Highlights
-
•
Development of a reliable and cost-effective tool for IBV Mass-type and QX-like genotyping.
-
•
IBV QX vaccine markers enable vaccine differentiation via melting curve analysis.
-
•
SYBRgreen RT-qPCR assay supports multiplexing and enables IBV genotyping.
Keywords: Infectious bronchitis virus, Genotyping, Vaccine differentiation, RT-qPCR, Melting curve
Abstract
Infectious Bronchitis Virus (IBV) is a highly contagious virus of chicken, causing huge economic losses in the poultry industry. Many genotypes circulate in a given area, and optimal protection relies on vaccination with live attenuated vaccines of the same genotype. As these live vaccines are derived from field viruses and circulate, understanding the prevalence of different IBV genotypes in any area is complex. In a recent study, the genome comparison of an IBV QX vaccine and its progenitor field strain led to the identification of vaccine markers. Here we developed a simplex SYBRgreen RT-qPCR assay for differentiation between QX-like field and vaccine strains and a multiplex SYBRgreen RT-qPCR assay for IBV genotyping with melting curve analysis, as each virus produced distinct and reliable melting peaks. Both the simplex and the multiplex assays showed excellent efficiency, sensitivity and specificity representing a low cost diagnostic tool for IBV genotyping and vaccine differentiation.
1. Introduction
Infectious Bronchitis Virus (IBV) is a highly contagious avian coronavirus belonging to the family Coronaviridae, genus Gammacoronavirus. IBV infects primarily domestic fowl causing disease associated with respiratory signs, and in some cases with kidney lesions and proventriculitis, resulting in increased mortality in unprotected birds and in serious economic losses (Cook et al., 2012). There is a wide variety of genetically distinct IBV genotypes; some have been continuously detectable since suitable typing techniques were developed, some have appeared, briefly caused a problem and then disappeared, some proved restrained to a particular geographical area, some widespread (Cook et al., 2012; Jackwood et al., 2012). The control of the disease occurs mainly by vaccination using live attenuated homologous vaccines (Geerligs et al., 2011; Jackwood et al., 2003), or a combination of two antigenically distinct live attenuated vaccines (Jackwood et al., 2012; Jordan, 2017; Terregino et al., 2008). Effective knowledge of the prevalence and circulation of different IBV genotypes is needed to decide upon the most effective vaccination protocol. However, this decision is complicated by the circulation and thus presence of the very live vaccines applied for disease control. For example, withdrawal of IBV 793/B (lineage GI-13) live vaccine in Northern Italy led to the disappearance of this previously thought ubiquitous genotype, suggesting that IBV 793/B virulent strains were absent in the area (Franzo et al., 2014a).
IBV detection is commonly performed using TaqMan® RT-qPCR assays (Callison et al., 2006). IBVs’ differentiation and genotyping rely on serological analyses and/or on RT-PCR followed by sequencing of full or partial S1 gene, which, due to its variability and biological function, is the region commonly targeted for this purpose (Cavanagh et al., 1999; Worthington et al., 2008). However, they both represent expensive and/or time-consuming methodologies. SYBRgreen RT-qPCR paired with melting curve analysis represents a more economic and time-effective strategy for IBV genotyping and for the differentiation between vaccine and field strains in a single analysis. Using this approach genotyping and/or vaccine differentiation relies on sequence variants identification based on their individual melting temperatures, a strategy successfully applied for typing of other avian viruses (Franzo et al., 2014b; Pham et al., 2005; Steer et al., 2011). However, due to the high number of IBV genotypes (or lineages) (Valastro et al., 2016) and the lack of vaccine markers for most of IBV vaccines (Listorti et al., 2017) the development of effective SYBRgreen RT-qPCR assays represents a challenge for any research group embarking in this task.
Here we explored SYBRgreen RT-qPCR paired with melting curve analysis to a) distinguish between IBV field strains and homologous attenuated live vaccine and b) support multiplexing towards the differentiation between IBV genotypes. Comparison of the full genome sequences of vaccine Poulvac IB QX (Zoetis) and its progenitor 1148-A identified several vaccine markers, including three consecutive nucleotide changes in the S1 gene (Listorti et al., 2017). Those unique vaccine markers affecting the GC content of the target region were targeted to develop a SYBRgreen RT-qPCR assay enabling the differentiation of vaccine Poulvac IB QX from QX-like field strains via melting curve analysis. Next we developed a multiplex assay for the simultaneous genotyping of IBV Mass-type (lineage GI-1) and IBV QX-like (lineage GI-19) strains, as those two genotypes represent a major threat for the poultry industry worldwide (Cook et al., 2012; de Wit et al., 2018; Jackwood et al., 2012). To achieve this, specific primer sets were designed and RT-qPCR and SYBRgreen analysis settings were optimized in order to detect and differentiate the sequence variants of interest. Both the simplex and the multiplex approaches proved reliable and effective diagnostic tools, showing excellent sensitivity, specificity and repeatability within and between runs. The application of these protocols will aid in the understanding of the molecular epidemiology of IBV and they represent proves of principle for future developments and applications of melting curve analyses towards IBV diagnosis.
