Highlights
-
•
Feline coronavirus (FCoV) causes feline infectious peritonitis (FIP).
-
•
Repeated RT-PCR assays are necessary to identify FCoV shedders.
-
•
The identification of FCoV genome may support a diagnosis of FIP.
-
•
RT-LAMP assay for feline coronavirus was developed in this study.
-
•
RT-LAMP can be used to confirm the infection but not to exclude it.
Keywords: Feline infectious peritonitis, Isothermal gene amplification, Hydroxynaphtol blue
Abstract
The Feline coronavirus (FCoV) is the etiological agent of feline infectious peritonitis (FIP), a lethal disease of felids. The role of molecular methods is controversial for the diagnosis of FIP, while essential for the identification of the shedders. Thus, a fast and inexpensive method for the detection of FCoV could be beneficial, especially in multicat environments. A reverse transcription loop mediated isothermal amplification (RT-LAMP) assay was developed. RNA extraction and RT-nPCR for FCoV were performed on thirty-two samples (11 faeces, 9 blood, 8 effusions, and 4 lymph nodes) collected from 27 cats. Six RT-LAMP primers were designed from the same conserved region of RT-nPCR, and the assay was run at 63 °C for one hour. Results were evaluated through both agarose gel run and hydroxynapthol blue (HNB) dye and then compared with RT-nPCR results for the assessment of sensitivity and specificity. The overall specificity was 100%, but the sensitivity was 50% and 54.5% for agarose gel and HNB respectively. Therefore, RT-LAMP seems optimal to confirm the presence of the virus, but not applicable to exclude it.
Feline coronaviruses (family Coronaviridae, order Nidovirales) are enveloped, single-stranded positive sense RNA viruses belonging to the species Alphacoronavirus 1, genus Alphacoronavirus of the sub family Coronavirinae (Gonzalez et al., 2003). The feline coronavirus (FCoV) possesses a large genome (almost 30 kb) organized in 11 putative open reading frames (ORFs) encoding 16 nonstructural proteins involved in virus replication as well as four structural proteins (Spike, S; Membrane, M; Nucleocapsid, N; Envelope, E) and five accessory proteins (3 a-c, 7a and b) (Kipar and Meli, 2014). Not yet well characterized viral mutations, along with an inadequate immune response of the host, lead to the inevitably deadly disease of felids called feline infectious peritonitis (FIP) (Pedersen, 2009, Porter et al., 2014). The diagnosis of FIP is challenging in vivo and must rely on several clinico-pathological tests (e.g. serum protein electrophoresis, AGP measurement, effusion analysis) and only the immunohistochemical demonstration of the viral antigen inside the typical pyogranulomatous lesions can be considered as a gold standard (Pedersen, 2014). The FCoV is faecally-orally transmitted, worldwide distributed and highly prevalent in feline populations (Addie et al., 2009). Since most of the molecular and serological tools available up to date are not able to distinguish between the not mutated and the mutated pathogenic form of the FCoV, the use of reverse transcriptase polymerase chain reaction (RT-PCR) for the diagnosis of FIP is still extensively discussed (Doenges et al., 2015, Doenges et al., 2016, Felten et al., 2015, Longstaff et al., 2015). On the other hand, PCR is an extremely useful tool for the diagnosis of FCoV infection and shedding and consequently for the identification of shedders, when performed on faeces (Pedersen, 2014). Since shedders can spread the virus in the environment for months, RT-PCR should be repeatedly performed to identify both shedders and cats free from the infection (Addie and Jarrett, 2001).
Loop-mediated isothermal amplification (LAMP) is an amplification gene technique developed some years ago (Notomi et al., 2000) which allows to amplify nucleic acids in an hour and under isothermal conditions, and to evaluate the results by observation of the turbidity of the reaction or using different dyes. Reverse transcriptase LAMP (RT-LAMP) has been used to amplify the genome of several coronaviruses of both humans and animals (Amer et al., 2013, Bhadra et al., 2015, Cardoso et al., 2010, Hanaki et al., 2013, Nemoto et al., 2015, Pyrc et al., 2011, Thai et al., 2004, Yu et al., 2015). To our knowledge, RT-LAMP has never been used for the identification of FCoVs.
The aim of this study was to develop a RT-LAMP for the detection of feline coronavirus in the specimens most frequently used in clinical practice for both screening and diagnostic purposes.
