Highlights
-
•
Two Multiplex PCR-DNA microarray assays (BRDC and BED) were developed and validated.
-
•
The BRDC assay simultaneously detects 4 bacteria and 5 viruses.
-
•
The BED assay simultaneously detects 4 bacteria, 3 protozoa and 4 viruses.
-
•
Both assays were validated using laboratory and clinical samples.
Abbreviations: BRDC, bovine respiratory disease complex; BED, bovine enteric diseases; BRSV, bovine respiratory syncytial virus; BPI-3, bovine parainfluenza virus 3; BVDV, bovine viral diarrhea virus; BoHV-1, Bovine herpesvirus 1; BCoV, bovine corona virus; BRoV, bovine rotavirus; BToV, bovine torovirus; DENV, dengue virus; lktA, leukotoxin-A; E.coli, Escherichia coli; ETEC, enterotoxigenicE. coli; EHEC, enterohemorrhagic E. coli; EPEC, enteropathogenic E. coli; FI, fluorescence intensity; GI, Geninfo identifier; Lt, heat labile toxin; LOD, limit of detection; LSB, low salt buffer; LNA, lock nucleic acid; NTC, no-template control; nmaA, UDP-N-acetyle-D-glucosamine-2-epimerase gene; PCR, polymerase chain reaction; RT-PCR, reverse transcription polymerase chain reaction; stx-1, shiga-like toxin 1; tbpB, transferring-binding protein B gene; 5′UTR, 5′ untranslated region
Keywords: Bovine enteric diseases, Bovine respiratory disease complex, Multiplex PCR, DNA microarray
Abstract
Respiratory and enteric diseases continue to be two major causes of economic losses to the cattle industry worldwide. Despite their multifactorial etiology, the currently available diagnostic tests for bovine respiratory disease complex (BRDC) and bovine enteric disease (BED) are single-pathogen-tests. DNA microarray when combined with multiplex polymerase chain reaction (PCR) is a powerful tool in detection and differentiation of multiple pathogens in a single sample. This study reports development and initial validation of two independent highly sensitive and specific multiplex PCR-electronic microarray assays, one for the detection and differentiation of pathogens of the BRDC and the other for detection and differentiation of pathogens of the BED. The BRDC multiplex PCR-microarray assay was able to detect and differentiate four bacteria (Mannheimia haemolytica, Histophilus somni, Pasteurella multocida, and Mycoplasma bovis) and five viruses [bovine parainfluenza virus-3, bovine respiratory syncytial virus, bovine herpesvirus-1, bovine coronavirus (BCoV), and bovine viral diarrhea virus (BVDV)] associated with BRDC. The BED multiplex PCR- microarray- assay was able to detect and differentiate four bacteria (Clostridium perfringens, Escherichia coli, Salmonella enterica Dublin, and Salmonella enterica Typhimurium), three protozoa (Eimeria zuernii, Eimeria bovis, and Cryptosporidium parvum), and four viruses (bovine torovirus, bovine rotavirus, BCoV, and BVDV) associated with the BED. Both assays detected their respective targets individually or in combination when present. The limit-of-detection of each assay at the PCR amplification and DNA microarray levels was determined using previously titrated laboratory amplified target pathogens or using quantified synthetic nucleotides. Both assays showed very high analytical sensitivity and specificity, and were validated using a limited number of clinical samples. The BRDC and BED multiplex PCR- microarray-assays developed in this study, with further clinical validation, could be used in veterinary diagnostic laboratories for the rapid and simultaneous identification of pathogens to facilitate quick and accurate decision making for the control and treatment of these two economically important disease complexes. Furthermore, these assays could be very effective tools in epidemiological studies as well as for screening of healthy animals to identify carriers that may potentially develop BRDC or BED.
1. Introduction
Respiratory and enteric diseases are the two most common and costly diseases in cattle industry worldwide. In the USA, respiratory ailments account for 40–50% of feedlot deaths (Hilton, 2014; Snowder et al., 2006). Based on a National Animal Health Monitoring System’s (NAHMS) Beef Feedlot study conducted in 2011, approximately 2.29 million US beef cattle in feedlots are affected by respiratory diseases costing around 54.12 million US dollars annually for the treatments (USDA–APHIS–VS, 2013; Johnson and Pendell, 2017). In the UK, an estimated 1.9 million animals are affected by bovine respiratory disease each year costing the UK beef industry approximately £60 million (NADIS, 2007). Enteric diseases are responsible for more than a half of deaths among pre-weaned dairy calves in the USA and other countries (Gomez and Weese, 2017; Cho and Yoon, 2014; Hur et al., 2013; Azizzadeh et al., 2012). Both respiratory and enteric diseases continue to be major health threats to the global cattle industry despite many intervention strategies such as vaccination, medications, and herd management.
Bovine respiratory disease complex (BRDC) is multi-factorial in origin and involves a complex interaction between host, pathogen and environmental factors that makes its diagnosis, prevention and control a challenging task (Mosier, 2014). Several viruses and bacteria, including mycoplasma are associated with BRDC. The most common viral pathogens associated with BRDC are bovine respiratory syncytial virus (BRSV), bovine parainfluenza virus-3 (BPI-3), bovine viral diarrhea virus (BVDV), and bovine herpesvirus 1 (BoHV-1). The role of some viruses such as bovine coronavirus (BCoV) and bovine rhinitis A virus, in BRDC pathogenesis, is being increasingly recognized (Murray et al., 2016). Some of these viral infections alone may not result in any clinical disease, but lead to secondary bacterial infections resulting in severe respiratory disease. Bacterial species that are commonly associated with BRDC are Mannheimia haemolytica, Mycoplasma bovis, Histophilus somni, and Pasteurella multocida (reviewed in Griffin et al., 2010).
Like BRDC, bovine enteric disease (BED) in neonatal calves is often caused by concurrent infection of multiple pathogens of viral, bacterial and/or protozoan origin (Foster and Smith, 2009). Establishment of pathogenic infection in the alimentary epithelium leads to secretion, malabsorption or maldigestion leading to enteritis and scours. Among viral agents, group A bovine rotaviruses (BRoV) and enteropathogenic BCoV are the leading causes of diarrhea in young calves (Dhama et al., 2009; Boileau and Kapil, 2010). Additionally, BVDV and BRoV group B and C are also considered as causative agents of scours. Besides viruses, several bacteria including pathogenic strains of Escherichia coli, Clostridium perfringens, and Salmonella are associated with BED. Eimeria species and Cryptosporidium parvum are the major parasitic protozoans that cause diarrhea in young calves.
Given the multiple etiological nature of BRDC and BED, accurate and rapid identification of pathogen(s) involved is crucial towards effective treatment and prevention of such diseases. The current diagnostic assays for BRDC and BED include pathogen isolation, serology and detection of pathogen-specific nucleic acids using polymerase chain reaction (PCR) based methods. While many of such assays target single pathogens, few assays have been developed for simultaneous detection of multiple pathogens in a single reaction (Vilcek et al., 1994; Decaro et al., 2012; Angen et al., 1998). These multiplex assays depend on pathogen differentiation based on the size of PCR amplicons, and therefore requires gel-electrophoresis following PCR amplification. Recently several multiplex real-time PCR assays have been developed to detect pathogens associated with BRDC and BED (Horwood and Mahony, 2011; Marley et al., 2008; Thonur et al., 2012; Cho et al., 2010). Alternatively, a real-time PCR based system was described that includes multiple reactions in one-run with similar thermal cycling conditions for detection of BRDC and BED pathogens (Kishimoto et al., 2017; Tsuchiaka et al., 2016). Although these real-time PCR assays are rapid and highly sensitive, their multiplexing capability at present is restricted by the availability of limited number of compatible fluorescent dyes.
The DNA microarray platform is suitable for multiplex detection of many targets in a single sample and has been widely used for multiplex pathogen detection (Kostrzynska and Bachand, 2006; Yadav et al., 2015). Conventional microarray requires a substantial amount of manual processing, multiple pieces of equipment for array preparation, and longer time for passive target-probe hybridization. Meanwhile, the new automated electronic microarray platform is rapid, user-friendly, and automates all steps from microarray capture probe printing to all post-PCR processing steps including electronically driven hybridization, washing and reporting (Lung et al., 2012). Each electronic microarray cartridge consists of 400 individually activated probe binding sites that allow the probe layout to be customizable during each run based on the needs of the end user, in contrast to conventional glass slide microarray, where changes to the array layout is not possible once the arrays are printed. This allows the user to make changes to the detection probe panel if necessary and eliminates anticipation of the future needs before finalizing the array layout, which is the case with conventional microarray slide production. Furthermore, depending on the number of samples to be tested, not all probe binding sites on an electronic microarray cartridge need to be used on the initial run, and unused sites can be used in subsequent sessions (up to 10 times per cartridge). Electronic microarray based assays for detection of several avian, bovine, and swine viruses and bacteria have been described previously (Lung et al., 2012; Lung et al., 2015; Lung et al., 2016).
This study describes the development and initial validation of multiplex PCR and oligonucleotide electronic microarray assays that can detect a total of seven respiratory pathogens, nine enteric pathogens and two pathogens that cause both respiratory and enteric diseases in a single run.
2. Materials and methods
2.1. Sequence databases
Sequence databases were created for selected target genomic regions of four BRDC-associated bacteria (M. haemolytica, H. somni, P. multocida, and M. bovis), three BRDC-associated viruses (BPI-3, BRSV, and BoHV-1), four BED-associated bacteria (C. perfringens, E. coli, S. e. Typhimurium, and S. e. Dublin), three BED-associated protozoa (E. zuernii, E. bovis, and C. parvum), two BED-associated viruses (BoRV and BoTV), and two viruses that are associated with both BRDC and BED (BVDV and BCoV) in cattle. Highly conserved genomic regions of these pathogens were identified through literature search, and all available full and partial nucleic acid sequences for each target were downloaded from the nucleotide database of the National Center for Biotechnology Information (NCBI). Upon removal of duplicated sequences with identical GenInfo identifiers (GI), multiple sequence alignments for each genetic target were created with MAFFT v.7.0 (Katoh and Standley, 2013) using default settings. Databases were maintained with BioEdit Sequence Alignment Editor v.7.1.9 (Hall, 1999).