2. Materials and methods
2.1. Viruses and viral RNA isolation
IBV-D388 was isolated by GD Animal Health (Deventer, The Netherlands) in March 2004 from 19-day-old broiler breeders with respiratory signs and increased mortality due to renal failure and typed as IBV QX-like (Lineage GI-19) (de Wit et al., 2011). Poulvac IB QX (Zoetis) is a live attenuated avian infectious bronchitis virus derived from QX-like field strain IB 1148-A after serial passages in embryonated chicken eggs (ECEs) (Geerligs et al., 2011). IBV M41 strain was kindly provide by GD Animal Health (Deventer, The Netherlands); the virus was isolated from birds showing respiratory sings and drop in egg production. Nobilis® IB H120 (MSD/Animal Health, Boxmeer, The Netherlands) is a live attenuated avian infectious bronchitis Mass-type vaccine. For each virus a stock was produced. Briefly, ten 8-day-old specific pathogens free (SPF) ECEs were inoculated with a 50 % embryonic infectious dose (EID50) of 100, incubated for 48 h, and cooled at 4 °C for 24 h prior to harvesting and pooling of the allantoic fluid (AF). Viruses titration was performed in 8-day-old ECEs by determining the EID50 per ml at 7 days post infection (dpi) according to the Reed and Muench method (Reed and Muench, 1938). RNA was extracted from the allantoic fluid using QIAamp viral RNA minikit (QIAGEN, Hilden, Germany) following the manufacturer’s protocol.
2.2. Oligo design
All available IBV QX-like S1 sequences (96) deposited on ViPR (https://www.viprbrc.org) were downloaded on 30 November 2018 and aligned using MAFFT version 7 (https://mafft.cbrc.jp/alignment/software/). QX-specific oligo pairs were designed to amplify the portion of S1 gene containing the two transversion (G→T) affecting the GC content of the gene, hence affecting also the melting temperature of the amplicon. To confirm the higher GC content in QX-like field strains detected worldwide, the multiple sequence alignment was trimmed to include solely the target region. For easiness of analysis, the dataset was processed with cd‐hit‐est test of the CD‐HIT Suite (http://weizhong-cluster.ucsd.edu/cdhit_suite/cgi-bin/index.cgi?cmd = cd-hit-est) (Li and Godzik, 2006) to cluster sequences that shared 100 % identity, such as each cluster represents a unique prototype sequence. The resulting unique prototype sequences were processed using uMELT Melting Curve Predictions Software (https://www.dna.utah.edu/umelt/umelt.html) to compare the predicted melting curve of vaccine Poulvac IB QX to those of the field strains. Eventually, the specificity of the oligo pairs was assessed using Primer-BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) (Jian et al., 2012). Similarly, all available Mass-type S1 sequences (166) deposited on ViPR (https://www.viprbrc.org) were downloaded on 30 November 2018. Using the same approach Mass-type specific oligo sets were designed, with the exception that after clustering a web-based Shannon Entropy calculator (https://www.hiv.lanl.gov/content/sequence/ENTROPY) was used to identify conserved region among the dataset.
2.3. Simplex and multiplex RT-qPCR optimization
All RT-qPCRs were carried out in a Bio-Rad CFX Connect real-time PCR system using the iTaq universal SYBRgreen one-step kit (Bio-Rad Laboratories, Hercules, California, USA). Optimization of the assays was performed using ten-fold viral dilutions of Poulvac IB QX, IBV-D388, IBV M41 GD and Nobilis® IB H120, different primer concentrations, annealing temperatures, extension step lengths and temperature increment during the melting curve analysis.
2.4. Analytical validation
The efficiency of the simplex RT-qPCR was determined using serial dilution of Poulvac IB QX ranging from 107,83 EID50 to 100,83 EID50 and of IBV-D388 ranging from 107,31 EID50 to 100,31 EID50. Similarly the efficiency of the multiplex assay was determined using the same dilution panels for the QX-like viruses, plus dilution panels for IBV M41 GD (107,63 EID50-100,63 EID50) and Nobilis® IB H120 (107,75 EID50-100,75 EID50). Two operators tested the lowest dilution detected as positive by the assays, using nine independent replicates; the lowest viral amount that can be detected in at least 50 % of replicates was considered the assays’ limit of detection (LoD).
2.5. Repeatability
Two operators using three viral dilutions for each virus, corresponding to high, medium and low dilution (one log higher than LoD) during three different experiments performed at weekly intervals, evaluated the repeatability. Inter and intra-run coefficient of variations (CV) were calculated to assess the robustness of the assay. The significance of Tm difference between vaccine and field strains and between genotypes among different runs and operators were evaluated by T-test using GraphPad Prism 7 (https://www.graphpad.com/scientific-software/prism/).
2.6. Analytical specificity
Specificity of the assays was assessed against other IBV’s genotype (IBV M41 GD, Nobilis® IB H120, H52 BI, IBV B1648, IBV 4/91 GD, Nobilis® IB IBV 4/91) and a panel of avian viruses, including Avian Influenza H9N2 (A/Chicken/Saudi Arabia/SP02525/3AAV/2000), Avian Metapneumovirus subtype A (Nobilis® TRT), Newcastle Disease Virus (Nobilis® ND LaSota), and Infectious Laryngotracheitis Virus (Nobilis® ILT).
2.7. Diagnostic validation
Differentiation of Poulvac IB QX and IBV-D388 by melting curve analysis using the simplex assay was performed testing three panels of 20 samples of known infectious status (allantoic fluid obtained from IBV, non-IBV and mock infected ECEs) prepared by one operator and blindly tested by two different operators at weekly intervals. The diagnostic sensitivity (DSe), the diagnostic specificity (DSp), the overall accuracy (Acc) and the vaccine differentiation were calculated. The ability of the multiplex RT-qPCR to differentiate between the two genotypes and identified vaccines Poulvac IB QX by melting curve analysis was carried out following the experiment set up described above.
