Highlights
-
•
Diagnosis of RV is extremely challenging due to genetic and serological variability within the RV serotypes.
-
•
We report a novel isothermal nucleic acid amplification method for the detection of RV.
-
•
The method, RT-SIBA, detected RV in clinical specimens with high analytical sensitivity and specificity.
-
•
The method will facilitate prompt diagnosis of infection and thereby improving patient care.
Keywords: Rhinovirus, Virus, RT-SIBA, Diagnostics, Isothermal, Amplification, Point-of-care
Abstract
Background: Rhinovirus (RV), a major cause of respiratory infection in humans, imposes an enormous economic burden due to the direct and indirect costs associated with the illness. Accurate and timely diagnosis is crucial for deciding the appropriate clinical approach and minimizing unnecessary prescription of antibiotics. Diagnosis of RV is extremely challenging due to genetic and serological variability among its numerous types and their similarity to enteroviruses.
Objective: We sought to develop a rapid nucleic acid test that can be used for the detection of Rhinovirus within both laboratory and near patient settings.
Study design: We developed and evaluated a novel isothermal nucleic acid amplification method called Reverse Transcription Strand Invasion-Based Amplification (RT-SIBA) to rapidly detect Rhinovirus from clinical specimens.
Result: The method, RT-SIBA, detected RV in clinical specimens with high analytical sensitivity (96%) and specificity (100%). The time to positive result was significantly shorter for the RV RT-SIBA assay than for a reference RV nucleic acid amplification method (RT-qPCR).
Conclusion: The rapid detection time of the RV SIBA assay, as well as its compatibility with portable instruments, will facilitate prompt diagnosis of infection and thereby improve patient care.
1. Background
Upper respiratory tract infection (URTI), which includes the common cold, is the most prevalent human illness. Rhinoviruses (RVs) are responsible for more than one-half of URTIs (Jacobs et al., 2013; Megremis et al., 2012). RVs are positive-sense, single-stranded RNA viruses that belong to genus Enterovirus of family Picornaviridae. RVs comprise three species, Rhinovirus A, B, and C, and types within are commonly referred to by species letter and number (e.g. rhinovirus A2; RV-A2). To date, over 170 rhinovirus types have been identified, displaying high genetic and antigenic variability (Megremis et al., 2012; Tapparel et al., 2013). RV infection carries an enormous economic burden due to the direct and indirect costs associated with the illness. In addition, to causing common cold, these viruses are the causative agents of many cases of severe pneumonia in the elderly and immunocompromised patients, as well as exacerbation of chronic obstructive pulmonary disease and asthma (Jacobs et al., 2013; Tan et al., 2003; Wark et al., 2013). Moreover, rhinovirus infection increases susceptibility to bacterial infection, e.g., by disrupting epithelial cell barrier function and promoting bacterial adhesion and internalization into epithelia (Sajjan et al., 2008; Unger et al., 2014).
Because respiratory infections can be caused by a variety of viral or bacterial pathogens, accurate and timely diagnosis is crucial for optimal management and treatment. Diagnosis of RVs is challenging due to genetic and antigenic variability among the types, as well as presentation of similar signs or symptoms in patients infected by bacterial or other viral respiratory pathogens. Furthermore, rhinoviruses grow slowly in cell culture, and RV-C is currently uncultivable (Price, 1956; Lau et al., 2007). Development of serology- and antibody-based detection methods has been hampered due to structural diversity of RV types and slowness of the immune response to generate specific antibodies. Thus, real-time reverse transcription–polymerase chain reaction (RT-PCR) remains the only sensitive method for diagnosis of RVs (Österback et al., 2013; Dagher et al., 2004). RT-PCR is often targeting the highly conserved 5′ noncoding region (NCR) of RV types (RV NCR). However, because the RV NCR region is highly similar between entero- and rhinoviruses, most RT-PCR methods cannot distinguish between rhinoviruses and other members of the Enterovirus genus (Cassandra et al., 2011). Consequently, commercial RV RT-PCR tests report combined results as rhinovirus + enterovirus. Furthermore, RT-qPCR requires thermal cyclers and skilled personnel, which are costly and require laboratory environments, limiting its use in field or point-of-care applications. Isothermal nucleic acid amplification methods obviate the need for thermal cyclers because the reactions are performed at low and constant temperature.
