Abstract
Interleukin-22 (IL-22), a member of the IL-10 superfamily, plays essential roles in fighting against mucosal microbial infection and maintaining mucosal barrier integrity within the intestine. However, little knowledge exists on the ability of porcine IL-22 (pIL-22) to fight against viral infection in the gut. In this study, we found that recombinant mature pIL-22 (mpIL-22) inhibited the infection of multiple diarrhea viruses, including alpha coronavirus, porcine epidemic diarrhea virus (PEDV), transmissible gastroenteritis virus (TGEV), and porcine rotavirus (PoRV), in the intestinal porcine epithelial cell line J2 (IPEC-J2) cells. mpIL-22 up-regulated the expression of the antimicrobial peptide beta-defensin (BD-2), cytokine IL-18 and IFN-λ. Furthermore, we found that mpIL-22 induced phosphorylation of STAT3 on Ser727 and Tyr705 in IPEC-J2 cells. Inhibition of STAT3 phosphorylation by S3I-201 abrogated the antiviral ability of mpIL-22 and the mpIL-22-induced expression of BD-2, IL-18, and IFN-λ. Together, mpIL-22 inhibited the infection of PoRV and enteric coronaviruses, and up-regulated the expression of antimicrobial genes in IPEC-J2, which were mediated by the activation of the STAT3 signal pathway. The significant antiviral activity of IL-22 to curtail multiple enteric diarrhea viruses in vitro suggests that pIL-22 could be a novel therapeutic against devastating viral diarrhea in piglets.
Keywords: Interleukin 22, Porcine rotavirus, Porcine epidemic diarrhea virus, Transmissible gastroenteritis virus, Intestinal epithelia, STAT3
Highlights
-
•
Porcine IL-22 inhibits the most common three porcine diarrhea viruses (rotavirus, PEDV and TGEV) in vitro.
-
•
STAT3 signaling by IL-22 accounts for the antiviral activity and the enhanced expression of protective genes of mpIL-22.
-
•
IL-22 could provide a novel approach to control devastating viral diarrhea in piglets.
1. Introduction
Interleukin-22 (IL-22), an IL-10 family-related cytokine, is primarily secreted by T helper 17 cells, innate lymphoid cells (ILC), innate natural killer (INK) cells, and epithelial cells (Dudakov et al., 2015, Sonnenberg et al., 2011, Wolk et al., 2004). Unlike most other IL-10-related cytokines, IL-22 primarily targets nonhematopoietic epithelial cells and fibroblasts (Ouyang and Valdez, 2008, Sonnenberg et al., 2011, Wolk et al., 2004). The IL-22 receptor is composed of two heterodimeric subunits, IL-22R1 and a common IL-10R2 of the IL-10 family (Xie et al., 2000). While the IL-10R2, a shared receptor for IL-10, IL-26, IL-28, and IL-29, is expressed in various tissues, IL-22R1 is primarily expressed in mucosal epithelia such as the intestinal epithelial cells (IEC) (Eidenschenk et al., 2014, Ouyang and Valdez, 2008).
The growing evidence demonstrates that IL-22 is essential to host defense against invading pathogens and inflammatory response, especially in the mucosal tissue (Ouyang et al., 2011, Witte et al., 2010, Wolk et al., 2006). Initially, studies determined that IL-22 is essential for the host to control the infection of extracellular bacteria in the lung or gut (Eidenschenk et al., 2014, Ivanov et al., 2013, Zheng et al., 2008). Recent evidence indicates that IL-22 has a substantial role in host defense against mucosal viral infection, including human immunodeficiency virus (HIV-1), influenza virus, and rotavirus (Hernandez et al., 2015, Ivanov et al., 2013, Kim et al., 2012, Kumar et al., 2013). The mechanism of IL-22 protection against infection is elusive and includes the direct promotion of epithelial survival and regeneration and the maintenance of the tissue barrier, but IL-22 also induces epithelial-secreting antimicrobials such as defensins and antiviral cytokines such as IL-18 (Hernandez et al., 2015, Munoz et al., 2015, Ouyang and Valdez, 2008). Furthermore, IL-22 increases its protective role when combined with IL-18 or IFN-λ (Hernandez et al., 2015, Zhang et al., 2014). Given the critical role of IL-22 in epithelial regeneration, host defense, and pathology, IL-22 is an attractive target for clinical development in animals and humans. However, little knowledge exists regarding porcine IL-22 (pIL-22).
Porcine diarrhea caused by viruses is one of the major problems affecting suckling piglets and causes substantial economic loss in the pork industry. The most common pathogens of porcine viral diarrhea are porcine epidemic diarrhea virus (PEDV), transmissible gastroenteritis virus (TGEV), and porcine rotavirus (PoRV) (Wang et al., 2016, Zhang et al., 2013, Zhao et al., 2016). PEDV and TGEV are alpha coronaviruses (Brian and Baric, 2005), and PoRV is a member of the genus Rotavirus within the Reoviridae family (Taniguchi and Komoto, 2012). All three diarrhea viruses primarily infect villous epithelial cells throughout the small intestine and cause serious injury of IEC, including severe diffuse atrophic enteritis and superficial villous enterocyte swelling (Wang et al., 2016). The manifestations of the infection of the three viruses are indistinguishable and characterized by vomiting, anorexia, watery diarrhea, dehydration, and weight loss, with a high morbidity and mortality in suckling piglets (Wang et al., 2016, Zhang et al., 2013). The co-infection of two or three viruses of PEDV, TGEV, and PoRV in the field is frequent and makes the development of therapeutic and prophylactic strategies to protect suckling pigs against diarrhea complications (Zhang et al., 2013, Zhao et al., 2016).
In this study, we found that the recombinant mature porcine IL-22 (mpIL-22) prepared using a prokaryotic expression system broadly inhibited the infection of alpha coronaviruses, PEDV and TGEV, and PoRV in intestinal epithelial cells. The antiviral activity of mpIL-22 was through the activation of STAT3 signaling and the up-regulated expression of antimicrobials and antiviral cytokines.
