Abstract
The disease outbreak caused by Middle East respiratory syndrome coronavirus (MERS-CoV) is still ongoing in the Middle East. Over 1700 people have been infected since it was first reported in September 2012. Despite great efforts, licensed vaccines or therapeutics against MERS-CoV remain unavailable. The MERS-CoV spike (S) protein is an important viral antigen known to mediate host-receptor binding and virus entry, as well as induce robust humoral and cell-mediated responses in humans during infection. In this review, we highlight the importance of the S protein in the MERS-CoV life cycle, summarize recent advances in the development of vaccines and therapeutics based on the S protein, and discuss strategies that can be explored to develop new medical countermeasures against MERS-CoV.
Keywords: Coronavirus, MERS-CoV, Spike protein, Vaccines, Therapeutics, Animal models
Highlights
-
•
A licensed vaccine or therapeutic against MERS-CoV remains unavailable to date.
-
•
The S protein plays a pivotal role for virus entry and thus is an ideal target for vaccine and antiviral development.
-
•
DNA vaccines expressing the S protein merit further development for potential human application.
-
•
nAbs and peptides targeting the S protein needs to be evaluated in NHPs before clinical trials.
1. Introduction
Coronaviruses (CoVs) are spherical and enveloped viruses with large, unsegmented, single positive RNA genomes that are 26.2–31.7 kb in length. The two-thirds of the genome at the 5′ end are translated into two overlapping polyproteins, pp1a and pp1ab, that are later cleaved by 3CLpro and PLpro to yield nonstructural proteins (nsps). The remaining one-third of the genome is responsible for coding the structural proteins: spike (S) glycoprotein, small envelope protein (E), integral membrane protein (M), and genome-associated nucleocapsid protein (N). Some CoVs also contain a hemagglutinin esterase (HE) (van Boheemen et al., 2012). Interspersed between these genes in the coronavirus genome are several genes coding for accessory proteins. Although dispensable for virus growth in vitro, as determined using reverse genetics and targeted mutagenesis analyses, most of them are involved in regulating the host immune system (Narayanan et al., 2008, Niemeyer et al., 2013, Yang et al., 2013). The proteins E, M, and N are mainly responsible for the assembly of the virions, while the S protein is involved in receptor binding and bears membrane fusion capabilities during CoVs infection (Saif, 1993). Thus, the S protein has an essential role in virus entry and determines tissue and cell tropism, as well as host range (Lu et al., 2015).
CoVs infect both avian and mammalian species (Schmidt et al., 2005), with examples such as the infectious bronchitis virus (IBV) infecting chickens, porcine epidemic diarrhea virus (PEDV), porcine respiratory coronavirus (PRCV), and transmissible gastroenteric virus (TGEV) infecting pigs, bovine coronavirus (BCoV) infecting bovines, feline infectious peritonitis virus (FIPV) infecting cats, and canine coronavirus (CCoV) infecting dogs. These viruses have resulted in significant economic losses for the animal industry worldwide. Commonly, veterinary CoV infections are prevented through either modified live or inactivated CoV vaccines (Carmichael, 1999, Cavanagh, 2007, Hick et al., 2012, Olsen, 1993, Welter et al., 1993). The attenuated live viruses (BCoV, TGEV, and IBV) and a temperature-sensitive mutant (FIPV) are usually administered orally or intranasally to induce active mucosal immunity. In contrast, inactivated vaccines administered parenterally with adjuvant are available to induce active or passive immunity to BCoV, TGEV, CCoV, and IBV.
CoVs can also cause disease in humans (Fig. 1 ). In the 1960s, two human coronaviruses, human coronavirus-229E (HCoV-229E) and OC43 (HCoV-OC43), were isolated from volunteers at the Common Cold Unit in Salisbury, United Kingdom (Almeida and Tyrrell, 1967, Hamre and Procknow, 1966, Hendley et al., 1972, McIntosh et al., 1967, Tyrrell and Bynoe, 1965). In 2004, human coronavirus-NL63 (HCoV-NL63) was isolated from a 7-month-old child suffering from bronchiolitis and conjunctivitis in The Netherlands (van der Hoek et al., 2004). In the following year, another CoV, human coronavirus-HKU1 (HCoV-HKU1), was identified in a patient with pneumonia in Hong Kong (Woo et al., 2005). These human coronaviruses mainly cause subclinical or very mild infections and account for 10-15% of common colds (Heikkinen and Jarvinen, 2003). However, more severe lower respiratory tract-related cases have been reported in infants and the elderly (Falsey et al., 1997, McIntosh et al., 1974, Nokso-Koivisto et al., 2000, Pene et al., 2003, Vabret et al., 2003).
Fig. 1.
Timeline for the discovery of different human CoVs.
Human coronaviruses can also be life-threatening and have pandemic potential. During 2002–2003, the severe acute respiratory syndrome coronavirus (SARS-CoV) caused > 8000 infections and 774 related deaths worldwide (WHO, 2004). A decade later, the epidemic outbreak of Middle East respiratory syndrome (MERS) reminds us of the possibility for another potential pandemic on the scale of that of SARS. The first case was reported in the Kingdom of Saudi Arabia (KSA) in September 2012 (Bermingham et al., 2012, Zaki et al., 2012), and since then, the MERS-CoV (i.e., the etiological agent behind MERS (de Groot et al., 2013)) has spread to 25 other countries, totaling 1782 laboratory-confirmed infections and 634 related deaths (case fatality rate = 35.6%) (WHO, 2016). The majority of human cases is reported in the Middle East, likely due to the presence of dromedary camels, which have been confirmed to carry live MERS-CoV and are a potential source of human infections (Azhar et al., 2014, Memish et al., 2014). However, whether dromedary camels are intermediary hosts or the original reservoir remains open at present.
In May 2015, MERS-CoV spread to South Korea via a traveler from the Middle East and caused several infection clusters, resulting in a total of 186 confirmed cases, with evidence of virus super-spreading in hospitals and clinics (Wong et al., 2015). One of the subsequent patients traveled to China and fell ill, marking the first instance of an imported MERS-CoV case that did not spread directly from the Middle East (Su et al., 2015). These events are a reminder that past geographical barriers for disease can be easily bypassed due to the increasingly globalized nature of the world. Unlike SARS-CoV, which after great disease control efforts by the Chinese government suddenly disappeared, the number of MERS cases is still on the rise, reinforcing the urgent need to develop vaccines or specific drugs targeted at CoVs, especially anti-epidemic MERS-CoV measures.
Although more than 10 years have passed since the initial outbreak, there are presently still no available anti-SARS drugs on the market. Nonetheless, many antiviral agents have been identified, which have been summarized in several excellent reviews (Adedeji and Sarafianos, 2014, Keum and Jeong, 2012, Kumar et al., 2013, Liang, 2006, Ramajayam et al., 2011, Yang et al., 2006). Just as for SARS-CoV and other viruses (Liang, 2006, Lu et al., 2011, Yang et al., 2003), the 3C-like protease (3CLpro or Mpro) and papain-like protease (PLpro) in MERS-CoV are responsible for processing the huge polyproteins into mature nsps (Baez-Santos et al., 2014, Kilianski et al., 2013, Yang et al., 2014) and are deemed to be a viable target for inhibitor screening. Small molecules, such as 5-chloropyridine ester, 6-mercaptopurine, and 6-thioguanine targeting MERS-CoV 3CLpro or PLpro have shown efficacy in inhibiting MERS-CoV replication in vitro (Chan et al., 2013, Cheng et al., 2015, Kilianski et al., 2013), though their activity and cytotoxicity needs to be further assessed in vivo.
In addition, existing drugs have been screened to identify potential inhibitors, and several demonstrate clinical relevance, with mild adverse effects and low half-maximal effective concentrations (EC50). Among these, ribavirin and interferon-α display some effects in vitro (Chan et al., 2013, de Wilde et al., 2014) and in MERS-CoV infected rhesus macaques with mild diseases (Munster et al., 2013), but their benefits in MERS patients are not obvious. Fortunately, lopinavir/ritonavir and interferon-β1b have been shown to improve the outcome of MERS-CoV-infected common marmosets (Chan et al., 2015), which represents the severe manifestation of MERS patients (Falzarano et al., 2014) and have demonstrated the most potential for clinical use. Recently, miRNAs and siRNAs targeting MERS-CoV have also been predicted by utilizing a series of bioinformatics tools (Hasan et al., 2014, Nur et al., 2014), but further evaluation in vivo and in vitro is required before any clinical trials may begin.
In this review, we first describe the chemical and structural features of the S protein, summarize the current published vaccine and therapeutic research against MERS-CoV based on the S protein, and highlight the suitability of the S protein as a target for the design of medical countermeasures against MERS-CoV.
2. S protein: a mediator of virus entry and its structure
The MERS-CoV genome is assembled similar to other CoVs, and the gene coding for S is located in the one-third of the genome at the 3′ end (Fig. 2 A). S is a type I, trimeric, transmembrane protein located at the surface of the viral envelope, giving rise to spike-shaped protrusions from the virion. S is 1353 amino acids in length, heavily glycosylated (with 21 predicted N-linked glycosylation sites), and consists of a large ectodomain and a short cytosolic tail (Fig. 2B). The S proteins of CoVs can be divided into two functional subunits (Xia et al., 2014): the N-terminal S1 subunit forms the globular head, and the membrane-embedded C-terminal S2 region forms the stalk region. S1 is involved in receptor recognition and binding and is less conserved compared to the stalk. Two discrete, independently folded domains are located at the N- and C-termini of S1 and are able to bind receptors.
Fig. 2.
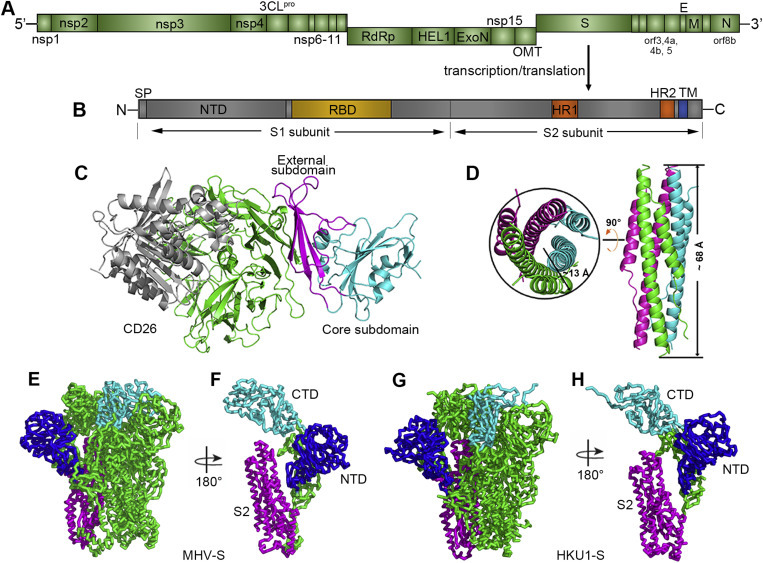
Genome arrangement of MERS-CoV and structure of the spike protein-receptor complex. (A) Schematic representation of the MERS-CoV genome. Abbreviations: nsp, nonstructural protein; 3CLpro, 3C-like protease; RdRp, RNA-dependent RNA polymerase; HEL1, superfamily 1 helicase; ExoN, 5′-3′ exonuclease; OMT, S-adenosylmethionine-dependent ribose 2′-O-methyltransferase; S, spike protein; orf, open reading frame; E, envelope protein; M, membrane protein; and N, nucleocapsid. In addition, PLpro (papain-like protease) is located in nsp3, and NendoU (nidoviral endoribonuclease specific for U) is located in nsp15. The gene coding for accessory protein orf8b overlaps with the N-coding gene (Cotten et al., 2013). (B) Schematic representation of the MERS-CoV spike protein. Abbreviations: SP, signal peptide; NTD, N-terminal domain; RBD, receptor binding domain; HR1/2, heptad repeat 1/2; and TM, transmembrane domain. (C) Complex structure between the MERS-RBD and its receptor CD26. The core and external subdomains are highlighted in cyan and magenta, respectively, while the receptor is colored in green for the β-propeller domain and in gray for the α/β-hydrolase domain, respectively. (D) Crystal structure of the HR1/HR2 fusion core. The three HR1/HR2 chains are colored in green, cyan, and magenta, respectively. The approximate size of the bundle is indicated. The left panel represents the top view, and the right panel represents the side view. The figure was used upon approval of the authors in Gao et al., 2013. (E) and (G) Ribbon diagrams showing the overall structures of MHV-S and HKU1-S trimers, respectively. (F) and (H) Ribbon diagrams showing one MHV-S and HKU1-S molecule, respectively. NTD, CTD and S2 are colored in blue, cyan, and magenta, respectively in E-H.