2. Objective
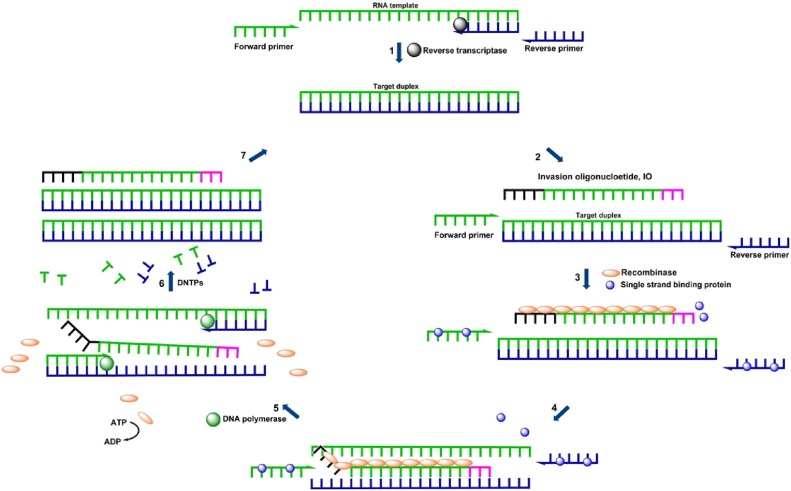
In this study, we developed an isothermal nucleic acid amplification method for the detection of RVs. This method, reverse-transcription strand invasion–based amplification (RT-SIBA), was previously shown to be useful for the diagnosis of infectious diseases (Eboigbodin et al., 2016a,b, 2017). In RT-SIBA reactions, RNA is first reverse transcribed into cDNA, and then immediately amplified and detected under isothermal reaction conditions (Fig. 1 ). The SIBA method relies on separation of the target duplex by a recombinase-coated invasion oligonucleotide (IO), allowing the primers to bind and DNA polymerase to extend the target sequence. The continuous strand separation and primer extension lead to exponential amplification of the target RNA under constant temperature. RT-SIBA detected RV in clinical samples with high analytical sensitivity and specificity comparable to diagnostic RT-qPCR methods.
Fig. 1.
Rhinovirus amplification by reverse-transcription strand invasion–based amplification (RT-SIBA). 1) Rhinovirus RNA is reverse transcribed to cDNA by the reverse transcriptase enzyme 2) SIBA amplification requires an invasion oligonucleotide (IO) and two target-specific primers. 3) Single strand binding protein, Gp32 binds to oligonucleotides in order to reduce the formation of secondary structures. The recombinase protein, UvsX, coats the IO displacing the bound Gp32. 4) The recombinase-IO complex invades and separates the target duplex. 5) This allows target-specific primers to bind and extend the target via the action of a DNA polymerase. 6) This leads to the synthesis of two copies of the target duplex. 7) The continuous recombinase-mediated target duplex separation and DNA polymerase extension process leads to an exponential amplification under isothermal conditions.
3. Study design
3.1. Microbial strains and clinical specimens
RV-A60 and RV-B17 were used as positive controls to establish the analytical sensitivity of the RT-SIBA assay. The 600-bp of the 5′ noncoding region (NCR) of RV-A60 and RV-B17 were cloned into their respective pEX-A2 plasmids. (Eurofins, Germany) These plasmid were used as controls for the quantification of RV-A60 and RV-B17 viral RNA using qRT-PCR. A total of 19 microbes (other than rhinoviruses) commonly found in respiratory specimens were used to determine the analytical specificity of the assay. A total of 26 respiratory clinical isolates (21 rhinovirus, 3 coronavirus, and 2 enterovirus) were from collections of University of Turku (Turku, Finland). A total of 50 retrospective nasopharyngeal (NP) swab specimens were obtained from Helsinki University Hospital (Finland), 26 of which were positive for RV. The specimens were handled and tested anonymously. The ethical approvals as well as patients' consent were obtained to collect and use the specimens for development of diagnostic methods and applications. The clinical isolates and the specimens were used to evaluate clinical performance of both the RV SIBA and RV RT-PCR assays. All specimens were obtained and used in accordance with the bioethics policies of Helsinki University Hospital and University of Turku.
