Highlights
-
•
This paper presents an in-depth review of coronavirus Orf1b enzymes.
-
•
It is organized in sections on RNA synthesis, RNA degradation, and RNA capping.
-
•
It discusses integration of these functions into the coronavirus replication/transcription complex.
-
•
It reviews published inhibitors of any of these functions.
Keywords: Coronavirus, SARS-CoV, Replication, Transcription, Nonstructural proteins
Abstract
The SARS (severe acute respiratory syndrome) pandemic caused ten years ago by the SARS-coronavirus (SARS-CoV) has stimulated a number of studies on the molecular biology of coronaviruses. This research has provided significant new insight into many mechanisms used by the coronavirus replication-transcription complex (RTC). The RTC directs and coordinates processes in order to replicate and transcribe the coronavirus genome, a single-stranded, positive-sense RNA of outstanding length (∼27–32 kilobases). Here, we review the up-to-date knowledge on SARS-CoV replicative enzymes encoded in the ORF1b, i.e., the main RNA-dependent RNA polymerase (nsp12), the helicase/triphosphatase (nsp13), two unusual ribonucleases (nsp14, nsp15) and RNA-cap methyltransferases (nsp14, nsp16). We also review how these enzymes co-operate with other viral co-factors (nsp7, nsp8, and nsp10) to regulate their activity. These last ten years of research on SARS-CoV have considerably contributed to unravel structural and functional details of one of the most fascinating replication/transcription machineries of the RNA virus world. This paper forms part of a series of invited articles in Antiviral Research on “From SARS to MERS: 10 years of research on highly pathogenic human coronaviruses”.
1. Introduction
Coronaviruses are ubiquitous viruses infecting a large variety of hosts. In humans, they were mainly responsible for mild respiratory diseases until 2002/2003 when a novel coronavirus was identified as the cause of a severe acute respiratory syndrome (SARS). The SARS-coronavirus (SARS-CoV) infected ∼8400 persons in the world, and differs from other human coronaviruses by its case/fatality rate (∼10%) (Cheng et al., 2013). Its discovery greatly renewed interest in coronavirus research (Hilgenfeld and Peiris, 2013). SARS-CoV may have evolved from coronaviruses infecting bats (SARS-like CoVs) (Li et al., 2005). The virus became infectious in humans probably through an intermediate passage into civets and mutations at least in the structural protein S recognized by the host receptor (Wang and Eaton, 2007, Li, 2013).
Since April, 2012, a novel human pathogenic coronavirus of zoonotic origin, the Middle East respiratory syndrome coronavirus (MERS-CoV) has emerged, causing severe pneumonia as well as kidney failure in some cases (De Groot et al., 2013, Zaki et al., 2012). As of November, 2013, more than 130 MERS-CoV-infected people have been identified, with a case fatality rate of ∼50%. All are related in some way to travel to the Arabian Peninsula. MERS-CoV is a close relative of bat coronaviruses HKU4 and HKU5 (Drexler et al., 2013). The ongoing outbreak confirms that coronaviruses constitute a threat to public health at world level.
The Coronavirinae subfamily contains 4 genera (alpha, beta, gamma and deltacoronaviruses) and, together with the Torovirinae subfamily, form the Coronaviridae family. Nidovirales order is formed by Coronaviridae, Roniviridae (also named long genome nidoviruses), and the Arteriviridae (short genome nidoviruses) families. Recently, a new family of viruses infecting insects was created, in this viral order. It was named Mesoniviridae in reference to their middle-sized genomes between long and short genome nidoviruses (Nga et al., 2011).
Nidoviruses are positive-sense, single-stranded RNA viruses ((+) ssRNA), whose distinctive feature is to produce a set of subgenomic mRNAs (sg mRNAs) coding for structural and accessory proteins. Coronaviruses have a long genome that carries a cap structure at 5′ end and a polyA tail at the 3′ end. Genomic RNA can be directly translated into viral proteins after cell infection. The main characteristic that makes coronaviruses (and long-genome nidoviruses in general) of outstanding interest is the unusual length of their RNA genomes (27–32 kb). It is two to three time larger than that of other RNA viruses, but is thought to be associated with a higher genetic stability (Belshaw et al., 2008, Jenkins et al., 2002).