2. Material and methods
2.1. Cells and viruses
The IPEC-J2 cell line was kindly provided by Dr. Anthony Blikslager (North Carolina State University, Raleigh, NC). The cells were grown in Dulbecco's modified Eagle's medium (DMEM): nutrient mixture F-12 (Ham) (1:1) with Gluta AMAX_-I (DMEM = F12) (Gibco), supplemented with 5% fetal bovine serum (FBS), 5 μg/ml insulin-transferring-selenium supplements (Life Technologies), 5 ng/ml epidermal growth factor (Life Technologies), and 1% penicillin-streptomycin in a humidified atmosphere of 5% CO2 at 37 °C. Cell culture media were changed every 2 days, and the cells were passaged every 4–5 days by trypsinization with 0.25% trypsin-EDTA. African green monkey kidney cells (Vero E6) were grown and maintained in DMEM supplemented with 10% heat-inactivated FBS and penicillin-streptomycin and incubated at 37 °C with 5% CO2.
The Vero cell-adapted PEDV CV777 strain, kindly provided by Maurice Pensaert at Ghent University (Merelbeke, Belgium), was propagated as previously described (Hofmann and Wyler, 1988, Sun et al., 2015). Briefly, Vero cells were inoculated with the virus at a multiplicity of infection (MOI) of 1 and cultured in serum-free DMEM for 72 h at 37 °C with 5% CO2. The progeny virions were filtered and titrated using the TCID50 method. TGEV Hua isolate H87 was derived from the virulent strain H16 by serial passage in PK-15 cells in our laboratory (Wang et al., 2010). The PoRV OSU strain was propagated in MA104 cells, and titers were determined by a plaque assay as previously described (Feng et al., 2009).
2.2. Phylogenetic analysis
An alignment of pIL-22 sequence with other mammalian IL-22 molecules (human, mouse, bovine, sheep, goat) was carried out using the Clustal W program. The amino acid (aa) sequence homologies among the species were analyzed using the MegAlign program of DNAstar (DNAstar Inc., Madison, Wis.). The phylogenetic tree based on the nucleotide sequence of the IL-22 genes was constructed by the neighbor-joining method, using DNAstar MegAlign Version 8.1.2. by Clustal W. The sequence data for phylogenetic analysis were taken from the GenBank nucleotide sequence database with the following accession numbers: pig IL-22, XM_001926156; human IL-22, NM_020525; mouse IL-22, NM_016971; bovine IL-22, EF560596; sheep IL-22, HE617662 and goat IL-22, HM542482.
2.3. Clone and prokaryotic expression of pIL-22
To clone pIL-22, total cellular mRNA was isolated from IPEC-J2 cells, and 1 μg total mRNA was used to prepare cDNA using a PrimeScript™ II 1st strand cDNA synthesis kit (Takara, Dalian, China). Primers used for pIL-22 are summarized in Table 1 . Amplification was performed using PrimeSTAR HS DNA polymerase (Takara, Dalian, China) under the following conditions: 1 cycle of 94 °C for 5 min, 35 cycles of 98 °C for 10 s, 60 °C for 15 s, 72 °C for 1 min and 1 cycle of 72 °C for 10 min. The cDNA fragment encoding the predicted mpIL-22 was cloned into a pET-30 vector containing a 6-histidine tag at the N-terminus by EcoRI and SalI, resulting in the plasmid pET-30a-IL-22, which was transformed into BL21 (DE3). The recombinant mpIL-22 was purified by a Ni–NTA column according to the manufacturer's protocol.
Table 1.
Real-time Q-PCR primers used for quantification of virus genomes and host cell gene expression.
| Target | Sequence |
|---|---|
| IL-18 | Forward: 5′-TCT ACT CTC TCC TGT AAG AAC-3′ |
| Reverse: 5′-CTT ATC ATG TCC AGG AAC-3′ | |
| IFN-λ | Forward: 5′-CCACGTCGAACTTCAGGCTT-3′ |
| Reverse: 5′-ATGTGCAAGTCTCCACTGGT-3′ | |
| BD-2 | Forward: 5′-CCAGAGGTCCGACCACTA-3′ |
| Reverse: 5′-GGTCCCTTCAATCCTGTT-3′ | |
| GAPDH | Forward: 5′-CCTTCCGTGTCCCTACTGCCAAC-3′ |
| Reverse: 5′-GACGCCTGCTTCACCACCTTCT-3′ | |
| β-actin | Forward: 5′-GTGCGGGACATCAAGGAGAAG-3′ |
| Reverse: 5′-CGTAGCTCTTCATCCAGGGAG-3′ | |
| Survivin | Forward: 5′-CCGATTTGGCTCAATGTTTC-3′ |
| Reverse: 5′-GACAGAAAGGAAAGCACAACC-3′ | |
| PEDV-ORF3 | Forward: 5′-GCACTTATTGGCAGGCTTTGT-3′ |
| Reverse: 5′-CCATTGAGAAAAGAAAGTGTCGTAG-3′ | |
| TGEV-S | Forward: 5′-GCTTGATGAATTGAGTGCTGATG-3′ |
| Reverse: 5′-CCTAACCTCGGCTTGTCTGG-3′ | |
| PoRV-VP6 | Forward: 5′-CAACTGCACCACAAACTGAAAGA-3′ |
| Reverse: 5′-CTCGGTAATAAAAGGCAGCAGAA-3′ |
2.4. Stimulation of IPEC-J2 by mpIL-22 and in vitro virus infection
IPEC-J2 cells were initially plated into 6-wells for 2 days to grow a confluent monolayer. The cells were stimulated in the presence of DMEM F12 (as control) or a range of concentrations (4, 40 and 400 ng/ml) of mpIL-22 protein for 24 h. For the kinetics experiment, IPEC-J2 cells were stimulated with 40 ng/ml mpIL-22 and then harvested at the indicated times post treatment. The cells were washed and harvested for total cellular RNA extraction and quantification.