3. Results
3.1. In silico analysis
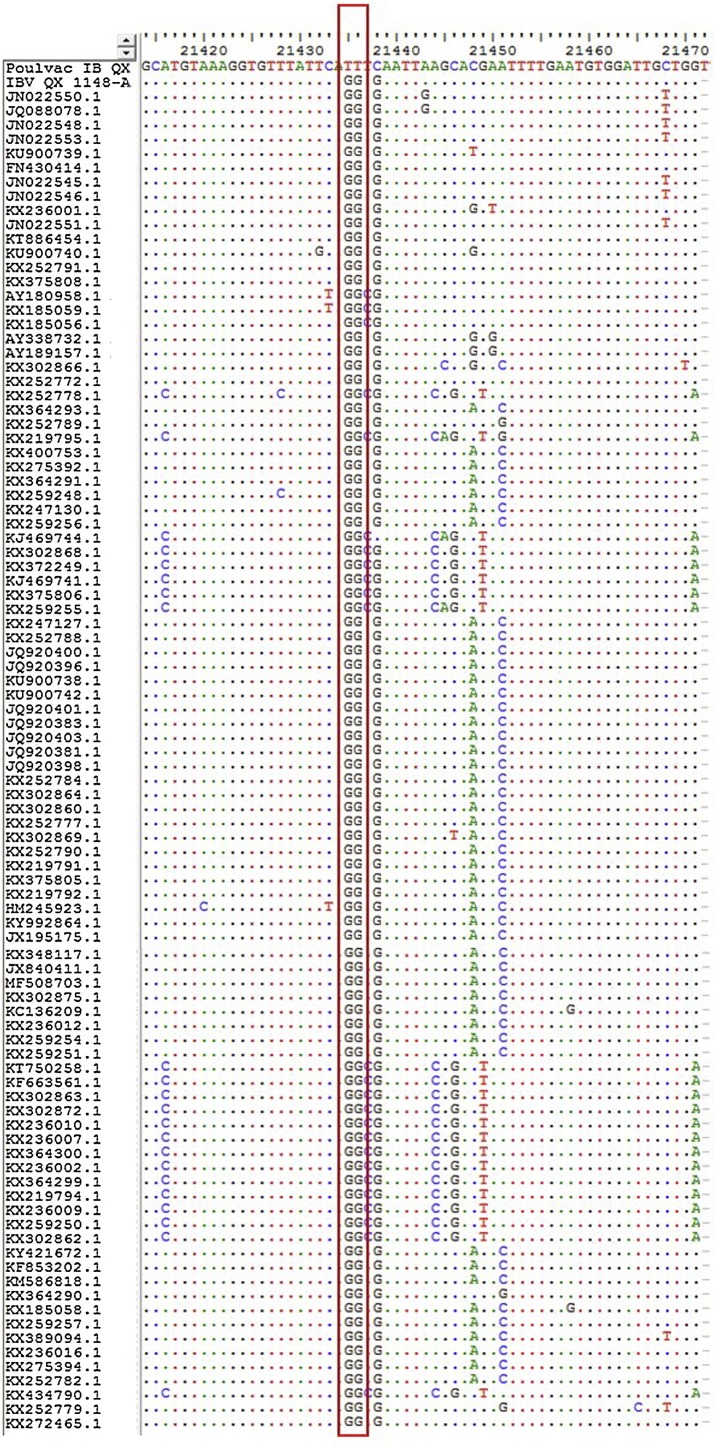
In silico analysis confirmed the distinctiveness of the two transversion (G→T) occurred during the attenuation process of vaccine Poulvac IB QX (Fig. 1 ); as a consequence the target region of the vaccine strain has a GC content of 34,48 %, while for the field strains it ranged from 36.62 % to 41.38 % (supplementary materials 1). The predicted differences in Tm between the vaccine and the field strains based on uMelt analysis ranged from 1 °C to 3 °C. The GC content of the QX field strain used in this study was 37.93 %, and the predicted Tm difference between Poulvac IB QX and IBV-D388 was 1.5 °C. The Mass-type specific oligo were designed to amplify a region longer than the QX-like one, characterized by a GC content ranging from 40.00 % of IBV M41 GD to 45.71 % of Nobilis® IB H120 (supplementary materials 1). uMelt analysis predicted a Tm difference between the two genotypes ranging from 3 °C to 5 °C, while the predicted Tm difference between the two Mass-type viruses used in this study was 1.5 °C.
Fig. 1.
Alignment of the S1 gene portion targeted for QX vaccine differentiation. The red box highlighted the distinctive transverstions (G→T) occurred at genome positions 21435 and 21436 affecting the GC content of Poulvac IB QX (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).
3.2. Assays optimization
The oligo pairs, their concentrations, and cycling protocol maximizing RT-qPCR performances, avoiding nonspecific amplification, was defined. Sequences of the selected oligo pairs, their concentration and the thermal profile are reported in Table 1 .
Table 1.