Thirty-two samples from 27 cats (11 faeces, 9 blood, 8 effusions, and 4 lymph nodes) submitted to our institution as part of a diagnostic panel for the clinical suspicion of FIP or, regarding faeces, for screening purposes, were used. All the specimens were subjected to RNA extraction using a NucleoSpin RNA Isolation Kit (Macherey-Nagel Bethlehem, PA). Whole blood and effusion samples were centrifuged (5 min at 3500 × g) and the obtained pellets were suspended in 200 μL of phosphate buffered saline (PBS) by vigorous vortexing and stored at −20 °C for further RNA extraction. Faecal samples were suspended at a final concentration of 10% (wt/vol) in PBS by vigorous vortexing. The supernatant was cleared by centrifugation for 4 min at 5000 × g and stored at −20 °C for further RNA extraction. For tissues, approximately 20 mg of sample were thinly shredded on sterile plates using sterile scalpels, followed by vigorous vortexing in RNA lysis buffer until complete disruption of the sample. All the further steps were performed according to the manufacturer’s instruction.
The extracted RNA samples were tested for the presence of FCoV using a reverse transcription nested PCR (RT-nPCR) targeting a 177 bp product of the highly conserved 3′ untranslated region (3′ UTR) of the genome of both type I and type II FCoV (Herrewegh et al., 1995). RT-nPCR positive FCoV RNA from a cat with FIP was used as positive control and RNase-free water as negative control. PCR products were visualized under UV transilluminator on a 2% agarose gel stained with ethidium bromide.
The RT-LAMP primers targeting the 3′UTR of the FCoVs were designed using the Primer Explorer V4 software (http://primerexplorer.jp/elamp4.0.0/index.html) based on a 296 nucleotides sequence (28981–29277 bp) of the FCoV C1Je strain (accession number: DQ848678) (Table 1 ). A Loopamp RNA amplification kit (RT-LAMP, New England Biolabs, UK) was used to perform RT-PCR LAMP and the reaction mixture was set up as follows: 1× Isothermal amplification buffer, 6 mM MgSO4, 1.4 mM dNTPs, 320 U/mL of warm start DNA, 300 U/mL of warm start RTx Reverse Transcriptase, 2 μM of both forward inner primer (FIP) and backward inner primer (BIP), 0.2 μM of both F3 and B3 primers, 1 μM of both Loop F and Loop B primers, 120 μM of Hydroxynaphtol blue (HNB) dye (Sigma-Aldrich®) and 5 μL of RNA template. The reaction mixture was then made up to 25 μL with RNase-free water and incubated in a thermal cycler (MyCycler, Bio-Rad Laboratories, Hercules, CA, USA) for 1 h at 63 °C followed by 10 min at 80 °C for heat inactivation. The same positive control used for traditional RT-nPCR, which tested positive also on the first RT-LAMP assay, was then used as a positive control in the following RT-LAMP assays, while RNase-free water was used as negative control. The products of the reaction were then inspected both by eye, in order to detect the color turning from violet to sky blue in case of positive results with HNB (Goto et al., 2009), and under UV transilluminator on a 2% agarose gel stained with ethidium bromide in order to detect a ladder-like pattern in case of positive result (Parida et al., 2008) (Fig. 1 ). Results obtained with RT-LAMP were then compared with those obtained with the RT-nPCR and the sensitivity and specificity of RT-LAMP obtained with both HNB and agarose gel were calculated.
Table 1.
List of the primers (Primm Biotech, Italy) used in this study, based on the FCoV strain C1Je (GeneBank accession number: DQ848678).