Following mpIL-22 stimulation for 24 h, IPEC-J2 cells were washed with PBS twice and inoculated with the Group A PoRV OSU strain, PEDV CV777 strain, or TGEV H87 strain at an MOI of 1. For S3I-201 inhibition experiment, IPEC-J2 cells were stimulated with 40 ng/ml mpIL-22 for 24 h, followed by incubation with 20 μM S3I-201 for 24 h before virus inoculation. After 2-h incubation, IPEC-J2 cells were washed and continued to culture with serum-free media for 0 to 48 h. The cells were washed and harvested for total RNA extraction and the quantification of viral genomes.
2.5. Quantification of virus genomes and host cell gene expression
Total RNA was isolated from cells using an RNeasy mini kit (Qiagen Sciences, Hilden, Germany) and reverse transcribed using a PrimeScript™ II 1st strand cDNA synthesis kit (Takara, Dalian, China). Quantitative PCR (Q-PCR) was performed in triplicate using the Power SYBR Green PCR master mix (Takara, Dalian, China) to quantify virus genomes and detect the expression of beta-defensin (BD-2), IL-18 and IFN-λ. The expression of β-actin was used as control whose expression does not change with treatment, and STAT3 dependent transcript survivin was used as control to demonstrate the specific effects of both mpIL-22 and the STAT3 inhibitor used (Naher et al., 2012). The results were normalized to those of the control housekeeping gene GADPH (encoding glyceraldehyde-3-phosphate dehydrogenase). All data were acquired with LightCycler 480 real time PCR machines (Roche) and analyzed with LightCycler 480 software 1.5 based on the cycle threshold (ΔΔCT) method.
For the quantification of PEDV, TGEV, or PoRV replication, the ORF3 gene of PEDV, the S gene of TGEV and the VP6 gene of PoRV were used as standards for the virus genome. Each pair of primers was synthesized for quantification of the PEDV, TGEV, and PoRV genome in Q-PCR. Quantification of viral RNA was calculated based on a standard curve with known amounts. All primers are listed in Table 1.
2.6. Western blotting analysis
The proteins were separated on 10% SDS-PAGE and transferred to polyvinylidene fluoride (PVDF) membranes (Millipore, Bedford, MA). Antibodies against total STAT3, p-STAT3 (S727), p-STAT3 (Y705) or β-actin were purchased from Cell Signaling Technology (Beverly, MA, USA). Monoclonal antibodies against TGEV N (mAb 3D3), PEDV N (mAb 2G3), and PoRV VP6 (mAb 1E5) were prepared by our team. After washing three times with TBST, the membrane was incubated with 1:10,000-diluted IRDye800 conjugated anti-mouse IgG or Alexa Fluor 680 goat anti-rabbit IgG (Invitrogen, USA) in blocking buffer for 1 h at room temperature. After washing three times with TBST, the membrane was scanned in an Odyssey infrared imaging system (LI-COR Biosciences). The fluorescence intensity of each band was measured using Odyssey 2.1 software (LI-COR Biosciences).
2.7. ELISA assay for the cytokines
The medium from the cell cultures was collected, pooled, and stored in aliquots at −70 °C until assayed. IL-22 in supernatants from PEDV, TGEV and PoRV-infected IPEC-J2 was measured using a porcine IL-22 ELISA kit (Bio Swamp, Shanghai, China) according to the manufacturer's instructions. The range of detection for the IL-22 ELISA is 62.5–1000 pg/ml.
2.8. Cytotoxicity assay
The cytotoxicity of purified mpIL-22 or S3I-201 in IPEC-J2 cells was measured using a Cell Counting Kit-8 (CCK-8) (Dojindo, China) according to the kit instructions. Briefly, IPEC-J2 cells were trypsinized and resuspended in fresh media at a concentration of 50,000 cells/ml. 200 μl of cell suspensions was then pipetted into 96-well microplates. Cells were allowed to grow overnight and were then treated with various concentrations of purified mpIL-22 or mpIL-22 following 20 μM S3I-201 for another 24 h in the incubator. The cells were quantified using a cell counting kit, CCK-8. Here, 20 μl of CCK-8 solution was added directly to cell cultures in 96-well plates with 200 μl of media per well. Cells were incubated in a humidified environment. After 2 h at 37 °C with 5% CO2, the absorbance at 450 nm was determined using an Elx808 microplate reader (BioTek Instruments). The assay for each concentration of purified mpIL-22 or S3I-201 was repeated in 4 wells.
2.9. Statistical analysis
All results in figures are presented, where appropriate, as the mean ± the standard error of the mean (SEM) from three independent experiments and were analyzed in GraphPad Prism with Student's t-test or a 2-way ANOVA test as indicated (GraphPad Software, Inc.). Differences were considered significant if the P value was <0.05. P values are indicated as follows: *P < 0.05; **P < 0.01; ***P < 0.001.
3. Results
3.1. Clone and expression of pIL-22
Growing evidence demonstrates that IL-22 plays critical roles in the host's fight against pathogens and inflammatory reactions in mucosal tissue. However, there is no report on pIL-22. We first tried to amplify the pIL-22 gene from IPEC-J2 cells. The full-length cDNA of pIL-22 containing a 570 bp was successfully amplified specifically from mRNA of IPEC-J2 cells infected by PoRV. Analysis of the aa homologies between species revealed that pIL-22 shared 65.4–75% aa identity with other mammalian IL-22 molecules (human, mouse, bovine, sheep, goat), with the highest identity seen with human (Fig. 1 A).
Fig. 1.