Sequences of the oligo, their final concentration and thermal cycling profile used.
| Oligo | Sequence (5’ – 3’) | Position (5’-3’) | Concentration (μM) | |
|---|---|---|---|---|
| QX-like_Forward | GCATGTAAAGGTGTTTA | 21628-21644 | 0,25 | |
| QX-like_Reverse | CCAGCAATCCACATTC | 21673-21685 | 0,25 | |
| Mass-type_Forward | GATGGGTGTCCTATAAC | 20742-20758 | 0,20 | |
| Mass-type_Reverse | GCTGGCCATTTTTCATAGCAGAAAC | 20787-20811 | 0,15 | |
| Thermal cycling profile | Temperature | Duration | N° cycles | |
| Reverse transcription reaction | 50 °C | 10 min | ||
| Polymerase Activation | 95 °C | 1 min | ||
| Denaturation | 95 °C | 10 sec | X40 | |
| Annealing | 55 °C | 10 sec | ||
| Extension | 60 °C | 20 sec | ||
| Melt-Curve Analysis | 60-90 °C – 0,2 °C increment 5 sec/step | |||
3.3. SYBRgreen RT-qPCR followed by melting curve analysis enables the differentiation of Poulvac IB QX from QX-like field strains
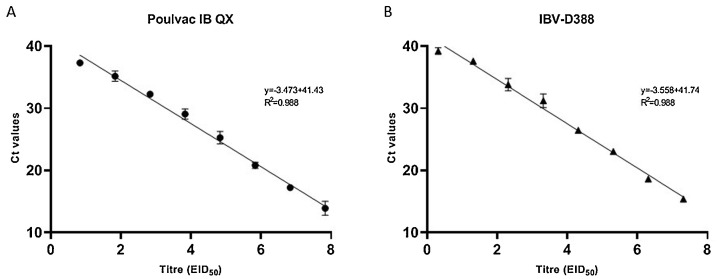
The detection range of the RT-qPCR assay was from 107,83 EID50 to 101,83 EID50 for Poulvac IB QX and 107,31 EID50 to 101,31 EID50 for IBV-D388, showing excellent efficiency and linearity for both the viruses up to the dilution above the limit of detection (Fig. 2 ), paired with excellent robustness, as CVs were consistently low both within and between runs (Table 2 ).
Fig. 2.
Standard curves for QX vaccine differentiation using serial ten-fold dilutions of vaccine Poulvac IB QX (A) and field strain IBV-D388 (B). Each point of the curves represents the mean of three replicates; branches represent standard deviation. Linear and R2 are reported for each standard curve.
Table 2.
Repeatability of the RT-qPCR assay for differentiation between vaccine Poulvac IB QX and QX-like field strain. Mean Ct (three replicates), standard deviation (SD) and CV are reported for each virus according to virus, dilution, operator and week of experiment.
| Week 1 |
Week 2 |
Week 3 |
Total |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Virus | Titre | Operator | Ct mean ± SD | CV | Ct mean ± SD | CV | Ct mean ± SD | CV | Ct mean ± SD | CV |
| Poulvac IB QX | 106.83 | 1 | 17,00 ± 0,14 | 0,80 | 16,93 ± 0,13 | 0,77 | 16,89 ± 0,15 | 0,90 | 16,77 ± 0,30 | 1,79 |
| 2 | 16,82 ± 0,20 | 1,21 | 16,66 ± 0,63 | 2,18 | 16,34 ± 0,31 | 1,89 | ||||
| 104.83 | 1 | 24,65 ± 0,12 | 0,85 | 23,88 ± 0,25 | 1,03 | 24,52 ± 0,07 | 0,29 | 24,36 ± 0,49 | 2,01 | |
| 2 | 24,81 ± 0,45 | 1,82 | 23,67 ± 0,17 | 0,72 | 24,66 ± 0,21 | 0,86 | ||||
| 102.83 | 1 | 32,33 ± 0,53 | 1,64 | 33,61 ± 0,37 | 1,11 | 32,12 ± 0,18 | 0,55 | 32,05 ± 0,93 | 2,90 | |
| 2 | 31,70 ± 0,53 | 1,68 | 31,20 ± 0,87 | 2,79 | 31,31 ± 0,03 | 0,08 | ||||
| IBV-D388 | 106.31 | 1 | 17,75 ± 0,23 | 1,30 | 17,72 ± 0,17 | 0,95 | 17,57 ± 0,16 | 0,93 | 17,70 ± 0,38 | 2,14 |
| 2 | 17,50 ± 0,28 | 1,58 | 17,37 ± 0,39 | 2,25 | 18,31 ± 0,26 | 1,40 | ||||
| 104.31 | 1 | 25,26 ± 0,42 | 1,64 | 24,63 ± 0,45 | 1,82 | 25,71 ± 0,47 | 1,86 | 24,98 ± 0,43 | 1,72 | |
| 2 | 25,26 ± 0,38 | 1,51 | 24,77 ± 0,25 | 1,01 | 24,82 ± 0,44 | 1,79 | ||||
| 102.31 | 1 | 33,26 ± 0,14 | 0,42 | 32,80 ± 0,30 | 0,91 | 31,39 ± 0,72 | 2,30 | 32,18 ± 0,93 | 2,90 | |
| 2 | 32,36 ± 0,81 | 2,51 | 31,35 ± 0,89 | 2,85 | 32,23 ± 0,89 | 2,77 | ||||
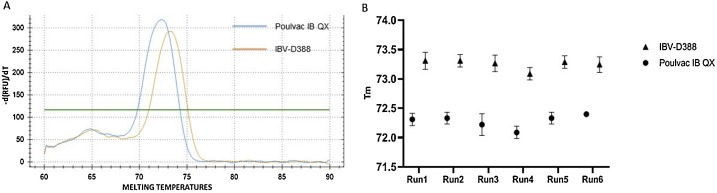
The assay produced two distinguishable melting temperatures between the vaccine and the field strain, 72.2 °C (±0,2 °C) and 73.2 °C (±0,2 °C) respectively (Fig. 3 A). The melting temperatures observed were constant and reproducible among different replicates, operators and experiments, as depicted in Fig. 3B. T-tests confirmed the presence of a significant (P < 0.001) difference in the Tm between the two viruses, with a mean Tm of 72,28 °C and of 73,25 °C for Poulvac IB QX and IBV-D388, respectively. DSe was equal to 98.41 %, Dsp to 100 %, Acc to 98,86 % and vaccine differentiation accuracy to 100 %. To conclude, no increase in fluorescence associated with sigmoidal amplification curve was observed (data not shown) for no template controls, non QX-like IBVs nor other avian pathogens as listed in Materials and Methods.