2.2. Primer design
Generated sequence databases were used to design four panels of multiplex PCR primers specific for BRDC viruses, BRDC bacteria, BED viruses, and BED bacteria and protozoa (Table 1 ). When possible, primers were sourced from literature. New primers were designed using either AlleleID (Premier Biosoft International, Palo Alto, CA) or Primer3web version 4.0.0 (Koressaar and Remm, 2007; Untergasser et al., 2012). Except for a few instances, a single gene from each pathogen was targeted for PCR amplification. For M. haemolytica, the leukotoxin A (lktA) gene was selected to detect all serotypes, nmaA and tbpB genes were targeted to differentiate pathogenic serotypes 1 and 6 from commensal serotypes. For E. coli, heat-labile toxin (Lt) genes for identification of enterotoxigenic (ETEC) strains, Intimin (eae) and Shiga-like toxin-1 (stx-1) genes for identification of enterohemorrhagic (EHEC) strains, and K99 gene for identification of enteropathogenic (EPEC) strains were selected. One pair of primers (a forward and a reverse primer) was used for amplification of each target virus except for 5′-untranslated region (5′UTR) of bovine viral diarrhea virus (BVDV). To cover the sequence diversity in the 5′ UTR region of BVDV and HoBi viruses, two forward primers and one reverse primer were used. All reverse primers were synthesized with the reverse complementary sequence of the red universal reporter probe at the 5′ end (Huang et al., 2009). In addition to pathogen-specific primers, each PCR primer panel contained primers for a selected 229 bp 3′ untranslated region of dengue virus (DENV) that was used as the internal control. Primer lengths and melting temperatures ranged from 19–29 bp and 45°-60 °C, respectively (Table 1). The best combination of primers for each multiplex panel was selected upon examining various combinations of primers using AlleleID with default settings.
Table 1.
Multiplex PCR assays, target pathogens, genomic regions and primers. All reverse primers were synthesized with 5′GCA GTA TAT CGC TTG ACA 3′, the reverse complementary sequence of the Red Universal Reporter Probe, at the 5′ end. In addition to the pathogen specific primers, each assay consisted of DENV specific primers, (F) AAACCGTGCTGCCTGTAG/(R) TCTCTCCCAGCGTCAA (Sudiro et al., 1997) as internal control primers. †includes two primers.
| Organism | Genomic region | Primer Sequence (5'->3') | Amplicon size (bp) | Reference | |
|---|---|---|---|---|---|
| Assay for BRDC associated bacteria | |||||
| H. somni | 16S ribosomal DNA (rDNA) | F | GTGATGAGGAAGGCGATTAGT | 416 | Angen et al., 2009 (modified) |
| R | TTCGGGCACCAAGTRTTCA | ||||
| P. multocida | Putative transcriptional regulator gene, Pm0762 | F | TTGTGCAGTTCCGCAAATAA | 567 | Liu et al., 2004 |
| R | TTCACCTGCAACAGCAAGAC | ||||
| M. bovis | 16S rDNA | F | CCTTTTAGATTGGGATAGCGGATG | 360 | Wiggins et al., 2007 |
| R | CCGTCAAGGTAGCATCATTTCCTAT | ||||
| M. haemolytica (all serotypes) | Leukotoxin A (lktA) | F | GTCCCTGTGTTTTCATTATAAG | 385 | Klima et al., 2014a, b |
| R | CACTCGATAATTATTCTAAATTAG | ||||
| M. haemoytica (pathogenic serotypes) | Transferring-binding protein B (tbpB) | F | CTACTTGCTGCTTGTTCCTC | 452 | Klima et al., 2014a, b |
| R | CCATGTGCACCTGTTCTCAAA | This study | |||
| UDP-N-acetyl-D-glucosamine- 2-epimerase (nmaA) | F | AAGCCGTTTCAACATTAGCGT | 396 | This study | |
| R | CATCGCCATAAGGGTTGTGA | Klima et al., 2014a, b | |||
| Assay for BRDC associated viruses | |||||
| BoHV-1 | Glycoprotein B | F | TGAGGCCTATGTATGGGCAGTT | 432 | Klima et al., 2014a, b |
| R | GGACACAACAAACAATGCGG | ||||
| BRSV | G attachment glycoprotein | F | ACACATCAATYCAAAGCACCACAC | 374 | This study |
| R | GCTRGTTCTGTGGTGGRTTGTTGTC | Vilcek et al., 1994 (modified) | |||
| BPI-3 | M gene | F | TGTCTTCCACTMGATAGAGGGATAAAATT | 203 | Klima et al., 2014a, b |
| R† | CCTTTTTCATCTAGAATCTGAACTACTCC/ CCTTTCTCATCTAAGATCTGGACMACC | This study | |||
| BVDV/Hobi | 5’ untranslated region (UTR) | F† | CATACCTTCAGTAGGACGAGC/ CATGCCCRYAGTAGGACTAGC | 280 | This study/ Deregt et al., 2006 |
| R | ATGTGCCATGTACAGCAGAG | ||||
| BCoV | N | F | GCCGATCAGTCCGACCAATC | 407 | Tsunemitsu et al., 1999 |
| R | AGAATGTCAGCCGGGGTAT | ||||
| Assay for BED associated bacteria/protozoa | |||||
| C. perfringens | Alpha toxin (plC) | F | CTATGGGCTGGGGCATCAACT | 480 | This study |
| R | AACATGTCCTGCGCTATCAAC | ||||
| E. bovis/ E. zuernii | Internal transcribed spacer-1 (ITS-1) | F | AAAGGATGCAAAAGTCGTAACAC | 535-514 | Kawahara et al., 2010 (modified) |
| R | TGCAATTCACAATGCGTATCG | ||||
| C. parvum | Cryptosporidium oocyst wall protein (COWP) | F | CAAATTGATACCGTTTGTCCTTCTG | 348 | Guy et al., 2003 |
| R | AGGGCAGACAGGTTGAGTTGG | This study | |||
| S. e. Dublin | Hypothetical protein coding sequence SeD_A1118 | F | CCGACCTTGCCGTGTATCTCA | 408 | This study |
| R | GCTGCCGACTGTTACCGAGAA | ||||
| S. e. Typhimurium | Putative cytoplasmic protein coding sequence STM4493 | F | GAGCGTGCGGCGATGTTAGT | 572 | This study |
| R | CGGGTACTGCTGGTTGAAGGT | ||||
| E coli (EHEC) | Intimin (eaeA) | F | GACCCGGCACAAGCATAAGC | 384 | Blais et al., 2012 |
| R | CCACCTGCAGCAACAAGAGG | ||||
| Shiga like toxin-1 (stx-1) | F | GTCATTCGCTCTGCAATAGGTA | 559 | Frank et al., 1998 (modified) | |
| R | TTCCCCAGTTCAATGTAAGATCA | Ok et al., 2009 (modified) | |||
| E. coli (ETEC) | K99 antigen coding gene (K99) | F | CGACTACCAATGCTTCTGCGAA | 439 | Cho et al., 2010 (modified) |
| R | TTAGACGGAGCGCGGTCATC | This study | |||
| E. coli (EPEC) | Heat-labile toxin (Lt) | F | CGGAGGTCTTATGCCCAGAGG | 481 | This study |
| R | CCAGGGTTCTTCTCTCCAAGCT | West et al., 2007 (Modified) | |||
| Assay for BED associated viruses | |||||
| BRoV A/C | Outer capsid protein VP6 | F | GCATGGATGAAATGGTTAGAGA | 467 | This study |
| R | TGGATTGAAGTACCATGTAGT | ||||
| BRoV B | VP6 | F | ACAGTGAATGCTTGCGTCAG | 584 | This study |
| R | GCTTCCATGCCTGAAACACA | ||||
| BToV | Nucleocapsid phosphoprotein gene (N) | F | CCAAATGCTATGCCATTTCAGC | 297 | This study |
| R | TGCAGTCTCATTTGCCATCAT | ||||
| BVDV/Hobi | 5’ untranslated region (UTR) | F† | CATACCTTCAGTAGGACGAGC/ CATGCCCRYAGTAGGACTAGC | 280 | This study/ Deregt et al., 2006 |
| R | ATGTGCCATGTACAGCAGAG | ||||
| BCoV | N | F | GCCGATCAGTCCGACCAATC | 407 | Tsunemitsu et al., 1999 |
| R | AGAATGTCAGCCGGGGTAT | ||||
2.3. Probe design
Except for BVDV and pestivirus probes, which were adapted from previously published work, all microarray capture probes were newly designed using Primer3web version 4.0.0 (Table 2 ). The probes contained minimal secondary structures (dG ≥ −8.0 kcal/mol) and their lengths, GC contents, and melting temperatures ranged from 20–45 bp, 22–59%, and 53°–72 °C, respectively. Specificity of each designed primer and probe were evaluated in silico by Nucleotide Basic Local Alignment Search Tool (BLASTn) homology search. Primers or probes that showed significant homology to any non-target species were excluded from further investigation.
Table 2.