3.2. Nucleic acid extraction
RNA was extracted from viral strains, clinical isolates, and NPS specimens using the QIAamp Viral RNA Mini Kit (Qiagen, Germany). DNA was extracted from microbial strains using the QIAamp DNA Mini Kit (Qiagen). All extractions were conducted in accordance with the manufacturer’s protocols.
3.3. RT-SIBA Rhino A and B assay design
RV 5′ NCR sequences were retrieved from GenBank and aligned to identify conserved regions. The RT-SIBA assay was designed to detect approximately 65 nucleotides in the conserved 5′ NCR of the rhinovirus genome. The conserved 5′ NCR RV used for the RT-SIBA assay was also selected to have sufficient mismatches with respect to enterovirus (EV) 5′ NCR sequences allowing better differentiation of EV and RV. A set of primers and invasion oligonucleotides (IOs) were designed to amplify this target region. The RT-SIBA assay used one forward primer, one reverse primer, and one IO each to detect serotypes A and B. The serotype RT-SIBA oligonucleotides were approximately 95% identical to those for serotype C (rhinovirus C26). The 5′-end of the IO includes poly-cytosine nucleotides (poly-C) that are non-complementary to the target region (seeding region), which promotes optimal coating of the IO by the recombinase (Eboigbodin et al., 2016a; Hoser et al., 2014; Formosa and Alberts, 1986). Furthermore, the 3′-end of the IO are modified with 2′-O-methyl RNA nucleotides in order to prevent IO DNA polymerase extension. The oligonucleotides used in this study were purchased from Integrated DNA Technologies (Leuven, Belgium). Primers and IOs were purified by reverse-phase HPLC and PAGE, respectively. The sequences of oligonucleotide used in this study are provided in Table 1 .
Table 1.
Oligonucleotides used for RV SIBA assay.
| Name | Sequence 5'→3' |
|---|---|
| RV-A F-primer | TGCACTAGCTGCAGGGTTA |
| RV-A R-primer | GTGTGCTCACTTTGAG |
| RV-B F-primer | GTCTCAAGGCTCCAGGGTTT |
| RV-B R-primer | GTGTGCTTAATTCTGAG |
| RV-A IO | CCCCCCCCCCCCCCCAGGGTTAAGGTTAGCCACATTCAGGGGmCmCmGmGmAmGmGmAmCmUmCmA |
| RV-B IO | CCCCCCCCCCCCCCCAGGGTTTAGGTTAGCCGCATTCAGGGGmCmCmGmGmAmGmGmAmCmUmCmA |
For invasion oligonucleotide (IO), bold sequences denote non-homologous seeding regions. mA, mC, mG, and mU denote 2′-O-methyl RNA nucleotides. F, forward; R, reverse; RV, human Rhinovirus; SIBA, strand invasion–based amplification; RV-A assay is designed to detect the sequence between position 421 and 483 within the genome of the human rhinovirus 60 strain ATCC VR-1473 (GenBank: FJ445133.1); RV-B assay is designed to detect the sequence between position 429 and 493 within the genome of the human rhinovirus 17 (GenBank: AF542419.1).
3.4. RV SIBA reaction
RT-SIBA reactions were performed using the SIBA reagent kit (Orion Diagnostica Oy, Espoo, Finland) with the addition of 16 U of GoScript Reverse Transcriptase (Promega, Madison, USA). UvsX and Gp32 were used at 0.25 mg/ml and 0.4 mg/ml, respectively. The reactions were initiated with 10 mM magnesium acetate. Forward and reverse primers were used at final concentrations of 200 and 400 nM, respectively. SIBA reactions were optimized for rapid detection of types A and B by determining the optimal IO concentration and ratio of A and B IOs. Both IOs were tested at final concentrations of 100, 200, 300, and 400 nM. The optimal ratio of IOs was used for subsequent RV RT-SIBA experiments.
SIBA products were detected using SYBR Green 1 (dilution, 1:100,000; Thermo Fisher Scientific, Waltham, MA, USA). The standard SIBA reaction volume was 20 μl, including 2 μl of template. Reactions were incubated at 41 °C for 60 min, and fluorescence readings were taken at 60 s intervals on a Bio-Rad CFX 96 (Bio-Rad Laboratories). A melting curve profile (40–95 °C) was generated after each amplification reaction to further verify that the reaction products were specific.