Fig. 3.
Melting curves observed for Poulvac IB QX and IBV-D388 (A) using the simplex RT-qPCR assay (A). Melting temperatures among six independent runs (B). Each point represent the mean of nine replicates, branches represent standard deviation (SD) (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).
3.4. SYBRgreen RT-qPCR followed by melting curve analysis supports multiplexing and enables IBV genotyping
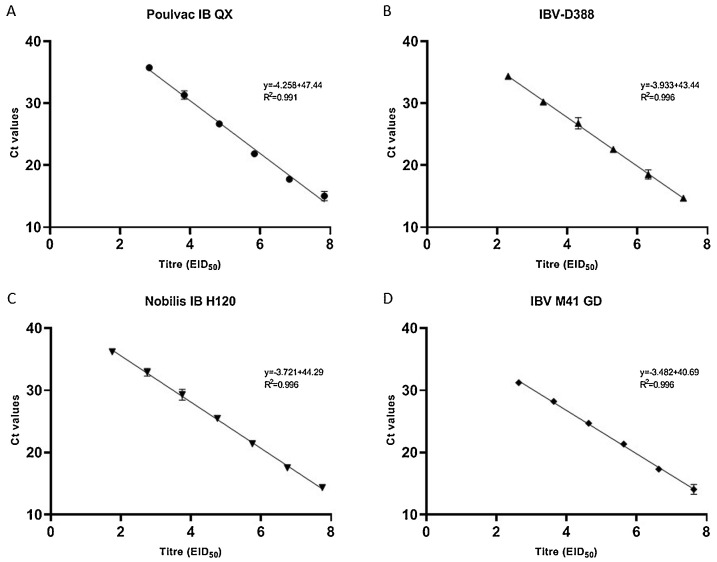
The detection range of the multiplex RT-qPCR was from 107,83 EID50 to 102,83 EID50 for Poulvac IB QX, from 107,31 EID50 to 102,31 EID50 for IBV-D388, from 107,63 EID50 to 102,83 EID50 for IBV M41 GD and 107,75 EID50 to 101,75 EID50 for Nobilis® IB H120. Furthermore, the assay proved linear when tested against the four dilution panels up to the lowest dilution detected (Fig. 4 ) and robust, as CVs were consistently low both within and between runs (Table 3 ).
Fig. 4.
Standard curves obtained for the multiplex assay using serial ten-fold dilutions of vaccine Poulvac IB QX (A), field strain IBV D388 (B), Nobilis® IB H120 (C) and IBV M41 GD (D). Each point of the curves represents the mean of three replicates; branches represent standard deviation. Linear and R2 are reported for each standard curve.
Table 3.
Repeatability of the multiplex RT-qPCR assay for IBV genotyping. Mean Ct (three replicates), standard deviation (SD) and CV are reported for each virus according to virus, dilution, operator and week of experiment.