The prokaryotic expression of mpIL-22. (A) The amino acid (aa) sequence homologies and phylogenetic analysis of pIL-22 sequence with other mammalian IL-22 molecules (human, mouse, bovine, sheep, goat). The amino acid (aa) sequence homologies among the species were analyzed using the MegAlign program of DNAstar (DNAstar Inc., Madison, Wis.). The phylogenetic tree based on the nucleotide sequence of the IL-22 genes was constructed by the neighbor-joining method, using DNAstar MegAlign Version 8.1.2. by Clustal W. The sequence data for phylogenetic analysis were taken from the GenBank nucleotide sequence database. (B) SDS-PAGE analysis of the expression and purification of mpIL-22 from the E. coli BL21(DE3)/pET-30a. 1, affinity-purified mpIL-22-his protein; 2, the total protein extract of mpIL-22-his inclusion body pellet following 0.5 mM IPTG induction at 37 °C for 4 h; 3, the total protein of empty pET-30a vector; 4, unstained protein marker. (C) Purified mpIL-22-his was western blotted with mouse anti-His antibody. 1, the purified mpIL-22-his; 2, the total protein extract of pET-30a vector; 3, prestained protein marker.
To identify the presence and location of signal peptide cleavage sites in precursor pIL-22, the aa sequences of precursor pIL-22 were analyzed using the SignalP4.1 server program (http://www.cbs.dtu.dk/services/SignalP/). The most likely signal peptidase cleavage site found by the SignalP4.1 server program is located between aa positions 33 and 34, which would yield a mature protein of 156 aa and a signal peptide of 33 aa. The mpIL-22 containing 471 bp was amplified using precursor IL-22 as a template and cloned into the E. coli-expressing vector pET-30a. A fusion protein of the expected size at approximately 22 kDa was expressed after IPTG 0.5 mM induction at 37 °C for 4 h and was not present in the lysate of IPTG-induced bacterial cells harboring the pET-30a empty vector. The denatured recombinant mpIL-22 proteins in inclusion body isolates were refolded by dialysis and purified by Ni–NTA Affinity Chromatography (Fig. 1B). The expression of mpIL-22-his was further confirmed by western blotting with mouse anti-his monoclonal antibodies (Fig. 1C).
3.2. Recombinant mpIL-22 inhibits PoRV infection in small intestinal epithelial cells
Recently, two studies discovered that IL-22, in collaboration with other mucosal cytokines, exerts substantial roles in controlling rotavirus infection in vivo in a mouse model (Hernandez et al., 2015, Zhang et al., 2014). To test whether pIL-22 inhibits porcine rotavirus in small intestinal epithelial cells, we treated IPEC-J2 cells with 40 ng/ml purified mpIL-22 for 24 h before infection of the PoRV OSU strain. The mpIL-22 treatment resulted in more than 90% reduction in the viral infection (8.40 ± 4.0% for mpIL-22) (Fig. 2 A). The viral RNA quantification was consistent with the results of viral protein western blotting that the levels of PoRV VP6 protein expression were decreased about 97.22% in IL-22-treated cells at 48 hpi compared to those in the control cells (Fig. 2B). The results demonstrate that mpIL-22 also exhibits significant anti-porcine rotavirus activity in IPEC-J2 cells.
Fig. 2.
mpIL-22 inhibited PoRV infection in IPEC-J2 cells. Following mpIL-22 stimulation for 24 h, IPEC-J2 cells were washed with PBS twice and inoculated with the PoRV OSU strain at an MOI of 1. After 2-h incubation, IPEC-J2 cells were washed and continued to culture with serum-free media for 48 h. (A) The total amount of viral RNA was quantified by Q-PCR. The % virus infection compared to the mock treatment was calculated. The results represent the mean ± SEM of three independent experiments. (B) The PoRV infection of IPEC-J2 cells at 48 hpi was determined by PORV VP6 protein western blotting following pretreatment with mpIL22.
3.3. Recombinant mpIL-22 inhibits alpha coronavirus infection in small intestinal epithelial cells
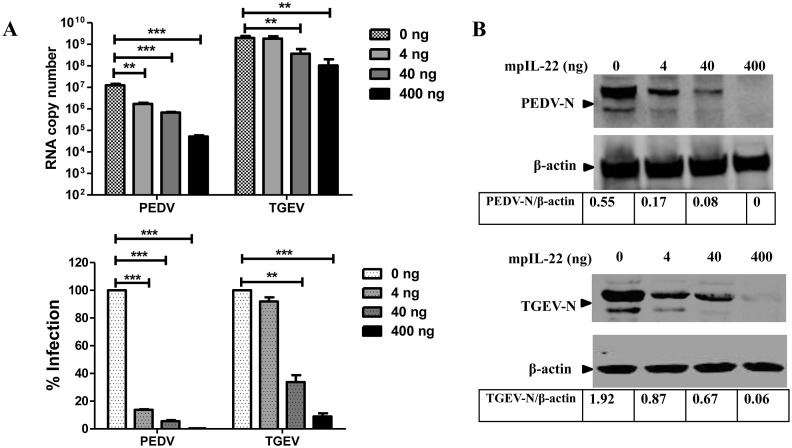
Alpha coronaviruses PEDV and TGEV are the causative pathogens of one of the most economically devastating viral diseases of swine in the world. Like PoRV, PEDV and TGEV primarily infect small intestinal epithelial cells and cause a similar injury of the intestines with similar clinical signs. We reasoned that mpIL-22 is also capable of curtailing PEDV and TGEV infection in IPEC-J2, as it does in rotavirus infection. Similar to the rotavirus experiment, we stimulated IPEC-J2 cells with mpIL-22 for 24 h before inoculating with PEDV or TGEV. As we expected, mpIL-22 treatment inhibited PEDV and TGEV infection in a dose-dependent manner (Fig. 3 A). Pretreatment with 400 ng/ml of mpIL-22 resulted in a more than 90% reduction in infection of PEDV and TGEV, whereas 40 ng/ml mpIL-22 caused a 93.82 ± 0.34% and 68.13 ± 7.34% inhibition of PEDV and TGEV infection, respectively. The anti-coronavirus activity of mpIL-22 was further confirmed by western blotting the N protein expression levels and exhibited a dose-dependent manner for mpIL-22 treatment at 48 hpi compared to those in the control cells (Fig. 3B). PEDV and TGEV showed a different sensitivity to the antiviral activity of mpIL-22; mpIL-22 exhibited a more efficient inhibition of the PEDV infection in IPEC-J2 than TGEV (Fig. 3). mpIL-22 exhibited a significant suppression of PEDV infection from 1 ng/ml (Fig. S2). These results show that IL-22 is a significant innate antiviral cytokine against coronavirus infection within the intestine cells.