3.5. Rhinovirus RT-PCR
The performance of the RT-SIBA assay was compared to highly sensitive RT-PCR assay for the detection of entero- and rhinoviruses (Österback et al., 2013). This assay uses entero + rhinovirus-specific 5′ NCR primers for RT-PCR followed by rhinovirus-specific LNA probes for specific detection of rhinoviruses. One-step RT-PCR reactions were performed using the Express One-Step SuperScript® qRT-PCR SuperMix Kit (Thermo Fisher Scientific). Primers and probes were used at 600 nM and 100 nM, respectively. Reaction products were detected on a CFX 95 PCR instrument (Bio-Rad Laboratories, Finland). The following thermal cycling protocol was used: 50 °C for 15 min (cDNA synthesis), 95 °C for 2 min (reverse transcription and UDG inactivation), and 50 cycles of 95 °C for 15 s and 60 °C for 60 s (PCR amplification).
4. Results
4.1. Optimization of the RV SIBA assay
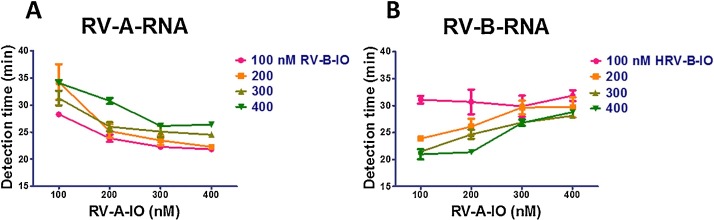
The rhino RT-SIBA uses two sets of oligonucleotides (one for RV-A and one for RV-B), allowing the detection of the members of species Rhinovirus A and or Rhinovirus B within the same reaction tube. We reasoned that these sets of oligonucleotides, particularly the IOs, could compete among themselves, resulting in sub-optimal amplification conditions. Therefore, we sought to determine the optimal concentrations of RV-A and RV-B IO (RV-A-IO and RV-B-IO) that would efficiently amplify both RV-A and RV-B RNA in the same reaction tube. To this end, we conducted the RV RT-SIBA assay using 100–400 nM of each IO, and determined the effect of IO concentration on the detection time of RV A and B types.
The amounts of time required to obtain a positive result from 1000 copies of RV-A and RV-B RNA per reaction are shown in Fig. 2 . A result was considered positive when the fluorescence signal exceeded the background signal. The amount of time to obtain a positive result for RV-A decreased (i.e., the amplification rate was faster) as the concentration of RV-A-IO increased from 100 to 400 nM. However, an increase in the concentration of RV-A-IO also led to a dramatic increase in the time taken to detect RV-B virus (i.e., the amplification rate for RV-B RNA was slower). Similarly, increasing the RV-B-IO concentration from 100 to 400 nM decreased the detection time for RV-B RNA but increased the detection time for RV-A RNA. These results suggested that RV-A-IO and RV-B-IO compete for reaction components. Based on optimization results, we performed subsequent experiments using RV-A-IO and RV-B-IO at final concentrations of 200 and 300 nM, respectively. These concentrations allowed for optimal detection of RV-A and -B in the same reaction tube.
Fig. 2.
Optimization of RV SIBA reaction conditions using different invasion oligonucleotide (IO) concentrations. (A) Amplification of 1000 copies of rhinovirus A60 RNA. (B) amplification of 1000 copies of rhinovirus B17 RNA; RV-A-IO, rhinovirus A invasion oligonucleotide; RV-B-IO, rhinovirus B invasion oligonucleotide.