Four Microarray panels and the capture probes used in identification of each target pathogen. Unless referred, probes were designed for the current study.
| Organism | Genomic Region | Probe name | Sequence |
|---|---|---|---|
| BRDC Bacteria Panel | |||
| H. somni | 16S rDNA | Hsom-16S-P629 | GAACAGCATTTCAGACTGGGTGAC |
| Hsom-16S-P648 | GAGATTAATTGATTGACGATAATCACAGAAG | ||
| P. multocida | Pm0762 | Pmul-ulaR-P448 | AGAGCCACAGGTCAGAATCACACT |
| Pmul-ulaR-P531 | TATTGATGGCGAGCGGTTGAGA | ||
| M. bovis | 16S rDNA | Mbov-16S-P208 | GTTTGCTTCGCTAAAAGATCGGAGTG |
| Mbov-16S-P284 | GATGAAGGCCCTATGGGTTGTAAACTG | ||
| M. haemolytica | lktA | Mhae-Lkt-P194 | TCTCAATCCTCTTGATTCCTCTATCTCA |
| tbpB | Mhae-tbpB-P138 | ACAAGATGATAATAGTAACGCAAGACG | |
| nmaA | Mhae-nmaA-P801 | GCCTTGCGACTATCTCTCCTTTGTT | |
| Mhae-nmaA-P937 | GAAGCTGTAGAAGCCGGAACAGTA | ||
|
BRDC Virus Panel BoHV-1 |
Adjacent gB | BHV-glyco b Pb | CCACAAAGCACATTTGACCC |
| BHV-2 probe | GCGCGAATCTTATTTAAGTGCACACCGTGTTATTT | ||
| gB-BHV-2mod | GCGAATCTTATTTAAGTGCACACC | ||
| BRSV | G | BRSV-G-P1 | GAAAACCACCAAGACCACAACAAC |
| BRSV-G-P2 | ACCCAAAATAGAAAAAACAAAAGCCAATC | ||
| BRSV-G-Pmod | CAAGCAGAGCCCCCACAATCA | ||
| BPI-3 | M | BPI3-M-Pmod | TGCACAGCAATTGGATCAATAAC |
| BPI3-M-P2a | CCCAAATCCATGGCATTGTTATCATTGCC | ||
| BPI3-M-P2b | CCCAAATCTATGGCTTTGCTATCATTGCC | ||
| BPI3-M-P3a | ACAATATCAATAAATCTACAAGTACATATCAAAAC | ||
| BPI3-M-P3b | ACGATTTCAATAAACTTGCAAGTACACATTAAAAC | ||
| BPI3-M-P3c | ACCATATCAATAAACTTACAGGTACACATAAAAAC | ||
| BVDV -1 | 5’UTR | BVDV-5'UTR P1 | CTGCAGAGGCCCACTGTATTGCTAC1 |
| BVDV 1 | CAGGTAAAAGCAGTTTTAACCGACTGTTACGAATACAGCCTGATA1 | ||
| BVDV -2 | 5’UTR | BVDV-5'UTR P2 | CAGTTGAGGAGTCTCGAGATGCCATG1 |
| BVDV 2 | GACACTCCATTAGTCGAGGAGTCTCGAGATGCC1 | ||
| Hobi virus | 5’UTR | BVDV-5'UTR P3 | CGACGCATCAAGGAATGCCTCG2 |
| Pestivirus common | 5’UTR | BVDV com | GGGTAGCAACAGTGGTGAGTTCGTTGGATGGCT2 |
| Pesti-167R, Common B | CCTGAGTACAGGGCAGTCGTCAGTGGTTC1 | ||
| Pesti-215, Common A | CTCGAGATGCCATGTGGACGAGGGCATGCCCAAG1 | ||
| BCoV | N | BCoV-N-P1 | GTTGTACCCTACTATTCTTGGTTCTCTGG1 |
| BCoV-N-P2 | GTTCAAACCAGGGGTAGAAGAGCTC1 | ||
|
BED Bacteria/protozoa Panel C. perfringens |
plC | Cper-plC-P397 | GCTAGATATGAATGGCAAAGAGGAA |
| Cper-plC-P436 | ACATTCTATCTTGGAGAGGCTATGC | ||
| E. zuernii | ITS1 | Ezu-ITS1-P117 | TGTTTCTACCCACTACATCCAAC |
| E. bovis | ITS1 | Ebov-ITS1-P199 | GGTCTCATAAAACATCACCTCCAA |
| C. parvum | COWP | Cpar-COWP-P636 | CAGGACAACAATGTATGGCACCA |
| Cpar-COWP-P735 | CTGCCCACCAGGATATACAGA | ||
| S.e. Dublin | A1118 | SeD-A1118-P633 | AAATTACGCAGGGGTGGAAATAG |
| S.e. Typhimurium | STM4493 | SeT-STM4493-P257 | CGTAAGCATTGTTCAACTTCAGCAT |
| SeT-STM4493-P695 | CTGAGGAAATTCTGGAAAGCCGT | ||
| E. coli | eaeA | Ecol-eae-P318 | TGGTCAGCAGATCATTTTGCCACTC |
| Ecol-eae-P66 | GCTTAGTGCTGGTTTAGGATTGTTT | ||
| K99 | Ecol-K99-P252 | TGCTCGTATTGACTGGTCTGGTTC | |
| Ecol-K99-P398 | TCACTACGGCTGAATACACTCAC | ||
| stx-1 | Ecol-stx1-P782 | GCGTTCTTATGTAATGACTGCTG | |
| Ecol-stx1-P681 | GTGGCAAGAGCGATGTTACGGTTTG | ||
| Lt | Ecol-Lt-P680 | CCGGCAGAGGATGGTTACAGATT | |
| Ecol-Lt-P545 | GAGGTTTCTGCGTTAGGTGGAATA | ||
|
BED Virus Panel BRoV A |
VP6 | BRoV-VP6-PA1 | AGTTTTCCAAGAGTGATTAATTCAGC |
| BRoV-VP6-PA2 | GGATCAGAAATTCAGGTCGCTGGATT | ||
| BRoV C | VP6 | BRoV-VP6-PC1 | AATGGAATGCAACCTCAATCACCAAC |
| BRoV-VP6-PC2 | ACAGTTTGGAAATGCTATGGGATTGAG | ||
| BRoV B | VP6 | BRoV-VP6-PB1 | TTGTGAGACAACCAGCTAGTCT |
| BRoV-VP6-PB2a | ATTGAACGTCGGGTTGTGGAACC | ||
| BRoV-VP6-PB2b | GAAGGCTCGAGACCAGTTAGAGTG | ||
| BToV | N | BToV-N-P1-uni | CAGCAGTCACTATCTTTGCCATTTG |
| BToV-N-P2-bre | AATAGGCGGCGGAATAATAATGG | ||
| BVDV -1 | 5’UTR | BVDV-5'UTR P1 | CTGCAGAGGCCCACTGTATTGCTAC1 |
| BVDV 1 | CAGGTAAAAGCAGTTTTAACCGACTGTTACGAATACAGCCTGATA1 | ||
| BVDV -2 | 5’UTR | BVDV-5'UTR P2 | CAGTTGAGGAGTCTCGAGATGCCATG1 |
| BVDV 2 | GACACTCCATTAGTCGAGGAGTCTCGAGATGCC1 | ||
| Hobi virus | 5’UTR | BVDV-5'UTR P3 | CGACGCATCAAGGAATGCCTCG2 |
| Pestivirus common | 5’UTR | BVDV com | GGGTAGCAACAGTGGTGAGTTCGTTGGATGGCT2 |
| Pesti-167R, Common B | CCTGAGTACAGGGCAGTCGTCAGTGGTTC1 | ||
| Pesti-215, Common A | CTCGAGATGCCATGTGGACGAGGGCATGCCCAAG1 | ||
| BCoV | N | BCoV-N-P1 | GTTGTACCCTACTATTCTTGGTTCTCTGG1 |
| BCoV-N-P2 | GTTCAAACCAGGGGTAGAAGAGCTC1 | ||
Oliver Lung, unpublished. BVDV 1, 2, Hobi virus, pestivirus common and BCoV probes are present in both Respiratory and Enteric Virus panels.
2.4. Laboratory isolates, synthetic constructs, and nucleic acid extraction
A list of laboratory isolates and synthetic genetic materials used in the evaluation (specificity and sensitivity) and initial validation of these assays and their sources are listed in Table 3 . Specificity of each assay was further evaluated using nucleic acids from a comprehensive panel of non-target pathogens (Supplementary Table 1). Target sequences of Breda virus, a BToV prevalent in North America or any other BToV strains or genomic RNA, were synthesized as gBlocks® gene fragments (IDT, Coralville, IA), cloned into pCR-Blunt II-TOPO vector (Life Technologies Inc., Carlsbad, CA, USA) and propagated in One Shot TOP10 chemically competent E. coli (Life Technologies Inc.) following the manufacturer’s protocol. The plasmids were extracted from overnight liquid cultures using QIAprep Miniprep kit (Qiagen Inc., Hilden Germany) and the sequence information and the orientation of the insert were confirmed by DNA sequencing using the Eurofins sequencing services (Eurofins MWG, Louisville, KY). Extracted plasmids were used as templates for PCR amplification of the T7 promoter and the cloned gene target using M13 forward and reverse primers. The amplicons were purified using QIAquick PCR purification kit (Qiagen Inc.) according to manufacturer’s recommendations and subjected to in vitro RNA transcription using MEGAscript T7 Transcription Kit (Thermo Fisher Scientific, Waltham, MA, USA). The in vitro synthesized BToV RNA was treated with TURBO DNase (Life Technologies Inc.) to remove any template DNA, quantified using Qubit RNA BR Assay kit on Qubit 2.0 fluorometer, (both from Life Technologies Inc.) and used in multiplex PCR and microarray assay evaluation and validation.
Table 3.
Number of laboratory isolates and/or synthetic constructs used in evaluation of multiplex PCR and electronic microarray.
| Target pathogen | Number of isolates tested by multiplex PCR | Number of isolates tested by electronic microarray | Reference to the heat map (Fig. 2) |
|---|---|---|---|
| Viruses associated with bovine respiratory disease complex | |||
| BVDV-1 | 4 | 3 | 1-3 |
| BVDV-2 | 4 | 3 | 4-6 |
| Hobi virus | 1 | 1 | 7 |
| BoHV-1 | 7 | 4 | 8-11 |
| BCoV | 3 | 3 | 12-14 |
| BPI3 | 3 | 2 | 15,16 |
| BRSV | 2 | 1 | 17 |
| Bacteria associated with bovine respiratory disease complex | |||
| M. haemolytica type 1 | 6 | 2 | 18, 19 |
| M. haemolytica type 2 | 6 | 1 | 20 |
| M. haemolytica type 6 | 6 | 3 | 21-23 |
| P. multocida | 6 | 6 | 24-29 |
| H. somni | 1 | 1 | 30 |
| M. bovis | 5 | 3 | 31-33 |
| Viruses associated with bovine enteric diseases | |||
| BVDV-1 | 4 | 3 | 34-36 |
| BVDV-2 | 4 | 3 | 37-39 |
| Atypical pestivirus | 1 | 1 | 40 |
| BRoV -A | 2 | 2 | 41, 42 |
| BRoV-B | 4 | 1 | 43 |
| BRoV-C | 1 | 1 | 44 |
| BToV | 2 | 2 | 45, 46 |
| BCoV | 3 | 3 | 47-49 |
| Bacteria and protozoa associated with bovine enteric diseases | |||
| C. perfringens | 1 | 1 | 50 |
| E. zuernii | 1 | 1 | 51 |
| E. bovis | 1 | 1 | 52 |
| C. parvum | 2 | 1 | 53 |
| S.e. Dublin | 3 | 3 | 54-56 |
| S.e. Typhimurium | 4 | 4 | 57-60 |
| E. coli | 5 | 4 | 61-64 |
QIAamp Viral RNA mini and DNeasy Blood and Tissue kits (both from Qiagen Inc.) were selected to extract nucleic acids from viral and bacterial pathogens respectively, based on the results from previous studies (Lung et al., 2015) following manufacturer’s recommendations.