Nonstructural proteins forming the replication/transcription complex (RTC) are coded by the genomic mRNA, where two overlapping ORFs are found. The ORF1a encodes for polyprotein 1a (pp1a) cleaved by viral proteases into 11 nonstructural protein (nsp1–nsp11). The second (ORF1b) produces 5 additional nonstructural proteins (nsp12–16) upon processing by the viral proteases contained into polyprotein 1ab (pp1ab) (Fig. 1 ). ORF1ab is translated upon a −1 ribosomal frameshift inside ORF1a, implying that nsp12–nsp16 are produced at significantly lower levels than ORF1a-encoded products. The nsp12–nsp16 proteins and their known co-factors/partners are the subject of this review. In an attempt to document their potential as drug targets, we report the present state of the art on the enzymatic machinery formed by the SARS-CoV ORF1b-encoded nonstructural proteins.
Fig. 1.
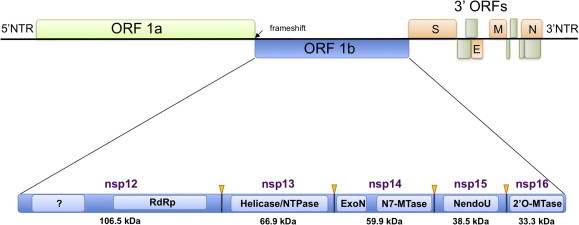
Coronavirus genome organization and expression. Coronavirus genome is depicted and divided into five main segments: 5′NTR (nontranslated region), ORF1a (light green), ORF1b (blue), 3′ORFs, corresponding to all open reading frames coding for structural (orange) and accessory proteins (dark green), and 3′NTR. The ORF1b is zoomed and the derived nonstructural proteins are shown. The nsp5 protease cleavage sites (yellow arrow), the domains harbouring enzymatic activities (soft blue) and the size of each nsp are also depicted. ?: predicted domain of unknown function; RdRp, RNA-dependent RNA polymerase; NTPase, nucleoside triphosphatase, also capable of hydrolysizing 5′-triphosphate-RNA to 5′-diphosphate-RNA); ExoN, 3′ to 5′ exonuclease; N7-MTase, guanine-N7-methyltransferase; NendoU, endoribonuclease; 2′-O-MTase, 2′-O-methyltransferase. Protein size is indicated below each nsp.
2. Nsp12 and nsp13, the presumed replication/transcription complex catalytic core
2.1. The RNA-dependent RNA polymerase nsp12
Nsp12 (102 kDa) is the most conserved protein in coronaviruses, and thus a central enzyme in the viral replication/transcription complex. It is an RNA-dependent RNA polymerase (RdRp) presenting all conserved motifs of canonical RdRps. The polymerase active site (Ser-Asp-Asp within motif C) is conserved in all nidoviruses. More intriguing is the presence of motif G (Gorbalenya et al., 2002), which is a signature of RdRps that initiate RNA synthesis in a primer-dependent manner (te Velthuis et al., 2010). Nsp12 of all coronaviruses also carries a ∼42 kDa amino-terminal extension whose function is yet unknown. Even if bioinformatics studies did not reveal any specific signature sequence, this extension is necessary for RdRp activity (Cheng et al., 2005).
Although it plays a central role in viral replication, nsp12 RdRp is poorly characterized to date, certainly due to the difficulty to express and purify satisfactory amounts of protein. SARS-CoV nsp12 fused to a GST-tag exhibits weak polymerase activity in vitro using poly (rA)/oligo dT12–18 as a template (Cheng et al., 2005). Neither metal requirements nor the ability to initiate RNA synthesis with or without an RNA primer are clearly established, as reported by several authors which used full-length histidine-tagged nsp12 (Ahn et al., 2012, Cheng et al., 2005, Te Velthuis et al., 2010). In any case, the in vitro nsp12 polymerase activity was weak, contrasting with the replication of ∼30 kb-long coronavirus RNA genomes.
Several protein/protein interaction studies of the SARS CoV ORFeome were initiated and showed a validated direct interaction between nsp8 and nsp12 (Imbert et al., 2008, Von Brunn et al., 2007). We have recently found that the association of SARS-CoV nsp7 and nsp8 confers processivity of RNA synthesis to nsp12 (Subissi et al., unpublished data). The nsp7/nsp8 complex increases binding of nsp12 to RNA, resulting in a larger stretch of nucleotide synthesized per binding event. Nsp8 bears a second, non canonical RdRp activity for it does not have any characteristic signature sequence of “classic” RdRps (Imbert et al., 2006, Te Velthuis et al., 2012, Xiao et al., 2012). Nsp8 is able to polymerize small oligomers in a sequence-specific fashion and was therefore proposed to act as an RNA primase for nsp12. The nsp7/nsp8/nsp12 polymerase complex catalyzes long RNA synthesis de novo but the role of nsp8 in this complex is unclear (Subissi et al. unpublished data).