| Week 1 |
Week 2 |
Week 3 |
Total |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Virus | Titre | Operator | Ct mean ± SD | CV | Ct mean ± SD | CV | Ct mean ± SD | CV | Ct mean ± SD | CV |
| Poulvac IB QX | 107.83 | 1 | 15,71 ± 0,38 | 2,43 | 15,70 ± 0,11 | 0,73 | 15,53 ± 0,13 | 0,89 | 15,44 ± 0,29 | 1,92 |
| 2 | 15,26 ± 0,01 | 0,11 | 15,14 ± 0,34 | 2,29 | 15,29 ± 0,11 | 0,76 | ||||
| 105,83 | 1 | 23,53 ± 0,44 | 1,88 | 23,40 ± 0,19 | 0,85 | 23,17 ± 0,32 | 1,41 | 23,37 ± 0,48 | 2,05 | |
| 2 | 23,11 ± 0,22 | 0,98 | 24,11 ± 0,21 | 0,89 | 22,85 ± 0,18 | 0,82 | ||||
| 103.83 | 1 | 32,22 ± 0,75 | 2,34 | 31,00 ± 0,02 | 0,08 | 32,32 ± 0,42 | 1,30 | 31,66 ± 0,78 | 2,49 | |
| 2 | 31,56 ± 0,18 | 0,59 | 32,33 ± 0,22 | 0,68 | 30,56 ± 0,43 | 1,44 | ||||
| IBV-D388 | 10731 | 1 | 15,85 ± 0,43 | 2,75 | 16,13 ± 0,40 | 2,53 | 15,59 ± 0,28 | 1,83 | 15,82 ± 0,34 | 2,17 |
| 2 | 15,73 ± 0,30 | 1,95 | 15,67 ± 0,25 | 1,62 | 15,93 ± 0,35 | 2,20 | ||||
| 105.31 | 1 | 22,97 ± 0,47 | 2,07 | 23,05 ± 0,34 | 1,50 | 24,12 ± 0,57 | 2,38 | 23,40 ± 0,59 | 2,56 | |
| 2 | 23,75 ± 0,41 | 1,74 | 23,77 ± 0,19 | 0,80 | 22,84 ± 0,37 | 1,64 | ||||
| 103,31 | 1 | 31,51 ± 0,92 | 2,93 | 30,54 ± 0,53 | 1,74 | 32,14 ± 0,24 | 0,75 | 31,28 ± 0,76 | 2,45 | |
| 2 | 31,52 ± 0,48 | 1,52 | 31,57 ± 0,22 | 0,73 | 30,40 ± 0,34 | 1,14 | ||||
| Nobilis® IB H120 | 107.75 | 1 | 16,58 ± 0,26 | 1,62 | 16,93 ± 0,17 | 1,08 | 16,71 ± 0,22 | 1,37 | 16,51 ± 0,22 | 1,31 |
| 2 | 16,58 ± 0,12 | 0,73 | 16,25 ± 0,08 | 0,53 | 16,56 ± 0,11 | 0,66 | ||||
| 105.75 | 1 | 24,12 ± 0,30 | 1,28 | 23,10 ± 0,02 | 0,11 | 24,05 ± 0,10 | 0,42 | 23,66 ± 0,45 | 1,90 | |
| 2 | 24,02 ± 0,04 | 0,20 | 23,39 ± 0,20 | 0,89 | 23,29 ± 0,21 | 0,91 | ||||
| 103,75 | 1 | 29,93 ± 0,62 | 2,09 | 30,71 ± 0,61 | 2,01 | 31,55 ± 0,81 | 2,58 | 30,85 ± 0,75 | 2,43 | |
| 2 | 31,52 ± 0,46 | 1,46 | 30,85 ± 0,22 | 0,71 | 30,53 ± 0,44 | 1,45 | ||||
| IBV M41 GD | 107.63 | 1 | 16,17 ± 0,20 | 1,27 | 15,84 ± 0,12 | 0,77 | 16,13 ± 0,07 | 0,46 | 15,91 ± 0,32 | 2,04 |
| 2 | 15,34 ± 0,21 | 1,39 | 16,15 ± 0,09 | 0,58 | 15,83 ± 0,12 | 0,81 | ||||
| 104.63 | 1 | 23,11 ± 0,03 | 0,15 | 22,45 ± 0,30 | 1,32 | 23,47 ± 0,03 | 0,16 | 22,86 ± 0,43 | 1,91 | |
| 2 | 22,43 ± 0,11 | 0,51 | 23,03 ± 0,07 | 0,33 | 22,46 ± 0,19 | 0,85 | ||||
| 103.63 | 1 | 30,68 ± 0,47 | 1,53 | 29,58 ± 0,16 | 0,54 | 30,45 ± 0,13 | 0,43 | 30,36 ± 0,54 | 1,80 | |
| 2 | 29,90 ± 0,37 | 1,23 | 30,67 ± 0,32 | 1,05 | 30,90 ± 0,24 | 0,77 | ||||
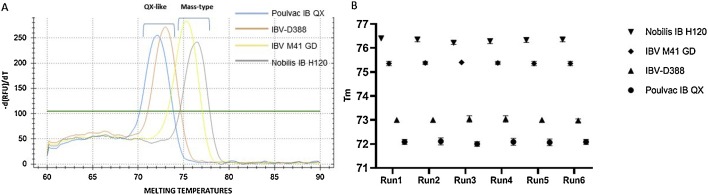
The melting curve analysis confirmed the Tm previously observed for QX-like viruses, while the Tm detected for IBV M41 GD and Nobilis® IB H120 were 75.4 °C (±0,2 °C) and 76.4 °C (±0,2 °C) respectively (Fig. 5 A). The Tm proved to be constant and reproducible among different replicates, operators and experiment as depicted in Fig. 5B. T-test confirmed the presence of significant difference (P < 0.001) between the Tm of the two genotypes, with a mean Tm of 72,54 °C and of 75,84 °C for QX-like and Mass-type IBVs respectively. Furthermore, T-test showed a significant difference (P < 0.001) between the Tm of vaccine Poulvac IB QX and its progenitor, and between vaccine Nobilis® IB H120 and Mass-type field strain IBV M41 GD. No increase in fluorescence associated with a sigmoidal amplification curve was observed for no template controls, non-QX-like and non-Mass-type IBVs nor other avian pathogens (data not shown). A slight increase in fluorescence was detected when processing an IBV B1648 (GI-14 lineage) isolate; however, it was associated with a non-sigmoidal amplification curve (linear shape) at Ct values above 38 and paired with no melting curves. In the diagnostic setting used, the assay showed DSe of 98,11 %, DSp of 100 %, Acc of 98,31 % and genotyping accuracy of 100 %.
Fig. 5.
Melting curves observed for Poulvac IB QX, IBV-D388, Nobilis IB H120 and IBV M41 GD when tested using the multiplex approach for IBV genotyping (A). Melting temperatures among six independent runs (B). Each point represent the mean of nine replicates, branches represent standard deviation (SD) (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).