4.2. Analytical sensitivity and specificity of the RV SIBA and RT-PCR assay
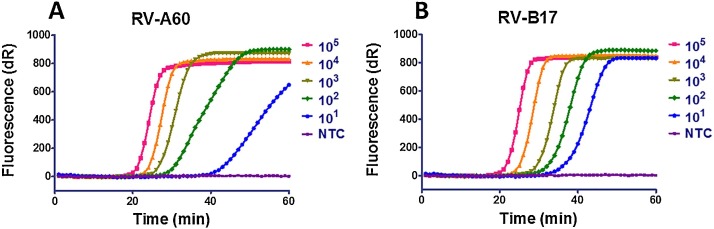
The analytical sensitivities of the RV SIBA and RT-PCR assays were elucidated using quantified RNA extracted from rhinovirus A60 and B17. These experiments were performed in three independent replicates by adding serial dilutions of RNA (from 1 to 105 copies per reaction). Each RNA dilution was performed in triplicate, and the results are shown in Fig. 3 and Table 2 . The RV SIBA assay detected as little as 10 copies of either rhinovirus A or B RNA. The RV SIBA assay detected 1000 copies of RV RNA in less than 30 min. In comparison, the lowest concentration of RV-A and RV-B RNA detected by RT-PCR was 1000 and 100 copies, respectively. Furthermore, we performed Probit regression analysis to determine the limited of detection (LOD) for both RV RT-SIBA and RV RT-PCR, which is the concentration of RV RNA that were detected 95% of the time. Serial dilutions of quantified RV-A RNA and RV-B RNA from 104 to 100 copies were used. Each RNA dilution were performed using eight replicates. The LODs of RV RT-SIBA, for RV-A RNA and RV-B RNA were 31 and 15 copies per reaction, respectively. Meanwhile, The LODs of RV RT-PCR, for RV-A RNA and RV-B RNA were 662 and 17 copies per reaction, respectively. Thus, the RV RT-SIBA assay was more sensitive than the RV RT-PCR for detection of purified RV RNA particularly RV-A RNA. Furthermore, the time to positive results was significantly shorter for RV RT-SIBA than for RV RT-PCR, which took approximately 2 h.
Fig. 3.
Sensitivity of RV SIBA assay for detection of rhinoviruses (RVs). (A) rhinovirus A60 RNA. (B) rhinovirus B17 RNA; NTC, no template control.
Table 2.
Analytical sensitivity and average detection time of RT-SIBA vs. RT-PCR for the detection of RV.
| RNA copy number/ reaction | RT-SIBA |
RT-PCR |
||
|---|---|---|---|---|
| Average amount of time taken to achieve positive results (min) |
Average cycle threshold to achieve positive results (Ct) |
|||
| RV-A | RV-B | RV-A | RV-B | |
| 105 | 20 | 20 | 25 | 22 |
| 104 | 23 | 24 | 29 | 26 |
| 103 | 25 | 28 | 34 | 29 |
| 102 | 28 | 31 | ND | 33 |
| 101 | 40 | 34 | ND | ND |
| 0 | ND | ND | ND | ND |
The analytical specificities of the RV RT-SIBA and RV-RT-PCR assays were determined by challenging the assays with DNA or RNA extracted from 19 microbial strains commonly found in respiratory specimens (Table 2). None of these 19 microbes were detected by either assay, indicating that both assays are highly specific for the detection of RV.
4.3. Evaluation of RV SIBA and RT-PCR assays using clinical isolates and NPS specimens
We compared the performance of RV SIBA and RT-PCR assays using RNA extracted from 26 respiratory clinical isolates containing 21 rhinoviruses, 3 coronaviruses and 2 enteroviruses (Table 3 ). Both the RV SIBA and RT-PCR assays only detected RV clinical isolates, and did not cross-react with coronaviruses or enteroviruses. The RV SIBA assay detected 20 of 21 RV isolates, whereas RT-PCR detected 21 of 21 RV isolates.
Table 3.
List of microbes used for cross-reactivity testing.