Fig. 3.
mpIL-22 inhibited PEDV and TGEV infection in IPEC-J2 cells. IPEC-J2 cells were washed with PBS twice and inoculated with PEDV CV777 strain or TGEV H87 strain at an MOI of 1 following mpIL-22 stimulation for 24 h. After 2-h incubation, IPEC-J2 cells were washed and continued to culture with serum-free media for 48 h. (A) The total amount of PEDV and TGEV viral RNA was quantified by Q-PCR. The % virus infection compared to the mock treatment was calculated. The results represent the mean ± SEM of three independent experiments. (B) The infection of PEDV or TGEV in IPEC-J2 cells at 48 hpi was determined by western blotting for the N protein after treatment with mpIL-22.
3.4. Recombinant mpIL-22 elicits an up-regulation of expression of BD-2, IL-18, and IFN-λ in small intestinal epithelial cells
IL-22 has been demonstrated to combat a broad range of infectious agents in the gut through the up-regulation of antimicrobial peptides or cytokines (Ouyang and Valdez, 2008, Sonnenberg et al., 2011). Beta-defensin-2 (BD-2), IL-18 and IFN-λ are among the antimicrobials and antiviral cytokines in small intestinal epithelial cells that are well studied (Chen et al., 2009, Lazear et al., 2015, Schroder and Harder, 1999, Zhang et al., 2014). We initially investigated whether mpIL-22 induced the expression of BD-2, IL-18, and IFN-λ in IPEC-J2. mpIL-22 stimulation increased the transcriptional expression of IL-22-induced gene survivin (Naher et al., 2012) instead of the house-keeping gene β-actin, indicating that mpIL-22 is biological active and its activity is specific (Fig. 4 ). mpIL-22 pretreatment specifically enhanced the expression of BD-2, IL-18 and IFN-λ in IPEC-J2 cells and exhibited a dose response (Fig. 4A). Next, we determined the time kinetics of the up-regulation of BD-2, IL-18 and IFN-λ mRNA following 40 ng/ml mpIL-22 stimulation. The levels of mRNA of the protective genes instead of β-actin increased with the time post stimulation and peaked at 48 h post stimulation, with fold increases of 6.50 ± 0.65, 3.48 ± 0.83, and 7.37 ± 4.84 relative to mock control for BD-2, IL-18, and IFN-λ, respectively (Fig. 4B).
Fig. 4.
The up-regulated expression of BD-2 and cytokines in IPEC-J2 cells following stimulation with recombinant mpIL-22. (A) The transcriptional mRNA of BD-2, IL-18 and IFN-λ by IPEC-J2 cells exhibited a dose response to mpIL-22. IPEC-J2 cells were stimulated with various concentrations of mpIL-22. The total cellular RNA was extracted 24 h post treatment. The mRNA of cytokines and BD-2 was quantified by real time Q-PCR as described in the Methods section. The expression of β-actin and survivin was used as controls. (B) Kinetics of the mRNA of cytokines and BD-2 in IPEC-J2 cells following mpIL-22 stimulation. The expression of β-actin and survivin was used as controls. IPEC-J2 cells were stimulated with 40 ng/ml mpIL-22 and harvested at the indicated time following treatment. The results are represented as the mean ± SEM of three independent experiments.
To exclude the possibility that the cellular proliferation and cytotoxicity of mpIL-22 caused the increased expression of BD-2 and cytokines, we performed the mpIL-22 cytotoxicity assay with a CCK-8 kit, a routine assay of cellular proliferation and cytotoxicity based on dehydrogenase activity detection in viable cells. There were no significant differences in the absorbance among cells following treatment with purified recombinant mpIL-22 ranging from 4 to 400 ng/ml (from 1.25 ± 0.04 to 1.32 ± 0.14 for mpIL-22 versus 1.36 ± 0.04 for mock), indicating that no proliferation and cytotoxicity was observed at the concentrations used in this study (Supplementary Fig. S3).
3.5. IL-22 inhibits virus replication by activation of STAT3
Most of the biological effects of IL-22 are primarily mediated by the activation of STAT3 signaling downstream of the IL-22R in epithelial cells. To address the potential role of STAT3 signaling in the IL-22 antiviral activity against the infection of porcine enteric coronavirus and rotavirus in IPEC-J2, the effects of IL-22 on STAT3 activation in IPEC-J2 cells were analyzed. We found that IL-22 induced phosphorylation of STAT3 on Ser727 and Tyr705 in IPEC-J2 cells (Fig. 5 A). To further confirm STAT3 activation specifically induced by mpIL-22, S3I-201, a STAT3-specific inhibitor, was used. The treatment of S3I-201 completely inhibited STAT3 phosphorylation induced by mpIL-22 stimulation.