Inhibition studies supposedly directed at the nsp12 RdRp are emerging, as only a handful of reports have appeared in the literature. The inhibitory activity of some nucleoside analogues was tested against SARS-CoV in African green monkey kidney (Vero) cells. Most of the hits were cytotoxic, however the D-isomer of a thymine analogue with a substituted ribose exhibited good activity (EC50 = 20 μM) and did not show toxicity at dosages up to 100 μM (Chu et al., 2006). In addition to reporting a poor effect of Ribavirin (IC50 = 82 μM), authors have shown that 6-chloropurine nucleosides may exhibit interesting anti-SARS properties (Ikejiri et al., 2007). However, most interesting compounds have a poor selectivity index (<10), and the most active is protected in 5′, indicating that it might not be phosphorylated to the 5′-triphosphate, Thus, the target might not necessarily be nsp12. Others (Barnard et al., 2006, Pyrc et al., 2006) reported that beta-D-N4 hydroxycytidine was active against SARS-CoV and human coronavirus NL63, and these two groups agree on the lack of activity of ribavirin.
As of today, to the best of our knowledge, there are no studies showing inhibition of purified nsp12 RdRp in vitro. Thus, it remains to be demonstrated that nsp12 is the actual target of these poorly active nucleoside analogues.
2.2. The NTPase/helicase nsp13
CoV nsp13 (66.5 kDa) is a multi-functional protein with a zinc-binding domain in N-terminus while a helicase domain featuring the typical conserved motifs of superfamily 1 helicases is present in the C-terminal half (Gorbalenya and Koonin, 1989). Initially biochemical studies with SARS-CoV nsp13 expressed from bacteria have demonstrated that nsp13 can unwind both double-stranded (ds) DNA and RNA in a 5′–3′ direction, fueled by hydrolysis of all deoxyribonucleotide and ribonucleotide triphosphates (Ivanov and Ziebuhr, 2004, Ivanov et al., 2004b, Lee et al., 2010, Tanner et al., 2003), for review (Keum and Jeong, 2012). The nsp13 N-terminus contains 26 cysteine residues, 14 of which are highly conserved and predicted to form a binuclear Zn2+-binding cluster (Seybert et al., 2005). This cluster is conserved in all nidoviruses and is critical for helicase activity in vitro. Moreover, some of these cysteine residues may participate in disulfide bonds, and the latter seem to increase helicase activity (Seybert et al., 2005). Nsp13 helicase activity is also stimulated (2-fold) in the presence of nsp12 by a direct interaction between both proteins (Adedeji et al., 2012a). 5′-end single-stranded overhangs of at least five nucleotides are sufficient to promote unwinding of double-stranded nucleic acids with a estimated rate of ∼ 280 base-pairs per second, in the same range as other helicases (Adedeji et al., 2012a).
Among the nonstructural proteins carried by pp1ab, nsp13 is carrying essential functions for viral replication (Fang et al., 2007, Seybert et al., 2005) and it has been presented as a possible antiviral target. Natural flavonoids, myricetin and scultellarein, are able the inhibit the ATPase activity of purified SARS-CoV nsp13 in vitro (Yu et al., 2012). Bismuth complexes (Yang et al., 2007a, Yang et al., 2007b) dihydroxychromones and aryl diketoacids (Lee et al., 2009a, Lee et al., 2009b) have been reported to target nsp13. The compound 3-[(2-nitrophenyl)sulphanylmethyl]-4prop-2-enyl-1H-1,2,4-triazole-5-thione inhibits the nucleic acid unwinding activity of nsp13 with an IC50 around 5 μM, the compound showing also activity in SARS-CoV infected cells without significant toxicity (Adedeji et al., 2012b). The latter compound is the only one for which inhibition of the nsp13 helicase activity is observed both on purified enzyme and infected cells. Adamantane-related molecules are also targeting the ATPase activity of nsp13, leading to EC50 below 10 μM in infected cell based assays (Tanner et al., 2005). Interestingly, escape mutants were obtained, leading to the substitution of Serine 259 to Leucine, far from the catalytic pocket (Wang et al., 2011).