| Microbial strains | RV SIBA result |
RV PCR result |
|---|---|---|
| Rhinovirus A60 | + | + |
| Rhinovirus B17 VR-1663 | + | + |
| Enterovirus 71 ATCC VR-1432 | – | – |
| Streptococcus pyogenes NCTC 9994 | – | – |
| Streptococcus dysgalactiae ATCC 12388 | – | – |
| Streptococcus pneumoniae ATCC 6305 | – | – |
| Streptococcus agalactiae ATCC 12386 | – | – |
| Escherichia coli ATCC 25922 | – | – |
| Klebsiella pneumoniae | – | – |
| Neisseria sicca 29193 | – | – |
| Neisseria meningitides BAA 335 | – | – |
| Staphylococcus aureus ATCC 6538 | – | – |
| Staphylococcus epidermidis 2954 | – | – |
| Parainfluenza virus 1 ATCC VR-94 | – | – |
| Coronavirus ATCC-VR-740 | – | – |
| Adenovirus 1 ATCC VR-1 | – | – |
| Adenovirus 7 ATCC VR-7 | – | – |
| Human respiratory syncytial virus A ATCC VR-1540 | – | – |
| Human respiratory syncytial virus B ATCC VR-1400 | – | – |
| Influenza A VR-1736 | – | – |
| Influenza B VR-1813 | – | – |
We further evaluated the performance of both the RV SIBA and RT-PCR assays using 50 retrospective NPS specimens previously determined to be positive(N = 26) or negative (N = 24) for RV. The results are shown in Table 4 . The RV SIBA assay detected 25 of 26 RV-positive specimens, whereas RV RT-PCR detected 26 of 26 RV-positive specimens. None of the negative RV NPS specimens was detected by the RV SIBA assay. Based on comparisons with the previous results, the sensitivities and specificities of RV detection were 96% (95% CI: 81–100%) and 100% (95% CI: 86–100%), respectively, for the RV SIBA assay, and 100% (95% CI: 87–100%) and 100% (95% CI: 96–100%), respectively, for the RV RT-PCR assay. This suggests that the RV SIBA assay is more specific, but slightly less sensitive, than RV RT-PCR for the detection of NPS specimens. However, the RV SIBA assay had a significantly shorter time to positive result than the RV RT-PCR assay (Table 5 ).
Table 4.
Detection of RV clinical isolates using RV RT-SIBA and RT-PCR.
| Microbial strainsa | RV SIBA result |
RV PCR result |
|---|---|---|
| RV-A1b | + | + |
| RV-A2 | – | + |
| RV-A12 | + | + |
| RV-A16 | + | + |
| RV-A20 | + | + |
| RV-A30 | + | + |
| RV-A34 | + | + |
| RV-A40 | + | + |
| RV-A44 | + | + |
| RV-A58 | + | + |
| RV-A66 | + | + |
| RV-A74 | + | + |
| RV-A78 | + | + |
| RV-A80 | + | + |
| RV-A85 | + | + |
| RV-A90 | + | + |
| RV-A95 | + | + |
| RV-B14 | + | + |
| RV-B26 | + | + |
| RV-B52 | + | + |
| RV-B99 | + | + |
| CV-B3 | – | – |
| CV-B4 | – | – |
| CV-A9 | – | – |
| EV-11 | – | – |
| EV-30 | – | – |
RV, rhinovirus; CV, coronavirus; EV, enterovirus.
Table 5.
Clinical performance of RV SIBA assay vs. RT-PCR assay for detection of RV virus types.
| RV SIBA |
RV PCR |
|||
|---|---|---|---|---|
| Positive | Negative | Positive | Negative | |
| Positive | 25 | 1 | 26 | 0 |
| Negative | 0 | 24 | 1 | 23 |
| Total no. of samples | 50 | 50 | ||
| Sensitivity (95% CI) | 96% (81–100) | 100% (87–100.0) | ||
| Specificity (95% CI) | 100 % (86–100) | 100 % (86–100) | ||
5. Discussion
Diagnosis of RV is extremely challenging due to genetic and serological variability among its numerous types and their similarity to enteroviruses (Price, 1956; Lau et al., 2007). Nucleic acid amplification test (NAAT), i.e., RT-PCR, remains the most sensitive method for rhinovirus detection. These phenomena are responsible for inconsistent performance of most previously reported RV RT-PCR assays (Cassandra et al., 2011). Furthermore, due to the requirement for thermal cycling instruments and skilled personnel, RT-PCR reactions are costly. Alternative NAAT techniques, such as isothermal nucleic acid amplification test (iNAAT), aim to overcome the problem of expensive instrumentation.
In this study, we developed and evaluated an iNAAT, RT-SIBA, for rapid detection of RV. RT-SIBA uses a reverse transcriptase, recombinase, and recombinase-coated oligonucleotide to catalyze separation of a specific target sequence within the RV genome. This setup enables target-specific RV primers to exponentially amplify the target sequence under isothermal conditions. This method was previously shown to be a rapid and highly sensitive technique for the detection of infectious diseases (Eboigbodin et al., 2016a,b, 2017; Liikonen et al., 2018; Buissonnière et al., 2018). The RV SIBA assay uses two sets of oligonucleotides, allowing for detection of RV-A and RV-B in a single reaction tube.