Fig. 5.
mpIL-22 activated the STAT3 signaling pathway in IPEC-J2 cells. (A) IL-22 induced pY705 and pS727 phosphorylation of STAT3 in IPEC-J2 cells. IPEC-J2 cells were stimulated with mpIL-22 and harvested 24 h post treatment, 30 μg of protein lysates were analyzed for P-STAT3 (Y705 and S727) and total STAT3 by western blot analysis. (B) Inhibition of STAT3 phosphorylation by S3I-201 down-regulated BD-2, IL-18, and IFN-λ expression. IPEC-J2 cells were stimulated with 40 ng/ml mpIL-22 for 24 h and then treated with 20 μM STAT3 phosphorylation inhibitor (S3I-201) for 24 h; the expression levels of BD-2, IL-18, and IFN-λ were quantified by real time Q-PCR as described in the Methods section. The expression of β-actin and survivin was used as controls. (C) Inhibition of STAT3 phosphorylation by S3I-201 abolished the anti-PEDV, TGEV, or PoRV activity of mpIL-22. The PEDV, TGEV, or PoRV replication was measured by viral RNA copies.
Next, we investigated whether the production of antiviral genes induced by mpIL-22 was mediated through STAT3 signaling. PEDV infection alone elicited the expression of BD-2 and IFN-λ instead of IL-18 in IPEC-J2 cells, and the pre-stimulation of mpIL-22 enhanced the expression of BD-2, IFN-λ, and IL-18 following PEDV infection (Fig. 5B). Inhibition of STAT3 phosphorylation by S3I-201 almost completely abrogated the up-regulated expression of BD-2, IL-18, and IFN-λ by mpIL-22 and PEDV infection but had no effect on β-actin expression, indicating that the expression of these protective genes elicited by mpIL-22 is through the STAT3 signaling pathway. In concordance with these results, the presence of S3I-201 completely abolished the antiviral activity of mpIL-22 against PEDV, TGEV and PoRV (Fig. 5C). These effects did not result from the S3I-201 cellular cytotoxicity since no changes of cellular viability following treatment with 20 μM S3I-201 were observed in comparison with mock control (Fig. S4). These data indicate that mpIL-22 inhibits porcine enteric coronavirus and rotavirus infection via STAT3 signaling.
3.6. IL-22 was induced in PEDV, TGEV and PoRV-infected IPEC-J2 cells
We next investigated whether PEDV, TGEV, and PoRV infection elicited the production of pIL-22 in IPEC-J2 cells. The expression of IL-22 in the supernatants at different times after virus infection was determined by ELISA. The uninfected IPEC-J2 cells in vitro constitutively produced low levels of IL-22. The increased IL-22 was detected in the supernatants of the PEDV, TGEV, and PoRV-infected IPEC-J2 cells from 6 to 48 h post infection (Fig. 6 ). Following infection with PEDV, TGEV, or PoRV, IPEC-J2 cells secreted markedly higher amounts of pIL-22 than the uninfected IPEC-J2 cells. However, the kinetics profile of pIL-22 production was variable following different virus infection. The production of pIL-22 was maintained at a plateau level during the entire PEDV infection period while pIL-22 declined gradually after peaking at 6 hpi for TGEV infection. For PoRV, pIL-22 was induced and increased steadily from 6 to 48 hpi. These results demonstrate that the infection of IPEC-J2 with enteric coronavirus or rotavirus stimulates the production of pIL-22.
Fig. 6.
IL-22 was induced in PEDV, TGEV and PoRV-infected IPEC-J2 cells. IPEC-J2 cells were infected with PEDV CV777, TGEV H87, or PoRV OSU strain. After a 2-h infection, the unabsorbed virus was removed, and fresh medium was added. The cells were cultured further, and the supernatants were collected at different times after infection. The expression of pIL-22 in the supernatants was determined by porcine IL-22 ELISA. The concentration of the supernatant-secreted pIL-22 was calculated based on the standards of the IL-22 ELISA kits.
4. Discussion
IL-22 has recently been found to primarily target epithelial cells and exerts critical protective roles against various bacterial or viral infections (Ouyang and Valdez, 2008, Sonnenberg et al., 2011). The diarrhea of suckling piglets, primarily caused by PEDV, TGEV, or PoRV, is a major concern for the pig industry and results in substantial economic loss (Wang et al., 2016, Zhang et al., 2013). In this study, we demonstrated that mpIL-22 broadly inhibited swine enteric coronaviruses (PEDV and TGEV) and porcine rotavirus. To our knowledge, this is the first report that shows that IL-22 inhibits coronavirus infection in epithelia. Given that most coronaviruses infect epithelial cells in the respiratory and/or enteric tracts and that coronaviruses such as SARS-CoV, MERS-CoV, and PEDV are important for human health and livestock, it is worthwhile to further develop IL-22 as a potential therapeutic agent against the infection of coronaviruses.
The mechanism of the antiviral activity of IL-22 remains elusive. IL-22 has been linked to elicit the expression of antimicrobial defensins, synergistically cooperate with another mucosal antiviral cytokine IFN-λ, and promote the tissue barrier in IEC to curtail mucosal viral infection. We herein found that porcine IL-22 induced the expression of protective genes BD-2, IFN-λ, and IL-18 in IPEC-J2, especially IFN-λ, which primarily acts on mucosal epithelial cells and exhibits robust antiviral activity within the mucosal surface. Our previous results have demonstrated that porcine IFN-λ substantially curtailed PEDV infection in IPEC-J2 and Vero E6 cells in vitro (Li et al., 2016). Previous studies of rotavirus in mice have shown that IL-22 enhances the antiviral activity of IFN-λ to synergistically amplify the STAT1 signal pathway induced by IFN-λ (Hernandez et al., 2015). Here we reasoned that IL-22 may boost the antiviral activity of IFN-λ by promoting the production of IFN-λ. IL-18, a member of the IL-1 family, is induced by TLR5 activation in IEC and mutually enhanced the antiviral activity of IL-22 (Zhang et al., 2014). IL-22 is indispensable for the induction and maintenance of the expression of IL-18 by IEC during infection with Toxoplasma gondii (Munoz et al., 2015). Our results further reinforce the notion that the synergetic feedback loop between IL-22 and IFN-λ or IL-18 acts on IEC for the protection of the intestine against infection. Therefore, the positive feedback loop among IL-22, IL-18, and IFN-λ consists of a complex mutual regulating cytokine network in the gut and provides synergistic innate immunity to combat mucosal virus infection. Our results and the findings from other groups indicate that the combination of IL-22 with IL-18 or IFN-λ is a potential therapeutic strategy to fight against gut viral pathogens.