| PRIMER | GENOME TARGETING POSITION | SEQUENCE (5′-3′) |
|---|---|---|
| F3 | 28982–29002 | GCAACCCGATGTTTAAAACTG |
| B3 | 29179–29162 | CCATTGTTGGCTCGTCAT |
| FIP (F1c + F2) | F1c, 29065-29043; F2, 29003–29023 | ACACGTGCTTACCATTCTGTACA-GTCTTTCCGAGGAATTACTGG |
| BIP (B1c + B2) | B1c, 29077–29101; B2, 29161-29141 | CAAGCAACCCTATTGCATATTAGGA-AGCGGATCTTTAAACTTCTCT |
| LOOP F | 29042–29024 | AGAGTAGACAGCGCGATGA |
| LOOP B | 29103–29128 | GTTTAGATTTGATTTGGCAATGCTAG |
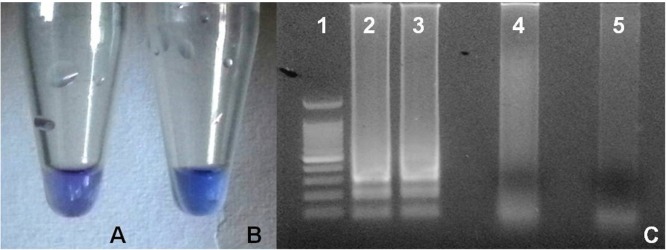
Fig. 1.
Evaluation methods of RT-LAMP for feline coronavirus. A. Negative result (violet) with hydroxynaphtol blue. B. Positive result (light blue) with hydroxynapthol blue. C. Agarose gel electrophoresis run showing 100 bp Ladder (lane 1); ladder like patterns typical of positive reaction (lanes 2 and 3); negative results (lanes 4 and 5). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Results are reported in Table 2 . All the negative samples using RT-nPCR were also negative by RT-LAMP, using both gel electrophoresis and HNB for the visualization of the results, leading to a 100% specificity.
Table 2.
Results obtained on the samples tested with RT-nPCR (PCR) and LAMP and evaluated with agarose gel electrophoresis (LAMP GEL) and hydroxynaphtol blue dye (LAMP HNB).
| ID | Specimen | PCR | LAMP GEL | LAMP HNB |
|---|---|---|---|---|
| 1 | Faeces | POS | POS | POS |
| 2 | Faeces | POS | POS | POS |
| 3 | Faeces | POS | POS | POS |
| 4 | Faeces | POS | POS | NEG |
| 5 | Faeces | POS | POS | NEG |
| 6 | Faeces | POS | NEG | POS |
| 7 | Faeces | POS | NEG | NEG |
| 8 | Faeces | POS | NEG | NEG |
| 9 | Faeces | POS | NEG | NEG |
| 10 | Faeces | POS | NEG | NEG |
| 11 | Faeces | NEG | NEG | NEG |
| TOT POS | 10/11 | 5/11 | 4/11 | |
| 12 | Blood | POS | POS | POS |
| 13 | Blood | POS | NEG | POS |
| 14 | Blood | POS | NEG | NEG |
| 15 | Blood | POS | NEG | NEG |
| 16 | Blood | NEG | NEG | NEG |
| 17 | Blood | NEG | NEG | NEG |
| 18 | Blood | NEG | NEG | NEG |
| 19 | Blood | NEG | NEG | NEG |
| 20 | Blood | NEG | NEG | NEG |
| TOT POS | 4/9 | 1/9 | 2/9 | |
| 21 | Effusion | POS | POS | POS |
| 22 | Effusion | POS | POS | POS |
| 23 | Effusion | POS | NEG | POS |
| 24 | Effusion | POS | NEG | NEG |
| 25 | Effusion | POS | NEG | NEG |
| 26 | Effusion | NEG | NEG | NEG |
| 27 | Effusion | NEG | NEG | NEG |
| 28 | Effusion | NEG | NEG | NEG |
| TOT POS | 5/8 | 2/8 | 3/8 | |
| 29 | Tissue | POS | POS | POS |
| 30 | Tissue | POS | POS | POS |
| 31 | Tissue | POS | POS | POS |
| 32 | Tissue | NEG | NEG | NEG |
| TOT POS | 3/4 | 3/4 | 3/4 | |
On the other hand, a conspicuous number of false negative results was recorded (11/22 for agarose gel and 10/22 for HNB) and the overall sensitivity was 50% and 54.5% using gel electrophoresis and HNB, respectively. Positive samples were characterized by a slight difference in the intensity of coloration, as shown in Fig. 1. The sensitivity of RT-LAMP was also different according to specimens: faeces showed a sensitivity of 50% and 40%, with gel electrophoresis and HNB respectively. On blood, the sensitivity was 25% and 50%, with gel electrophoresis and HNB respectively while on effusions the sensitivity was 40% with both the visualization methods. Only on tissues, the sensitivity resulted to be absolute, but the number of tested samples was too low to be discussed in terms of diagnostic accuracy. Based on the results of this pilot study, RT-LAMP for FCoV, due to its high specificity, appears to be a solid molecular test to confirm the diagnosis of FCoV infection, and eventually to support a clinical diagnosis of FIP, when performed on specimens from cats with an high pre-test probability of FIP (e.g. cats with clinical signs and laboratory findings consistent with FIP, like effusions with physico-chemical and cytological features consistent with FIP) but its low sensitivity makes this test not reliable in case of negative results (Pedersen, 2009). The design of this study did not allow us to understand the possible mechanisms responsible for the low sensitivity recorded. Technical problems are unlikely since the method described in this study has been developed after testing different primers, working conditions or temperatures (data not included in this short communication). Possible explanations of the high rate of false negative results compared with conventional nested RT-PCR include the lower analytical sensitivity of RT-LAMP, as previously reported for other coronaviruses when compared with real time RT-PCR (Bhadra et al., 2015), that allow to obtain positive results only in samples with a high viral burden.