The analytical sensitivity of each multiplex PCR assay was determined using nucleic acids extracted from serially diluted laboratory-amplified isolates of each pathogen with known titers, when available. Titrated virus stocks of BVDV 1 and 2, BoHV1, BRSV, and BPI3 were from Prairie Diagnostic Services, Saskatoon, SK, and bacterial cultures of S. enterica Dublin and Typhimurium strains, and E. coli O101, O111 and O149 strains were kindly provided by Dr. John Fairbrother, Faculty of Veterinary Medicine, University of Montreal, QC. Cultures of M. haemolytica type 1, 2 and 6 were from the Lethbridge Research and Development Centre, Lethbridge, AB and P. multocida was from Manitoba Agriculture, Food and Rural Initiative, Winnipeg, MB. Titrated C. parvum oocysts were purchased from Creative Sciences, Scotland, UK.
Nucleic acids from viruses were extracted from 10-fold dilutions of titrated virus stocks. With respect to bacteria, frozen stocks of M. haemolytica strains, S. e. Dublin, S. e. Typhimurium, and E. coli strains were streaked onto LB agar plates and P. multocida onto 5% sheep blood agar plates. Following overnight incubation at 37 °C, single colonies of each bacterium was harvested and inoculated into 5 ml of Miller’s LB broth and incubated overnight at 37 °C on a shaking incubator (150 rpm). On the next day, the cultures were enumerated using viable plate count method (Miller, 1972). The cultures were standardized to 3.33 × 10^6 cfu/ml so that a 30 μl aliquot of each 10x serial dilution for each pathogen yielded CFUs to the nearest power of base 10 (i.e. 1 × 106, 1 × 105, etc.). Thirty microliters of each dilution were used for nucleic acid extraction.
Titrated stocks were not available for M. bovis, H. somni, E. bovis, E. zuernii, C. perfringens, BRoV A, B and C strains, and BCoV. Therefore synthetic gene fragments targeting the genes of interest of these pathogens were artificially synthesized and individually cloned into pBlueScript SK(+) expression vector (GenScript, Piscataway, NJ). For DNA targets, cloned plasmids were amplified, quantified and directly used in limit-of-detection experiments. For RNA targets, the plasmids with target sequences were subjected to in vitro RNA transcription using MEGAscript T7 transcription kit as described above (Thermo Fisher Scientific). Based on the DNA/RNA yield and the length of each target sequence, copy numbers were calculated using a copy number calculator (http://www.endmemo.com/bio/dnacopynum.php) and used to determine the analytical sensitivities of multiplex PCR assays.
2.5. Clinical samples and nucleic acid extraction
Clinical samples (Table 4 ) used in validation of the assays were from Prairie Diagnostic Services, Saskatoon, SK and Animal Health Centre, Ministry of Agriculture, Abbotsford, BC. The clinical samples for BRDC-associated pathogens included nasal swabs (n = 12) and lung tissues (n = 2), whereas for BED-associated pathogens, the clinical sample were feces (n = 13), intestinal tissues (n = 1) and rectal swabs. Nasal swabs (n = 10) and fecal swabs (n = 8) from clinically healthy cattle were obtained from the herd at the Lethbridge Laboratory of the Canadian Food Inspection Agency. DNeasy Blood & Tissue kit and QIAamp DNA Stool Mini kit (both from Qiagen inc.) were used to extract nucleic acid from BRDC clinical samples and BED clinical samples, respectively, according to the manufacturer’s recommendations.
Table 4.
Validation of current multiplex PCR and capture probes in electronic microarray using clinical samples. Genetic material from each sample was amplified using BRDC (a) or BED (b) viral and bacterial multiplex PCR/RT-PCR assays. +: microarray detectable reaction, §initial results from provincial diagnostic laboratories. ¶ Initial diagnosis at the species level (without genotype).
| (a) Sample ID | Sample type | BoHV-1 | BRSV | BPI3 | BCoV | M. bovis | P. multocida | M. haemolytica |
H. somni | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| nmaA | tbpB | lktA | |||||||||
| 16-02999-2 | Swab | +§ | + | + | + | ||||||
| 16-03583-248 | Swab | + | + | +§ | |||||||
| 16-3747-1 | Swab | + | +§ | ||||||||
| 15-18333 | Lung tissue | + | +¶ | + | + | ||||||
| 15-18345 | Lung tissue | +¶ | + | + | |||||||
| 16-03583-242 | Swab | + | +¶ | ||||||||
| 16-03583-243 | Swab | +§ | + | ||||||||
| 16-03583-245 | Swab | +§ | |||||||||
| 16-03747-4 | Swab | + | § | + | |||||||
| 14-34524-3 | Swab | +§ | +§ | +§ | +§ | + | + | ||||
| 14-34524-7 | Swab | § | +§ | +§ | +§ | +§ | + | + | + | ||
| 14-34524-22 | Swab | +§ | +§ | + | + | + | |||||
| 14-34524-23 | Swab | § | § | +§ | +§ | + | |||||
| 14-34524-34 | Swab | § | § | +§ | +§ | + | + | ||||
| (b) Sample ID | Sample type | BRoV A | C. perfringens | C. parvum | E. zuernii |
E. coli |
|
|---|---|---|---|---|---|---|---|
| eaeA | stx-1 | ||||||
| 16-02423 | Intestinal tissue | +§ | + | + | |||
| 16-03954 | Feces | +§ | |||||
| 16-04427 | Feces | § | |||||
| 16-04443 | Feces | + | +§ | ||||
| 15-14192 | Feces | + | +§ | ||||
| 15-14530 | Feces | + | +§ | ||||
| 15-15110-318 | Feces | + | +§ | ||||
| 15-15110-221 | Feces | + | +§ | ||||
| 15-15473 | Feces | +§ | |||||
| 15-13531 | Feces | + | + | + | |||
| 15-11157 | Feces | +§ | |||||
| 15-11906 | Feces | +§ | + | + | + | ||
| Rota+ | Feces | +§ | |||||
| 03-9822 | Feces | +§ | + | + | |||
2.6. PCR and RT-PCR conditions
Initially, four pools of PCR primers were prepared (75 μM final concentration of each primer) to use in their respective multiplex PCR assays i.e. BRDC virus, BRDC bacteria, BED virus, and BED bacteria/protozoa assays. As presented in Table 1, the BRDC virus primer panel consisted of 12 primers targeting various genomic regions of five viruses. The BRDC bacteria panel consisted of 14 primers targeting four pathogenic bacterial species. The BED virus primer panel contained 13 primers targeting four viruses, and the BED bacteria/protozoa panel contained 20 primers targeting various genomic regions in seven pathogenic bacteria and protozoa. Forward and reverse primers for the DENV internal control were included in the total number of primers in each pool.
All four multiplex PCR assays were optimized for buffer and magnesium concentration, annealing temperature, cycle number, and internal control concentration. All reactions were performed on a Veriti® 96-Well Fast Thermal Cycler (Applied Biosystems, Foster City, CA). For virus targets, both singleplex and multiplex reverse transcription (RT)-PCR assays were performed using Superscript III One-step RT-PCR kit with Platinum Taq DNA polymerase (Thermo Fisher Scientific). The finalized RT-PCR mixtures consisted of 1 μl of nucleic acid, 0.001 pg of internal control, 1 μl of enzyme mix, 2 μl of pooled primers (0.25 μM final concentration) and 12.5 μl of 2 x reaction buffer in a final reaction volume of 25 μl. For bacterial and protozoal targets, singleplex PCR assays were performed using Platinum Taq DNA Polymerase (Invitrogen) according to the manufacturer’s instructions and multiplex PCR were performed using Platinum® Multiplex PCR Master Mix (Applied Biosystems). Finalized multiplex PCR mixtures consisted of 1 μl of nucleic acid, 0.001 pg of internal control, 12.5 μl of 2x Platinum® Multiplex PCR Master Mix, and 2 μl of pooled primers to a final volume of 25 μl. PCR cycling conditions were 2 min of initial denaturation at 94 °C followed by 40 cycles of 30 s at 94 °C, 30 s at 55 °C, and 40 s at 68 °C, and 5 min at 68 °C for final extension. The RT-PCR cycling conditions included an initial RT step of 30 min at 50 °C followed by the same cycling conditions described above. Amplified products were visualized by gel electrophoresis using the QIAxcel Advanced System (Qiagen). The minimum number of target copies in a sample that could produce a detectable band by electrophoresis upon PCR amplification was considered as the limit-of-detection (LOD) for each multiplex PCR. Any loss of sensitivity due to multiplexing was evaluated by comparison of LODs between singleplex and multiplex PCRs for each target.
