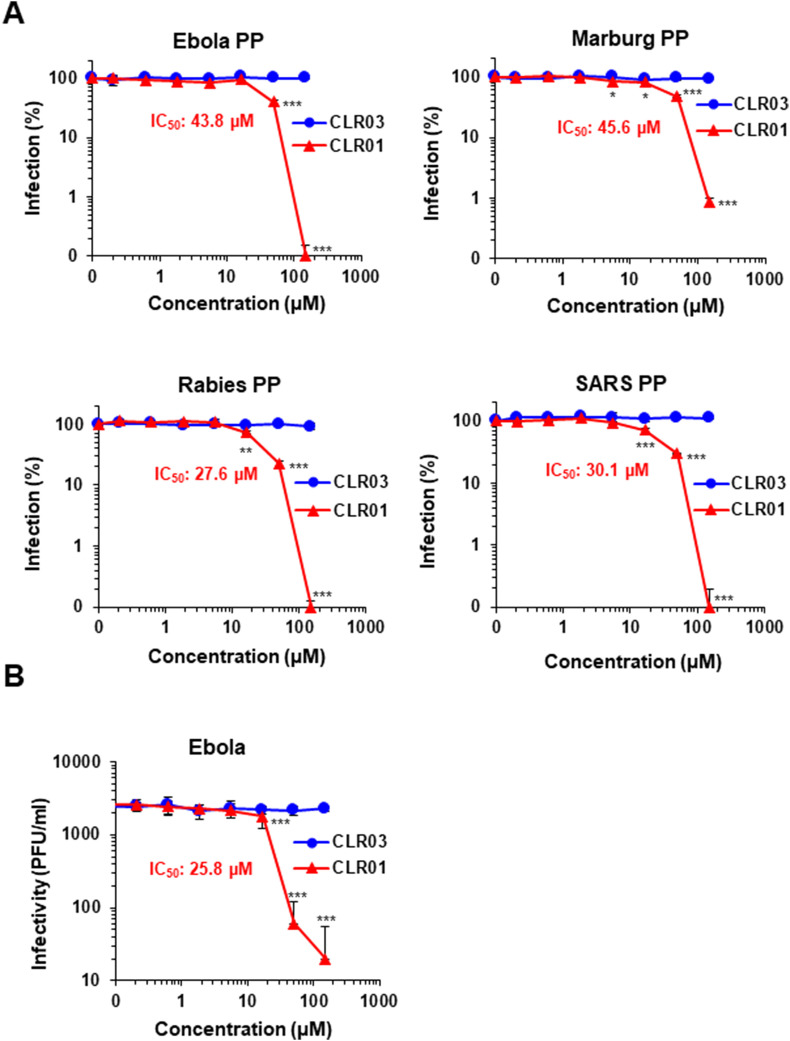
We have shown previously that CLR01 inhibits HIV-1 infection by disrupting the viral membrane (Lump et al., 2015), suggesting that the antiviral activity of the tweezer is independent of the presence of the viral glycoproteins. To test this hypothesis, we generated luciferase-encoding retroviral particles harboring glycoproteins derived from EBOV, the closely related Filovirus Marburg virus, Rabies virus (Rhabdoviridae) or SARS-CoV (Coronaviridae). These pseudoparticles were exposed to CLR01 and then added to Huh-7 cells. CLR03, which lacks hydrophobic side arms and has no anti-HIV-1 activity (Lump et al., 2015), served as a negative control. Infection rates were determined three days later by quantifying luciferase activity and showed that CLR01, in contrast to CLR03, efficiently blocked infection by all tested pseudoparticles (Fig. 1 A). The half-maximal inhibitory concentrations (IC50) of CLR01 against the four pseudoparticles were similar and ranged between 27.6 μM for Rabies and 45.6 μM for Marburg virus (Fig. 1A). These data corroborate our previous findings that CLR01 targets the viral membrane rather than a viral glycoprotein (Lump et al., 2015). We tested next whether CLR01 blocked infection by the replication-competent BSL4 pathogen EBOV. Vero E6 cells were inoculated with GFP-encoding EBOV (Ebihara et al., 2007, Hoenen et al., 2013) that had been exposed to CLR01 or CLR03. After 7 days, the number of virus-induced plaques revealed that CLR01, but not CLR03, reduced the infectious titer in a dose-dependent manner with an IC50 of 25.8 μM (Fig. 1B).
Fig. 1.
CLR01 inhibits lentiviral infection independently of the viral glycoprotein and prevents EBOV plaque formation. (A) Luciferase-encoding lentiviral particles pseudotyped with glycoproteins of viruses of the Filoviridae (EBOV, Marburg virus),Rhabdoviridae (Rabies virus) or the Coronaviridae family (SARS-CoV), were exposed to indicated concentrations of CLR01 or CLR03 and then used to infect Huh-7 cells. After three days, infection rates were determined by quantifying firefly luciferase activity and subtracting background activity derived from uninfected cells. Values represent % reporter gene activities ± SD derived from triplicate infections, normalized to values obtained for cells infected in the absence of tweezers. (B) Analysis of replication competent EBOV was performed using confluent Vero E6 cells in 24-well plates. rgEBOV-eGFP was preincubated with CLR01 or CLR03 and mixtures were added to the cells. After 7 days, samples were analyzed by counting the number of plaques per well and calculating the corresponding plaque forming units per milliliter (PFU/ml) for each inhibitor and dilution. Displayed values represent the mean of three independent experiments ± SD. IC50 values were calculated by GraphPad Prism. One-way ANOVA (non-parametric, grouped), followed by Bonferroni's multiple comparison tests were applied to compare the different CLR01/CLR03 concentrations to cells infected in the absence of compound (* denotes p < 0.01; ** denotes p < 0.001; *** denotes p < 0.0001).