4. Discussion
In the present study, we developed two SYBRgreen RT-qPCR assays paired with melting curve analysis for the differentiation between IBV vaccine and field strains and for genotyping. To this purpose we first developed and validated a protocol to differentiate vaccine Poulvac IB QX from QX-like field strains using three distinct vaccine makers arose during the attenuation process (Listorti et al., 2017). Then we developed a multiplex protocol for IBV genotyping, focusing on QX-like (GI-19) and Mass-type (GI-1) strains, being two of the most widespread IBV genotypes (Cook et al., 2012; de Wit et al., 2018; Jackwood et al., 2012).
Both the assays showed high analytical sensitivity, however the limit of detection estimated for Poulvac IB QX and IBV-D388 differs between the two assays, with the simplex proving to be more sensitive than the multiplex approach. It is possible that the lower sensitivity of the multiplex assay is due to heterodimers assembling between QX-like and Mass-type oligo, reducing the availability of the QX-specific oligo during the annealing phase of the thermal cycle.
The good efficiency and linearity at least up to one dilution above the limit of detection, together with the high repeatability observed for both the assays, suggest their suitability for viral quantification not only for diagnostic, but also for research purposes. To support, the simplex RT-qPCR has been successfully used in a study aiming to assess the tissue distribution of IBV-D388 at early time points after infection (manuscript in preparation). The same study showed that the efficiency of the protocol is not affected by the sample’s matrix, as viral RNA was detected and quantified in tracheal and cloacal swabs, lungs and kidneys. The optimization of the designed oligo sets, reaction chemistries, thermal protocol and melting curve analyses avoided the presence of nonspecific amplifications, which, coupled with the in silico evaluation performed, ensured the high specificity of the assays, as demonstrated when tested against others IBV genotypes and different avian pathogens.
Implementation of the melting curve analysis enables differentiation between vaccine Poulvac IB QX and field strains IBV-D388, as the melting temperatures of each virus proved constant within not only the same run, but also among different experiments and operators, and no overlaps between the melting temperatures of the two viruses were observed throughout the study. Noteworthy, the melting curve analysis proved extremely accurate, being able to identify sequence variants that differ only by three nucleotides, of which just two affect the GC content of the amplicon. QX-like field viruses will continuously evolve and the persistence in the field of Poulvac IB QX might lead to the introduction of mutations in the latter; however, the chance of the emergence of field variants showing a sequence identical to Poulvac IB QX by random mutations, or back mutations in the applied vaccine lessening the strength of the protocol, are extremely low (Laconi et al., 2019; Oade et al., 2019).
A constant difference in melting temperature was also observed between vaccine Nobilis® IB H120 and field strain IBV M41 GD; however, the unavailability of vaccine markers for Nobilis® IB H120 hinders to confidently identify it. In our study, the melting curve analysis towards vaccine differentiation proved an efficient and reliable tool, remarking the urgency of the availability of vaccine and progenitor sequences to determine markers for all IBV vaccines. This will be needed to obtain a precise knowledge of field viruses circulating in a certain area.
Melting curve analysis was also successfully used to genotype QX-like and Mass-type strains in a single run using a multiplex RT-qPCR approach. These two genotypes were chosen as target representing a major threat worldwide; however, this multiplex approach might be easily improved and adapted, via designing of specific oligo, to identify other IBV genotypes depending on the geographical area of interest and the epidemiological data available.
Taken as a whole, the data gathered in the present study clearly demonstrate that melting curve analysis enables IBV genotyping and vaccine differentiation, representing a cost and time effective method in comparison to those in use in diagnostic laboratories worldwide.
Funding
This research was financially supported by the research institute.
Declaration of Competing Interest
The authors have no competing interests to declare.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.jviromet.2019.113771.