We compared the performance of our RV SIBA assay with that of a previously published RV RT-PCR method (Österback et al., 2013). We found that the RV SIBA assay was more sensitive for the detection of low copy numbers of quantified RV RNA than the RV RT-PCR assay. In addition, we compared the performance of the RV SIBA and RV RT-PCR assays using clinical isolates and NPS specimens. On the NPS specimens, the sensitivity and specificity of the RV SIBA assay were 96% and 100%, respectively, whereas those of the RV RT-PCR assay were 100% and 100%, respectively. Thus, RV SIBA assay exhibited slightly lower sensitivity than RV RT-PCR for the detection of RV from NPS specimens. The time to positive results was significantly shorter for RV SIBA than for the RV RT-PCR assay. A loop-mediated isothermal amplification method (LAMP) for the detection of RV had previously been reported (Guan et al., 2016). Both SIBA and LAMP displayed similar performance with respect to the detection of RV from clinical specimens. However, SIBA displayed a faster detection time than LAMP, particularly for the detection of rhinovirus B virus.
The results of RV SIBA and RV RT-PCR were well correlated. However, to further evaluate the performance of both RV assays, it will be necessary to test a larger set of clinical specimens. Another limitation of our RV SIBA assay is that it was not tested with RV-C, mainly due to the unavailability of well-characterized RV-C types. The oligonucleotides used in our assay share approximately 95% identity with RV-C26, suggesting that the assay could also detect RV-C types. However, it may be necessary to include additional oligonucleotides to facilitate optimum detection of RV-C. Because the RV SIBA assay can be performed at low and constant temperature, the reactions can be performed using relatively small and inexpensive fluorescence readers. The shorter detection time of the RV SIBA assay, along with its compatibility with portable devices, will facilitate timely diagnosis of infection, thereby improving patient care and helping in avoiding unnecessary use of antibiotics.
Funding
This work is funded by Orion Diagnostica Oy, Salwe Research program for GET IT DONE (Finnish funding agency for technology and innovation Grant 534/14) and Academy of Finland: For financial support from Academy of Finland for VK (grant number 285632).
Conflict of interest
SE, MM and KE are employees of Orion Diagnostica Oy. All SIBA patents/patent applications are owned by Orion Diagnostica Oy. SE and KE are named inventors in SIBA patents applications. VK, PS, AP, JK, RK declare no conflict of interest.
Ethical approval
The ethical approvals as well as patients' consent were obtained to collect and use the specimens for development of diagnostic methods and applications.
Acknowledgments
The authors will also like to thank Amanda Raitosalo and members of the Research and Development team, Orion Diagnostica Oy for their support.
References
- Buissonnière A., Bénéjat L., Charron P., Bessède E., Lehours P., Valdenaire G. A new kit to detect Campylobacter species in stool specimens: the Orion GenRead Campylobacter®. Eur. J. Clin. Microbiol. Infect. Dis. 2018;37(8):1585–1587. doi: 10.1007/s10096-018-3288-5. [DOI] [PubMed] [Google Scholar]
- Cassandra E.F., Katherine E.A., Stephen B.L., Michael D.N., Terry M.N., Anne B.C. Usefulness of published PCR primers in detecting human rhinovirus infection. Emerg. Infect. Dis. J. 2011;17:296. doi: 10.3201/eid1702.101123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagher H., Donninger H., Hutchinson P., Ghildyal R., Bardin P. Rhinovirus detection: comparison of real-time and conventional PCR. J. Virol. Methods. 2004;117:113–121. doi: 10.1016/j.jviromet.2004.01.003. [DOI] [PubMed] [Google Scholar]
- Eboigbodin K., Filén S., Ojalehto T., Brummer M., Elf S., Pousi K. Reverse transcription strand invasion based amplification (RT-SIBA): a method for rapid detection of influenza A and B. Appl. Microbiol. Biotechnol. 2016;100:5559–5567. doi: 10.1007/s00253-016-7491-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eboigbodin K.E., Brummer M., Ojalehto T., Hoser M. Rapid molecular diagnostic test for Zika virus with low demands on sample preparation and instrumentation. Diagn. Microbiol. Infect. Dis. 2016;86:369–371. doi: 10.1016/j.diagmicrobio.2016.08.027. [DOI] [PubMed] [Google Scholar]