MERS-CoV utilizes the C-terminal domain, which spans ∼240 residues, as the receptor binding domain (RBD) (Du et al., 2013b, Lu et al., 2013a, Mou et al., 2013, Wang et al., 2013). Specifically, the MERS-RBD can be further divided into two parts, the core subdomain and external subdomain. Although sharing a low amino acid sequence identity, the core subdomain structures of SARS-RBD and MERS-RBD can be superimposed well (Li et al., 2005), indicating a conserved core structure among the CoVs. In contrast, the external subdomain structure of these two proteins differs greatly. Interestingly, it is the external subdomain that is mainly involved in receptor binding and thus results in the different receptor usage between CoVs. In the MERS-RBD, the four antiparallel β-strands comprising the external region bind to the blade IV and V in the β-propeller of human CD26 (hCD26, also known as dipeptidyl peptidase IV), the receptor for MERS-CoV (Fig. 2C) (Raj et al., 2013). Accordingly, blocking the binding between the MERS-RBD and hCD26 can prevent virus attachment and inhibit virus infection. Intriguingly, the MERS-RBD stimulates the production of robust antibodies in mice and rabbits that neutralize virus infection (Ma et al., 2014a, Mou et al., 2013), suggesting that the S protein may represent a promising immunogen for eliciting neutralizing antibodies (nAbs).
In addition to receptor recognition and binding, proteolysis between S1 and S2 is another critical determinant for CoV tropism and pathogenesis (Millet and Whittaker, 2015, Simmons et al., 2013). Similarly, MERS-S protein can be cleaved by different host proteases, which has previously been thoroughly reviewed (Lu et al., 2015). It is worth noting that MERS-CoV uses a two-step furin cleavage activation process to infect cells. Specifically, S is first cleaved during biosynthesis at position R751/S752. When MERS-CoV enters target cells, S is then further cleaved at R887/S888, the position adjacent to the fusion peptide within S2, and designated as S2' (Millet and Whittaker, 2014). The importance of the first cleavage has been confirmed by the analysis of bat coronavirus HKU4 (batCoV-HKU4), which is able to bind to hCD26 but is unable to infect cells due to the lack of the first proteolytic conserved motif recognized by furin. Introduction of a furin recognition site into S (Yang et al., 2015) or treatment of the pseudotyped HKU4 particles with trypsin confers particles with the ability to infect hCD26-expressing cells (Wang et al., 2014). Likewise, inhibition of S2′ proteolysis by furin inhibitors decreases MERS-CoV infection in susceptible cells in a concentration-dependent manner (Millet and Whittaker, 2014), indicating both cleavages are critical for virus entry.
Viral fusion proteins can be grouped into three classes. Class I fusion proteins, represented by influenza hemagglutinin, are trimers of a single-chain precursor and primed by the cleavage of a trimeric precursor, forming six-helix bundles. Class II fusion proteins, represented by flavivirus prM/E proteins, are icosahedrally symmetrically, assemble as dimers on the virion, and are primed by cleavage of heterodimeric partner proteins to form homotrimers during membrane fusion. Class III fusion proteins are more complicated, including one or several proteins, and undergo no obvious priming event (Earp et al., 2005, Gao, 2007, Harrison, 2015, Kielian and Rey, 2006, Melikyan, 2008, White et al., 2008). CoV-S protein is a typical Class I fusion protein. Recently, the cryo-electron microscopy structures of MHV-S and HKU1-S proteins were reported (Kirchdoerfer et al., 2016, Walls et al., 2016). Both structures reveal the overall pre-fusion architecture of full length S ectodomains (Fig. 2E and G), showing that the N-terminal domain (NTD) and C-terminal domain (CTD) present in a “V” shape and therefore contributing to the overall triangular appearance of the S trimer (Fig. 2F and H). The S2 subunit connects to the viral membrane and is characterized by the presence of long α-helices (Fig. 2F and H). This structural information can serve as a foundation for structure-based design of β-CoV vaccine immunogens.
After binding to hCD26, fusion between the MERS-CoV envelope and the host cell membrane occurs and is mediated by the S2 subunit (Belouzard et al., 2012). As a Class I fusion protein, MERS-CoV S2 contains multiple key components, including fusion peptides and conserved heptad repeats (HRs) (Fig. 2B). However, unlike the extensive studies of SARS-CoV fusion peptides, little experimental investigation has been undertaken concerning MERS-CoV, though a similar concentration of hydrophobic residues can be identified in the equivalent regions of MERS-S (Lu et al., 2015).
In contrast, the HRs containing heptad repeat 1 (HR1) and heptad repeat 2 (HR2) in the virus have been well studied (Gao et al., 2013, Lu et al., 2014). Consistent with other typical Class I fusion proteins, the HRs in MERS-CoV S2 assemble into a canonical six-helix bundle called the fusion core, with three HR1 helices forming the central coiled-coil core and three HR2 chains surrounding the core in the HR1 side grooves (Fig. 2D). It is believed that the fusion core represents the post-fusion state of the part of the envelope structure. Although HR1 and HR2 are assumed to be located distal from one another in the pre-fusion state, a series of conformational changes occur in the envelope structure after S1 binding to the receptor that result in the binding of the fusion peptide located at the N-terminus of HR1 to the cell membrane. Through a short-lived intermediate state, HR1 and HR2 bend to bind each other, moving the virus membrane and cell membrane into close proximity to enable the membrane fusion process. In the intermediate state, both HRs are exposed, and inhibitors blocking the formation of the fusion core or that maintain HRs at the intermediate state can be used as drugs to inhibit MERS-CoV infection.
3. Vaccines
The roles of S in receptor binding and membrane fusion make it a perfect target for vaccine and antiviral development. Previous studies on SARS-CoV reveal that vaccines based on the S protein can induce antibodies to block virus binding and fusion or neutralize virus infection (Du et al., 2009). Although there is no licensed MERS vaccines available, researchers are working to develop potential vaccines based on S protein, and several previous reviews have touched on this topic (Papaneri et al., 2015, Zhang et al., 2014). Here, we focus on the more recent progress on vaccines showing efficacy in mice and nonhuman primates (NHPs) and/or dromedaries, including replication-deficient virus vectors, DNA vaccines, and subunit vaccines, which are summarized in Table 1 .
Table 1.
S protein-based vaccines against MERS-CoV.
| Candidate | Antigen | Animal model | Vaccine regimens | Efficacy or observed effects | Reference |
|---|---|---|---|---|---|
| MVA-S | S | Mice | i.m. administered of 1 × 108 PFU twice at a 3-weeks interval | ND | (Song et al., 2013) |
| Ad5-hCD26-transduced mice | s.c. administered of 1 × 108 PFU twice at a 3-weeks interval | 100% protection (n = 5) | (Volz et al., 2015) | ||
| Ad5-hCD26-transduced mice | i.m. administered of 1 × 108 PFU twice at a 3-weeks interval | 100% protection (n = 5) | |||
| Dromedaries | i.n. administered of 2 × 108 PFU and i.m. Injection of 1 × 108 at the neck twice at a 4-weeks interval | Significant reduction of excreted virus (n = 4) | (Haagmans et al., 2016) | ||
| VRP-S | S | Ad5-hCD26-transduced mice | 1 × 105 IU delivered in footpad twice at 4-week interval | 100% protection (n = 3−4) | (Zhao et al., 2014) |
| DNA | S-DNA and S1-protein | Mice | 20 μg of S DNA were i.m. delivered followed by EP twice at 3-week interval and boosted with 10 μg of S1 protein plus Ribi 3 weeks later | ND | (Wang et al., 2015) |
| Rhesus macaque | 1 mg of S DNA were i.m. delivered followed by EP twice at 4-week interval and boosted with 100 μg of S1 protein plus AlPO4 4 weeks later | 100% protection (n = 6) | |||
| S | Mice | 25 μg of DNA i.m. delivered followed by EP three times at 2-week intervals | ND | (Muthumani et al., 2015) | |
| Dromedaries | EP-enhanced i.m. delivery three times at 4-week intervals | ND | |||
| Rhesus macaque | 0.5 mg or 2 mg of DNA i.m. delivered followed by EP three times at 3-week intervals | 100% protection (n = 4 for each dosage) | |||
| subunit | RBD-Fc | Ad5-hCD26-transduced mice | 10 μg/mice of RBD-Fc plus MF59 s.c. delivered three times at 3-week interval | 100% protection (n = 4) | (Zhang et al., 2015) |
Abbreviations: i.m. – intramuscular, s.c. – subcutaneous; i.n. – intranasal; ND – not determined; EP – electroporation.
3.1. Modified vaccinia Ankara (MVA)-based chimeric virus vaccines
MVA is a highly attenuated vaccinia virus strain that has lost ∼15% of the vaccinia genome and is replication-defective in mammalian cells (Ura et al., 2014). It has previously been safely administered to over 120,000 individuals as a vaccine against smallpox (Ura et al., 2014). Currently, MVA-based vaccines against several other diseases/conditions are undergoing clinical trials, including HIV-1, hepatitis, influenza, malaria, tuberculosis, and cancer (Gilbert, 2013). A similar approach has been adopted to develop vaccines against MERS-CoV.
Song et al. show that mice intramuscularly immunized with 1 × 108 plaque-forming units (PFU) of recombinant MVA-MERS-S twice within a 21-day interval elicit nAbs (Song et al., 2013). Volz et al. further dissect the immunogenicity of the same vaccine in mice (Volz et al., 2015), and in agreement with the data obtained with MVA used as smallpox vaccine, immunizations through subcutaneous and intramuscular routes induce equivalent humoral and cellular immunity in mice. In addition, intramuscular vaccination evokes equal amounts of MERS-S-specific CD8+ T cells across low (1 × 106 PFU) to high (1 × 108 PFU) doses. When challenged with MERS-CoV, vaccinated mice exhibit little or no virus replication, irrespective of the route or dose used (Volz et al., 2015), indicating the efficacy of this MVA-MERS-S in the mouse model. More recent studies by Haagmans et al. further reveal that this vaccine significantly reduces MERS-CoV shedding in dromedary camels (Haagmans et al., 2016). Four dromedaries were inoculated twice at four-week intervals with 2 × 108 PFU MVS-MERS-S administered in nostrils and 1 × 108 PFU MVA-MERS-S delivered intramuscularly in the neck. In addition to the systemic humoral and cellular immunity evoked by the vaccine, low levels of MERS-CoV nAbs (virus neutralization titer of 1:20 to 1:40) are detected 3 weeks after the boost immunization through the nostrils. When challenged with a 1 × 107 median tissue culture infectious dose (TCID50) of the virus via the intranasal route, the animals show only mild clinical signs and significantly reduced mean viral titers in the nasal respiratory tract compared to the control animals.
The advantage of this vaccine is its compatibility with clinical use and industrial-scale production. In addition, the vector can be grown in chicken embryo fibroblasts (CEFs) without the need for additional animal-derived components in culture, and S protein can be synthesized upon serial amplification at low multiplicities of infection. Although its efficacy in mice and dromedaries has clearly been shown, there is still no data demonstrating the protective effect of MVA-MERS-S in a NHP model. This is important for human application because in some studies, high doses of MVA vector (>1 × 108 PFU) result in severe adverse events in humans (Gilbert, 2013). Thus, strict evaluation needs to be performed before MVA-MERS-S application in humans. In addition, one immunized dromedary (1/4) developed undetectable nAbs at the nostrils at the time of challenge and excreted low levels of infectious virus at 6 day post infection (dpi) after virus challenge (Haagmans et al., 2016). Further sequencing of the spike gene of the infectious virus showed no changes in the RBD region, excluding the possibility of escape mutations under the stress of vaccine-induced antibodies. Consequently, the underlying mechanisms that resulted in the failure of MVA-MERS-S to boost immunity warrant further investigation.
3.2. Virus-like replicon particle (VRP)-based chimeric virus vaccines
Venezuelan equine encephalitis virus (VEEV) belongs to the family Alphaviridae (Lundstrom, 2014). The structure protein of VEEV can be replaced by a heterologous protein of interest, resulting in a recombinant RNA replicon that can then be “packaged” into VRPs using co-transfection with helper RNAs (Lundstrom, 2014). Due to the lack of structural protein gene(s), these replicons are replication-defective. VRP-based vaccines against prostate cancer and HIV-1 have been tested in Phase I clinical trials (Slovin et al., 2013, Wecker et al., 2012), with no toxicity associated with the immunization. In an astute study, Zhao et al. generated a vaccine candidate against MERS-CoV by inserting the S gene into the VRP (VRP-S) (Zhao et al., 2014). Mice were first immunized with 1 × 105 infectious units (IU) of VRP-S in the footpad and boosted with the same dose 4 weeks later. Mice were afterwards transduced with 2.5 × 108 PFU of Ad5-hCD26 and then infected with 1 × 105 PFU of MERS-CoV (EMC2012 strain). By day 1 post infection, the viral load in hCD26 transgenic mice decreased to nearly undetectable levels (Zhao et al., 2014), indicating its efficacy in the mouse model. However, further evaluation in NHPs is required before human application.