3.2. CLR01 prevents the formation of the ZIKV-induced CPE
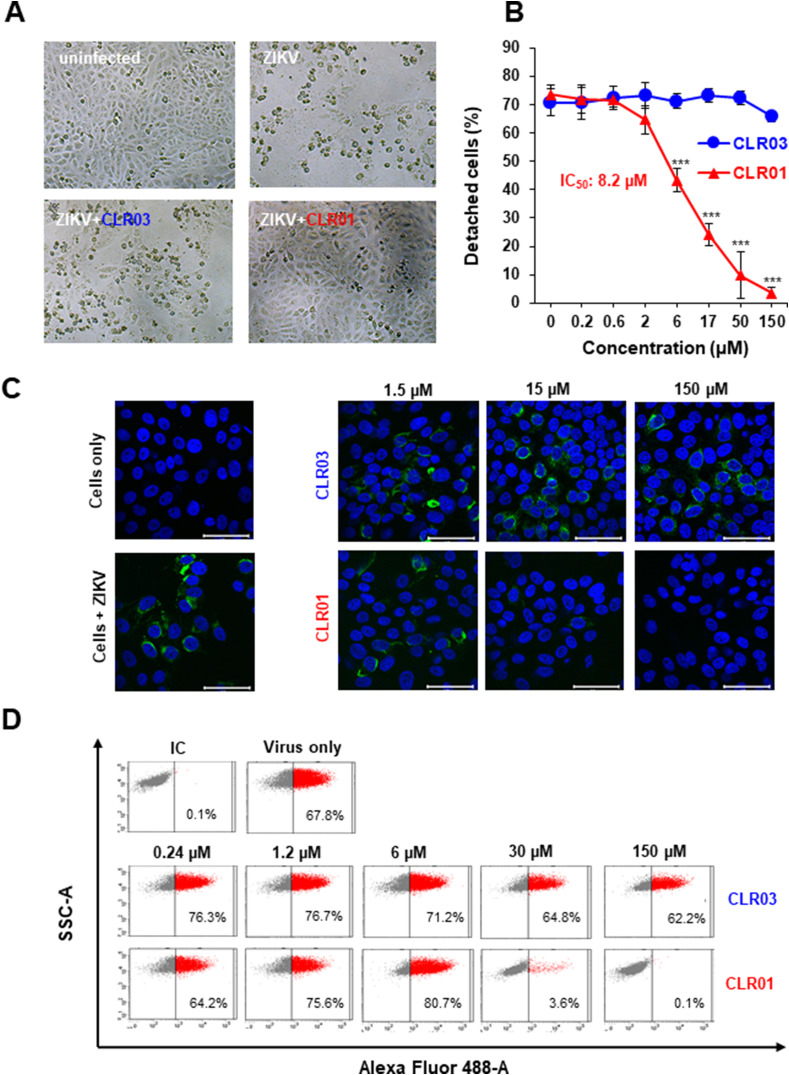
Next, we determined the effect of CLR01 on ZIKV infection. Vero E6 cells were inoculated with the African ZIKV MR766 strain that was isolated in 1947 from a sentinel rhesus macaque (Dick et al., 1952) in the absence or presence of CLR01 or CLR03 and viral replication was monitored by light microscopy. In the absence of CLR01, ZIKV caused a profound cytopathic effect (CPE) as evidenced by large plaques caused by detachment of cells. CLR03 had no effect on this phenotype (Fig. 2 A). However, exposure to 150 μM CLR01 prevented formation of the virus-induced CPE entirely. To quantitatively assess the antiviral activity, we used a colorimetric MTT assay measuring the virally induced CPE (Müller et al., 2016). In the absence of compounds or in the presence of CLR03, ZIKV resulted in more than 70% dead (detached) cells. In contrast, CLR01 suppressed CPE formation with an IC50 of 8.2 μM (Fig. 2B).
Fig. 2.
CLR01 inhibits ZIKV infection of Vero E6 cells. (A) Light microscopy images of Vero E6 cells infected with ZIKV MR766 in the absence or presence of 150 μM CLR01 or CLR03. Images were taken 4 days post infection (dpi). (B) ZIKV MR766 was incubated with 0.2-150 μM CLR01 or CLR03 before these mixtures were used to infect Vero E6 cells. After 4 days, when significant cytopathic effects were visible, the number of adherent, viable cells were determined using the MTT assay (Müller et al., 2016). Values represent mean ± SD of percentages derived from triplicate infections. IC50 was determined using GraphPad Prism. One-way ANOVA (non-parametric, grouped), followed by Bonferroni's multiple comparison tests were applied to compare the different CLR01/CLR03 concentrations to cells infected in the absence of compound (*** denotes p < 0.0001) (C) Confocal microscopy images of Vero E6 cells infected with CLR01- or CLR03-treated ZIKV at day 3 post infection. Cells were stained for ZIKV E protein (green) and nuclei (Hoechst, blue) and imaged using confocal microscopy. Scale bar: 50 μm. (D) Flow cytometry of infected Vero E6 cells. Virus was pretreated with PBS, CLR01 or CLR03 and added to cells. 48 h later, cells were fixed, permeabilized, and stained with an anti E protein antibody, and quantified using an Alexa 488-coupled secondary antibody. Percentages indicate the fraction of protein E positive cells. IC, isotype control.
3.3. CLR01 blocks ZIKV infection
The MTT assay allows an indirect measurement of ZIKV infection as the tetrazolium dye is metabolized only in living, non-infected cells. To evaluate the effect of CLR01 on ZIKV more directly, infected cells were stained for the viral E protein and analyzed by confocal microscopy (Schandock et al., 2017). Upon treatment with 15 or 150 μM of CLR01, E protein-specific fluorescence could not be detected, whereas CLR03 had no effect, as expected (Fig. 2C). Lack of E protein expression in cells infected with CLR01-treated virus was confirmed by flow cytometry (Fig. 2D, Supplementary Fig. S1A). Of note, CLR01 was not cytotoxic at concentrations active against ZIKV (Supplementary Fig. S1B), and did not cause alterations in cell morphology (compare confocal images and dot plots of uninfected cells vs. 150 μM CLR01 exposed cells in Fig. 2C and D). Thus, CLR01 prevents ZIKV infection of Vero E6 cells.
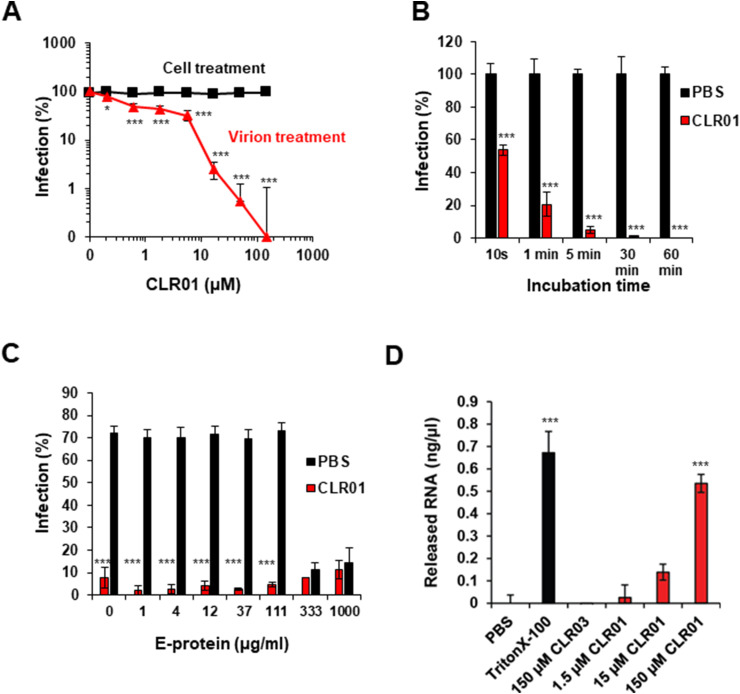
3.4. CLR01 targets the virion
We hypothesized that if CLR01 inhibited ZIKV infection by a similar mechanism to other enveloped viruses, its activity would be directed against the viral membrane (Lump et al., 2015). Indeed, when CLR01 was added to cells, rather than to the virus, and washed out prior to infection, no antiviral effect was observed in a cell-based ZIKV immunodetection assay (Fig. 3 A). We performed next a time-course analysis in which ZIKV was exposed to 10 μM CLR01 for 10 to 3600 s prior to infection. Already after exposure for only 10 s, CLR01 reduced infection by ∼43%. Infection declined further with increasing incubation time and was nearly at the level of uninfected cells after 1800 s incubation (Fig. 3B). In agreement with a direct activity against the virion, the IC50 of CLR01 increased from 4.3 μM to 32.3 μM when a 10-fold higher multiplicity of infection was applied (Supplementary Fig. S2). CLR01 was shown to destroy HIV-1 membrane integrity through interaction with raft-rich regions in the retroviral envelope (Lump et al., 2015). In contrast to HIV-1, the ZIKV particle is relatively densely packed with glycoproteins (Sirohi et al., 2016), which might hamper the interaction of the tweezer with the ZIKV membrane. To determine whether CLR01 might inhibit ZIKV infection through direct interaction with glycoproteins, increasing amounts of recombinant viral E protein were titrated to CLR01, and then these solutions were assayed for anti-ZIKV activity. As shown in Fig. 3C, even E-antigen concentrations of 1000 μg/ml did not affect the antiviral activity of CLR01, suggesting that the tweezer did not reduce ZIKV infectivity through binding to the viral E protein. Interestingly, we also observed that elevated E-antigen concentrations of 333 and 1000 μg/ml reduced infection in the absence of CLR01 (Fig. 3C), which is likely due to the competition between the virion-associated E protein and the recombinant E protein for cellular receptors.