- Eboigbodin K.E., Moilanen K., Elf S., Hoser M. Rapid and sensitive real-time assay for the detection of respiratory syncytial virus using RT-SIBA®. BMC Infect. Dis. 2017;17:134. doi: 10.1186/s12879-017-2227-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formosa T., Alberts B.M. Purification and characterization of the T4 bacteriophage uvsX protein. J. Biol. Chem. 1986;261:6107–6118. [PubMed] [Google Scholar]
- Guan L., Zhao L.-Q., Zhou H.-Y., Nie K., Li X.-N., Zhang D. Reverse transcription genome exponential amplification reaction assay for rapid and universal detection of human rhinoviruses. Arch. Virol. 2016;161:1891–1898. doi: 10.1007/s00705-016-2858-z. [DOI] [PubMed] [Google Scholar]
- Hoser M.J., Mansukoski H.K., Morrical S.W., Eboigbodin K.E. Strand invasion based amplification (SIBA®): a novel isothermal DNA amplification technology demonstrating high specificity and sensitivity for a single molecule of target analyte. PLoS One. 2014;9 doi: 10.1371/journal.pone.0112656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs S.E., Lamson D.M., St, George K., Walsh T.J. Human rhinoviruses. Clin. Microbiol. Rev. 2013;26:135–162. doi: 10.1128/CMR.00077-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.K.P., Yip C.C.Y., H-w Tsoi, Lee R.A., L-y So, Y-l Lau. Clinical features and complete genome characterization of a distinct human rhinovirus (HRV) genetic cluster, probably representing a previously undetected HRV species, HRV-C, associated with acute respiratory illness in children. J. Clin. Microbiol. 2007;45:3655–3664. doi: 10.1128/JCM.01254-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liikonen K., Ojalehto T., Elf S., Mäki M., Matero P., Eboigbodin K.E. Clinical evaluation of a novel and simple-to-use molecular platform for diagnosis of respiratory syncytial virus. Anal. Biochem. 2018;551:4–6. doi: 10.1016/j.ab.2018.04.017. [DOI] [PubMed] [Google Scholar]
- Megremis S., Demetriou P., Makrinioti H., Manoussaki A.E., Papadopoulos N.G. The genomic signature of human rhinoviruses A, B and C. PLoS One. 2012;7 doi: 10.1371/journal.pone.0044557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Österback R., Tevaluoto T., Ylinen T., Peltola V., Susi P., Hyypiä T. Simultaneous detection and differentiation of human rhino- and enteroviruses in clinical specimens by real-time PCR with locked nucleic acid probes. J. Clin. Microbiol. 2013;51:3960–3967. doi: 10.1128/JCM.01646-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price W.H. The isolation of a new virus associated with respiratory clinical disease in humans. Proc. Natl. Acad. Sci. U. S. A. 1956;42:892–896. doi: 10.1073/pnas.42.12.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajjan U., Wang Q., Zhao Y., Gruenert D.C., Hershenson M.B. Rhinovirus disrupts the barrier function of polarized airway epithelial cells. Am. J. Respir. Crit. Care Med. 2008;178:1271–1281. doi: 10.1164/rccm.200801-136OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan W.C., Xiang X., Qiu D., Ng T.P., Lam S.F., Hegele R.G. Epidemiology of respiratory viruses in patients hospitalized with near-fatal asthma, acute exacerbations of asthma, or chronic obstructive pulmonary disease. Am. J. Med. 2003;115:272–277. doi: 10.1016/s0002-9343(03)00353-x. [DOI] [PubMed] [Google Scholar]
- Tapparel C., Siegrist F., Petty T.J., Kaiser L. Picornavirus and enterovirus diversity with associated human diseases. Infect. Genet. Evol. 2013;14:282–293. doi: 10.1016/j.meegid.2012.10.016. [DOI] [PubMed] [Google Scholar]
- Unger B.L., Ganesan S., Comstock A.T., Faris A.N., Hershenson M.B., Sajjan U.S. Nod-like receptor X-1 is required for rhinovirus-induced barrier dysfunction in airway epithelial cells. J. Virol. 2014;88:3705–3718. doi: 10.1128/JVI.03039-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wark P.A.B., Tooze M., Powell H., Parsons K. Viral and bacterial infection in acute asthma and chronic obstructive pulmonary disease increases the risk of readmission. Respirology. 2013;18:996–1002. doi: 10.1111/resp.12099. [DOI] [PMC free article] [PubMed] [Google Scholar]