In the case the sensitivity of the test might be ameliorated through further studies, the RT-LAMP could be extremely useful, due to its low costs and rapidity, in those situations where the detection of FCoV must be repeated over time and on a high number of cats (e.g. breeding catteries). An additional future perspective would be the optimization of the test to obtain quantitative results, possibly by establishing a standard curve of color intensity using RNA samples with known viral load (e.g. quantification of RNA copies by quantitative PCR techniques). Moreover, the development of an internal control to assess the integrity of RNA in each sample would be advisable before the use of this test in field conditions.
Declaration of interest
The authors declared no conflicts of interest regarding the research, authorship, and/or publication of this article. The authors received no financial support for the research, authorship, and/or publication of this article.
Acknowledgment
None.
References
- Addie D.D., Jarrett O. Use of a reverse-transcriptase polymerase chain reaction for monitoring the shedding of feline coronavirus by healthy cats. Vet. Rec. 2001;148(21):649–653. doi: 10.1136/vr.148.21.649. [DOI] [PubMed] [Google Scholar]
- Addie D., Belák S., Boucraut-Baralon C., Egberink H., Frymus T., Gruffydd-Jones T., Hartmann K., Hosie M.J., Lloret A., Lutz H., Marsilio F. Feline infectious peritonitis. ABCD guidelines on prevention and management. J. Feline Med. Surg. 2009;11(7):594–604. doi: 10.1016/j.jfms.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amer H.M., El Wahed A.A., Shalaby M.A., Almajhdi F.N., Hufert F.T., Weidmann M. A new approach for diagnosis of bovine coronavirus using a reverse transcription recombinase polymerase amplification assay. J. Virol. Methods. 2013;193(2):337–340. doi: 10.1016/j.jviromet.2013.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhadra S., Jiang Y.S., Kumar M.R., Johnson R.F., Hensley L.E., Ellington A.D. Real-time sequence-validated loop-mediated isothermal amplification assays for detection of Middle East respiratory syndrome coronavirus (MERS-CoV) PLoS One. 2015;10(4):e0123126. doi: 10.1371/journal.pone.0123126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso T.C., Ferrari H.F., Bregano L.C., Silva-Frade C., Rosa A.C.G., Andrade A.L. Visual detection of turkey coronavirus RNA in tissues and feces by reverse-transcription loop-mediated isothermal amplification (RT-LAMP) with hydroxynaphthol blue dye. Mol. Cell. Probes. 2010;24(6):415–417. doi: 10.1016/j.mcp.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doenges S.J., Weber K., Dorsch R., Fux R., Fischer A., Matiasek L.A., Matiasek K., Hartmann K. Detection of feline coronavirus in cerebrospinal fluid for diagnosis of feline infectious peritonitis in cats with and without neurological signs. J. Feline Med. Surg. 2015;18(2):104–109. doi: 10.1177/1098612X15574757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doenges S.J., Weber K., Dorsch R., Fux R., Hartmann K. Comparison of real-time reverse transcriptase polymerase chain reaction of peripheral blood mononuclear cells, serum and cell-free body cavity effusion for the diagnosis of feline infectious peritonitis. J. Feline Med. Surg. 2016 doi: 10.1177/1098612X15625354. Epub ahead of print 1098612x15625354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felten S., Weider K., Doenges S., Gruendl S., Matiasek K., Hermanns W., Mueller E., Matiasek L., Fischer A., Weber K., Hirschberger J. Detection of feline coronavirus spike gene mutations as a tool to diagnose feline infectious peritonitis. J. Feline Med. Surg. 2015 doi: 10.1177/1098612X15623824. Epub ahead of print 1098612x15623824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez J.M., Gomez-Puertas P., Cavanagh D., Gorbalenya A., Enjuanes L. A comparative sequence analysis to revise the current taxonomy of the family Coronaviridae. Arch. Virol. 2003;148(11):2207–2235. doi: 10.1007/s00705-003-0162-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto M., Honda E., Ogura A., Nomoto A., Hanaki K.I. Short technical reports. Biotechniques. 2009;46:167–172. doi: 10.2144/000113072. [DOI] [PubMed] [Google Scholar]