Fig. 3.
Mechanism of CLR01 inhibition of ZIKV infectivity. (A) For virus treatment, ZIKV was incubated with CLR01 for 10 min at room temperature before the mixtures were added to Vero E6 cells. For cell treatment, CLR01 was added directly to the cells; after 2 h, the medium was replaced and the cells were infected with ZIKV MR766. 2 dpi, cell-based ZIKV immunodetection assay was performed. Values represent mean ± SD (n = 3). One-way ANOVA (non-parametric, grouped), followed by Bonferroni's multiple comparison tests were applied to compare the different CLR01/CLR03 concentrations to cells infected in the absence of compound (* denotes p < 0.01; *** denotes p < 0.0001) (B) ZIKV was incubated for the indicated times with PBS or 10 μM CLR01 before the mixture was added to Vero E6 cells. 2 dpi, cell-based ZIKV immunodetection assay was performed. Values represent mean ± SD (n = 3). One-way ANOVA (non-parametric, grouped), followed by Bonferroni's multiple comparison tests were applied to compare cells infected with CLR01-treated ZIKV to cells infected with ZIKV that had been incubated with PBS for the same time period (*** denotes p < 0.0001). (C) Indicated concentrations of ZIKV E protein were titrated to 100 μM CLR01 or PBS before ZIKV was added. Mixtures were used to inoculate Vero E6 cells. 3 dpi, the number of adherent, viable cells were determined using the MTT assay (Müller et al., 2016). The values shown are mean ± SD from triplicate infections. One-way ANOVA (non-parametric, grouped), followed by Bonferroni's multiple comparison tests were applied to compare cells treated with different concentrations of ZIKV E protein and CLR01 to cells that had been treated with the same concentrations of E protein and PBS (*** denotes p < 0.0001). (D) ZIKV MR766 was incubated with PBS, 150 μM Triton X-100, 1.5-150 μM CLR01 or 150 μM CLR03 for 30 min at 37 °C. Samples were inactivated by UV light of a laminar flow for 60 min. Then, 10 μl of the samples were used to determine RNA concentrations using the QuantiFluor® RNA System and a Quantus Fluorometer (Promega). Background values obtained from control samples using cell supernatant of uninfected cells were subtracted from the respective signals. RNA levels of virus stock incubated with PBS were subtracted from these values. Data points represent mean ± SD (n = 3). One-way ANOVA (non-parametric, grouped), followed by Bonferroni's multiple comparison tests were applied to compare samples treated with different CLR01 concentrations, CLR03 or Triton X-100 to PBS-treated virus (*** denotes p < 0.0001).
To test whether CLR01 interaction with the viral particle results in loss of membrane integrity, as was shown in the case of HIV-1, (Lump et al., 2015), we measured viral RNA genome release. Sucrose cushion purified ZIKV was incubated with CLR01, CLR03, PBS or Triton X-100, as a positive control, for 30 min at 37 °C and the amount of total RNA in the solution was determined fluorometrically. As expected, no viral RNA was detectable when ZIKV was incubated with buffer or CLR03 (Fig. 3D). In contrast, incubation with Triton X-100 resulted in readily detectable viral RNA suggesting that the detergent lysed the ZIKV particle. Similarly, elevated amounts of RNA were detected upon treatment of ZIKV with CLR01, demonstrating the physical destruction of the viral particle by the tweezer. Of note, CLR01 itself did not act as detergent but targets the viral membrane (Lump et al., 2015).
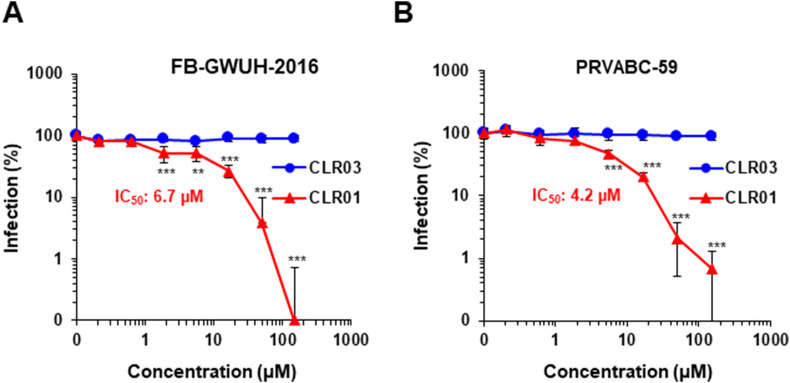
3.5. CLR01 blocks clinically relevant ZIKV strains
Next, we analyzed whether CLR01 was active against epidemic ZIKV strains. The FB-GWUH-2016 isolate was derived in 2016 from the fetus of a pregnant Finnish tourist returning from South America (Driggers et al., 2016), and the PRVABC-59 isolate represents the current American epidemic strain, isolated in 2015 from a Puerto Rican patient (Lanciotti et al., 2016). Both ZIKV strains were exposed to CLR01 upon infection of Vero E6 cells. Cell-based immunodetection assay and flow cytometry experiments demonstrated that the tweezer suppressed infection of both strains with IC50 values of 6.7 μM for FB-GWUH-2016 (Fig. 4 A and Supplementary Fig. S3), and 4.2 μM against PRVABC-59 (Fig. 4B), respectively.