2.7. Electronic microarray
Based on the evaluation of 76 microarray capture probes, 58 probes were selected to generate the four microarray panels to detect the BRDC and BED pathogens (Table 2). In addition to the pathogen-specific probes, each panel also consisted of a probe specific for the DENV internal control. The selected probes were validated on the NanoChip® 400 (NC400) electronic microarray platform (Nanogen, San Diego, CA), which is an “amplicon-to-answer” system that automates the post-PCR analytical steps i.e. array printing, electrophoretically-driven amplicon hybridization, washing, and reporting (Takahashi et al., 2008; Papatheodorou et al., 2010; Keen-Kim et al., 2006). The NC400 microarray protocol was carried out as previously described by (Lung et al., 2012) with modifications as described by (Lung et al., 2015). Briefly, biotinylated capture probes (250 nM) prepared in 50 mM histidine buffer containing 0.05% Proclin, were electronically addressed (“printed”) to selected electrodes (test sites) in duplicate on the NC 400 cartridge through the application of a positive 350 nA current for 30 s. After five washes of the cartridge with histidine buffer, unpurified PCR amplicons, diluted 1:8 in Cap-Down A Buffer (Nexogen Inc., San Diego, CA), were electronically addressed to selected test sites (800 nA for 120 s). After five more washes with histidine buffer, the cartridge was subsequently filled with low-salt buffer (LSB) (Nexogen Inc.). Hybridized amplicons were detected using 5′-Alexa Fluor 647 modified locked nucleic acid (LNA) variant (5′-/5Alex647 N/TGT + CA + AGCG + AT + AT + ACT + GC-3′) (IDT). The reporting protocol included washing of the microarray cartridge with LSB at 2 °C temperature increments between 24 °C and 60 °C. An image was taken at each temperature increment, and the raw fluorescent intensity (FI) data from all utilized electrodes at each temperature increment were obtained and analyzed using Microsoft Excel. For each probe, positive-to-no-template control (P:N) ratios were calculated by dividing the FI value from each analyte by the FI value produced by the no template control (NTC). For each assay, samples that produced P:N ratios of 2.0 or greater were considered positive. Average P:N data derived from the microarrays were visualized with a heat map generated using Microsoft Excel. Based on preliminary data, P:N ratios calculated using FI values at 50 °C were considered as optimum and used throughout this study.
Amplicons generated by multiplex PCR using 10-fold serial dilutions of each target were used to determine the detection limits of the microarray (Table 5). The highest dilution of the target that resulted in P:N ratio of 2 or higher was considered as the LOD of the electronic microarray for that target.
Table 5.
Limits of detection of each target used in this study are expressed as the minimum number of target gene copies in 25 μl PCR volume needed to produce a band visible by gel electrophoresis (detection limit of multiplex PCR) or to produce a reactivity of >2 PN ratio by electronic microarray (detection limit in electronic microarray).
| Target pathogen | Target gene | Detection limit by multiplex PCR/RT-PCR | Detection limit by electronic microarray |
|---|---|---|---|
| BRD virus multiplex RT-PCR | |||
| BoHV-1 | gB | 100 | 1000 |
| BRSV | G | 1 | 1 |
| BPI-3 | M | 1 | 10 |
| BVDV 1 | 5’UTR | 1 | 1 |
| BVDV 2 | 5’UTR | 1 | 10 |
| BRD bacteria multiplex PCR | |||
| H. somni | 16S rDNA | 1 | 1 |
| P. multocida | ulaR | 1 | 1 |
| M. bovis | 16S rDNA | 1 | 1 |
| M. haemolytica | nmaA | 10 | 10 |
| tbpB | 10 | 100 | |
| lktA | 100 | 1000 | |
| BED virus multiplex RT-PCR | |||
| BCoV | N | 1 | 1 |
| BRoV A | VP6 | 1 | 1 |
| BRoV C | VP6 | 100 | 1000 |
| BRoV B | VP6 | 1 | 10 |
| BToV (common) | N | 1000 | 10000 |
| BToV (Breda) | N | 1000 | 1000 |
| BVDV 1 | 5’UTR | 1 | 1 |
| BVDV 2 | 5’UTR | 1 | 10 |
| BED bacteria multiplex PCR | |||
| C. perfringens | plC | 10 | 10 |
| E. zuernii | ITS1 | 10 | 100 |
| E. bovis | ITS1 | 10 | 10 |
| C. parvum | COWP | 100 | 1000 |
| S. e. Dublin | A1118 | 10 | 100 |
| S. e. Typhimurium | STM4493 | 100 | 100 |
| E. coli | eaeA | 10 | 10 |
| stx-1 | 10 | 100 | |
| Lt | 1 | 10 | |
| K99 | 10 | 100 | |
3. Results
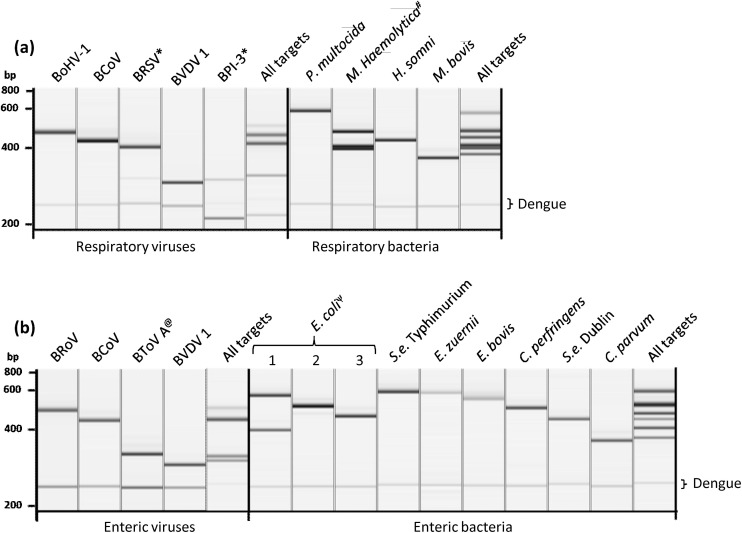
Four separate multiplex PCR assays were designed to amplify selected genetic targets from BRDC-associated viruses, BRDC-associated bacteria, BED-associated viruses, and BED-associated bacteria and protozoa, respectively (Table 1). Each assay was successfully validated using a panel of laboratory isolates of target (Table 3) and related non-target pathogens (Supplementary Table 1). PCR using each primer panel specifically amplified intended size PCR products that ranged from 203–584bp (Fig. 1 ). There was no detectable amplification when non-target pathogens were used as templates. In addition to the target-specific primers, primers for DENV internal control were also included in each panel (Fig. 1). Simultaneous inclusion of all relevant targets and the internal control in each assay generated multiple bands of expected sizes respectively (Fig. 1).
Fig. 1.
QIAxcel images of a representative set of BRDC (a) and BED (b) target pathogens amplified with multiplex PCR assays. In each multiplex PCR, primers specific for the 3′ untranslated region of the dengue virus were also included to detect dengue virus internal control (229bp). No amplification was observed in no template controls (data not shown). *BVDV contamination. # M. Haemolytica type 1 expressing nmaA, tbpB and Lkt genes. ψ lanes 1, 2, and 3 contains E. coli strains O157:H7 (expressing eae and stx-1), O149:H10 (expressing Lt), and O101 (expressing K99), respectively. @similar size bands were produced for different strains by PCR and subsequently were differentiated by electronic microarray.
3.1. Multiplex RT-PCR for BRDC and BED viruses
All respiratory and enteric viral pathogens targeted in this study are RNA viruses except for BoHV-1, which is a DNA virus. RT-PCR was performed to amplify BRDC and BED viral targets using respective multiplex primer panel. Optimized five-plex RT-PCR assay targeting BRDC viruses could successfully amplify genetic targets from all three BRDC-specific viruses (BoHV-1, BRSV, and BPI-3) and two viruses that are associated with both respiratory and enteric diseases (BCoV and BVDV). Primers targeting BVDV were able to amplify ∼280bp region of 5′UTR sequence of type 1 and 2 strains of BVDV as well as HoBi virus (a.k.a. BVDV type 3). Differentiation of each type of BVDV upon PCR amplification was achieved by the subsequent use of type-specific microarray detection probes (see below). With respect to the BED virus assay, the optimized RT-PCR assay included primers specific for BToV and BRoV along with primers specific for BVDV and BCoV. The BED primer panel consisted of two sets of BRoV-specific primers, one set targeting 467bp segment of VP6 gene of BRoV type A and C and another set targeting a 584bp segment of the same gene of BRoV type B. As observed by using laboratory strains of target pathogens or transcribed RNA (Table 3), both the BRDC and BED assays generated PCR products of expected sizes without any detectable cross reaction (Fig. 1).
3.2. Multiplex PCR for BRDC and BED bacteria/protozoa
Multiplex PCR assays for bacterial pathogens associated with BRDC and bacterial and protozoan pathogens associated with BED were successfully optimized and validated using laboratory isolates, when available, and synthetic genetic material (Table 3). With a view of potential simultaneous testing of a single sample for both RNA and DNA pathogens, PCR cycling conditions used for bacterial assays were the same as those used in the RT-PCR. The additional step of 50 °C for 30 min (i.e. conditions for RT step) before the PCR did not have any detectable negative impact on the amplification of DNA targets (data not shown). Optimized multiplex panels for both BRDC and BED pathogens targeted one gene from each pathogen except for M. haemolytica and E. coli. With respect to M. haemolytica, the assay consisted of three sets of primers targeting lktA, nmaA, and tbpB genes (Table 1). By doing so commensal M. haemolytica strain type 2 (lktA+, nmaA-, tbpB-) was successfully differentiated from potentially pathogenic type 1 and 6 strains (lktA+, nmaA+, tbpB+) (Klima et al., 2014a, b). Similarly, genes encoding four virulence factors in various strains of E. coli were targeted in the BED bacteria/protozoa assay. As expected, the assay could amplify respective target genes from each E. coli strain used, namely, O157:H7 and O111 (stx-1+, eaeA+, Lt-, K99-), O149:H10 (stx-1-, eaeA-, Lt+, K99-), and O101 (stx-1-, eaeA-, Lt-, K99+) (Fig. 1 and Table 3).