3.3. DNA vaccines
DNA vaccines induce protection by injecting genetically engineered DNA containing target antigen genes, and several DNA vaccines have been approved for veterinary use (Redding and Weiner, 2009). There are currently no DNA vaccines approved for human use, but >100 products are under clinical trials targeting a variety of diseases, including HIV, influenza, hepatitis B and C, human papillomavirus (HPV), and cancer (Ferraro et al., 2011).
Wang et al. first reported the development of DNA vaccines by inserting codon-optimized S (strain England1) coding sequences into mammalian expression vector VRC8400, which has since been used for HIV DNA vaccines and has been proven to be safe in mice and NHPs (Barouch et al., 2005, Catanzaro et al., 2007). Although three intramuscular immunizations at three-to-four week intervals with plasmid DNA followed by electroporation stimulate MERS-S-specific nAbs in mice and rhesus macaques, the immunogenicity is not as efficient as the combination of the same plasmid DNA with S1 protein (Wang et al., 2015). Both mice and NHPs develop higher titer of nAbs when immunized with plasmid DNA twice within three-to-four weeks followed by S1 protein mixed with adjuvant than using either DNA vaccine or S1 protein alone. When challenged with approximately 3 × 108 PFU of MERS-CoV (Strain JordanN3), NHPs immunized with S DNA/S1 protein experience a lower peak volume of pulmonary disease than the S1 protein group and clear pulmonary infiltrates more rapidly (Wang et al., 2015), indicating that this strategy is a promising approach for MERS vaccine development.
Another independent group has also developed DNA-based MERS vaccines. To increase the immunogenicity, Muthumani et al. optimized the expression of S (England/2/2013) in mammalian expression plasmid pVax1 through codon and RNA optimization, as well as the addition of a highly efficient immunoglobulin E (IgE) signal peptide instead of the N-terminal methionine. By EP-enhanced delivery three times at two-to-four-week intervals, the optimized DNA vaccine induces potent cellular immunity and antigen-specific nAbs in mice, macaques, and camels. Notably, in NHPs, two doses (0.5 mg vs. 2 mg per immunization) were assessed for immunogenicity, and both groups developed similar humoral immunity after three immunizations. However, the high-dose group produced more cellular immunity than the low-dose group. Four weeks after the final immunization, 7 × 106 TCID50 of MERS-CoV (EMC/2012) was applied to inoculate NHPs. Compared to the control animals, both groups of vaccinated rhesus macaques exhibited significantly lower viral loads. Interestingly, all four NHPs immunized with lower doses of DNA vaccines failed to demonstrate radiographic evidence of infiltration at any time point, whereas half of animals vaccinated with higher doses showed evidence of minor infiltration that resolved by day 5 after infection. Whether this phenomenon is associated with the vaccine dose needs to be ascertained before human usage. Moreover, when this vaccine was used to immunize dromedaries, one of three animals failed to develop robust nAbs, pointing to the need for further research before veterinary use.
DNA vaccines have the characteristics of simplicity, versatility, and the absence of pre-existing immunity against vectors and, thus, can be used repeatedly, which has sparked extensive studies into their development. However, an initial trial failed to efficiently stimulate recipients’ immune responses and may in part be due to insufficient exposure to the antigens (Ferraro et al., 2011). Wang et al. and Muthumani et al. apply different strategies to solve this problem. Combining these two strategies, DNA-subunit vaccine regimens would stimulate more robust immunoreactions in recipients, if possible, to the level to decrease times of vaccination, which would save time, cost, and effort.
3.4. Subunit vaccines
MERS-CoV relies on the binding between its RBD region and the hCD26 receptor to initiate infection, and blocking this interaction is speculated to abolish virus entry. Therefore, the RBD is the main immunogen for eliciting specific nAbs. The MERS-RBD has previously been shown to efficiently induce nAbs in mice and rabbits (Du et al., 2013a, Du et al., 2013b, Ma et al., 2014b, Mou et al., 2013). Following subcutaneous vaccination with 10 μg/mouse of RBD-Fc in the presence of an equal volume of MF59 and following two boost doses with the same immunogen and adjuvants at three-week intervals, mice develop systemic humoral and cellular immunity against MERS-CoV. Ten days after the last immunization, mice were transduced with Ad5-hCD26, and then intranasally challenged with 1 × 105 PFU of MERS-CoV (EMC-2012) 5 days later. No MERS-CoV was detected in the lungs of immunized mice 3 and 5 dpi, while high titers of the virus were found in control mice (Zhang et al., 2015), indicating that MERS-CoV RBD is highly effective in protecting mice from MERS-CoV challenge. S1 vaccines have also been tested in the immunization of mice and rhesus macaques, but they are not as efficient as the combination of S DNA/S1 protein in protection of NHPs from virus challenge (Wang et al., 2015).
Compared to other types of vaccines, subunit vaccines have defined components and can be produced consistently. More importantly, the vaccines are designed based on the knowledge of viral pathogenesis and only contain synthetic immunogenic fragments of a pathogen, which consequently avoids allergenic and/or reactogenic sequences (Li et al., 2014). Considering the aforementioned features, subunit vaccines represent attractive vaccine candidates. However, compared to inactivated and live-attenuated vaccines, protein-based subunit vaccines still exhibit a relatively low immunogenicity (De Gregorio et al., 2013, Naz and Dabir, 2007, Podda and Del Giudice, 2003), which may be improved by several strategies, such as the addition of suitable adjuvant, increasing vaccination times, and being combined with other types of vaccines.
4. Therapeutics targeting the S protein
To date, there are no licensed anti-MERS-CoV therapeutics. However, several strategies are currently being investigated to identify various specific antivirals targeting S, including MERS-RBD-targeted nAbs to block viral attachment, peptide inhibitors targeting S2 to prevent formation of the fusion core, and small molecules without defined mechanisms.
4.1. nAbs
A large number of humanized monoclonal antibodies (mAbs) and human mAbs have been characterized and licensed against a number of diseases (Carter, 2006, Dimitrov, 2012), but they are only approved against respiratory syncytial virus (RSV) in terms of viral infectious disease (Shadman and Wald, 2011). Experimental mAb therapies have been developed against HIV-1, influenza, and Ebola virus, and they are currently undergoing clinical trials (Bossart et al., 2009, Bossart et al., 2011, Lingwood et al., 2012, Mascola and Haynes, 2013, Qiu et al., 2014, Zhu et al., 2006). Against SARS-CoV, convalescent sera has so far been tested, in which most antibodies target the RBD or S2 regions, thus constituting the most promising targets for the design of mAb therapeutics. After the outbreak of MERS, the International Severe Acute Respiratory and Emerging Infection Consortium also recommended the use of convalescent sera for MERS treatment. Thus, a vast effort has been devoted to identify nAbs targeting S through a variety of strategies, such as mouse hybridomas (Li et al., 2015, Wang et al., 2015), screening libraries (Jiang et al., 2014, Tang et al., 2014), and isolation from humanized mice (Pascal et al., 2015). Here, we only focus on those proven to be efficient in vivo, as summarized in Table 2 .
Table 2.
Properties of monoclonal antibodies against MERS-CoV.
| Source | Ab | Binding affinity (Kd, nM) | IC50 (μg/ml) |
Efficacy | Reference | |
|---|---|---|---|---|---|---|
| Pseudotyped virus | Live virus | |||||
| Mouse hybridoma | 4C2 | 162 | ∼0.71 | 6.25 | ND | (Li et al., 2015) |
| 2E6 | 60.3 | ∼0.29 | 1.56 | ND | ||
| Humanized | 4C2h | 217 | ∼1.8 | 6.25 | 100% protection of Ad5-hCD26-transduced mice against virus infection | |
| Humanized mice | REGN3051 | 0.0433 | 0.0098 | 0.069 | 100% protection hCD26 mice from virus infection | (Pascal et al., 2015) |
| REGN3048 | 0.0485 | 0.011 | 0.027 | |||
| Phage-display (Fab) | m336 | 4.27 | 0.005 | 0.07 | 100% survival in rabbits when administered before challenge; no protection observed when given after challenge | (Houser et al., 2016; Ying et al., 2014) |
Kd, the equilibrium dissociation constant; IC50, 50% inhibitory concentration.
4.1.1. nAbs elicited in mice
In a recent study, two mouse-derived nAbs (4C2 and 2E6) against MERS-CoV were generated from mice immunized with the MERS-RBD (Li et al., 2015). Both nAbs specifically bind to the immunogen with sub-micromolar binding affinity. A competition assay revealed that the two nAbs likely recognize proximate or overlapping epitopes and neutralize the virus by interfering with S protein binding to hCD26. The 4C2 nAb was then humanized (4C2h) to circumvent adverse effects associated with using mAbs of foreign origin, such as serum sickness. Based on the structure of 4C2 bound to the MERS-RBD (Li et al., 2015), only the paratope residues are preserved, and the remaining amino acids are substituted with counterparts from human immunoglobulins. The resulting 4C2h exhibits similar binding affinity and neutralizing activity to the parental mouse antibody, and thus, further in vivo evaluation was performed.
For control Ad5-hCD26-transduced mice intranasally infected with 1 × 105 PFU MERS-CoV (EMC/2012), the viral load plateaus at ∼106 PFU/g tissue at 3 dpi and then decreases to ∼104 PFU/g tissue at 5 dpi. Whereas either intravenous administration of 200 μg/mouse 4C2h one day before or after challenge efficiently decreases the viral load in the lungs of transduced mice. In each case, the viral load decreases by approximately two orders of magnitude to ∼104.5 PFU/g tissue at 3 dpi and to < 102 (which is below the limit of detection) PFU/g tissue at 5 dpi, indicating the ability of prophylaxis and therapy of this antibody against MERS-CoV infection.
4.1.2. Human antibodies generated in transgenic mice
VeloImmune mice are a type of transgenic mice that include DNA encoding human Ig heavy and κ light-chain variable regions instead of the corresponding mouse DNA, while leaving the mouse constant regions intact (Murphy et al., 2014, Valenzuela et al., 2003). Antibodies generated in these mice are essentially indistinguishable from naturally occurring human antibodies, and hundreds of thousands of doses of VeloImmune-derived antibodies have been safely administered to humans (Bruggemann et al., 2015, DiGiandomenico and Sellman, 2015, Economides et al., 2013, Murphy et al., 2014). Pascal et al. immunize VeloImmune mice with DNA encoding MERS-S and recombinant purified MERS-RBD (Pascal et al., 2015). Two fully humanized antibodies (REGN3051 and REGN3048) that bind to the MERS-RBD and block the interaction of S protein with hCD26, which potently neutralizes MERS-CoV infectivity in vitro, were identified. Specifically, the two nAbs do not cross-compete for binding on MERS-RBD and therefore may potentially be used together as a cocktail to reduce the occurrence of viral escape mutants.
The in vivo efficacy of the two mAbs was also assessed in a transgenic mouse model generated by replacing the mouse CD26 coding sequence with the human counterpart (Pascal et al., 2015). For prophylaxis tests, low to high doses of mAbs were intraperitoneally injected 24 h before intranasal infection with 1 × 105 PFU of MERS-CoV. Both nAbs are able to significantly decrease MERS-CoV-specific RNA levels in the lungs at 4 dpi by over two logs at the 200 μg/mouse dose, compared to the isotype control antibody. More strikingly, the 20-μg dose of REGN3051 reduces virus levels as efficiently as the 200-μg dose, to near the level of detection in the assay. In addition, REGN3051 also exhibits potential for disease treatment. When the mAbs are administered 1 day after virus challenge, virus titers in the lungs of the mice are significant abated, with a greater than two log reduction at day 4 dpi, indicating pre- and post-exposure efficacy of the human mAbs in vivo.
4.1.3. Human antibodies isolated through screening of phage-displayed library
Aside from the nAbs generated from wild-type or transgenic mice, human nAbs were also isolated by screening the phage-displayed IgM Fab library (Ying et al., 2014). Using MERS-RBD as the antigen, Ying et al. identified a panel of nAbs against MERS-CoV, amongst which m336 showed the highest affinity to the antigen and neutralized both pseudotyped and live MERS-CoV (50% neutralization at 0.005 and 0.07 μg/ml m336, respectively). When m336 (1 mg/kg or 10 mg/kg) were injected intravenously into New Zealand white rabbits followed by intranasal infection with 1 × 105 TCID50 of MERS-CoV, complete protection was observed. In contrast, rabbits injected with control antibodies developed pulmonary disease characterized by mild perivascular and peribronchiolar inflammation, with viral titers peaking at about 3 dpi. Viral load was significantly reduced in rabbits administered with m336 at both doses and through both routes (intravenous or intranasal). In addition, evidence of inflammation or virus antigen were not observed in rabbits administered with m336. Although m336 completely protected rabbit against MERS-CoV in a prophylactic setting, post-exposure treatment with this nAb did not lead to a significant reduction in viral RNA titers (Houser et al., 2016).