Appendix A. Supplementary data
The following is supplementary data to this article:
References
- Callison S.A., Hilt D.A., Boynton T.O., Sample B.F., Robison R., Swayne D.E., Jackwood M.W. Development and evaluation of a real-time Taqman RT-PCR assay for the detection of infectious bronchitis virus from infected chickens. J. Virol. Methods. 2006;138:60–65. doi: 10.1016/j.jviromet.2006.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh D., Mawditt K., Britton P., Naylor C.J. Longitudinal field studies of infectious bronchitis virus and avian pneumovirus in broilers using type-specific polymerase chain reactions. Avian Pathol. 1999;28:593–605. doi: 10.1080/03079459994399. [DOI] [PubMed] [Google Scholar]
- Cook J.K.A., Jackwood M., Jones R.C. The long view: 40 years of infectious bronchitis research. Avian Pathol. 2012;41:239–250. doi: 10.1080/03079457.2012.680432. [DOI] [PubMed] [Google Scholar]
- de Wit J.J., Cazaban C., Dijkman R., Ramon G., Gardin Y. Detection of different genotypes of infectious bronchitis virus and of infectious bursal disease virus in European broilers during an epidemiological study in 2013 and the consequences for the diagnostic approach. Avian Pathol. 2018;47:140–151. doi: 10.1080/03079457.2017.1387231. [DOI] [PubMed] [Google Scholar]
- de Wit J.J., Nieuwenhuisen-van Wilgen J., Hoogkamer A., vande Sande H., Zuidam G.J., Fabri T.H.F. Induction of cystic oviducts and protection against early challenge with infectious bronchitis virus serotype D388 (genotype QX) by maternally derived antibodies and by early vaccination. Avian Pathol. 2011;40:463–471. doi: 10.1080/03079457.2011.599060. [DOI] [PubMed] [Google Scholar]
- Franzo G., Drigo M., Lupini C., Catelli E., Laconi A., Listorti V., Bonci M., Naylor C.J., Martini M., Cecchinato M. A sensitive, reproducible, and economic real-time reverse transcription PCR detecting avian metapneumovirus subtypes A and B. Avian Dis. 2014;58 doi: 10.1637/10676-092413-Reg.1. [DOI] [PubMed] [Google Scholar]
- Franzo G., Naylor C.J., Lupini C., Drigo M., Catelli E., Listorti V., Pesente P., Giovanardi D., Morandini E., Cecchinato M. Continued use of IBV 793B vaccine needs reassessment after its withdrawal led to the genotype’s disappearance. Vaccine. 2014;32:6765–6767. doi: 10.1016/j.vaccine.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geerligs H.J., Tarres-Call J., Stuurman B.G.E., Bru T., Wijmenga W., Symons J., Meinders C.A.M., Karaca K., Boelm G.-J., Kumar M., Vila R., Mombarg M. Efficacy and safety of an attenuated live QX-like infectious bronchitis virus strain as a vaccine for chickens. Avian Pathol. 2011;40:93–102. doi: 10.1080/03079457.2010.542742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackwood M.W., Hall D., Handel A. Molecular evolution and emergence of avian gammacoronaviruses. Infect. Genet. Evol. 2012;12:1305–1311. doi: 10.1016/j.meegid.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackwood M.W., Hilt D.A., Brown T.P. Attenuation, safety, and efficacy of an infectious bronchitis virus GA98 serotype vaccine author (s) In: Jackwood Mark W., Hilt Deborah A., Thomas P., editors. Brown Published by : American Association of Avian Pathologists Stable. 2003. pp. 627–632.http://www.jstor.org47 URL: [DOI] [PubMed] [Google Scholar]
- Jian Y., George C., Irena Z., Ioana C., Steve R., Madden Thomas L. Primer- BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012;13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan B. Vaccination against infectious bronchitis virus: a continuous challenge. Vet. Microbiol. 2017;206:137–143. doi: 10.1016/j.vetmic.2017.01.002. [DOI] [PubMed] [Google Scholar]
- Laconi A., Catelli E., Cecchinato M., Naylor C.J. Two similar commercial live attenuated AMPV vaccines prepared by random passage of the identical field isolate, have unrelated sequences. Vaccine. 2019 doi: 10.1016/j.vaccine.2019.04.036. [DOI] [PubMed] [Google Scholar]
- Li W., Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- Listorti V., Laconi A., Catelli E., Cecchinato M., Lupini C., Naylor C.J. Identification of IBV QX vaccine markers: should vaccine acceptance by authorities require similar identifications for all live IBV vaccines? Vaccine. 2017:35. doi: 10.1016/j.vaccine.2017.06.021. [DOI] [PubMed] [Google Scholar]
- Oade M.S., Keep S., Freimanis G.L., Orton R.J., Britton P., Hammond J.A., Bickerton E. Crossm attenuation of infectious bronchitis virus in eggs results in different patterns of genomic variation across multiple replicates. J. Virol. 2019;93:1–14. doi: 10.1128/JVI.00492-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham H.M., Konnai S., Usui T., Chang K.S., Murata S., Mase M., Ohashi K., Onuma M. Rapid detection and differentiation of Newcastle disease virus by real-time PCR with melting-curve analysis. Arch. Virol. 2005;150:2429–2438. doi: 10.1007/s00705-005-0603-0. [DOI] [PubMed] [Google Scholar]
- Reed L.J., Muench H. A simple method of estimating fifty per cent endpoints. Am. J. Hygene. 1938;27:493–497. [Google Scholar]
- Steer P.A., O’Rourke D., Ghorashi S.A., Noormohammadi A.H. Application of high-resolution melting curve analysis for typing of fowl adenoviruses in field cases of inclusion body hepatitis. Aust. Vet. J. 2011;89:184–192. doi: 10.1111/j.1751-0813.2011.00695.x. [DOI] [PubMed] [Google Scholar]
- Terregino C., Toffan, AnnaBeato M., Serena Beato M.S., De Nardi R., Meini A., Ortali G., Capua I., Vascellari M., Mancin M. Pathogenicity of a QX strain of infectious bronchitis virus in specific pathogen free and commercial broiler chickens, and evaluation of protection induced by a vaccination programme based on the Ma5 and 4/91 serotypes. Avian Pathol. 2008;37:487–493. doi: 10.1080/03079450802356938. [DOI] [PubMed] [Google Scholar]
- Valastro V., Holmes E.C., Britton P., Fusaro A., Jackwood M.W., Cattoli G., Monne I. S1 gene-based phylogeny of infectious bronchitis virus: an attempt to harmonize virus classification. Infect. Genet. Evol. 2016;39:349–364. doi: 10.1016/j.meegid.2016.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worthington K.J., Currie R.J.W., Jones R.C. A reverse transcriptase-polymerase chain reaction survey of infectious bronchitis virus genotypes in Western Europe from 2002 to 2006. Avian Pathol. 2008;37:247–257. doi: 10.1080/03079450801986529. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.