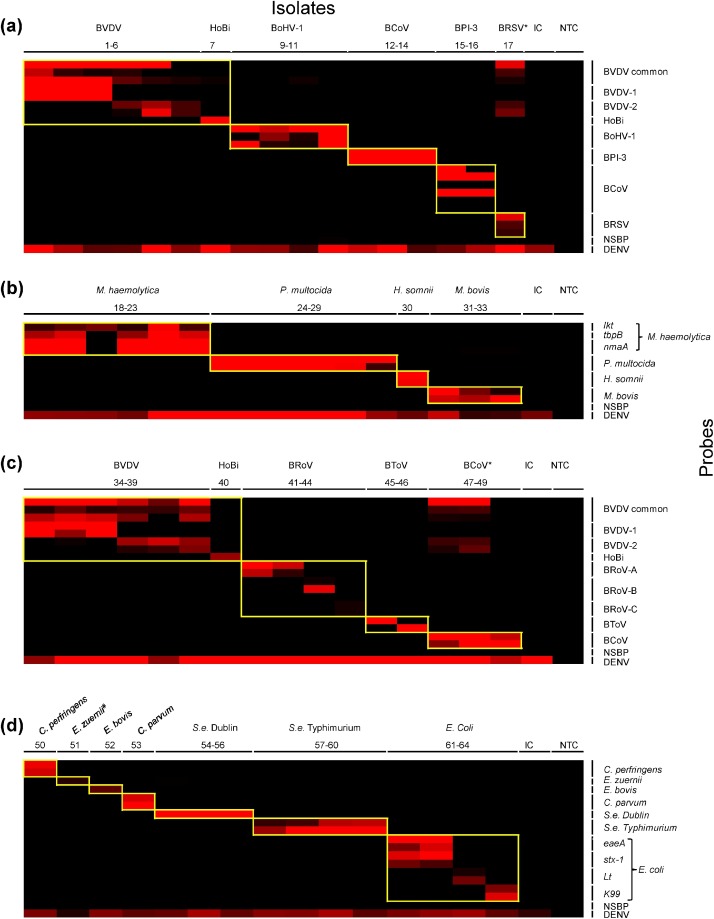
3.3. Electronic microarray
A total of 58 microarray detection probes targeting BRDC and BED pathogens were selected based on their reactivity on the microarray through preliminary screening (Table 2) and successfully validated on NC400 electronic microarray platform using multiplex PCR amplified products generated using laboratory isolates or synthetic genetic material (Table 3 and Fig. 2 a–d). Panels specific for BRDC-associated pathogens (Fig. 2a and b) consisted of 32 probes targeting five viruses and four bacteria. Meanwhile, there were 36 probes in the finalized BED panels (Fig. 2c and d) that were designed to detect four viruses, four bacteria and three protozoa that are associated with neonatal bovine diarrhea. As BCoV and BVDV are associated with both respiratory and enteric ailments in cattle, probes specific for BCoV and for different strains of BVDV were included in both BRDC and BED panels. In addition to the pathogen-specific probes, each panel consisted of two control probes, a negative probe and a probe specific for the dengue virus internal control.
Fig. 2.
Summary of NanoChip 400 electronic microarray results representing BRDC associated viruses (a) and bacteria (b), and BED associated viruses (c) and bacteria/protozoa (d). The heat maps depict positive to negative (PN) fluorescent ratios where PN ratios ≥2 are shown in red and PN ratios <2 are in black. The reactivity of specific reactions between targets and each pathogen-specific probes is outlined in yellow. NTC = No template control, NSBP = Non-specific binding probe negative control, IC = internal control (DENV), *BVDV contamination. # Sample 51 was weakly positive (just above the cut-off) for E. zuernii and dengue armored RNA and therefore represented as faint red bands in the heat map (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).
All target pathogens were accurately detected by their respective probes without any cross reactivity. In the BED viral assay, upon amplification of VP6 gene of BRoV A and C strains using the same set of PCR primers, two strain-specific detection probes for each strain could differentiate between them. Similarly, the Breda virus, a BToV found in North America (Hoet and Saif, 2004) was accurately differentiated from the other bovine toroviruses by using species-specific detection probes (Table 3). In both the BRDC and BED viral assays, while BVDV 5′UTR was amplified in multiplex RT-PCR using common primers, each strain, namely BVDV 1 and 2, and HoBi virus, was accurately differentiated in the electronic microarray using strain-specific probes alongside the BVDV common probes (Table 3). Two laboratory isolates of BCoV and BRSV produced an additional PCR product corresponding to the size of BVDV amplicon by RT-PCR and reacted with BVDV detection probes (Fig. 2a and c) suggesting a possible contamination of the original isolates with BVDV, which was confirmed by subsequent sequencing of PCR amplicons (data not shown). In order to exclude potential contamination of the samples during the assay, the presence of BVDV genomes in the original BCoV and BRSV isolates was confirmed by PCR and sequencing. The source of BVDV contamination in these samples was not investigated but could be non-irradiated fetal bovine serum used in cell culture during virus isolation.
3.4. Analytical sensitivity and specificity
Analytical sensitivities of multiplex PCR and subsequent microarray detection for all 23 pathogens (27 gene targets) were determined using series of dilutions of each target (Table 5 ). All multiplex PCR assays could detect as few as 1–10 copies of most target genes (n = 21). The lowest sensitivity, 1000 copies/reaction, was observed for the BToV N gene in vitro transcribed RNA. The analytical sensitivities of the microarray capture probes were comparable to those of the multiplex PCR assays for most targets. Several targets including BPI-3, BVDV-2, BRoV B, BRoV C, BToV, M. haemolytica (tbpB and lktA), E. zuernii, C. parvum, S.e. Dublin, and E. coli (stx-1, Lt, and K99), showed a 10-fold drop in microarray sensitivity as compared to the corresponding multiplex PCR assay (Table 5).
The specificity of each assay was demonstrated by testing a panel of related non-target pathogens including other bovine pathogens and coronaviruses, herpesviruses and pestiviruses from other species (Supplementary Table 1). As there was no detectable amplification with any of the related non-target pathogens by multiplex PCR, only a few randomly selected reactions were tested on the electronic microarray and observed no positive reactivity, as expected (data not shown).
3.5. Clinical validation of the finalized multiplex PCR-electronic microarray assay
Performance of the multiplex PCR-electronic microarray assay was evaluated using a panel of clinical samples collected from cattle with respiratory and/or enteric ailments (Table 4). According to the diagnostic records from provincial laboratories, most of the samples had tested for a single pathogen only. However, while confirming the original detection, we could demonstrate the presence of multiple pathogens in most samples. Where multiple pathogens were detected, any false positive reactivity was ruled out by demonstrating amplicons of the respective sizes by singleplex PCR using pathogen-specific primers, followed by Sanger sequencing. Among samples collected from clinically healthy animals, several produced faint non-specific bands of various sizes by multiplex PCR. However, such samples did not generate any positive signal on any target specific probe on the microarray (data not shown).
4. Discussion
Identification of causative pathogens by isolation is the gold standard in affirmative diagnoses of infectious diseases. However, isolation of pathogens is difficult in many routine diagnostics due to long turnaround times and for practical reasons such as the requirement of specialized facilities, culture conditions, and high level of technical expertise as well as sensitivity to sample degradation. In contrast, detection of pathogen-specific nucleic acid by PCR-based assays is a rapid, highly sensitive and cost effective alternative method for detection of infectious agents in clinical samples. In many cases, PCR-based assays also show significantly higher sensitivity than conventional culture methods (Bell et al., 2014; Shanthalingam et al., 2014).
Several previous studies documented the development of PCR-based assays for detection of pathogens associated with bovine respiratory and enteric diseases. However, most studies target either a single or a limited number of pathogens. Given the multifactorial nature and frequent involvement of multiple pathogens, a comprehensive assay that could simultaneously detect multiple pathogens is essential for rapid diagnosis and effective control of these economically significant diseases.
This study describes the development and initial validation of four highly sensitive multiplex PCR assays and their respective electronic microarray assays for detection and differentiation of a total of 22 important viral, bacterial, and protozoal pathogens associated with respiratory and enteric diseases in cattle. The newly developed BRDC assays could successfully detect and differentiate four bacteria and five viruses, while the BED assays could detect and differentiate six viruses, four bacteria, and three protozoa. As demonstrated with laboratory strains as well as clinical samples, both the BRDC and BED assays could simultaneously amplify and detect multiple pathogens present in a single sample.
As demonstrated using a limited number of clinical samples collected from the field, both BRDC bacterial and viral assays could successfully detect and differentiate multiple pathogens in a single sample (Table 4) it is notable that the initial diagnosis of clinical samples by corresponding laboratory was reproduced in most cases by the current multiplex assay. However, despite the demonstration of the presence of multiple pathogens, we were unable to reproduce the historically reported detections in a few cases, potentially due to possible sample degradation. While limited access to clinical samples is a hurdle, this highlights the need for use of an adequate number of freshly collected samples for assay validation. BRDC-associated viruses such as BoHV-1, BVDV, and BPI-3 alone can be primary pathogens, and often lead to BRDC of multiple etiologies through immunosuppression and damage to the respiratory epithelium (Srikumaran et al., 2007). Determination of the non-viral etiological organism(s) of BRDC is often challenging as many of the bacterial agents such as H. somni, P. multocida, M. bovis, M. haemolytica causing BRDC are part of the commensal flora of the upper respiratory tract of cattle (Angen et al., 2009). These opportunistic microbes increase in numbers and/or migrate to the lower respiratory tract (trachea and lungs) when the host is stressed or immunocompromised due to concurrent viral infections. Although detection of these pathogens in nasal or oral swabs may have less diagnostic value, the presence of these bacteria in tracheal swabs or lung tissues is indicative of their role in BRDC.
Although several molecular assays have been developed to detect multiple BED pathogens (Fukuda et al., 2012; Asano et al., 2010; Loa et al., 2006; Tsuchiaka et al., 2016), to our knowledge, the viral and bacterial/protozoa assays described here are the most comprehensive assays published to date for detection and differentiation of BED causative pathogens in a single reaction. A multiplex PCR assay for detection of five BED-associated viruses, namely BCoV, BRoV (strains A, B, and C) and BToV has been described previously (Fukuda et al., 2012). In addition to those pathogens, the NC400 BED viral assay could detect and differentiate BVDV 1, 2, and HoBi virus and differentiate Breda virus, which is prevalent in the North America from other toroviruses. E. coli is commensal bacteria in the gut. Therefore, virulence factors associated with pathogenic strains of E. coli have been used to differentiate commensal strains from those that cause disease in calves (West et al., 2007; Sharma and Dean-Nystrom, 2003; Sharma, 2006). By including a combination of virulence genes that are associated with pathogenic E. coli strains, the NC400 BED viral assay could differentiate pathotypes such as enterohemorrhagic (stx-1+, eaeA+), Enterotoxigenic (Lt+), and enteropathogenic (K99+) E. coli strains from those which are commensal inhabitants of the normal gut microflora.