3. Nsp10, a molecular switch
Because of its central regulatory role, described below, it appears important to present this small protein (18 kDa) encoded by ORF1a. Nsp10 is well conserved among coronaviruses. In the polyprotein 1a, nsp10 is located upstream of nsp11, a short 13-residue peptide encoded by the 3′-terminal nucleotides of ORF1a. Nsp10 has long been known to have an essential role in viral replication. It was shown that a murine hepatitis virus (MHV) temperature-sensitive mutant carrying a non-synonymous mutation in the nsp10 coding sequence had a defect in minus-strand RNA synthesis at non-permissive temperatures (Sawicki et al., 2005, Siddell et al., 2001). Crystal structures of nsp10 revealed monomers and homodimers (Joseph et al., 2006) whereas a complex dodecameric structure was observed when nsp10 was expressed and crystallized as a fusion with nsp11 (Su et al., 2006). The nsp10 monomer consists of a pair of antiparallel N-terminal helices stacked against a β-sheet, a coil-rich C terminus, and two zinc fingers. Nsp10 represents a novel fold and is the first structural representative of this family of zinc-finger proteins. Zinc-finger motifs are strictly conserved in coronaviruses, supporting their essential role in viral replication. Gel shift assays indicate that nsp10 binds single- and double-stranded RNA and DNA without obvious sequence specificity (Joseph et al., 2006, Matthes et al., 2006). Interactome analysis showed that nsp10 also strongly interact with different nsps from ORF1b (see below) (Imbert et al., 2008, Von Brunn et al., 2007). Therefore, it is likely that nsp10 is mediating interactions with viral proteins and RNA (Matthes et al., 2006).
It is currently unclear whether or not the nsp10 dodecameric structure seen in one of the structural studies is of biological relevance. The relevance has been questioned for several reasons. First, site-directed mutagenesis of the nsp10–nsp11 cleavage site in the MHV genome generated nonviable viruses, indicating that 3CLpro-mediated cleavage at this site is essential for viral replication (Deming et al., 2007). Second, the monomer structure has an intact second zinc-finger which appears to stabilize the structure of the C-terminus tail of nsp10. By contrast, in the dodecamer structure, the second zinc-finger lacks the C-proximal cysteine residue, resulting in local disorder at the nsp10 carboxyl terminus. Finally, substitutions of residues predicted to be crucial for the dodecamer formation did not cause a lethal phenotype in MHV (Donaldson et al., 2007).
4. ORF1b nuclease activities
4.1. The 3′–5′ exoribonuclease nsp14 is involved in a replicative mismatch repair system
Nsp14 is a 60 kDa bi-functional enzyme. It carries at its N-terminus a 3′–5′ exonuclease (ExoN), that is unique to nidoviruses with a large genome (Minskaia et al., 2006). ExoN is related to the DEDD superfamily of exonucleases (Zuo and Deutscher, 2001) and has been shown to hydrolyze single-stranded and double-stranded RNAs to final products of 8–12 and 5–7 nt, respectively. How coronaviruses have been able to enlarge and maintain their exceptionally large RNA genomes is a fundamental question that has been recently addressed (Lauber et al., 2013). Viral RNA high mutational rates, partly due to the lack of mismatch repair systems (Holland et al., 1982), has restricted RNA genome size (Belshaw et al., 2008). The nsp14-ExoN domain is exclusively encoded by nidoviruses with genomes larger than 20 kb (Lauber et al., 2013). This observation has strongly supported that nsp14-ExoN enzymatic activity is improving fidelity of coronavirus replication through RNA proofreading. SARS-CoV as well as MHV-infected cells with mutated ExoN active site (ExoN-) were shown to still replicate but exhibit a mutator phenotype, with an overall 12- to 20-fold increase in mutation frequency and up to 14-fold increase in mutator rate compared to wt (Eckerle et al., 2007, Eckerle et al., 2010). Moreover SARS-CoV and MHV ExoN-viruses were susceptible to lethal mutagenesis and they were found to incorporate the mutagenic agent 5′-fluorouracil in their genomes, driving increased mutagenesis in ExoN-viruses (Smith et al., 2013).
Recently, an in vitro study showed that the weak nsp14 ExoN activity is strongly enhanced (>35-fold) by the addition of the nsp10 protein (Bouvet et al., 2012). The nsp10/nsp14 ExoN complex was shown to be dsRNA-specific, but also able to excise one 3′ mismatched nucleotide mimicking a polymerase-mediated misincorporation product. This finding further strengthens the idea that nsp14 is involved in RNA synthesis proofreading.