PEDV, TGEV, or PoRV infection enhanced the endogenous production of IL-22 by IPEC-J2 (Fig. 6). This indicates that pIL-22 potentially exerts a protective role in the control of swine enteric coronaviruses (PEDV and TGEV) and PoRV in the host given the significant viral suppression of mpIL-22 in vitro. The previous results of the rotavirus in vivo mouse model have demonstrated that the IL-22 endogenously produced by IEC has a critical role in restricting enteric virus infection in vivo (Hernandez et al., 2015, Zhang et al., 2014). Given the frequent co-infection of the three porcine diarrhea viruses in the field (Zhang et al., 2013, Zhao et al., 2016), pIL-22 could provide a novel approach to control the devastating diarrhea of suckling piglets. We are investigating the therapeutic benefits of pIL-22 alone or combined with IFN-λ against viral enteric infection in vivo.
In conclusion, we demonstrated that the prokaryotic mpIL-22 is capable of inhibiting the infection of the most common three different porcine diarrhea viruses (PoRV, PEDV and TGEV) in vitro. The antiviral activity of mpIL-22 is through the activation of STAT3 signaling, which results in the up-regulated expression of protective genes.
Acknowledgments
This work was supported by grants from the National Key R & D Program (2016YFD0500100) and the China Postdoctoral Science Foundation (2015M570185).
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.antiviral.2017.03.006.
Contributor Information
Li Feng, Email: fengli_h@163.com.
Pinghuang Liu, Email: liupinghuang@caas.cn.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- Brian D.A., Baric R.S. Coronavirus genome structure and replication. Curr. Top. Microbiol. Immunol. 2005;287:1–30. doi: 10.1007/3-540-26765-4_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.Y., Zheng L.L., Li X.S., Wei Z.Y., Cui B.A., Li X.K., Liu J.P., Yin H.Z., Meng J.T., Zhang Y., Li S.M. Cloning, in vitro expression, and bioactivity of interleukin-18 isolated from a domestic porcine breed found in Henan. Fems. Immunol. Med. Mic. 2009;57:129–135. doi: 10.1111/j.1574-695X.2009.00589.x. [DOI] [PubMed] [Google Scholar]
- Dudakov J.A., Hanash A.M., van den Brink M.R. Interleukin-22: immunobiology and pathology. Annu. Rev. Immunol. 2015;33:747–785. doi: 10.1146/annurev-immunol-032414-112123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eidenschenk C., Rutz S., Liesenfeld O., Ouyang W. Role of IL-22 in microbial host defense. Curr. Top. Microbiol. Immunol. 2014;380:213–236. doi: 10.1007/978-3-662-43492-5_10. [DOI] [PubMed] [Google Scholar]
- Feng N., Sen A., Nguyen H., Vo P., Hoshino Y., Deal E.M., Greenberg H.B. Variation in antagonism of the interferon response to rotavirus NSP1 results in differential infectivity in mouse embryonic fibroblasts. J. Virol. 2009;83(14):6987–6994. doi: 10.1128/JVI.00585-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez P.P., Mahlakoiv T., Yang I., Schwierzeck V., Nguyen N., Guendel F., Gronke K., Ryffel B., Holscher C., Dumoutier L., Renauld J.C., Suerbaum S., Staeheli P., Diefenbach A. Interferon-lambda and interleukin 22 act synergistically for the induction of interferon-stimulated genes and control of rotavirus infection. Nat. Immunol. 2015;16(7):698–707. doi: 10.1038/ni.3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann M., Wyler R. Propagation of the virus of porcine epidemic diarrhea in cell culture. J. Clin. Microbiol. 1988;26(11):2235–2239. doi: 10.1128/jcm.26.11.2235-2239.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov S., Renneson J., Fontaine J., Barthelemy A., Paget C., Fernandez E.M., Blanc F., De Trez C., Van Maele L., Dumoutier L., Huerre M.R., Eberl G., Si-Tahar M., Gosset P., Renauld J.C., Sirard J.C., Faveeuw C., Trottein F. Interleukin-22 reduces lung inflammation during influenza A virus infection and protects against secondary bacterial infection. J. Virol. 2013;87(12):6911–6924. doi: 10.1128/JVI.02943-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C.J., Nazli A., Rojas O.L., Chege D., Alidina Z., Huibner S., Mujib S., Benko E., Kovacs C., Shin L.Y., Grin A., Kandel G., Loutfy M., Ostrowski M., Gommerman J.L., Kaushic C., Kaul R. A role for mucosal IL-22 production and Th22 cells in HIV-associated mucosal immunopathogenesis. Mucosal. Immunol. 2012;5(6):670–680. doi: 10.1038/mi.2012.72. [DOI] [PubMed] [Google Scholar]
- Kumar P., Thakar M.S., Ouyang W., Malarkannan S. IL-22 from conventional NK cells is epithelial regenerative and inflammation protective during influenza infection. Mucosal. Immunol. 2013;6(1):69–82. doi: 10.1038/mi.2012.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazear H.M., Nice T.J., Diamond M.S. Interferon-lambda: immune functions at barrier surfaces and beyond. Immunity. 2015;43(1):15–28. doi: 10.1016/j.immuni.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Fu F., Xue M., Chen W.Y., Liu J., Shi H.Y., Chen J.F., Feng L., Bu Z.G. IFN-lambda preferably inhibits PEDV infection of porcine intestinal epithelial cells compared with IFN-alpha. Antivir. Res. 2016;140:76–82. doi: 10.1016/j.antiviral.2017.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz M., Eidenschenk C., Ota N., Wong K., Lohmann U., Kuhl A.A., Wang X., Manzanillo P., Li Y., Rutz S., Zheng Y., Diehl L., Kayagaki N., van Lookeren-Campagne M., Liesenfeld O., Heimesaat M., Ouyang W. Interleukin-22 induces interleukin-18 expression from epithelial cells during intestinal infection. Immunity. 2015;42(2):321–331. doi: 10.1016/j.immuni.2015.01.011. [DOI] [PubMed] [Google Scholar]