The current assays showed an overall high level of sensitivity for the detection of target pathogens however, when field-derived clinical samples are used, potential PCR inhibitors in the samples may reduce the sensitivity of these assays. When the multiplex assays were combined with microarray detection a slight reduction (1/10 drop) in sensitivity was observed for several targets. However, given the potential high number of target pathogen particles present in clinical samples, the reduced sensitivity observed may not have a significant impact on the performance of these assays. Nevertheless, these assays should be further investigated through a comprehensive clinical validation, before applying to routine diagnostics.
5. Conclusions
In summary, new pathogen detection assays based on multiplex PCR and automated electronic microarray have been developed for comprehensive detection of bacterial, viral, and protozoal pathogens that are associated with BRDC and BED. With further clinical validation and subsequent deployment to veterinary diagnostic laboratories, these assays could be used for the rapid identification of pathogens in disease outbreaks and facilitate quick and accurate decision making for the control and treatment of economically important diseases of cattle. Furthermore, these assays could be very effective tools in epidemiological studies as well as screening of healthy animals to identify carriers, and those that may potentially develop BRD or BED. These assays are currently being adapted for use on a fully-integrated and fully-automated “sample-to-answer” system which reduces costs associated with labour, decreases turnaround time, and minimizes potential cross-contamination due to user intervention.
Acknowledgements
Authors would like to thank Drs. Carlos Hermosilla at University of Geisen for Eimeria samples, Dr. Jeremie Diedhiou for his work on development of BRDC and BED sequence database, Dr. Noriko Goji at the CFIA-Lethbridge Laboratory and the two anonymous reviewers for their critical review of this manuscript and valuable suggestions. This study was funded by the Beef Science Cluster project ANH.13.13 (Development of a fully-automated DNA microarray chip for multiplex detection of bovine pathogens) from the Beef Cattle Research Council, Calgary, AB, Canada.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.jviromet.2018.08.010.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- Angen O., Ahrens P., Tegtmeier C. Development of a PCR test for identification of Haemophilus somnus in pure and mixed cultures. Vet. Microbiol. 1998;63:39–48. doi: 10.1016/s0378-1135(98)00222-3. [DOI] [PubMed] [Google Scholar]
- Angen O., Thomsen J., Larsen L.E., Larsen J., Kokotovic B., Heegaard P.M., Enemark J.M. Respiratory disease in calves: microbiological investigations on trans-tracheally aspirated bronchoalveolar fluid and acute phase protein response. Vet. Microbiol. 2009;137:165–171. doi: 10.1016/j.vetmic.2008.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano K.M., de Souza S.P., de Barros I.N., Ayres G.R., Silva S.O., Richtzenhain L.J., Brandao P.E. Multiplex semi-nested RT-PCR with exogenous internal control for simultaneous detection of bovine coronavirus and group A rotavirus. J. Virol. Methods. 2010;169:375–379. doi: 10.1016/j.jviromet.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azizzadeh M., Shooroki H.F., Kamalabadi A.S., Stevenson M.A. Factors affecting calf mortality in Iranian Holstein dairy herds. Prev. Vet. Med. 2012;104:335–340. doi: 10.1016/j.prevetmed.2011.12.007. [DOI] [PubMed] [Google Scholar]
- Bell C.J., Blackburn P., Elliott M., Patterson T.I., Ellison S., Lahuerta-Marin A., Ball H.J. Investigation of polymerase chain reaction assays to improve detection of bacterial involvement in bovine respiratory disease. J. Vet. Diagn. Invest. 2014;26:631–634. doi: 10.1177/1040638714540166. [DOI] [PubMed] [Google Scholar]
- Blais B.W., Gauthier M., Descheenes M., Huszczynski G. Polyester cloth-based hybridization array system for identification of enterohemorrhagic Escherichia coli serogroups O26, O45, O103, O111, O121, O145, and O157. J. Food Prot. 2012;75:1691–1697. doi: 10.4315/0362-028X.JFP-12-116. [DOI] [PubMed] [Google Scholar]
- Boileau M.J., Kapil S. Bovine coronavirus associated syndromes. Vet. Clin. North Am. Food Anim. Pract. 2010;26:123–146. doi: 10.1016/j.cvfa.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y.-i., Yoon K.-J. An overview of calf diarrhea - infectious etiology, diagnosis, and intervention. J. Vet. Sci. 2014;15:1. doi: 10.4142/jvs.2014.15.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y.I., Kim W.I., Liu S., Kinyon J.M., Yoon K.J. Development of a panel of multiplex real-time polymerase chain reaction assays for simultaneous detection of major agents causing calf diarrhea in feces. J. Vet. Diagn. Invest. 2010;22:509–517. doi: 10.1177/104063871002200403. [DOI] [PubMed] [Google Scholar]
- Decaro N., Sciarretta R., Lucente M.S., Mari V., Amorisco F., Colaianni M.L., Cordioli P., Parisi A., Lelli R., Buonavoglia C. A nested PCR approach for unambiguous typing of pestiviruses infecting cattle. Mol. Cell. Probes. 2012;26:42–46. doi: 10.1016/j.mcp.2011.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deregt D., Gilbert S.A., Dudas S., Pasick J., Baxi S., Burton K.M., Baxi M.K. A multiplex DNA suspension microarray for simultaneous detection and differentiation of classical swine fever virus and other pestiviruses. J. Virol. Methods. 2006;136:17–23. doi: 10.1016/j.jviromet.2006.03.025. [DOI] [PubMed] [Google Scholar]
- Dhama K., Chauhan R.S., Mahendran M., Malik S.V. Rotavirus diarrhea in bovines and other domestic animals. Vet. Res. Commun. 2009;33:1–23. doi: 10.1007/s11259-008-9070-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster D.M., Smith G.W. Pathophysiology of diarrhea in calves. Vet. Clin. North Am. Food Anim. Pract. 2009;25:13–36. doi: 10.1016/j.cvfa.2008.10.013. xi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank S.M., Bosworth B.T., Moon H.W. Multiplex PCR for enterotoxigenic, attaching and effacing, and Shiga toxin-producing Escherichia coli strains from calves. J. Clin. Microbiol. 1998;36:1795–1797. doi: 10.1128/jcm.36.6.1795-1797.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda M., Kuga K., Miyazaki A., Suzuki T., Tasei K., Aita T., Mase M., Sugiyama M., Tsunemitsu H. Development and application of one-step multiplex reverse transcription PCR for simultaneous detection of five diarrheal viruses in adult cattle. Arch. Virol. 2012;157:1063–1069. doi: 10.1007/s00705-012-1271-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez D.E., Weese J.S. Viral enteritis in calves. Can. Vet. J. 2017;58(December (12)):1267–1274. [PMC free article] [PubMed] [Google Scholar]
- Griffin D., Chengappa M.M., Kuszak J., McVey D.S. Bacterial pathogens of the bovine respiratory disease complex. Vet. Clin. North Am. Food Anim. Pract. 2010;26:381–394. doi: 10.1016/j.cvfa.2010.04.004. [DOI] [PubMed] [Google Scholar]
- Guy R.A., Payment P., Krull U.J., Horgen P.A. Real-time PCR for quantification of Giardia and Cryptosporidium in environmental water samples and sewage. Appl. Environ. Microbiol. 2003;69:5178–5185. doi: 10.1128/AEM.69.9.5178-5185.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall T.A. Nucleic Acids Symposium Series. Information Retrieval Ltd.; London: 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT; pp. c1979–c2000. [Google Scholar]
- Hilton W.M. BRD in 2014: where have we been, where are we now, and where do we want to go? Anim. Health Res. Rev. 2014;15:120–122. doi: 10.1017/S1466252314000115. [DOI] [PubMed] [Google Scholar]
- Hoet A.E., Saif L.J. Bovine torovirus (Breda virus) revisited. Anim. Health Res. Rev. 2004;5:157–171. doi: 10.1079/ahr200498. [DOI] [PubMed] [Google Scholar]
- Horwood P.F., Mahony T.J. Multiplex real-time RT-PCR detection of three viruses associated with the bovine respiratory disease complex. J. Virol. Methods. 2011;171:360–363. doi: 10.1016/j.jviromet.2010.11.020. [DOI] [PubMed] [Google Scholar]
- Huang Y., Tang H., Duffy S., Hong Y., Norman S., Ghosh M., He J., Bose M., Henrickson K.J., Fan J., Kraft A.J., Weisburg W.G., Mather E.L. Multiplex assay for simultaneously typing and subtyping influenza viruses by use of an electronic microarray. J. Clin. Microbiol. 2009;47:390–396. doi: 10.1128/JCM.01807-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur T.Y., Jung Y.H., Choe C.Y. The dairy calf mortality: the causes of calf death during ten years at a large dairy farm in Korea. Korean J. Vet. Res. 2013;53:103–108. [Google Scholar]
- Johnson K.K., Pendell D.L. Market impacts of reducing the prevalence of bovine respiratory disease in United States beef cattle feedlots. Front. Vet. Sci. 2017;4:189. doi: 10.3389/fvets.2017.00189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara F., Zhang G., Mingala C.N., Tamura Y., Koiwa M., Onuma M., Nunoya T. Genetic analysis and development of species-specific PCR assays based on ITS-1 region of rRNA in bovine Eimeria parasites. Vet. Parasitol. 2010;174:49–57. doi: 10.1016/j.vetpar.2010.08.001. [DOI] [PubMed] [Google Scholar]
- Keen-Kim D., Grody W.W., Richards C.S. Microelectronic array system for molecular diagnostic genotyping: nanogen NanoChip 400 and molecular biology workstation. Expert Rev. Mol. Diagn. 2006;6:287–294. doi: 10.1586/14737159.6.3.287. [DOI] [PubMed] [Google Scholar]