4.1.4. Neutralizing mechanism
Although the mechanism of nAbs can be suggested by competition binding with the host receptor to the MERS-RBD, direct evidence comes from the structures of nAbs binding to the ligand. Several complex structures of nAbs bound to the MERS-RBD have been solved, and recognized epitopes are well defined (Li et al., 2015, Wang et al., 2015, Ying et al., 2015, Yu et al., 2015). Overall, both the heavy-chain variable domain (VH) and light-chain variable domain (VL) of the four nAbs are involved in the interaction, albeit the VHs contribute 50–85% of the total binding surface. Reciprocally, the major elements in the MERS-RBD recognized by four nAbs overlap and are focused on the inter-loop (the η3/β8 loop) connecting the η3 310 helix and the β8 strand, which is also involved in hCD26 binding.
Further superimposition of the mAbs/MERS-RBD structure with a previously reported structure of the hCD26/MERS-RBD complex (PDB code: 4KR0) (Lu et al., 2013a) indicates that three mAbs (D12, 4C2, and MERS-27) and hCD26 converge almost perpendicularly to recognize largely different surface patches on the viral ligand. However, the m336 mAb uses similar angles of approach as the host receptor to target the MERS-RBD (Fig. 3 ). The close proximity leads to strong steric clashes in one hot spot on the ligand, which is responsible for interaction with N229-linked carbohydrate moiety in the receptor, indicating that the protein-carbohydrate interaction might be important for hCD26 binding and the viral entry of MERS-CoV. In support of this observation, all N229 residues of CD26s from susceptible species are predicted to be glycosylated (Lu et al., 2015). In addition, the binding affinity of mutated hCD26 to the MERS-RBD is reduced by approximately 10-fold compared to wild-type hCD26 (Yu et al., 2015).
Fig. 3.
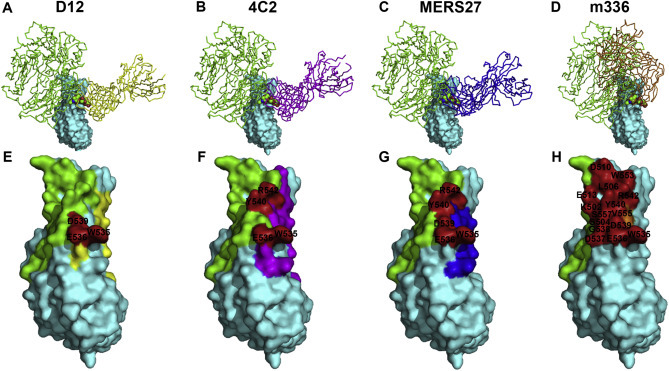
Comparison of the MERS-RBD binding sites among different antibodies. Top panels (A–D) show the superimposition of the structures between the indicated antibody (shown in ribbons) and the MERS-RBD (surface shown in cyan) with a previously reported structure of hCD26 (shown in green ribbons) bound to the MERS-RBD (Lu et al., 2013a). Antibodies D12 (Wang et al, 2015), 4C2 (Li et al., 2015), MERS27 (Yu et al. 2015), and m336 (Ying et al., 2015) are marked in yellow, magenta, blue, and orange, respectively. Bottom panels (E–H) indicate footprint overlaps in the MERS-RBD (surface shown in cyan) between the indicated antibody and hCD26. Residues recognized by the indicated antibody and hCD26 are represented in the same manner as the top panels. The overlapped interface residues are highlighted in red, and the amino acid identities/positions are labeled.
Footprints of four mAbs and hCD26 in the MERS-RBD were also compared by characterizing the interface residues located within 4.5 Å distance from the bound antibody or receptor. Though recognizing different surface patches in the viral ligand, two residues, W535 and E536, overlap in the five footprints. Hence, the four mAbs may interfere with the MERS-RBD/hCD26 interaction by both presenting strong steric hindrance and competing for the hCD26-interface residues, thereby neutralizing MERS-CoV infection.
4.2. Peptide inhibitors targeting the HRs of S2
Based on the fusion mechanism of Class I fusion proteins, soluble HR2 peptides display strong effective fusion-inhibiting potential. T20, derived from the HIV-1 gp41 HR2 region, was approved by the U.S. Food and Drug Administration as the first HIV fusion/entry inhibitor for treatment of HIV infection two decades ago (Jiang et al., 1993). Since then, many HR2-based viral fusion inhibitors against enveloped viruses with Class I membrane fusion proteins have been developed (Blacklow et al., 1995, Lombardi et al., 1996, Wang et al., 2003, Watanabe et al., 2000, Yu et al., 2002).
Following the same notion, HR2-based peptides that could potently inhibit MERS-CoV fusion and entry have been designed. Initially, two peptides based on HR2 were synthesized, and their inhibitory effects were tested in the pseudotyped virus system. Peptide P1, corresponding to the full-length HR2, is a potent inhibitor against viral infection, but a truncated peptide P2 is not (Gao et al., 2013). Similar correlations between the length of HR2 peptides and antiviral activity are observed in other HR2 peptides of Class I enveloped viruses (Jiang and Lin, 1995). The EC50 for inhibition of MERS-CoV infection by P1 is calculated to be ∼3 μM. Lu et al. design a similar HR2-based peptide inhibitor (HR2P), with EC50 values ranging from 0.6 to 13.9 μM, depending on the cell line used for the assay. By introducing hydrophilic residues to increase salt-bridge interactions, one HR2P mutant (HR2P-M2) shows significant improvements in stability, solubility, and antiviral activity. The inhibitory activity against MERS-S-mediated cell-cell fusion is increased by approximately 69% (Gao et al., 2013, Lu et al., 2014). Further in vivo studies demonstrate that intranasally administered HR2P-M2 effectively protect hCD26 transgenic mice from MERS-CoV infection, with a >1000-fold reduction of viral titers in the lungs, and protection is enhanced by combining HR2P-M2 with interferon β (Channappanavar et al., 2015).
4.3. Small molecules targeting S
By using a pseudovirus bearing the full-length S protein of MERS-CoV in inhibition assays, Zhao et al. assess several HIV entry inhibitors for their activity on MERS-CoV entry, among which ADS-J1 and 3-hydroxyphthalic anhydride-modified human serum albumin (HP-HSA) show antiviral activity (Zhao et al., 2013). Both compounds carry net negative charges and are predicted to interact with the positively charged residues in the S protein on the MERS-CoV virion, similar to those that inhibit HIV, SARS-CoV, and HPV entry (Chu et al., 2008, Debnath et al., 1999, Li et al., 2013, Lu et al., 2013b, Wang et al., 2009). The compounds also have a sub-micromolar IC50 and relative low cytotoxicity to the cells, but their efficacy and cytotoxicity need to be further evaluated in vivo.
5. Animal models for MERS-CoV to test the efficacy of vaccines and therapeutics
Development of suitable animal models for MERS-CoV is critical for understanding pathogenesis and for preclinical testing of vaccines and therapeutics. Unfortunately, commonly used laboratory animal species such as hamsters, mice, and ferrets are not susceptible to MERS-CoV (van Doremalen and Munster, 2015) because of the variation in CD26 among species. NHPs were initially used as animal models to test the efficacy of drugs, and subsequently, transgenic mice with hCD26 have been generated, which is summarized below.
5.1. Mice expressing hCD26
Although mice are not susceptible to MERS-CoV infection, three mouse models have been developed. In the first, a modified adenovirus expressing hCD26 is administrated intranasally to mice, leading to human receptor expression in all cells of the lung, including those negatively expressing mouse CD26 (Zhao et al., 2014). After infection with MERS-CoV, the virus can be detected in lungs but is cleared by days 6–8 in young mice and days 10–14 in old mice. Moderate weight loss is only observed in old mice. Further, virus clearance is delayed in several strains of immunocompromised mice compared to wild type mice. Infected mice develop a pneumonia characterized by extensive inflammatory cell infiltration, which may recapitulate the respiratory disease observed in mild or moderate human cases but not the fatal cases nor the occasionally occurring kidney disease. Accordingly, this model can be difficult as an evaluative model for therapeutics.
In the second model, transgenic mice globally expressing hCD26 were generated by microinjection by two independent groups (Agrawal et al., 2015, Zhao et al., 2015). When challenged with an overwhelming dose of MERS-CoV, the transgenic mice generated by Agrawal et al. develop severe pneumonia, leading to death in 4–6 days. Viral RNAs are detected in many organs, including the heart, spleen, and intestine, but not in the liver or kidney. In addition, infectious virus can be isolated from the lungs and brain (Agrawal et al., 2015). However, when infected with lower doses of virus, this mouse model exhibits early and persistent lung infection and delayed occurrence of brain infection. Persistent inflammatory infiltrates are seen in the lungs and brain, as well as focal infiltrates in the liver, but pathology was not seen in other tissues. The efficiency of the MERS-RBD-based subunit vaccine and peptide inhibitor HR2P-M6 was proven using this mouse model (Tao et al., 2015). Similarly, transgenic mice developed by Zhao et al. also exhibit high viral burdens and viral-positive neurons after infection with MERS-CoV. Systemic inflammation with mild-to-severe pneumonia accompanied by liver, kidney, and spleen injury are also observed (Zhao et al., 2015).
In the third model, the mouse CD26 gene was replaced with the human ortholog, which is expressed under the control of the mouse regulatory elements (Pascal et al., 2015). Unlike the transgenic mice expressing hCD26 in the whole body generated by Agrawal et al., the resulting mice designed by Pascal et al. preserve the proper expression regulation and protein tissue distribution of hCD26. When infected with MERS-CoV, transgenic mice resemble human patients in that robust virus replication in the lungs is evident, and interstitial pneumonia and significant lung disease are observed, indicating the potency of this mouse model to study both the host response to MERS-CoV and for the efficient testing of vaccines and drugs. By utilizing this model, two nAbs showed exceptional potency to inhibit pre- and post-exposed MERS-CoV infection.
5.2. NHPs
Two species of NHPs have been developed as MERS-CoV infection models: the rhesus macaque and common marmoset. MERS-CoV-infected rhesus macaques develop mild-to-moderate clinical signs within 24 h, including elevated body temperature, reduced appetite, increased respiratory rate, cough, pilo-erection, and hunched posture. Clinical signs are transient and last for a few days. Radiographic changes show varying degrees of localized infiltration and interstitial markings, but none of the animals reach a clinical score requiring euthanasia (de Wit et al., 2013, Munster et al., 2013). Similar symptoms in infected rhesus macaques have also been observed by another independent group (Yao et al., 2014). Using this animal model, Falzarano et al. evaluate the effectiveness of combination treatment with IFN-α2b and ribavirin, which are used to treat SARS patients (Stockman et al., 2006), and demonstrate anti-MERS-CoV effects in vitro (Falzarano et al., 2013a). Interestingly, the regimen reduces virus replication, moderates the host response, and improves clinical outcomes for infected animals (Falzarano et al., 2013b). This treatment strategy was further used for treatment of MERS-CoV-positive patients, but patients with multiple comorbidities who are diagnosed late in the course of their illness may unfortunately not benefit from this strategy (Al-Tawfiq et al., 2014).
The common marmoset model displays more severe clinical disease than rhesus macaques in that initial viral loads in the lungs are up to 1000 times higher than those in the rhesus macaque lungs, and several marmosets developed severe disease and had to be euthanized. Moreover, viral replication occurs both in the lower respiratory tract, which is also seen in rhesus macaque, and is detected in the blood and in nearly all tested tissues. This is associated with liver and/or kidney failure, as indicated by changes in blood chemistry. Taken together, these results suggest a more systemic dissemination in all infected common marmosets (Falzarano et al., 2014). Therefore, rhesus macaque appears to model the mild-to-moderate transient MERS cases, whereas the marmoset serves as a severe disease model. Both models provide important insights into the MERS-CoV infection mechanism and host immunity and are critical for antiviral therapeutics and vaccines evaluation.