Fig. 4.
CLR01 inactivates epidemic ZIKV isolates. (A) ZIKV FB-GWUH-2016 or (B) PRVABC-59 was incubated for 10 min at 37 °C with 0.2-150 μM CLR01 or CLR03. Thereafter, Vero E6 cells were infected and 2 days later, infection rates were determined via a cell-based ZIKV immunodetection assay. Data points represent mean ± SD (n = 3). IC50 values were determined with GraphPad Prism. One-way ANOVA (non-parametric, grouped), followed by Bonferroni's multiple comparison tests were applied to compare the different CLR01 concentrations to cells infected in the absence of compound (** denotes p < 0.001; *** denotes p < 0.0001).
3.6. CLR01 inhibits ZIKV infection of cells derived from the anogenital region and the brain
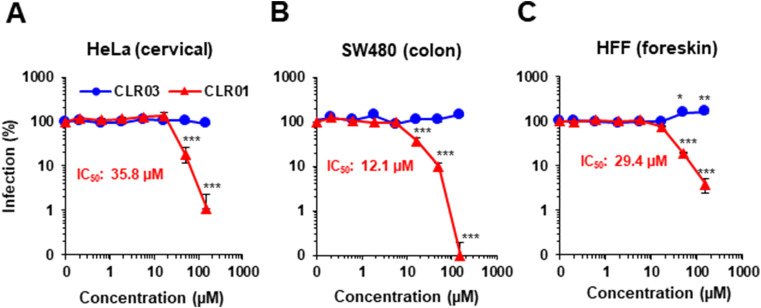
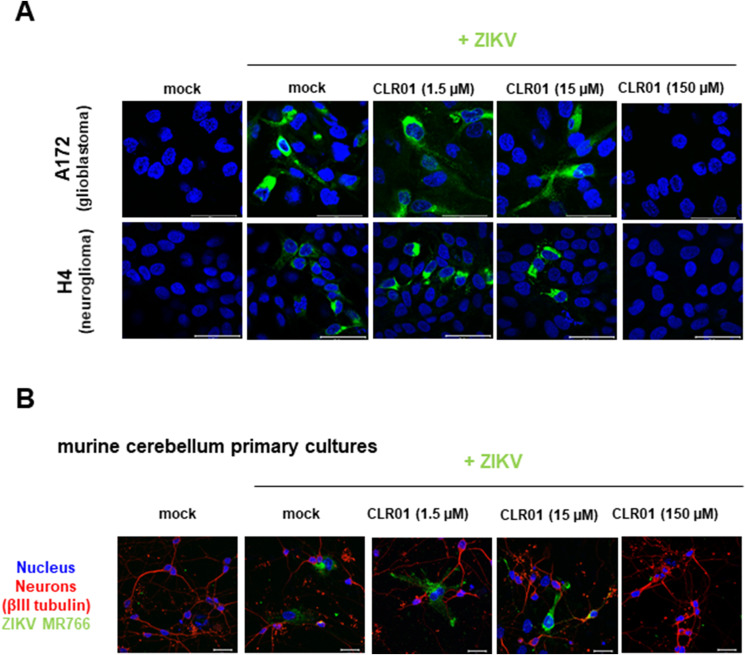
As ZIKV can be transmitted via sexual intercourse, we studied whether ZIKV infects cells derived from the anogenital region and if so, whether infection can be blocked by CLR01. ZIKV effectively infected cell lines derived from cervix (Fig. 5 A, Supplementary Fig. S4), colon (Fig. 5B, Supplementary Fig. S4), and primary foreskin cells (Fig. 5C, Supplementary Fig. S4). Viral infectivity was suppressed by CLR01 but not CLR03, with IC50 values in the μM range (Fig. 5 and Supplementary Fig. S4). Because ZIKV is neurotropic (Cao-Lormeau et al., 2016, Mlakar et al., 2016) and CLR01 has been shown previously to penetrate through the blood–brain barrier (BBB) when administered systemically (Attar et al., 2012, Attar et al., 2014, Richter et al., 2017), we also tested whether CLR01 blocked ZIKV infection of brain-derived cells. Both A172 glioblastoma and H4 neuroglioma cells were susceptible to ZIKV infection and CLR01 entirely abrogated infection at 150 μM (Fig. 6 A). Finally, we confirmed these findings obtained in human cell lines using primary murine cerebellar neurons. ZIKV infected neuronal bystander cells, and this was blocked by CLR01 (Fig. 6B). There was no apparent toxicity of CLR01 in the primary neurons at this concentration.
Fig. 5.
CLR01 abrogates ZIKV infection of cells derived from the anogenital region. ZIKV was incubated for 10 min at 37 °C with 0.2-150 μM CLR01 or CLR03. Next, mixtures were used to inoculate HeLa (A), SW480 (B), or HFF (C) cells. 3 days later, infection rates were determined via quantification of the viral E protein using a cell-based ZIKV immunodetection assay. Data points represent mean ± SEM (n = 6). One-way ANOVA (non-parametric, grouped), followed by Bonferroni's multiple comparison tests were applied to compare the different CLR01 concentrations to cells infected in the absence of compound (* denotes p < 0.01; ** denotes p < 0.001; *** denotes p < 0.0001).
Fig. 6.
CLR01 inhibits infection of human and murine brain cells. (A) Human glioblastoma (A172) or neuroglioma (H4) cells were infected with ZIKV in the presence or absence of 1.5-150 μM CLR01. Two days later, cells were stained for ZIKV E protein (green) and nuclei (with Hoechst; blue) and imaged by confocal microscopy. Scale bar: 50 μm. (B) Primary murine cerebellum cultures were infected with ZIKV MR766 that had been incubated with 1.5–150 μM CLR01 for 10 min at 37 °C. 3 dpi, cultures were fixed, permeabilized and stained for the neuronal protein βIII tubulin (red), ZIKV E protein (green) and nuclei (with Hoechst; blue). Scale bar: 20 μm.