- Kishimoto M., Tsuchiaka S., Rahpaya S.S., Hasebe A., Otsu K., Sugimura S., Kobayashi S., Komatsu N., Nagai M., Omatsu T., Naoi Y., Sano K., Okazaki-Terashima S., Oba M., Katayama Y., Sato R., Asai T., Mizutani T. Development of a one-run real-time PCR detection system for pathogens associated with bovine respiratory disease complex. J. Vet. Med. Sci. 2017;79:517–523. doi: 10.1292/jvms.16-0489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klima C.L., Alexander T.W., Hendrick S., McAllister T.A. Characterization of Mannheimia haemolytica isolated from feedlot cattle that were healthy or treated for bovine respiratory disease. Can. J. Vet. Res. 2014;78:38–45. [PMC free article] [PubMed] [Google Scholar]
- Klima C.L., Zaheer R., Cook S.R., Booker C.W., Hendrick S., Alexander T.W., McAllister T.A. Pathogens of bovine respiratory disease in North American feedlots conferring multidrug resistance via integrative conjugative elements. J. Clin. Microbiol. 2014;52:438–448. doi: 10.1128/JCM.02485-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koressaar T., Remm M. Enhancements and modifications of primer design program Primer3. Bioinformatics. 2007;23:1289–1291. doi: 10.1093/bioinformatics/btm091. [DOI] [PubMed] [Google Scholar]
- Kostrzynska M., Bachand A. Application of DNA microarray technology for detection, identification, and characterization of food-borne pathogens. Can. J. Microbiol. 2006;52:1–8. doi: 10.1139/w05-105. [DOI] [PubMed] [Google Scholar]
- Liu D., Lawrence M.L., Austin F.W. Specific PCR identification of Pasteurella multocida based on putative transcriptional regulator genes. J. Microbiol. Methods. 2004;58:263–267. doi: 10.1016/j.mimet.2004.04.008. [DOI] [PubMed] [Google Scholar]
- Loa C.C., Lin T.L., Wu C.C., Bryan T.A., Hooper T.A., Schrader D.L. Differential detection of turkey coronavirus, infectious bronchitis virus, and bovine coronavirus by a multiplex polymerase chain reaction. J. Virol. Methods. 2006;131:86–91. doi: 10.1016/j.jviromet.2005.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lung O., Beeston A., Ohene-Adjei S., Pasick J., Hodko D., Hughes K.B., Furukawa-Stoffer T., Fisher M., Deregt D. Electronic microarray assays for avian influenza and Newcastle disease virus. J. Virol. Methods. 2012;185:244–253. doi: 10.1016/j.jviromet.2012.07.005. [DOI] [PubMed] [Google Scholar]
- Lung O., Ohene-Adjei S., Buchanan C., Joseph T., King R., Erickson A., Detmer S., Ambagala A. Multiplex PCR and microarray for detection of swine respiratory pathogens. Transbound. Emerg. Dis. 2015;64:834–848. doi: 10.1111/tbed.12449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lung O., Furukawa-Stoffer T., Burton Hughes K., Pasick J., King D.P., Hodko D. Multiplex RT-PCR and automated microarray for detection of eight bovine viruses. Transbound. Emerg. Dis. 2016 doi: 10.1111/tbed.12591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marley M.S., Givens M.D., Galik P.K., Riddell K.P., Stringfellow D.A. Development of a duplex quantitative polymerase chain reaction assay for detection of bovine herpesvirus 1 and bovine viral diarrhea virus in bovine follicular fluid. Theriogenology. 2008;70:153–160. doi: 10.1016/j.theriogenology.2008.03.007. [DOI] [PubMed] [Google Scholar]
- Miller J.H. Determination of viable cell counts: bacterial growth curves. Experiments in molecular genetics. In: Miller J.H., editor. Experiments in Molecular Genetics. Cold Spring Harbor; New York: 1972. pp. 31–36. [Google Scholar]
- Mosier D. Review of BRD pathogenesis: the old and the new. Anim. Health Res. Rev. 2014;15:166–168. doi: 10.1017/S1466252314000176. [DOI] [PubMed] [Google Scholar]
- Murray G.M., O’Neill R.G., More S.J., McElroy M.C., Earley B., Cassidy J.P. Evolving views on bovine respiratory disease: an appraisal of selected key pathogens - Part 1. Vet. J. 2016;217:95–102. doi: 10.1016/j.tvjl.2016.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Animal Disease Information Service (NADIS) NADIS; UK: 2007. ). Health Bulletin: Respiratory Disease in Cattle; pp. 1–3. [Google Scholar]
- Ok M., Guler L., Turgut K., Ok U., Sen I., Gunduz I.K., Birdane M.F., Guzelbektes H. The studies on the aetiology of diarrhoea in neonatal calves and determination of virulence gene markers of Escherichia coli strains by multiplex PCR. Zoonoses Public Health. 2009;56:94–101. doi: 10.1111/j.1863-2378.2008.01156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papatheodorou A., Makrythanasis P., Kaliakatsos M., Dimakou A., Orfanidou D., Roussos C., Kanavakis E., Tzetis M. Development of novel microarray methodology for the study of mutations in the SERPINA1 and ADRB2 genes--their association with obstructive pulmonary disease and disseminated bronchiectasis in Greek patients. Clin. Biochem. 2010;43:43–50. doi: 10.1016/j.clinbiochem.2009.08.026. [DOI] [PubMed] [Google Scholar]
- Shanthalingam S., Goldy A., Bavananthasivam J., Subramaniam R., Batra S.A., Kugadas A., Raghavan B., Dassanayake R.P., Jennings-Gaines J.E., Killion H.J., Edwards W.H., Ramsey J.M., Anderson N.J., Wolff P.L., Mansfield K., Bruning D., Srikumaran S. PCR assay detects Mannheimia haemolytica in culture-negative pneumonic lung tissues of bighorn sheep (Ovis canadensis) from outbreaks in the western USA, 2009-2010. J. Wildl. Dis. 2014;50:1–10. doi: 10.7589/2012-09-225. [DOI] [PubMed] [Google Scholar]
- Sharma V.K. Real-time reverse transcription-multiplex PCR for simultaneous and specific detection of rfbE and eae genes of Escherichia coli O157:H7. Mol. Cell. Probes. 2006;20:298–306. doi: 10.1016/j.mcp.2006.03.001. [DOI] [PubMed] [Google Scholar]
- Sharma V.K., Dean-Nystrom E.A. Detection of enterohemorrhagic Escherichia coli O157:H7 by using a multiplex real-time PCR assay for genes encoding intimin and Shiga toxins. Vet. Microbiol. 2003;93:247–260. doi: 10.1016/s0378-1135(03)00039-7. [DOI] [PubMed] [Google Scholar]
- Snowder G.D., Van Vleck L.D., Cundiff L.V., Bennett G.L. Bovine respiratory disease in feedlot cattle: environmental, genetic, and economic factors. J. Anim. Sci. 2006;84:1999–2008. doi: 10.2527/jas.2006-046. [DOI] [PubMed] [Google Scholar]
- Srikumaran S., Kelling C.L., Ambagala A. Immune evasion by pathogens of bovine respiratory disease complex. Anim. Health Res. Rev. 2007;8:215–229. doi: 10.1017/S1466252307001326. [DOI] [PubMed] [Google Scholar]
- Sudiro T.M., Ishiko H., Green S., Vaughn D.W., Nisalak A., Kalayanarooj S., Rothman A.L., Raengsakulrach B., Janus J., Kurane I. Rapid diagnosis of dengue viremia by reverse transcriptase-polymerase chain reaction using 3′-noncoding region universal primers. Am. J. Trop. Med. Hyg. 1997;56:424–429. doi: 10.4269/ajtmh.1997.56.424. [DOI] [PubMed] [Google Scholar]
- Takahashi H., Norman S.A., Mather E.L., Patterson B.K. Evaluation of the NanoChip 400 system for detection of influenza A and B, respiratory syncytial, and parainfluenza viruses. J. Clin. Microbiol. 2008;46:1724–1727. doi: 10.1128/JCM.01947-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thonur L., Maley M., Gilray J., Crook T., Laming E., Turnbull D., Nath M., Willoughby K. One-step multiplex real time RT-PCR for the detection of bovine respiratory syncytial virus, bovine herpesvirus 1 and bovine parainfluenza virus 3. BMC Vet. Res. 2012;8:37. doi: 10.1186/1746-6148-8-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiaka S., Masuda T., Sugimura S., Kobayashi S., Komatsu N., Nagai M., Omatsu T., Furuya T., Oba M., Katayama Y., Kanda S., Yokoyama T., Mizutani T. Development of a novel detection system for microbes from bovine diarrhea by real-time PCR. J. Vet. Med. Sci. 2016;78:383–389. doi: 10.1292/jvms.15-0552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunemitsu H., Smith D., Saif L. Experimental inoculation of adult dairy cows with bovine coronavirus and detection of coronavirus in feces by RT-PCR. Arch. Virol. 1999;144:167–175. doi: 10.1007/s007050050493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Untergasser A., Cutcutache I., Koressaar T., Ye J., Faircloth B.C., Remm M., Rozen S.G. Primer3--new capabilities and interfaces. Nucleic Acids Res. 2012;40:e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- USDA–APHIS–VS . 2013. Part IV: Health and Health Management on U.S. Feedlots With a Capacity of 1,000 or More Head.https://www.aphis.usda.gov/animal_health/nahms/feedlot/downloads/feedlot2011/Feed11_dr_PartIV.pdf National Animal Health Monitoring System Beef Feedlot Study 2011. [Google Scholar]
- Vilcek S., Elvander M., Ballagi-Pordany A., Belak S. Development of nested PCR assays for detection of bovine respiratory syncytial virus in clinical samples. J. Clin. Microbiol. 1994;32:2225–2231. doi: 10.1128/jcm.32.9.2225-2231.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West D.M., Sprigings K.A., Cassar C., Wakeley P.R., Sawyer J., Davies R.H. Rapid detection of Escherichia coli virulence factor genes using multiplex real-time TaqMan PCR assays. Vet. Microbiol. 2007;122:323–331. doi: 10.1016/j.vetmic.2007.01.026. [DOI] [PubMed] [Google Scholar]
- Wiggins M.C., Woolums A.R., Sanchez S., Hurley D.J., Cole D.J., Ensley D.T., Pence M.E. Prevalence of Mycoplasma bovis in backgrounding and stocker cattle operations. J. Am. Vet. Med. Assoc. 2007;230:1514–1518. doi: 10.2460/javma.230.10.1514. [DOI] [PubMed] [Google Scholar]
- Yadav B.S., Pokhriyal M., Ratta B., Kumar A., Saxena M., Sharma B. Viral diagnosis in Indian livestock using customized microarray chips. Bioinformation. 2015;11:489–492. doi: 10.6026/97320630011489. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.