6. Concluding remarks
In conclusion, in spite of the efforts of many research groups, there are yet no licensed vaccines or therapeutics against MERS-CoV infection. Due to its susceptibility in the virus life cycle, vaccines targeting either full length or truncated S protein have been developed, among which S DNA and S DNA/S1 protein candidates show the most advanced progress to protect immunized mice and NHPs from virus challenge and merit further development as candidate vaccines against MERS-CoV for potential human application. It is worth noting that aside from direct human protection against virus infection, an effective dromedary vaccine is also needed to stop ongoing camel-to-human transmission. Both MVA-S and S DNA evoke nAbs in dromedaries. Regardless, although MERS-CoV infected dromedaries may show no severe clinical manifestations, they may carry MERS-CoV viral RNA in their nasal excretions even in the presence of specific antibodies (Alagaili et al., 2014, Hemida et al., 2014, Khalafalla et al., 2015). The MVA-MERS-S vaccines developed by Haagmans et al. evoke a local mucosal immune response in the nostrils and thus reduce virus excretion after MERS-CoV infection in dromedaries, which may therefore be the most promising candidate vaccine for dromedary application.
Therapeutics targeting S have also been developed. nAbs inhibit viral entry mainly through blocking virus attachment. Among the identified nAbs, two display efficacy in hCD26-expressing mice, exhibit protection against pre- and post-challenge infection in mice, and thus deserve further evaluation in NHPs models. Although escape mutations might occur under the selection of nAbs, usage of nAb cocktails that target different epitopes may alleviate this problem. Moreover, a peptide inhibitor that was designed based on the fusion core structure of S has fortunately shown benefit during post-challenge infection, and its protection is enhanced by combining with interferon β as assayed in Ad5-hCD26-tranduced mice (Channappanavar et al., 2015), which also merit further evaluation in NHPs. Although some small molecules have shown anti-MERS-CoV potential in vitro, their in vivo efficacy remains unclear.
To evaluate the efficacy of vaccines and therapeutics, appropriate animal models are needed. Among the developed animal models, Ad5-hCD26-transduced mice are the most widely used due to their low cost and ease of use. For further evaluation, the common marmoset appears to represent the most appropriate NHP model, as they recapitulate the severe manifestations of MERS patients.
Taken together, the current advancements in the development of MERS-CoV vaccines and therapeutics targeting S have been discussed in this review and have been shown to have important implications for tackling future CoV outbreaks.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (NSFC, Grant No. C180501 and 81461168030) and China Ministry of Science and Technology National 973 Project (Grant No. 2015CB910500). G.W. is the recipient of a Banting Postdoctoral Fellowship from the Canadian Institutes of Health Research (CIHR) and the President’s International Fellowship Initiative from the Chinese Academy of Sciences (CAS).
Contributor Information
Qihui Wang, Email: wangqihui@im.ac.cn.
George F. Gao, Email: gaof@im.ac.cn.
References
- Adedeji A.O., Sarafianos S.G. Antiviral drugs specific for coronaviruses in preclinical development. Curr. Opin. Virol. 2014;8:45–53. doi: 10.1016/j.coviro.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal A.S., Garron T., Tao X.R., Peng B.H., Wakamiya M., Chan T.S., Couch R.B., Tseng C.T.K. Generation of a transgenic mouse model of Middle East respiratory syndrome coronavirus infection and disease. J. Virol. 2015;89:3659–3670. doi: 10.1128/JVI.03427-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Tawfiq J.A., Momattin H., Dib J., Memish Z.A. Ribavirin and interferon therapy in patients infected with the Middle East respiratory syndrome coronavirus: an observational study. Int. J. Infect. Dis. 2014;20:42–46. doi: 10.1016/j.ijid.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alagaili A.N., Briese T., Mishra N., Kapoor V., Sameroff S.C., Burbelo P.D., de Wit E., Munster V.J., Hensley L.E., Zalmout I.S., Kapoor A., Epstein J.H., Karesh W.B., Daszak P., Mohammed O.B., Lipkin W.I. Middle East respiratory syndrome coronavirus infection in dromedary camels in Saudi Arabia. mBio. 2014;5 doi: 10.1128/mBio.00884-14. e00884-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida J.D., Tyrrell D.A. The morphology of three previously uncharacterized human respiratory viruses that grow in organ culture. J. Gen. Virol. 1967;1:175–178. doi: 10.1099/0022-1317-1-2-175. [DOI] [PubMed] [Google Scholar]
- Azhar E.I., El-Kafrawy S.A., Farraj S.A., Hassan A.M., Al-Saeed M.S., Hashem A.M., Madani T.A. Evidence for camel-to-human transmission of MERS coronavirus. N. Engl. J. Med. 2014;370:2499–2505. doi: 10.1056/NEJMoa1401505. [DOI] [PubMed] [Google Scholar]
- Baez-Santos Y.M., Mielech A.M., Deng X.F., Baker S., Mesecar A.D. Catalytic function and substrate specificity of the papain-like protease domain of nsp3 from the Middle East respiratory syndrome coronavirus. J. Virol. 2014;88:12511–12527. doi: 10.1128/JVI.01294-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barouch D.H., Yang Z.Y., Kong W.P., Korioth-Schmitz B., Sumida S.M., Truitt D.M., Kishko M.G., Arthur J.C., Miura A., Mascola J.R., Letvin N.L., Nabel G.J. A human T-cell leukemia virus type 1 regulatory element enhances the immunogenicity of human immunodeficiency virus type 1 DNA vaccines in mice and nonhuman primates. J. Virol. 2005;79:8828–8834. doi: 10.1128/JVI.79.14.8828-8834.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belouzard S., Millet J.K., Licitra B.N., Whittaker G.R. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4:1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermingham A., Chand M.A., Brown C.S., Aarons E., Tong C., Langrish C., Hoschler K., Brown K., Galiano M., Myers R., Pebody R.G., Green H.K., Boddington N.L., Gopal R., Price N., Newsholme W., Drosten C., Fouchier R.A., Zambon M. Severe respiratory illness caused by a novel coronavirus, in a patient transferred to the United Kingdom from the Middle East, September 2012. Euro. Surveill. 2012;17:20290–20294. [PubMed] [Google Scholar]
- Blacklow S.C., Lu M., Kim P.S. A trimeric subdomain of the simian immunodeficiency virus envelope glycoprotein. Biochemistry. 1995;34:14955–14962. doi: 10.1021/bi00046a001. [DOI] [PubMed] [Google Scholar]
- Bossart K.N., Geisbert T.W., Feldmann H., Zhu Z., Feldmann F., Geisbert J.B., Yan L., Feng Y.R., Brining D., Scott D., Wang Y., Dimitrov A.S., Callison J., Chan Y.P., Hickey A.C., Dimitrov D.S., Broder C.C., Rockx B. A neutralizing human monoclonal antibody protects african green monkeys from hendra virus challenge. Sci. Transl. Med. 2011;3:105ra103. doi: 10.1126/scitranslmed.3002901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossart K.N., Zhu Z., Middleton D., Klippel J., Crameri G., Bingham J., McEachern J.A., Green D., Hancock T.J., Chan Y.P., Hickey A.C., Dimitrov D.S., Wang L.F., Broder C.C. A neutralizing human monoclonal antibody protects against lethal disease in a new ferret model of acute nipah virus infection. PLoS Pathog. 2009;5:e1000642. doi: 10.1371/journal.ppat.1000642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruggemann M., Osborn M.J., Ma B., Hayre J., Avis S., Lundstrom B., Buelow R. Human antibody production in transgenic animals. Arch. Immunol. Ther. Exp. Warsz. 2015;63:101–108. doi: 10.1007/s00005-014-0322-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmichael L.E. Canine viral vaccines at a turning point - a personal perspective. Adv. Veter Med. Ap. 1999;41:289–307. doi: 10.1016/S0065-3519(99)80022-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter P.J. Potent antibody therapeutics by design. Nat. Rev. Immunol. 2006;6:343–357. doi: 10.1038/nri1837. [DOI] [PubMed] [Google Scholar]
- Catanzaro A.T., Roederer M., Koup R.A., Bailer R.T., Enama M.E., Nason M.C., Martin J.E., Rucker S., Andrews C.A., Gomez P.L., Mascola J.R., Nabel G.J., Graham B.S., Team V.R.C.S. Phase I clinical evaluation of a six-plasmid multiclade HIV-1 DNA candidate vaccine. Vaccine. 2007;25:4085–4092. doi: 10.1016/j.vaccine.2007.02.050. [DOI] [PubMed] [Google Scholar]
- Cavanagh D. Coronavirus avian infectious bronchitis virus. Vet. Res. 2007;38:281–297. doi: 10.1051/vetres:2006055. [DOI] [PubMed] [Google Scholar]
- Chan J.F., Chan K.H., Kao R.Y., To K.K., Zheng B.J., Li C.P., Li P.T., Dai J., Mok F.K., Chen H., Hayden F.G., Yuen K.Y. Broad-spectrum antivirals for the emerging Middle East respiratory syndrome coronavirus. J. Infect. 2013;67:606–616. doi: 10.1016/j.jinf.2013.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F., Yao Y., Yeung M.L., Deng W., Bao L., Jia L., Li F., Xiao C., Gao H., Yu P., Cai J.P., Chu H., Zhou J., Chen H., Qin C., Yuen K.Y. Treatment with lopinavir/ritonavir or interferon-beta1b improves outcome of MERS-CoV infection in a nonhuman primate model of common marmoset. J. Infect. Dis. 2015;212:1904–1913. doi: 10.1093/infdis/jiv392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channappanavar R., Lu L., Xia S., Du L., Meyerholz D.K., Perlman S., Jiang S. Protective effect of intranasal regimens containing peptidic Middle East respiratory syndrome coronavirus fusion inhibitor against MERS-CoV infection. J. Infect. Dis. 2015;212:1894–1903. doi: 10.1093/infdis/jiv325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng K.W., Cheng S.C., Chen W.Y., Lin M.H., Chuang S.J., Cheng I.H., Sun C.Y., Chou C.Y. Thiopurine analogs and mycophenolic acid synergistically inhibit the papain-like protease of Middle East respiratory syndrome coronavirus. Antivir. Res. 2015;115:9–16. doi: 10.1016/j.antiviral.2014.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu L.H., Chan S.H., Tsai S.N., Wang Y., Cheng C.H., Wong K.B., Waye M.M., Ngai S.M. Fusion core structure of the severe acute respiratory syndrome coronavirus (SARS-CoV): in search of potent SARS-CoV entry inhibitors. J. Cell Biochem. 2008;104:2335–2347. doi: 10.1002/jcb.21790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotten M., Lam T.T., Watson S.J., Palser A.L., Petrova V., Grant P., Pybus O.G., Rambaut A., Guan Y., Pillay D., Kellam P., Nastouli E. Full-genome deep sequencing and phylogenetic analysis of novel human betacoronavirus. Emerg. Infect. Dis. 2013;19:736–742B. doi: 10.3201/eid1905.130057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gregorio E., Caproni E., Ulmer J.B. Vaccine adjuvants: mode of action. Front. Immunol. 2013;4:214–219. doi: 10.3389/fimmu.2013.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot R.J., Baker S.C., Baric R.S., Brown C.S., Drosten C., Enjuanes L., Fouchier R.A.M., Galiano M., Gorbalenya A.E., Memish Z.A., Perlman S., Poon L.L.M., Snijder E.J., Stephens G.M., Woo P.C.Y., Zaki A.M., Zambon M., Ziebuhr J. Middle East respiratory syndrome coronavirus (MERS-CoV): announcement of the Coronavirus Study Group. J. Virol. 2013;87:7790–7792. doi: 10.1128/JVI.01244-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wilde A.H., Jochmans D., Posthuma C.C., Zevenhoven-Dobbe J.C., van Nieuwkoop S., Bestebroer T.M., van den Hoogen B.G., Neyts J., Snijder E.J. Screening of an FDA-approved compound library identifies four small-molecule inhibitors of Middle East respiratory syndrome coronavirus replication in cell culture. Antimicrob. Agents Ch. 2014;58:4875–4884. doi: 10.1128/AAC.03011-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E., Rasmussen A.L., Falzarano D., Bushmaker T., Feldmann F., Brining D.L., Fischer E.R., Martellaro C., Okumura A., Chang J., Scott D., Benecke A.G., Katze M.G., Feldmann H., Munster V.J. Middle East respiratory syndrome coronavirus (MERS-CoV) causes transient lower respiratory tract infection in rhesus macaques. Proc. Natl. Acad. Sci. U. S. A. 2013;110:16598–16603. doi: 10.1073/pnas.1310744110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debnath A.K., Radigan L., Jiang S. Structure-based identification of small molecule antiviral compounds targeted to the gp41 core structure of the human immunodeficiency virus type 1. J. Med. Chem. 1999;42:3203–3209. doi: 10.1021/jm990154t. [DOI] [PubMed] [Google Scholar]