3.7. The antiviral effect of CLR01 is abrogated by serum but not urine, saliva, semen, or CSF
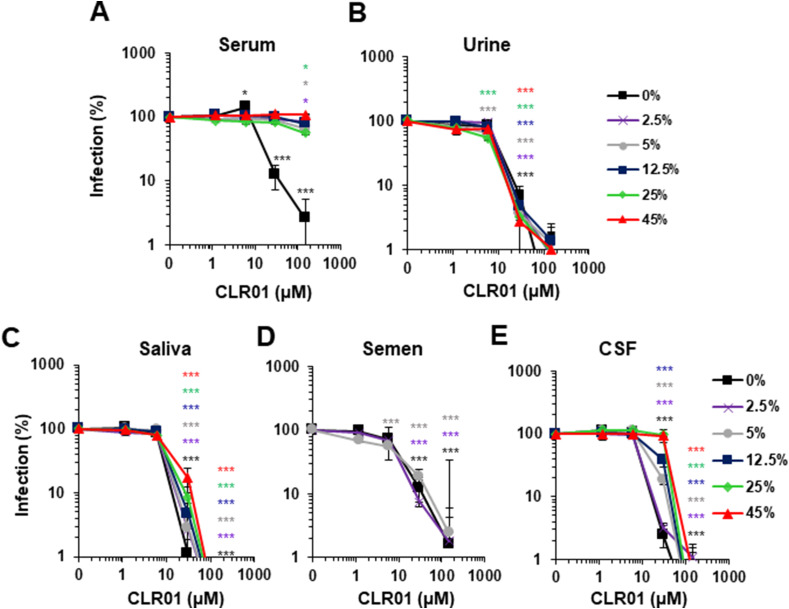
Its broad antiviral activity makes CLR01 an interesting lead compound for antiviral prevention or treatment. For a systemic application, CLR01 must retain antiviral activity in blood. To test whether this was the case, CLR01 was diluted in human serum, and then exposed to ZIKV. As shown in Fig. 7 A, serum concentrations of 2.5% and higher abrogated the anti-ZIKV activity of the tweezer. Similarly, serum also abrogated the anti-HIV-1 activity of CLR01 (Supplementary Fig. S5), precluding its use as a systemically applied antiviral drug. In contrast, CLR01 remained active and inhibited ZIKV infection in the presence of up to 45% of urine (Fig. 7B), saliva (Fig. 7C), cerebrospinal fluid (Fig. 7E) and 5% human semen (Fig. 7D). Thus, CLR01 inactivates ZIKV in the presence of body fluids associated with virus transmission, and could be administered locally to halt virus replication, or applied topically as a microbicide to prevent e.g. sexual virus transmission.
Fig. 7.
CLR01 loses antiviral activity in the presence of serum but is active in other body fluids. Indicated concentrations of CLR01 were incubated with ZIKV MR766 in the presence of increasing concentrations of human (A) serum, (B) urine, (C) saliva, (D) semen and (E) cerebrospinal fluid (CSF). After 10 min of incubation, the mixtures were used to infect Vero E6 cells. 2 days later, infection rates were determined via a cell-based ZIKV immunodetection assay. Data points represent mean ± SEM (n = 6), except for CSF and serum: mean ± SD (n = 3). One-way ANOVA (non-parametric, grouped), followed by Bonferroni's multiple comparison tests were applied to compare the different CLR01 concentrations to cells infected in the absence of CLR01 but in presence of the same respective body fluid concentration (* denotes p < 0.01; *** denotes p < 0.0001).
4. Discussion
4.1. CLR01 is a broadly active antiviral
Our results establish CLR01 as a broad-spectrum antiviral agent, which not only inhibits infection of established viral pathogens (Lump et al., 2015), but also of emerging viruses, such as EBOV and ZIKV. We characterized here mainly the effect of CLR01 on ZIKV, as there is currently no specific antiviral therapy nor a preventive vaccine available. CLR01 blocked ZIKV infection of primary brain-derived murine cells (Fig. 6B) as well as human cell lines derived from the anogenital region (Fig. 5, Supplementary Fig. S4) and the brain (Fig. 6A) with IC50 values between 5 and 50 μM. These values are in the same range as IC50 values obtained against EBOV (26 μM, Fig. 1B) and HIV-1 (14-20 μM) (Lump et al., 2015).
4.2. Mechanism of action
Several lines of evidence demonstrate that the antiviral activity of CLR01 is directed against the ZIKV particle itself. First, treatment of cells with CLR01 prior to infection had no antiviral effect (Fig. 3A). Second, the IC50 of CLR01 increased with increasing viral concentrations (Supplementary Fig. S2). Third, the integrity of the ZIKV particle was lost upon CLR01 treatment, as shown by the release of viral RNA from CLR01-treated virions (Fig. 3D). These findings were in agreement with our previous observations that CLR01 selectively disrupted viral membranes (Lump et al., 2015). Interestingly, the selectivity stems from CLR01's preferential interaction with membranes containing high levels of sphingomyelin and cholesterol, two lipids that are enriched in membranes of enveloped viruses, such as HIV-1 or EBOV (Bavari et al., 2002, Brügger et al., 2006, Chazal and Gerlier, 2003, Lorizate et al., 2013). The selective interaction of CLR01 with lipids that are enriched in the viral but not the cellular membrane may also explain its minimal effects on cell viability (Attar et al., 2012, Attar et al., 2014, Ferreira et al., 2014, Lopes et al., 2015, Prabhudesai et al., 2012, Sinha et al., 2011) (Fig. 2C, D, Supplementary Fig. S1B). CLR01 is slightly cytotoxic at concentrations of ∼1 mM, corresponding to selectivity indices of 50–100, which is in a reasonable range for drug development (Buschmann and Mannhold, 2017, Food and Drug Administration, 2006).
4.3. Antiviral activity in presence of body fluids
CLR01 lost antiviral activity in the presence of serum, precluding application as a systemic antiviral drug. This finding was unexpected because CLR01 has been found previously to be stable in mouse and human plasma, with a half-life of ∼2.5 h in mice after SC or IV injection (Attar et al., 2014). Moreover, CLR01 showed therapeutic effects in animal models of Alzheimer's and Parkinson's diseases (Attar et al., 2012, Malik et al., 2017, Prabhudesai et al., 2012, Richter et al., 2017). These data suggest that the loss of antiviral activity in the presence of serum likely was due to binding of CLR01 to serum proteins. The concentrations of CLR01 required to prevent amyloid formation in vitro and in vivo are in the sub-micromolar range, which is 2–3 orders of magnitude lower than those needed to block viral infection. Presumably, the vast majority of CLR01 was bound to serum proteins and hence unavailable to act on the virus, whereas the minority of the tweezer remains free at concentrations allowing to exert anti-amyloid activity. This interpretation also is supported by the observation that CLR01 retained anti-ZIKV activity in semen, urine, saliva, and CSF. The total protein concentrations in these body fluids are ∼ 1–3 orders of magnitude lower than in serum (Hu et al., 2006, Vibhakar et al., 2013). Thus, a larger percentage of CLR01 molecules likely are free in these biofluids and hence antivirally active.