- Hanaki K.I., Ike F., Hatakeyama R., Hirano N. Reverse transcription-loop-mediated isothermal amplification for the detection of rodent coronaviruses. J. Virol. Methods. 2013;187(2):222–227. doi: 10.1016/j.jviromet.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrewegh A.A.P.M., de Groot R.J., Cepica A., Egberink H.F., Horzinek M.C., Rottier P.J.M. Detection of feline coronavirus RNA in feces, tissues, and body fluids of naturally infected cats by reverse transcriptase PCR. J. Clin. Microbiol. 1995;33:684–689. doi: 10.1128/jcm.33.3.684-689.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipar A., Meli M.L. Feline infectious peritonitis: still an enigma? Vet. Pathol. 2014;51(2):505–526. doi: 10.1177/0300985814522077. [DOI] [PubMed] [Google Scholar]
- Longstaff L., Porter E., Crossley V.J., Hayhow S.E., Helps C.R., Tasker S. Feline coronavirus quantitative reverse transcriptase polymerase chain reaction on effusion samples in cats with and without feline infectious peritonitis. J. Feline Med. Surg. 2015 doi: 10.1177/1098612X15606957. Epub ahead of print 1098612x15606957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemoto M., Morita Y., Niwa H., Bannai H., Tsujimura K., Yamanaka T., Kondo T. Rapid detection of equine coronavirus by reverse transcription loop-mediated isothermal amplification. J. Virol. Methods. 2015;215:13–16. doi: 10.1016/j.jviromet.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28(12):E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parida M., Sannarangaiah S., Dash P.K., Rao P.V.L., Morita K. Loop mediated isothermal amplification (LAMP): a new generation of innovative gene amplification technique; perspectives in clinical diagnosis of infectious diseases. Rev. Med. Virol. 2008;18(6):407–421. doi: 10.1002/rmv.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen N.C. A review of feline infectious peritonitis virus infection: 1963–2008. J. Feline. Med. Surg. 2009;11(4):225–258. doi: 10.1016/j.jfms.2008.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen N.C. An update on feline infectious peritonitis: diagnostics and therapeutics. Vet. J. 2014;201(2):133–141. doi: 10.1016/j.tvjl.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter E., Tasker S., Day M.J., Harley R., Kipar A., Siddell S.G., Helps C.R. Amino acid changes in the spike protein of feline coronavirus correlate with systemic spread of virus from the intestine and not with feline infectious peritonitis. Vet. Res. 2014;45:1–11. doi: 10.1186/1297-9716-45-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyrc K., Milewska A., Potempa J. Development of loop-mediated isothermal amplification assay for detection of human coronavirus-NL63. J. Virol. Methods. 2011;175(1):133–136. doi: 10.1016/j.jviromet.2011.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thai H.T.C., Le M.Q., Vuong C.D., Parida M., Minekawa H., Notomi T., Hasebe F., Morita K. Development and evaluation of a novel loop-mediated isothermal amplification method for rapid detection of severe acute respiratory syndrome coronavirus. J. Clin. Microbiol. 2004;42(5):1956–1961. doi: 10.1128/JCM.42.5.1956-1961.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Shi L., Lv X., Yao W., Cao M., Yu H., Wang Z., Zheng S. Development of a real-time reverse transcription loop-mediated isothermal amplification method for the rapid detection of porcine epidemic diarrhea virus. Virol. J. 2015;12(1):1–8. doi: 10.1186/s12985-015-0297-1. [DOI] [PMC free article] [PubMed] [Google Scholar]