- DiGiandomenico A., Sellman B.R. Antibacterial monoclonal antibodies: the next generation? Curr. Opin. Microbiol. 2015;27:78–85. doi: 10.1016/j.mib.2015.07.014. [DOI] [PubMed] [Google Scholar]
- Dimitrov D.S. Therapeutic proteins. Methods Mol. Biol. 2012;899:1–26. doi: 10.1007/978-1-61779-921-1_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., Kou Z., Ma C., Tao X., Wang L., Zhao G., Chen Y., Yu F., Tseng C.T., Zhou Y., Jiang S. A truncated receptor-binding domain of MERS-CoV spike protein potently inhibits MERS-CoV infection and induces strong neutralizing antibody responses: implication for developing therapeutics and vaccines. PLoS One. 2013;8:e81587. doi: 10.1371/journal.pone.0081587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L.Y., He Y.X., Zhou Y.S., Liu S.W., Zheng B.J., Jiang S.B. The spike protein of SARS-CoV - a target for vaccine and therapeutic development. Nat. Rev. Microbiol. 2009;7:226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L.Y., Zhao G.Y., Kou Z.H., Ma C.Q., Sun S.H., Poon V.K.M., Lu L., Wang L.L., Debnath A.K., Zheng B.J., Zhou Y.S., Jiang S.B. Identification of a receptor-binding domain in the S protein of the novel human coronavirus Middle East respiratory syndrome coronavirus as an essential target for vaccine development. J. Virol. 2013;87:9939–9942. doi: 10.1128/JVI.01048-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earp L.J., Delos S.E., Park H.E., White J.M. The many mechanisms of viral membrane fusion proteins. Curr. Top. Microbiol. Immunol. 2005;285:25–66. doi: 10.1007/3-540-26764-6_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Economides A.N., Frendewey D., Yang P., Dominguez M.G., Dore A.T., Lobov I.B., Persaud T., Rojas J., McClain J., Lengyel P., Droguett G., Chernomorsky R., Stevens S., Auerbach W., Dechiara T.M., Pouyemirou W., Cruz J.M., Jr., Feeley K., Mellis I.A., Yasenchack J., Hatsell S.J., Xie L., Latres E., Huang L., Zhang Y., Pefanis E., Skokos D., Deckelbaum R.A., Croll S.D., Davis S., Valenzuela D.M., Gale N.W., Murphy A.J., Yancopoulos G.D. Conditionals by inversion provide a universal method for the generation of conditional alleles. Proc. Natl. Acad. Sci. U. S. A. 2013;110:E3179–E3188. doi: 10.1073/pnas.1217812110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falsey A.R., McCann R.M., Hall W.J., Criddle M.M., Formica M.A., Wycoff D., Kolassa J.E. The 'common cold' in frail older persons: impact of rhinovirus and coronavirus in a senior daycare center. J. Am. Geriatr. Soc. 1997;45:706–711. doi: 10.1111/j.1532-5415.1997.tb01474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falzarano D., de Wit E., Feldmann F., Rasmussen A.L., Okumura A., Peng X., Thomas M.J., van Doremalen N., Haddock E., Nagy L., LaCasse R., Liu T., Zhu J., McLellan J.S., Scott D.P., Katze M.G., Feldmann H., Munster V.J. Infection with MERS-CoV causes lethal pneumonia in the common marmoset. PLoS Pathog. 2014;10:e1004250. doi: 10.1371/journal.ppat.1004250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falzarano D., de Wit E., Martellaro C., Callison J., Munster V.J., Feldmann H. Inhibition of novel beta coronavirus replication by a combination of interferon-alpha2b and ribavirin. Sci. Rep. 2013;3:1686–1691. doi: 10.1038/srep01686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falzarano D., de Wit E., Rasmussen A.L., Feldmann F., Okumura A., Scott D.P., Brining D., Bushmaker T., Martellaro C., Baseler L., Benecke A.G., Katze M.G., Munster V.J., Feldmann H. Treatment with interferon-[alpha]2b and ribavirin improves outcome in MERS-CoV-infected rhesus macaques. Nat. Med. 2013;19:1313–1317. doi: 10.1038/nm.3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraro B., Morrow M.P., Hutnick N.A., Shin T.H., Lucke C.E., Weiner D.B. Clinical applications of DNA vaccines: current progress. Clin. Infect. Dis. 2011;53:296–302. doi: 10.1093/cid/cir334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G.F. Peptide inhibitors targeting virus-cell fusion in class I enveloped viruses. In: Torrence P.F., editor. Combating the Threat of Pandemic Influenza: Drug Discovery Approaches. John Wiley & Sons Inc; Hoboken, NJ: 2007. pp. 226–246. [Google Scholar]
- Gao J., Lu G., Qi J., Li Y., Wu Y., Deng Y., Geng H., Li H., Wang Q., Xiao H., Tan W., Yan J., Gao G.F. Structure of the fusion core and inhibition of fusion by a heptad repeat peptide derived from the S protein of Middle East respiratory syndrome coronavirus. J. Virol. 2013;87:13134–13140. doi: 10.1128/JVI.02433-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert S.C. Clinical development of modified vaccinia virus Ankara vaccines. Vaccine. 2013;31:4241–4246. doi: 10.1016/j.vaccine.2013.03.020. [DOI] [PubMed] [Google Scholar]
- Haagmans B.L., van den Brand J.M.A., Raj V.S., Volz A., Wohlsein P., Smits S.L., Schipper D., Bestebroer T.M., Okba N., Fux R., Bensaid A., Solanes Foz D., Kuiken T., Baumgärtner W., Segalés J., Sutter G., Osterhaus A.D.M.E. An orthopoxvirus-based vaccine reduces virus excretion after MERS-CoV infection in dromedary camels. Science. 2016;351:77–81. doi: 10.1126/science.aad1283. [DOI] [PubMed] [Google Scholar]
- Hamre D., Procknow J.J. A new virus isolated from the human respiratory tract. Proc. Soc. Exp. Biol. Med. 1966;121:190–193. doi: 10.3181/00379727-121-30734. [DOI] [PubMed] [Google Scholar]
- Harrison S.C. Viral membrane fusion. Virology. 2015;479–480:498–507. doi: 10.1016/j.virol.2015.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan M.M., Akter R., Ullah M.S., Abedin M.J., Ullah G.M., Hossain M.Z. A computational approach for predicting role of human microRNAs in MERS-CoV genome. Adv. Bioinforma. 2014;2014:967946–967953. doi: 10.1155/2014/967946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heikkinen T., Jarvinen A. The common cold. Lancet. 2003;361:51–59. doi: 10.1016/S0140-6736(03)12162-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemida M.G., Chu D.K., Poon L.L., Perera R.A., Alhammadi M.A., Ng H.Y., Siu L.Y., Guan Y., Alnaeem A., Peiris M. MERS coronavirus in dromedary camel herd, Saudi Arabia. Emerg. Infect. Dis. 2014;20:1231–1234. doi: 10.3201/eid2007.140571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendley J.O., Fishburne H.B., Gwaltney J.M., Jr. Coronavirus infections in working adults. Eight-year study with 229 E and OC 43. Am. Rev. Respir. Dis. 1972;105:805–811. doi: 10.1164/arrd.1972.105.5.805. [DOI] [PubMed] [Google Scholar]
- Hick P.M., Read A.J., Lugton I., Busfield F., Dawood K.E., Gabor L., Hornitzky M., Kirkland P.D. Coronavirus infection in intensively managed cattle with respiratory disease. Aust. Vet. J. 2012;90:381–386. doi: 10.1111/j.1751-0813.2012.00978.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houser K.V., Gretebeck L., Ying T., Wang Y., Vogel L., Lamirande E.W., Bock K.W., Moore I.N., Dimitrov D.S., Subbarao K. Prophylaxis With a Middle East Respiratory Syndrome Coronavirus (MERS-CoV) -Specific Human Monoclonal Antibody Protects Rabbits From MERS-CoV Infection. J. Infect. Dis. 2016;213:1557–1561. doi: 10.1093/infdis/jiw080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L.W., Wang N.S., Zuo T., Shi X.L., Poon K.M.V., Wu Y.K., Gao F., Li D.Y., Wang R.K., Guo J.Y., Fu L.L., Yuen K.Y., Zheng B.J., Wang X.Q., Zhang L.Q. Potent neutralization of MERS-CoV by human neutralizing monoclonal antibodies to the viral spike glycoprotein. Sci. Transl. Med. 2014;6 doi: 10.1126/scitranslmed.3008140. 234ra59. [DOI] [PubMed] [Google Scholar]
- Jiang S., Lin K. Effect of amino acid replacements, additions and deletions on the antiviral activity of a peptide derived from the HIV-1 GP41 sequence. Pept. Res. 1995;8:345–348. [PubMed] [Google Scholar]
- Jiang S., Lin K., Strick N., Neurath A.R. HIV-1 inhibition by a peptide. Nature. 1993;365:113. doi: 10.1038/365113a0. [DOI] [PubMed] [Google Scholar]
- Keum Y.S., Jeong Y.J. Development of chemical inhibitors of the SARS coronavirus: viral helicase as a potential target. Biochem. Pharmacol. 2012;84:1351–1358. doi: 10.1016/j.bcp.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalafalla A.I., Lu X., Al-Mubarak A.I., Dalab A.H., Al-Busadah K.A., Erdman D.D. MERS-CoV in upper respiratory tract and lungs of dromedary camels, Saudi Arabia, 2013-2014. Emerg. Infect. Dis. 2015;21:1153–1158. doi: 10.3201/eid2107.150070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kielian M., Rey F.A. Virus membrane-fusion proteins: more than one way to make a hairpin. Nat. Rev. Microbiol. 2006;4:67–76. doi: 10.1038/nrmicro1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilianski A., Mielech A.M., Deng X.F., Baker S.C. Assessing activity and inhibition of Middle East respiratory syndrome coronavirus papain-like and 3C-like proteases using luciferase-based biosensors. J. Virol. 2013;87:11955–11962. doi: 10.1128/JVI.02105-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchdoerfer R.N., Cottrell C.A., Wang N., Pallesen J., Yassine H.M., Turner H.L., Corbett K.S., Graham B.S., McLellan J.S., Ward A.B. Pre-fusion structure of a human coronavirus spike protein. Nature. 2016;531:118–121. doi: 10.1038/nature17200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V., Jung Y.S., Liang P.H. Anti-SARS coronavirus agents: a patent review (2008-present) Expert Opin. Ther. Pat. 2013;23:1337–1348. doi: 10.1517/13543776.2013.823159. [DOI] [PubMed] [Google Scholar]
- Li F., Li W., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- Li L., Qiu J., Lu L., An S., Qiao P., Jiang S., Liu S. 3-Hydroxyphthalic anhydride-modified human serum albumin as a microbicide candidate inhibits HIV infection by blocking viral entry. J. Antimicrob. Chemother. 2013;68:573–576. doi: 10.1093/jac/dks458. [DOI] [PubMed] [Google Scholar]
- Li W., Joshi M.D., Singhania S., Ramsey K.H., Murthy A.K. Peptide vaccine: progress and challenges. Vaccines (Basel) 2014;2:515–536. doi: 10.3390/vaccines2030515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Wan Y.H., Liu P.P., Zhao J.C., Lu G.W., Qi J.X., Wang Q.H., Lu X.C., Wu Y., Liu W.J., Zhang B.C., Yuen K.Y., Perlman S., Gao G.F., Yan J.H. A humanized neutralizing antibody against MERS-CoV targeting the receptor-binding domain of the spike protein. Cell Res. 2015;25:1237–1249. doi: 10.1038/cr.2015.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang P.H. Characterization and inhibition of SARS-coronavirus main protease. Curr. Top. Med. Chem. 2006;6:361–376. doi: 10.2174/156802606776287090. [DOI] [PubMed] [Google Scholar]
- Lingwood D., McTamney P.M., Yassine H.M., Whittle J.R., Guo X., Boyington J.C., Wei C.J., Nabel G.J. Structural and genetic basis for development of broadly neutralizing influenza antibodies. Nature. 2012;489:566–570. doi: 10.1038/nature11371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi S., Massi C., Indino E., La Rosa C., Mazzetti P., Falcone M.L., Rovero P., Fissi A., Pieroni O., Bandecchi P., Esposito F., Tozzini F., Bendinelli M., Garzelli C. Inhibition of feline immunodeficiency virus infection in vitro by envelope glycoprotein synthetic peptides. Virology. 1996;220:274–284. doi: 10.1006/viro.1996.0315. [DOI] [PubMed] [Google Scholar]