4.4. Future directions
CLR01 has been described previously as a novel bi-functional microbicide counteracting sexual HIV-1 transmission both through directly targeting virus infectivity and by inhibiting the infection-promoting activity of amyloids in semen (Lump et al., 2015). We show here that the tweezer also inhibits pathogenic ZIKV infection of cells derived from the anogenital region in the presence of semen. Interestingly, both ZIKV and EBOV are present in semen of infected individuals (Atkinson et al., 2016, Deen et al., 2017, Moreira et al., 2017) and can be transmitted by sexual intercourse, similar to HIV-1 (D'Ortenzio et al., 2016, Mate et al., 2015, Moreira et al., 2017). Thus, application of CLR01 as a microbicide in the vaginal or rectal tracts might protect from acquiring different viral pathogens. Interestingly, CLR01 has been shown to penetrate the BBB (Attar et al., 2012, Attar et al., 2014, Richter et al., 2017), where it may also block replication of neurotropic viruses such as Zika or Rabies (Cao-Lormeau et al., 2016, Ludlow et al., 2016, Mlakar et al., 2016). However, it needs to be considered that CLR01 may only block cell-free virus infection but not cell-to-cell viral spread, which may limit its utility as therapeutic agent. Therefore, the effect of CLR01 on cell-to-cell viral transmission needs to be explored. Another means of application of CLR01 is topical, e.g., by directly administering CLR01 on virus-infected body surfaces, such as the skin or mucous membranes, to treat, e.g., HSV-induced Herpes labialis or genitalis. Topical medications may also be inhalational, such as medications against Flu or Respiratory Syncytial Virus (RSV), or applied to the surface of tissues other than the skin, such as eye drops applied to the conjunctiva, for example, in CMV-induced retinitis.
4.5. Conclusions
To address the challenge of emerging and re-emerging viral pathogens, it is imperative to develop broad-spectrum classes of antiviral agents as the conventional one-bug-one-drug paradigm is insufficient (Zhu et al., 2015). Current direct-acting antiviral drugs are highly successful but have a narrow spectrum of coverage and are only available against a very limited number of viral pathogens. Moreover, drug development is slow and expensive, and it typically takes more than a decade to get market approval. Currently, no specific antiviral treatment exists against most, if not all, emerging and re-emerging viruses. Broad-spectrum antivirals may offer certain advantages as they reduce time and cost of drug development, allow off-label use, and could be applied even before a viral threat is diagnosed (Bekerman and Einav, 2015, Zhu et al., 2015). CLR01 is broadly active against enveloped viruses as it disrupts lipid bilayers containing elevated levels of sphingomyelin and cholesterol (Lump et al., 2015), which are typically enriched in viral membranes (Bavari et al., 2002, Brügger et al., 2006, Chan et al., 2008, Chazal and Gerlier, 2003, Lorizate et al., 2013). We have shown that CLR01 inactivates major viral pathogens such as HIV-1, HSV-2, CMV, and HCV, as well as ZIKV and EBOV. It is highly likely that CLR01 also blocks other enveloped viruses, such as Severe Acute Respiratory Syndrome (SARS) and Middle East respiratory syndrome (MERS) Corona Viruses, Influenza or RSV. Most notably, resistance development against CLR01 is very unlikely to occur, as this would require changes in the lipid composition of the cellular and the viral membrane, which is difficult to envisage. In conclusion, CLR01 represents a promising prototype of a broad-spectrum antiviral agent.
Statistical analysis
All data were analyzed using GraphPad Prism version 7.03 for Windows, GraphPad Software, La Jolla California USA (www.graphpad.com ). The significance level was calculated using one-way One-way analysis of variance (ANOVA) (non-parametric, grouped), followed by Bonferroni's multiple comparison test. P values of <0.01 were considered significant (*, p < 0.01 **, p < 0.001 ***, p < 0.0001).
Funding
J.M., T.S. and S.B. acknowledge DFG funding (CRC1279, CRC1921 and CRC1093, respectively). J.A.M. acknowledges funding by the Baden-Württemberg Stiftung.
Conflict of interests
Authors declare to have no conflict of interest.
Acknowledgements
A.E.R. is funded by a fellowship of the Landesgraduiertenförderung Baden-Württemberg and is part of the International Graduate School in Molecular Medicine Ulm. J.A.M. is indebted to the Baden-Württemberg Stiftung for the financial support of this research project by the Eliteprogramme for Postdocs.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.antiviral.2018.02.003.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
References
- Atkinson B., Hearn P., Afrough B., Lumley S., Carter D., Aarons E.J., Simpson A.J., Brooks T.J., Hewson R. Detection of Zika virus in semen. Emerg. Infect. Dis. 2016;22:940. doi: 10.3201/eid2205.160107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attar A., Chan W.-T.C., Klärner F.-G., Schrader T., Bitan G. Safety and pharmacological characterization of the molecular tweezer CLR01–a broad-spectrum inhibitor of amyloid proteins' toxicity. BMC Pharmacol. Toxicol. 2014;15:1. doi: 10.1186/2050-6511-15-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attar A., Ripoli C., Riccardi E., Maiti P., Puma D.D.L., Liu T., Hayes J., Jones M.R., Lichti-Kaiser K., Yang F. Protection of primary neurons and mouse brain from Alzheimer's pathology by molecular tweezers. Brain. 2012;135:3735–3748. doi: 10.1093/brain/aws289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrows N.J., Campos R.K., Powell S.T., Prasanth K.R., Schott-Lerner G., Soto-Acosta R., Galarza-Muñoz G., McGrath E.L., Urrabaz-Garza R., Gao J. A screen of FDA-approved drugs for inhibitors of Zika virus infection. Cell Host Microbe. 2016;20:259–270. doi: 10.1016/j.chom.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bavari S., Bosio C.M., Wiegand E., Ruthel G., Will A.B., Geisbert T.W., Hevey M., Schmaljohn C., Schmaljohn A., Aman M.J. Lipid raft microdomains. J. Exp. Med. 2002;195:593–602. doi: 10.1084/jem.20011500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekerman E., Einav S. Combating emerging viral threats. Science. 2015;348:282–283. doi: 10.1126/science.aaa3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brügger B., Glass B., Haberkant P., Leibrecht I., Wieland F.T., Kräusslich H.-G. The HIV lipidome: a raft with an unusual composition. Proc. Natl. Acad. Sci. U. S. A. 2006;103:2641–2646. doi: 10.1073/pnas.0511136103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschmann H., Mannhold R. John Wiley & Sons; 2017. Drug Selectivity: an Evolving Concept in Medicinal Chemistry. [Google Scholar]