- Naher L., Kiyoshima T., Kobayashi I., Wada H., Nagata K., Fujiwara H., Ookuma Y.F., Ozeki S., Nakamura S., Sakai H. STAT3 signal transduction through interleukin-22 in oral squamous cell carcinoma. Int. J. Oncol. 2012;41(5):1577–1586. doi: 10.3892/ijo.2012.1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang W., Rutz S., Crellin N.K., Valdez P.A., Hymowitz S.G. Regulation and functions of the IL-10 family of cytokines in inflammation and disease. Annu. Rev. Immunol. 2011;29:71–109. doi: 10.1146/annurev-immunol-031210-101312. [DOI] [PubMed] [Google Scholar]
- Ouyang W., Valdez P. IL-22 in mucosal immunity. Mucosal. Immunol. 2008;1(5):335–338. doi: 10.1038/mi.2008.26. [DOI] [PubMed] [Google Scholar]
- Schroder J.M., Harder J. Human beta-defensin-2. Int. J. Biochem. Cell. Biol. 1999;31(6):645–651. doi: 10.1016/s1357-2725(99)00013-8. [DOI] [PubMed] [Google Scholar]
- Sonnenberg G.F., Fouser L.A., Artis D. Border patrol: regulation of immunity, inflammation and tissue homeostasis at barrier surfaces by IL-22. Nat. Immunol. 2011;12(5):383–390. doi: 10.1038/ni.2025. [DOI] [PubMed] [Google Scholar]
- Sun D., Shi H., Guo D., Chen J., Shi D., Zhu Q., Zhang X., Feng L. Analysis of protein expression changes of the Vero E6 cells infected with classic PEDV strain CV777 by using quantitative proteomic technique. J. Virol. Methods. 2015;218:27–39. doi: 10.1016/j.jviromet.2015.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi K., Komoto S. Genetics and reverse genetics of rotavirus. Curr. Opin. Virol. 2012;2(4):399–407. doi: 10.1016/j.coviro.2012.06.001. [DOI] [PubMed] [Google Scholar]
- Wang C., Chen J., Shi H., Qiu H., Xue F., Liu C., Zhu Y., Liu S., Almazan F., Enjuanes L., Feng L. Molecular characterization of a Chinese vaccine strain of transmissible gastroenteritis virus: mutations that may contribute to attenuation. Virus Genes. 2010;40(3):403–409. doi: 10.1007/s11262-010-0467-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Fang L., Xiao S. Porcine epidemic diarrhea in China. Virus. Res. 2016;226:7–13. doi: 10.1016/j.virusres.2016.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte E., Witte K., Warszawska K., Sabat R., Wolk K. Interleukin-22: a cytokine produced by T, NK and NKT cell subsets, with importance in the innate immune defense and tissue protection. Cytokine Growth Factor Rev. 2010;21(5):365–379. doi: 10.1016/j.cytogfr.2010.08.002. [DOI] [PubMed] [Google Scholar]
- Wolk K., Kunz S., Witte E., Friedrich M., Asadullah K., Sabat R. IL-22 increases the innate immunity of tissues. Immunity. 2004;21(2):241–254. doi: 10.1016/j.immuni.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Wolk K., Witte E., Wallace E., Docke W.D., Kunz S., Asadullah K., Volk H.D., Sterry W., Sabat R. IL-22 regulates the expression of genes responsible for antimicrobial defense, cellular differentiation, and mobility in keratinocytes: a potential role in psoriasis. Eur. J. Immunol. 2006;36(5):1309–1323. doi: 10.1002/eji.200535503. [DOI] [PubMed] [Google Scholar]
- Xie M.H., Aggarwal S., Ho W.H., Foster J., Zhang Z., Stinson J., Wood W.I., Goddard A.D., Gurney A.L. Interleukin (IL)-22, a novel human cytokine that signals through the interferon receptor-related proteins CRF2-4 and IL-22R. J. Biol. Chem. 2000;275(40):31335–31339. doi: 10.1074/jbc.M005304200. [DOI] [PubMed] [Google Scholar]
- Zhang B., Chassaing B., Shi Z., Uchiyama R., Zhang Z., Denning T.L., Crawford S.E., Pruijssers A.J., Iskarpatyoti J.A., Estes M.K., Dermody T.S., Ouyang W., Williams I.R., Vijay-Kumar M., Gewirtz A.T. Viral infection. Prevention and cure of rotavirus infection via TLR5/NLRC4-mediated production of IL-22 and IL-18. Science. 2014;346(6211):861–865. doi: 10.1126/science.1256999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Hu R., Tang X., Wu C., He Q., Zhao Z., Chen H., Wu B. Occurrence and investigation of enteric viral infections in pigs with diarrhea in China. Arch. Virol. 2013;158(8):1631–1636. doi: 10.1007/s00705-013-1659-x. [DOI] [PubMed] [Google Scholar]
- Zhao Z.P., Yang Z., Lin W.D., Wang W.Y., Yang J., Jin W.J., Qin A.J. The rate of co-infection for piglet diarrhea viruses in China and the genetic characterization of porcine epidemic diarrhea virus and porcine kobuvirus. Acta. Virol. 2016;60(1):55–61. doi: 10.4149/av_2016_01_55. [DOI] [PubMed] [Google Scholar]
- Zheng Y., Valdez P.A., Danilenko D.M., Hu Y., Sa S.M., Gong Q., Abbas A.R., Modrusan Z., Ghilardi N., de Sauvage F.J., Ouyang W. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat. Med. 2008;14(3):282–289. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.