- Lu G., Hu Y., Wang Q., Qi J., Gao F., Li Y., Zhang Y., Zhang W., Yuan Y., Bao J., Zhang B., Shi Y., Yan J., Gao G.F. Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26. Nature. 2013;500:227–231. doi: 10.1038/nature12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G., Wang Q., Gao G.F. Bat-to-human: spike features determining 'host jump' of coronaviruses SARS-CoV, MERS-CoV, and beyond. Trends Microbiol. 2015;23:468–478. doi: 10.1016/j.tim.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G.W., Qi J.X., Chen Z.J., Xu X., Gao F., Lin D.Z., Qian W.K., Liu H., Jiang H.L., Yan J.H., Gao G.F. Enterovirus 71 and coxsackievirus A16 3C proteases: binding to rupintrivir and their substrates and anti-hand, foot, and mouth disease virus drug design. J. Virol. 2011;85:10319–10331. doi: 10.1128/JVI.00787-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L., Liu Q., Zhu Y., Chan K.H., Qin L., Li Y., Wang Q., Chan J.F., Du L., Yu F., Ma C., Ye S., Yuen K.Y., Zhang R., Jiang S. Structure-based discovery of Middle East respiratory syndrome coronavirus fusion inhibitor. Nat. Commun. 2014;5:3067. doi: 10.1038/ncomms4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L., Yang X., Li Y., Jiang S. Chemically modified bovine beta-lactoglobulin inhibits human papillomavirus infection. Microbes Infect. 2013;15:506–510. doi: 10.1016/j.micinf.2013.03.003. [DOI] [PubMed] [Google Scholar]
- Lundstrom K. Alphavirus-based vaccines. Viruses. 2014;6:2392–2415. doi: 10.3390/v6062392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C.Q., Li Y., Wang L.L., Zhao G.Y., Tao X.R., Tseng C.T.K., Zhou Y.S., Du L.Y., Jiang S.B. Intranasal vaccination with recombinant receptor-binding domain of MERS-CoV spike protein induces much stronger local mucosal immune responses than subcutaneous immunization: implication for designing novel mucosal MERS vaccines. Vaccine. 2014;32:2100–2108. doi: 10.1016/j.vaccine.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C.Q., Wang L.L., Tao X.R., Zhang N.R., Yang Y., Tseng C.T.K., Li F., Zhou Y.S., Jiang S.B., Du L.Y. Searching for an ideal vaccine candidate among different MERS coronavirus receptor-binding fragments-The importance of immunofocusing in subunit vaccine design. Vaccine. 2014;32:6170–6176. doi: 10.1016/j.vaccine.2014.08.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascola J.R., Haynes B.F. HIV-1 neutralizing antibodies: understanding nature's pathways. Immunol. Rev. 2013;254:225–244. doi: 10.1111/imr.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh K., Chao R.K., Krause H.E., Wasil R., Mocega H.E., Mufson M.A. Coronavirus infection in acute lower respiratory tract disease of infants. J. Infect. Dis. 1974;130:502–507. doi: 10.1093/infdis/130.5.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh K., Dees J.H., Becker W.B., Kapikian A.Z., Chanock R.M. Recovery in tracheal organ cultures of novel viruses from patients with respiratory disease. Proc. Natl. Acad. Sci. U. S. A. 1967;57:933–940. doi: 10.1073/pnas.57.4.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melikyan G.B. Common principles and intermediates of viral protein-mediated fusion: the HIV-1 paradigm. Retrovirology. 2008;5:111. doi: 10.1186/1742-4690-5-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memish Z.A., Cotten M., Meyer B., Watson S.J., Alsahafi A.J., Al Rabeeah A.A., Corman V.M., Sieberg A., Makhdoom H.Q., Assiri A., Al Masri M., Aldabbagh S., Bosch B.J., Beer M., Muller M.A., Kellam P., Drosten C. Human infection with MERS coronavirus after exposure to infected camels, Saudi Arabia, 2013. Emerg. Infect. Dis. 2014;20:1012–1015. doi: 10.3201/eid2006.140402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet J.K., Whittaker G.R. Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc. Natl. Acad. Sci. U. S. A. 2014;111:15214–15219. doi: 10.1073/pnas.1407087111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet J.K., Whittaker G.R. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015;202:120–134. doi: 10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou H., Raj V.S., van Kuppeveld F.J.M., Rottier P.J.M., Haagmans B.L., Bosch B.J. The receptor binding domain of the new Middle East respiratory syndrome coronavirus maps to a 231-residue region in the spike protein that efficiently elicits neutralizing antibodies. J. Virol. 2013;87:9379–9383. doi: 10.1128/JVI.01277-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munster V.J., de Wit E., Feldmann H. Pneumonia from human coronavirus in a macaque model. N. Engl. J. Med. 2013;368:1560–1562. doi: 10.1056/NEJMc1215691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy A.J., Macdonald L.E., Stevens S., Karow M., Dore A.T., Pobursky K., Huang T.T., Poueymirou W.T., Esau L., Meola M., Mikulka W., Krueger P., Fairhurst J., Valenzuela D.M., Papadopoulos N., Yancopoulos G.D. Mice with megabase humanization of their immunoglobulin genes generate antibodies as efficiently as normal mice. Proc. Natl. Acad. Sci. U. S. A. 2014;111:5153–5158. doi: 10.1073/pnas.1324022111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthumani K., Falzarano D., Reuschel E.L., Tingey C., Flingai S., Villarreal D.O., Wise M., Patel A., Izmirly A., Aljuaid A., Seliga A.M., Soule G., Morrow M., Kraynyak K.A., Khan A.S., Scott D.P., Feldmann F., LaCasse R., Meade-White K., Okumura A., Ugen K.E., Sardesai N.Y., Kim J.J., Kobinger G., Feldmann H., Weiner D.B. A synthetic consensus anti–spike protein DNA vaccine induces protective immunity against Middle East respiratory syndrome coronavirus in nonhuman primates. Sci. Transl. Med. 2015;7 doi: 10.1126/scitranslmed.aac7462. 301ra132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan K., Huang C., Makino S. SARS coronavirus accessory proteins. Virus Res. 2008;133:113–121. doi: 10.1016/j.virusres.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naz R.K., Dabir P. Peptide vaccines against cancer, infectious diseases, and conception. Front. Biosci. 2007;12:1833–1844. doi: 10.2741/2191. [DOI] [PubMed] [Google Scholar]
- Niemeyer D., Zillinger T., Muth D., Zielecki F., Horvath G., Suliman T., Barchet W., Weber F., Drosten C., Muller M.A. Middle East respiratory syndrome coronavirus accessory protein 4a is a Type I interferon antagonist. J. Virol. 2013;87:12489–12495. doi: 10.1128/JVI.01845-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nokso-Koivisto J., Pitkaranta A., Blomqvist S., Kilpi T., Hovi T. Respiratory coronavirus infections in children younger than two years of age. Pediatr. Infect. Dis. J. 2000;19:164–166. doi: 10.1097/00006454-200002000-00016. [DOI] [PubMed] [Google Scholar]
- Nur S.M., Hasan M.A., Amin M.A., Hossain M., Sharmin T. Design of potential RNAi (miRNA and siRNA) molecules for Middle East respiratory syndrome coronavirus (MERS-CoV) gene silencing by computational method. Interdiscip. Sci. 2014;7:257–265. doi: 10.1007/s12539-015-0266-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen C.W. A review of feline infectious peritonitis virus: molecular biology, immunopathogenesis, clinical aspects, and vaccination. Vet. Microbiol. 1993;36:1–37. doi: 10.1016/0378-1135(93)90126-R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaneri A.B., Johnson R.F., Wada J., Bollinger L., Jahrling P.B., Kuhn J.H. Middle East respiratory syndrome: obstacles and prospects for vaccine development. Expert Rev. Vaccines. 2015;14:949–962. doi: 10.1586/14760584.2015.1036033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascal K.E., Coleman C.M., Mujica A.O., Kamat V., Badithe A., Fairhurst J., Hunt C., Strein J., Berrebi A., Sisk J.M., Matthews K.L., Babb R., Chen G., Lai K.M.V., Huang T.T., Olson W., Yancopoulos G.D., Stahl N., Frieman M.B., Kyratsous C.A. Pre- and postexposure efficacy of fully human antibodies against Spike protein in a novel humanized mouse model of MERS-CoV infection. Proc. Natl. Acad. Sci. U. S. A. 2015;112:8738–8743. doi: 10.1073/pnas.1510830112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pene F., Merlat A., Vabret A., Rozenberg F., Buzyn A., Dreyfus F., Cariou A., Freymuth F., Lebon P. Coronavirus 229E-related pneumonia in immunocompromised patients. Clin. Infect. Dis. 2003;37:929–932. doi: 10.1086/377612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podda A., Del Giudice G. MF59-adjuvanted vaccines: increased immunogenicity with an optimal safety profile. Expert Rev. vaccines. 2003;2:197–203. doi: 10.1586/14760584.2.2.197. [DOI] [PubMed] [Google Scholar]
- Qiu X., Wong G., Audet J., Bello A., Fernando L., Alimonti J.B., Fausther-Bovendo H., Wei H., Aviles J., Hiatt E., Johnson A., Morton J., Swope K., Bohorov O., Bohorova N., Goodman C., Kim D., Pauly M.H., Velasco J., Pettitt J., Olinger G.G., Whaley K., Xu B., Strong J.E., Zeitlin L., Kobinger G.P. Reversion of advanced Ebola virus disease in nonhuman primates with ZMapp. Nature. 2014;514:47–53. doi: 10.1038/nature13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj V.S., Mou H., Smits S.L., Dekkers D.H.W., Muller M.A., Dijkman R., Muth D., Demmers J.A.A., Zaki A., Fouchier R.A.M., Thiel V., Drosten C., Rottier P.J.M., Osterhaus A.D.M.E., Bosch B.J., Haagmans B.L. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramajayam R., Tan K.P., Liang P.H. Recent development of 3C and 3CL protease inhibitors for anti-coronavirus and anti-picornavirus drug discovery. Biochem. Soc. Trans. 2011;39:1371–1375. doi: 10.1042/BST0391371. [DOI] [PubMed] [Google Scholar]
- Redding L., Weiner D.B. DNA vaccines in veterinary use. Expert Rev. Vaccines. 2009;8:1251–1276. doi: 10.1586/erv.09.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saif L.J. Coronavirus immunogens. Vet. Microbiol. 1993;37:285–297. doi: 10.1016/0378-1135(93)90030-B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A., Wolff M.H., Weber O.F. Birkhäuser Boston, Inc; 2005. Coronaviruses with Special Emphasis on First Insights Concerning SARS. [Google Scholar]
- Shadman K.A., Wald E.R. A review of palivizumab and emerging therapies for respiratory syncytial virus. Expert Opin. Biol. Ther. 2011;11:1455–1467. doi: 10.1517/14712598.2011.608062. [DOI] [PubMed] [Google Scholar]
- Simmons G., Zmora P., Gierer S., Heurich A., Pöhlmann S. Proteolytic activation of the SARS-coronavirus spike protein: cutting enzymes at the cutting edge of antiviral research. Antivir. Res. 2013;100:605–614. doi: 10.1016/j.antiviral.2013.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slovin S.F., Kehoe M., Durso R., Fernandez C., Olson W., Gao J.P., Israel R., Scher H.I., Morris S. A phase I dose escalation trial of vaccine replicon particles (VRP) expressing prostate-specific membrane antigen (PSMA) in subjects with prostate cancer. Vaccine. 2013;31:943–949. doi: 10.1016/j.vaccine.2012.11.096. [DOI] [PubMed] [Google Scholar]