- Cao-Lormeau V.-M., Blake A., Mons S., Lastère S., Roche C., Vanhomwegen J., Dub T., Baudouin L., Teissier A., Larre P. Guillain-Barré Syndrome outbreak associated with Zika virus infection in French Polynesia: a case-control study. The Lancet. 2016;387:1531–1539. doi: 10.1016/S0140-6736(16)00562-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (CDC) 2016. 2014 Ebola Outbreak in West Africa - Case Counts.https://www.cdc.gov/vhf/ebola/outbreaks/2014-west-africa/case-counts.html [Google Scholar]
- Chan R., Uchil P.D., Jin J., Shui G., Ott D.E., Mothes W., Wenk M.R. Retroviruses human immunodeficiency virus and murine leukemia virus are enriched in phosphoinositides. J. Virol. 2008;82:11228–11238. doi: 10.1128/JVI.00981-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chazal N., Gerlier D. Virus entry, assembly, budding, and membrane rafts. Microbiol. Mol. Biol. Rev. 2003;67:226–237. doi: 10.1128/MMBR.67.2.226-237.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deen G.F., Broutet N., Xu W., Knust B., Sesay F.R., McDonald S.L.R., Ervin E., Marrinan J.E., Gaillard P., Habib N., Liu H., Liu W., Thorson A.E., Yamba F., Massaquoi T.A., James F., Ariyarajah A., Ross C., Bernstein K., Coursier A., Klena J., Carino M., Wurie A.H., Zhang Y., Dumbuya M.S., Abad N., Idriss B., Wi T., Bennett S.D., Davies T., Ebrahim F.K., Meites E., Naidoo D., Smith S.J., Ongpin P., Malik T., Banerjee A., Erickson B.R., Liu Yongjian, Liu Yang, Xu K., Brault A., Durski K.N., Winter J., Sealy T., Nichol S.T., Lamunu M., Bangura J., Landoulsi S., Jambai A., Morgan O., Wu G., Liang M., Su Q., Lan Y., Hao Y., Formenty P., Ströher U., Sahr F. Ebola RNA persistence in semen of Ebola virus disease survivors - final report. N. Engl. J. Med. 2017;377:1428–1437. doi: 10.1056/NEJMoa1511410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick G.W.A., Kitchen S.F., Haddow A.J. Zika virus (I). Isolations and serological specificity. Trans. R. Soc. Trop. Med. Hyg. 1952;46:509–520. doi: 10.1016/0035-9203(52)90042-4. [DOI] [PubMed] [Google Scholar]
- D'Ortenzio E., Matheron S., de Lamballerie X., Hubert B., Piorkowski G., Maquart M., Descamps D., Damond F., Yazdanpanah Y., Leparc-Goffart I. Evidence of sexual transmission of Zika virus. N. Engl. J. Med. 2016;374:2195–2198. doi: 10.1056/NEJMc1604449. [DOI] [PubMed] [Google Scholar]
- Driggers R.W., Ho C.-Y., Korhonen E.M., Kuivanen S., Jääskeläinen A.J., Smura T., Rosenberg A., Hill D.A., DeBiasi R.L., Vezina G. Zika virus infection with prolonged maternal viremia and fetal brain abnormalities. N. Engl. J. Med. 2016;374:2142–2151. doi: 10.1056/NEJMoa1601824. [DOI] [PubMed] [Google Scholar]
- Ebihara H., Theriault S., Neumann G., Alimonti J.B., Geisbert J.B., Hensley L.E., Groseth A., Jones S.M., Geisbert T.W., Kawaoka Y. In vitro and in vivo characterization of recombinant Ebola viruses expressing enhanced green fluorescent protein. J. Infect. Dis. 2007;196:S313–S322. doi: 10.1086/520590. [DOI] [PubMed] [Google Scholar]
- Ferreira N., Pereira-Henriques A., Attar A., Klärner F.-G., Schrader T., Bitan G., Gales L., Saraiva M.J., Almeida M.R. Molecular tweezers targeting transthyretin amyloidosis. Neurotherapeutics. 2014;11:450–461. doi: 10.1007/s13311-013-0256-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fokkens M., Schrader T., Klärner F.-G. A molecular tweezer for lysine and arginine. J. Am. Chem. Soc. 2005;127:14415–14421. doi: 10.1021/ja052806a. [DOI] [PubMed] [Google Scholar]
- Food and Drug Administration . Department of Health and Human Services, FDA; June 2006. Guidance For Industry; Antiviral Product Development—Conducting and Submitting Virology Studies to the Agency. [Google Scholar]
- Haddow A.D., Schuh A.J., Yasuda C.Y., Kasper M.R., Heang V., Huy R., Guzman H., Tesh R.B., Weaver S.C. Genetic characterization of Zika virus strains: geographic expansion of the Asian lineage. PLoS Negl. Trop. Dis. 2012;6:e1477. doi: 10.1371/journal.pntd.0001477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes E.B. Zika virus outside Africa. Emerg. Infect. Dis. 2009;15:1347–1350. doi: 10.3201/eid1509.090442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoenen T., Groseth A., Callison J., Takada A., Feldmann H. A novel Ebola virus expressing luciferase allows for rapid and quantitative testing of antivirals. Antivir. Res. 2013;99:207–213. doi: 10.1016/j.antiviral.2013.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S., Loo J.A., Wong D.T. Human body fluid proteome analysis. Proteomics. 2006;6:6326–6353. doi: 10.1002/pmic.200600284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauer F., Riesen M., Reveiz L., Oladapo O.T., Martínez-Vega R., Porgo T.V., Haefliger A., Broutet N.J., Low N., Group W.Z.C.W. Zika virus infection as a cause of congenital brain abnormalities and Guillain–Barré syndrome: systematic review. PLoS Med. 2017;14 doi: 10.1371/journal.pmed.1002203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanciotti R.S., Lambert A.J., Holodniy M., Saavedra S., Signor L. del C.C. Phylogeny of Zika virus in western hemisphere, 2015. Emerg. Infect. Dis. 2016;22:933. doi: 10.3201/eid2205.160065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes D.H., Attar A., Nair G., Hayden E.Y., Du Z., McDaniel K., Dutt S., Bravo-Rodriguez K., Mittal S., Klärner F.-G. Molecular tweezers inhibit islet amyloid polypeptide assembly and toxicity by a new mechanism. ACS Chem. Biol. 2015;10:1555–1569. doi: 10.1021/acschembio.5b00146. [DOI] [PubMed] [Google Scholar]
- Lorizate M., Sachsenheimer T., Glass B., Habermann A., Gerl M.J., Kräusslich H.-G., Brügger B. Comparative lipidomics analysis of HIV-1 particles and their producer cell membrane in different cell lines. Cell. Microbiol. 2013;15:292–304. doi: 10.1111/cmi.12101. [DOI] [PubMed] [Google Scholar]