- Song F., Fux R., Provacia L.B., Volz A., Eickmann M., Becker S., Osterhaus A.D.M.E., Haagmans B.L., Sutter G. Middle East respiratory syndrome coronavirus spike protein delivered by modified vaccinia virus Ankara efficiently induces virus-neutralizing antibodies. J. Virol. 2013;87:11950–11954. doi: 10.1128/JVI.01672-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockman L.J., Bellamy R., Garner P. SARS: systematic review of treatment effects. PLoS Med. 2006;3:e343. doi: 10.1371/journal.pmed.0030343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S., Wong G., Liu Y., Gao G.F., Li S., Bi Y. MERS in South Korea and China: a potential outbreak threat? Lancet. 2015;385:2349–2350. doi: 10.1016/S0140-6736(15)60859-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X.C., Agnihothram S.S., Jiao Y.J., Stanhope J., Graham R.L., Peterson E.C., Avnir Y., Tallarico A.S., Sheehan J., Zhu Q., Baric R.S., Marasco W.A. Identification of human neutralizing antibodies against MERS-CoV and their role in virus adaptive evolution. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E2018–E2026. doi: 10.1073/pnas.1402074111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao X., Garron T., Agrawal A.S., Algaissi A., Peng B.H., Wakamiya M., Chan T.S., Lu L., Du L., Jiang S., Couch R.B., Tseng C.T. Characterization and demonstration of the value of a lethal mouse model of Middle East respiratory syndrome coronavirus infection and disease. J. Virol. 2015;90:57–67. doi: 10.1128/JVI.02009-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyrrell D.A., Bynoe M.L. Cultivation of a novel Type of common-cold virus in organ cultures. Br. Med. J. 1965;1:1467–1470. doi: 10.1136/bmj.1.5448.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ura T., Okuda K., Shimada M. Developments in viral vector-based vaccines. Vaccines (Basel) 2014;2:624–641. doi: 10.3390/vaccines2030624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vabret A., Mourez T., Gouarin S., Petitjean J., Freymuth F. An outbreak of coronavirus OC43 respiratory infection in Normandy, France. Clin. Infect. Dis. 2003;36:985–989. doi: 10.1086/374222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela D.M., Murphy A.J., Frendewey D., Gale N.W., Economides A.N., Auerbach W., Poueymirou W.T., Adams N.C., Rojas J., Yasenchak J., Chernomorsky R., Boucher M., Elsasser A.L., Esau L., Zheng J., Griffiths J.A., Wang X., Su H., Xue Y., Dominguez M.G., Noguera I., Torres R., Macdonald L.E., Stewart A.F., DeChiara T.M., Yancopoulos G.D. High-throughput engineering of the mouse genome coupled with high-resolution expression analysis. Nat. Biotechnol. 2003;21:652–659. doi: 10.1038/nbt822. [DOI] [PubMed] [Google Scholar]
- van Boheemen S., de Graaf M., Lauber C., Bestebroer T.M., Raj V.S., Zaki A.M., Osterhaus A.D., Haagmans B.L., Gorbalenya A.E., Snijder E.J., Fouchier R.A. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. mBio. 2012;3 doi: 10.1128/mBio.00473-12. e00473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoek L., Pyrc K., Jebbink M.F., Vermeulen-Oost W., Berkhout R.J., Wolthers K.C., Wertheim-van Dillen P.M., Kaandorp J., Spaargaren J., Berkhout B. Identification of a new human coronavirus. Nat. Med. 2004;10:368–373. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doremalen N., Munster V.J. Animal models of Middle East respiratory syndrome coronavirus infection. Antivir. Res. 2015;122:28–38. doi: 10.1016/j.antiviral.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volz A., Kupke A., Song F., Jany S., Fux R., Shams-Eldin H., Schmidt J., Becker C., Eickmann M., Becker S., Sutter G. Protective efficacy of recombinant modified vaccinia virus Ankara delivering Middle East respiratory syndrome coronavirus spike glycoprotein. J. Virol. 2015;89:8651–8656. doi: 10.1128/JVI.00614-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Tortorici M.A., Bosch B.J., Frenz B., Rottier P.J.M., DiMaio F., Rey F.A., Veesler D. Cryo-electron microscopy structure of a coronavirus spike glycoprotein trimer. Nature. 2016;531:114–117. doi: 10.1038/nature16988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E., Sun X., Qian Y., Zhao L., Tien P., Gao G.F. Both heptad repeats of human respiratory syncytial virus fusion protein are potent inhibitors of viral fusion. Biochem. Bioph Res. Co. 2003;302:469–475. doi: 10.1016/s0006-291x(03)00197-9. [DOI] [PubMed] [Google Scholar]
- Wang H., Qi Z., Guo A., Mao Q., Lu H., An X., Xia C., Li X., Debnath A.K., Wu S., Liu S., Jiang S. ADS-J1 inhibits human immunodeficiency virus type 1 entry by interacting with the gp41 pocket region and blocking fusion-active gp41 core formation. Antimicrob. Agents Ch. 2009;53:4987–4998. doi: 10.1128/AAC.00670-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Shi W., Joyce M.G., Modjarrad K., Zhang Y., Leung K., Lees C.R., Zhou T., Yassine H.M., Kanekiyo M., Yang Z.Y., Chen X., Becker M.M., Freeman M., Vogel L., Johnson J.C., Olinger G., Todd J.P., Bagci U., Solomon J., Mollura D.J., Hensley L., Jahrling P., Denison M.R., Rao S.S., Subbarao K., Kwong P.D., Mascola J.R., Kong W.P., Graham B.S. Evaluation of candidate vaccine approaches for MERS-CoV. Nat. Commun. 2015;6:7712. doi: 10.1038/ncomms8712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N., Shi X., Jiang L., Zhang S., Wang D., Tong P., Guo D., Fu L., Cui Y., Liu X., Arledge K.C., Chen Y.H., Zhang L., Wang X. Structure of MERS-CoV spike receptor-binding domain complexed with human receptor DPP4. Cell Res. 2013;23:986–993. doi: 10.1038/cr.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Qi J., Yuan Y., Xuan Y., Han P., Wan Y., Ji W., Li Y., Wu Y., Wang J., Iwamoto A., Woo P.C., Yuen K.Y., Yan J., Lu G., Gao G.F. Bat origins of MERS-CoV supported by bat coronavirus HKU4 usage of human receptor CD26. Cell Host Microbe. 2014;16:328–337. doi: 10.1016/j.chom.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S., Takada A., Watanabe T., Ito H., Kida H., Kawaoka Y. Functional importance of the coiled-coil of the Ebola virus glycoprotein. J. Virol. 2000;74:10194–10201. doi: 10.1128/jvi.74.21.10194-10201.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wecker M., Gilbert P., Russell N., Hural J., Allen M., Pensiero M., Chulay J., Chiu Y.L., Karim S.S.A., Burke D.S., Team H.P., Network N.H.V.T. Phase I safety and immunogenicity evaluations of an alphavirus replicon HIV-1 subtype C gag vaccine in healthy HIV-1-uninfected adults. Clin. Vaccine Immunol. 2012;19:1651–1660. doi: 10.1128/CVI.00258-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welter M.W., Horstman M.P., Welter C.J., Welter L.M. An overview of successful TGEV vaccination strategies and discussion on the interrelationship between TGEV and PRCV. Coronaviruses. 1993;342:463–468. doi: 10.1007/978-1-4615-2996-5_73. [DOI] [PubMed] [Google Scholar]
- White J.M., Delos S.E., Brecher M., Schornberg K. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme (vol. 43, pg 189, 2008) Crit. Rev. Biochem. Mol. 2008;43:287–288. doi: 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . 2004. Cumulative Number of Reported Probable Cases of Severe Acute Respiratory Syndrome (SARS)http://www.who.int/csr/sars/country/en/ [Google Scholar]
- WHO . 2016. Middle East Respiratory Syndrome Coronavirus (MERS-cov) – Update.http://www.who.int/csr/don/7-january-2016-mers-oman/en/ [Google Scholar]
- Wong G., Liu W., Liu Y., Zhou B., Bi Y., Gao G.F. MERS, SARS, and Ebola: The role of super-spreaders in infectious disease. Cell Host Microbe. 2015;18:398–401. doi: 10.1016/j.chom.2015.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Chu C.M., Chan K.H., Tsoi H.W., Huang Y., Wong B.H., Poon R.W., Cai J.J., Luk W.K., Poon L.L., Wong S.S., Guan Y., Peiris J.S., Yuen K.Y. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 2005;79:884–895. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S., Liu Q., Wang Q., Sun Z., Su S., Du L., Ying T., Lu L., Jiang S. Middle East respiratory syndrome coronavirus (MERS-CoV) entry inhibitors targeting spike protein. Virus Res. 2014;194:200–210. doi: 10.1016/j.virusres.2014.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Yang M., Ding Y., Liu Y., Lou Z., Zhou Z., Sun L., Mo L., Ye S., Pang H., Gao G.F., Anand K., Bartlam M., Hilgenfeld R., Rao Z. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc. Natl. Acad. Sci. U. S. A. 2003;100:13190–13195. doi: 10.1073/pnas.1835675100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H.T., Bartlam M., Rao Z.H. Drug design targeting the main protease, the Achilles' heel of coronaviruses. Curr. Pharm. Des. 2006;12:4573–4590. doi: 10.2174/138161206779010369. [DOI] [PubMed] [Google Scholar]
- Yang X., Chen X., Bian G., Tu J., Xing Y., Wang Y., Chen Z. Proteolytic processing, deubiquitinase and interferon antagonist activities of Middle East respiratory syndrome coronavirus papain-like protease. J. Gen. Virol. 2014;95:614–626. doi: 10.1099/vir.0.059014-0. [DOI] [PubMed] [Google Scholar]
- Yang Y., Liu C., Du L., Jiang S., Shi Z., Baric R.S., Li F. Two mutations were critical for bat-to-human transmission of Middle East respiratory syndrome coronavirus. J. Virol. 2015;89:9119–9123. doi: 10.1128/JVI.01279-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Zhang L., Geng H., Deng Y., Huang B., Guo Y., Zhao Z., Tan W. The structural and accessory proteins M, ORF 4a, ORF 4b, and ORF 5 of Middle East respiratory syndrome coronavirus (MERS-CoV) are potent interferon antagonists. Protein & Cell. 2013;4:951–961. doi: 10.1007/s13238-013-3096-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y., Bao L., Deng W., Xu L., Li F., Lv Q., Yu P., Chen T., Xu Y., Zhu H., Yuan J., Gu S., Wei Q., Chen H., Yuen K.Y., Qin C. An animal model of MERS produced by infection of rhesus macaques with MERS coronavirus. J. Infect. Dis. 2014;209:236–242. doi: 10.1093/infdis/jit590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying T.L., Prabakaran P., Du L.Y., Shi W., Feng Y., Wang Y.P., Wang L.S., Li W., Jiang S.B., Dimitrov D.S., Zhou T.Q. Junctional and allele-specific residues are critical for MERS-CoV neutralization by an exceptionally potent germline-like antibody. Nat. Commun. 2015;6:8223. doi: 10.1038/ncomms9223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M., Wang E., Liu Y., Cao D., Jin N., Zhang C.W., Bartlam M., Rao Z., Tien P., Gao G.F. Six-helix bundle assembly and characterization of heptad repeat regions from the F protein of Newcastle disease virus. J. Gen. Virol. 2002;83:623–629. doi: 10.1099/0022-1317-83-3-623. [DOI] [PubMed] [Google Scholar]
- Yu X.J., Zhang S.Y., Jiang L.W., Cui Y., Li D.X., Wang D.L., Wang N.S., Fu L.L., Shi X.L., Li Z.Q., Zhang L.Q., Wang X.Q. Structural basis for the neutralization of MERS-CoV by a human monoclonal antibody MERS-27. Sci. Rep. 2015;5:13133–13143. doi: 10.1038/srep13133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- Zhang N., Channappanavar R., Ma C., Wang L., Tang J., Garron T., Tao X., Tasneem S., Lu L., Tseng C.T., Zhou Y., Perlman S., Jiang S., Du L. Identification of an ideal adjuvant for receptor-binding domain-based subunit vaccines against Middle East respiratory syndrome coronavirus. Cell Mol. Immunol. 2016;13:180–190. doi: 10.1038/cmi.2015.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N., Jiang S., Du L. Current advancements and potential strategies in the development of MERS-CoV vaccines. Expert Rev. Vaccines. 2014;13:761–774. doi: 10.1586/14760584.2014.912134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G.Y., Du L.Y., Ma C.Q., Li Y., Li L., Poon V.K.M., Wang L.L., Yu F., Zheng B.J., Jiang S.B., Zhou Y.S. A safe and convenient pseudovirus-based inhibition assay to detect neutralizing antibodies and screen for viral entry inhibitors against the novel human coronavirus MERS-CoV. Virol. J. 2013;10:266. doi: 10.1186/1743-422X-10-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G.Y., Jiang Y.T., Qiu H.J., Gao T.T., Zeng Y., Guo Y., Yu H., Li J.F., Kou Z.H., Du L.Y., Tan W.J., Jiang S.B., Sun S.H., Zhou Y.S. Multi-organ damage in human dipeptidyl peptidase 4 transgenic mice infected with Middle East respiratory syndrome-coronavirus. PLoS One. 2015;10:e0145561. doi: 10.1371/journal.pone.0145561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Li K., Wohlford-Lenane C., Agnihothram S.S., Fett C., Zhao J., Gale M.J., Jr., Baric R.S., Enjuanes L., Gallagher T., McCray P.B., Jr., Perlman S. Rapid generation of a mouse model for Middle East respiratory syndrome. Proc. Natl. Acad. Sci. U. S. A. 2014;111:4970–4975. doi: 10.1073/pnas.1323279111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., Dimitrov A.S., Bossart K.N., Crameri G., Bishop K.A., Choudhry V., Mungall B.A., Feng Y.R., Choudhary A., Zhang M.Y., Feng Y., Wang L.F., Xiao X., Eaton B.T., Broder C.C., Dimitrov D.S. Potent neutralization of Hendra and Nipah viruses by human monoclonal antibodies. J. Virol. 2006;80:891–899. doi: 10.1128/JVI.80.2.891-899.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]