- Ludlow M., Kortekaas J., Herden C., Hoffmann B., Tappe D., Trebst C., Griffin D.E., Brindle H.E., Solomon T., Brown A.S. Neurotropic virus infections as the cause of immediate and delayed neuropathology. Acta Neuropathol. (Berl.) 2016;131:159–184. doi: 10.1007/s00401-015-1511-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lulla A. 2016. Neurotoxicity of the Parkinson's Disease-Associated Pesticide Ziram is Synuclein Dependent. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lump E., Castellano L.M., Meier C., Seeliger J., Erwin N., Sperlich B., Stürzel C.M., Usmani S., Hammond R.M., von Einem J. A molecular tweezer antagonizes seminal amyloids and HIV infection. Elife. 2015;4 doi: 10.7554/eLife.05397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik R., Di J., Nair G. Using molecular tweezers to remodel abnormal protein self-assembly and inhibit the toxicity of amyloidogenic proteins. Methods Mol. Biol. 2017 doi: 10.1007/978-1-4939-7811-3_24. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mate S.E., Kugelman J.R., Nyenswah T.G., Ladner J.T., Wiley M.R., Cordier-Lassalle T., Christie A., Schroth G.P., Gross S.M., Davies-Wayne G.J. Molecular evidence of sexual transmission of Ebola virus. N. Engl. J. Med. 2015;373:2448–2454. doi: 10.1056/NEJMoa1509773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlakar J., Korva M., Tul N., Popović M., Poljšak-Prijatelj M., Mraz J., Kolenc M., Resman Rus K., Vesnaver Vipotnik T., Fabjan Vodušek V. Zika virus associated with microcephaly. N. Engl. J. Med. 2016;374:951–958. doi: 10.1056/NEJMoa1600651. [DOI] [PubMed] [Google Scholar]
- Moreira J., Peixoto T.M., de Siqueira A.M., Lamas C.C. Sexually acquired Zika virus: a systematic review. Clin. Microbiol. Infect. 2017;23:296–305. doi: 10.1016/j.cmi.2016.12.027. [DOI] [PubMed] [Google Scholar]
- Müller J.A., Harms M., Schubert A., Jansen S., Michel D., Mertens T., Schmidt-Chanasit J., Münch J. Inactivation and environmental stability of Zika virus. Emerg. Infect. Dis. 2016;22:1685. doi: 10.3201/eid2209.160664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J.A., Harms M., Schubert A., Mayer B., Jansen S., Herbeuval J.-P., Michel D., Mertens T., Vapalahti O., Schmidt-Chanasit J. Development of a high-throughput colorimetric Zika virus infection assay. Med. Microbiol. Immunol. (Berl.) 2017;206:175–185. doi: 10.1007/s00430-017-0493-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münch J., Rücker E., Ständker L., Adermann K., Goffinet C., Schindler M., Wildum S., Chinnadurai R., Rajan D., Specht A. Semen-derived amyloid fibrils drastically enhance HIV infection. Cell. 2007;131:1059–1071. doi: 10.1016/j.cell.2007.10.014. [DOI] [PubMed] [Google Scholar]
- Paixão E.S., Barreto F., da Glória Teixeira M., da Conceição N., Costa M., Rodrigues L.C. History, epidemiology, and clinical manifestations of Zika: a systematic review. Am. J. Publ. Health. 2016;106:606–612. doi: 10.2105/AJPH.2016.303112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey A., Atkins K.E., Medlock J., Wenzel N., Townsend J.P., Childs J.E., Nyenswah T.G., Ndeffo-Mbah M.L., Galvani A.P. Strategies for containing Ebola in west Africa. Science. 2014;346:991–995. doi: 10.1126/science.1260612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen L.R., Jamieson D.J., Powers A.M., Honein M.A. Zika virus. N. Engl. J. Med. 2016;374:1552–1563. doi: 10.1056/NEJMra1602113. [DOI] [PubMed] [Google Scholar]
- Prabhudesai S., Sinha S., Attar A., Kotagiri A., Fitzmaurice A.G., Lakshmanan R., Ivanova M.I., Loo J.A., Klärner F.-G., Schrader T. A novel “molecular tweezer” inhibitor of α-synuclein neurotoxicity in vitro and in vivo. Neurotherapeutics. 2012;9:464–476. doi: 10.1007/s13311-012-0105-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen S.A., Jamieson D.J., Honein M.A., Petersen L.R. Zika virus and birth defects—reviewing the evidence for causality. N. Engl. J. Med. 2016:1981–1987. doi: 10.1056/NEJMsr1604338. [DOI] [PubMed] [Google Scholar]
- Richter F., Subramaniam S.R., Magen I., Lee P., Hayes J., Attar A., Zhu C., Franich N.R., Bove N., De La Rosa K. A molecular tweezer ameliorates motor deficits in mice overexpressing α-synuclein. Neurotherapeutics. 2017:1–13. doi: 10.1007/s13311-017-0544-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schandock F., Riber C.F., Röcker A., Müller J.A., Harms M., Gajda P., Zuwala K., Andersen A.H., Løvschall K.B., Tolstrup M. Macromolecular antiviral agents against Zika, Ebola, SARS, and other pathogenic viruses. Adv. Healthc. Mater. 2017;6(23) doi: 10.1002/adhm.201700748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader T., Klärner F.-G., Bitan G. Molecular tweezers for lysine and arginine–powerful inhibitors of pathologic protein aggregation. Chem. Commun. 2016;52(76):11318–11334. doi: 10.1039/c6cc04640a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma R., Ketki Jangid A. Ebola vaccine: how far are we? J. Clin. Diagn. Res. JCDR. 2017;11:DE01. doi: 10.7860/JCDR/2017/22184.9863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuaib W., Stanazai H., Abazid A.G., Mattar A.A. Re-emergence of Zika virus: a review on pathogenesis, clinical manifestations, diagnosis, treatment, and prevention. Am. J. Med. 2016;129 doi: 10.1016/j.amjmed.2016.02.027. 879–e7. [DOI] [PubMed] [Google Scholar]
- Sinha S., Lopes D.H., Du Z., Pang E.S., Shanmugam A., Lomakin A., Talbiersky P., Tennstaedt A., McDaniel K., Bakshi R. Lysine-specific molecular tweezers are broad-spectrum inhibitors of assembly and toxicity of amyloid proteins. J. Am. Chem. Soc. 2011;133:16958–16969. doi: 10.1021/ja206279b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirohi D., Chen Z., Sun L., Klose T., Pierson T.C., Rossmann M.G., Kuhn R.J. The 3.8 A resolution cryo-EM structure of Zika virus. Science. 2016;352:467–470. doi: 10.1126/science.aaf5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbiersky P., Bastkowski F., Klärner F.-G., Schrader T. Molecular clip and tweezer introduce new mechanisms of enzyme inhibition. J. Am. Chem. Soc. 2008;130:9824–9828. doi: 10.1021/ja801441j. [DOI] [PubMed] [Google Scholar]
- Usmani S.M., Zirafi O., Müller J., Sandi-Monroy N., Yadav J.K., Meier C., Weil T., Roan N.R., Greene W.C., Walther P. Direct visualization of HIV-enhancing endogenous amyloid fibrils in human semen. Nat. Commun. 2014;5:3508. doi: 10.1038/ncomms4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vibhakar P.A., Patankar S.R., Yadav M.R., Vibhakar P. Salivary total protein levels and their correlation to dental caries. Int. J. Oral Maxillofac. Pathol. 2013;4:13–16. [Google Scholar]
- Xu N., Bitan G., Schrader T., Klärner F.-G., Osinska H., Robbins J. Inhibition of mutant αB crystallin-induced protein aggregation by a molecular tweezer. J. Am. Heart Assoc. 2017;6 doi: 10.1161/JAHA.117.006182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J.-D., Meng W., Wang X.-J., Wang H.-C.R. Broad-spectrum antiviral agents. Front. Microbiol. 2015;6 doi: 10.3389/fmicb.2015.00517. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.