Coronavirus genome stability has been explored as a strategy for vaccine design (Graham et al., 2012). In mouse models, ExoN mutant viruses exhibit decreased virulence without reversion after more than 10 passages.
Thus, development of drugs blocking nsp14 exonuclease activity and acting as mutagenic agents may represent an antiviral strategy for viruses that have developed systems to maintain the integrity of their genome.
4.2. The still puzzling nsp15 endoribonuclease
All nidoviruses code for a uridine-specific endoribonuclease (called NendoU, nsp15 in coronaviruses) (Snijder et al., 2003). SARS-CoV nsp15 cleaves preferentially in 3′ of uridylates and generates 2′–3′ cyclic phosphate ends (Bhardwaj et al., 2004, Ivanov et al., 2004a). Mn2+ ions enhance NendoU activity. The crystal structure of SARS-CoV nsp15 shows a novel fold (Ricagno et al., 2006b). The structure of the monomeric protein (Fig. 2 A–C) consists of three extended domains: a small N-terminal domain (60 amino-acid residues), a central domain (128 amino-acid residues) sitting on a large C-terminal domain (153 amino-acid residues). The connectivity between domains is ensured by flexible linkers of 6–12 residues. The N-terminal domain is formed by 3 antiparallel β-strands packing against two perpendicular α-helices and its position is sideways in respect to the other two domains. The central domain is formed by a mix of α/β/α (11 β-strands and 3 α-helices) mainly organized as a central beta sheet of 7 strands reminiscent of a methyltransferase fold. Finally the C-terminal domain is formed by two sheets formed by 3 antiparallel β-strands and a segregated α helical sub-region (6 β-strands & 5 α-helices). The β-sheets serve as a space separator, one face of both sheets packs against a five α-helix subdomain, while the other side of the sheet is fully exposed. The endonuclease active site is carried by the C-terminal domain and involves the catalytic residues (His-His-Lys-Ser/Thr-Asp) forming a typical site signature. Despite low sequence similarity between nsp15 and other endonucleases the structure revealed that the catalytic residues (Lys/His) are superimposed with those of RNaseA, which also generates 2′–3′ cyclic phosphate ends (Ivanov et al., 2004a, Ricagno et al., 2006b). However, the proposed RNaseA-like catalytic mechanism does not involve metal ions and therefore, the Mn2+-stimulating effect on NendoU activity is not yet understood.
Fig. 2.
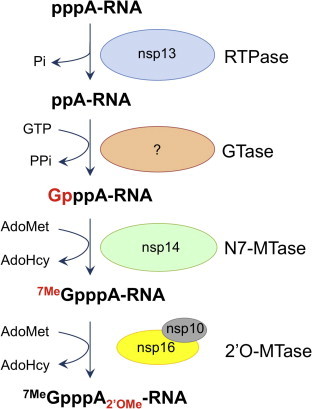
Coronavirus 5′ mRNA capping pathway. Coronaviruses use a conventional capping pathway made of four steps, as described in Decroly et al. (2011).
Crystals and solution studies showed that coronavirus NendoU forms an hexamer (Guarino et al., 2005, Ricagno et al., 2006a), composed of a dimer of trimers (Fig. 2, panel D) ensured mostly by the N-terminal domain, while on the other end, the C-terminal domains stabilise each other in a corolla like super structure (Fig. 2, panel E) concentrating the presence of six independent active sites. The biological relevance of the hexamer is however not yet established. It was shown that RNA binding and nuclease activity were affected in mutants altered in hexamer formation (Guarino et al., 2005). However, another study claimed that the nsp15 monomer had K m and K cat values similar to those of the nsp15 hexameric form, although this complex has 6 independent catalytic sites (Xu et al., 2006). Moreover, expression of nsp15 in fusion with the maltose binding protein (MBP) still exhibits endoribonuclease activity even though the MBP prevents hexamer formation (Ivanov et al., 2004a). Finally, the structure of SARS-CoV nsp15 with deleted N-terminal region was determined, revealing that without its adjacent monomer in the hexameric ring, nsp15 cannot be active (Joseph et al., 2007).
The role of NendoU in the viral cycle is still unknown. Reverse genetics experiments in MHV (Kang et al., 2007) and HCoV-229E (Ivanov et al., 2004a) indicate that NendoU activity is not essential for viral replication. Mutations of the nsp15 catalytic site His and Lys residues were shown in MHV to promote a subtle defect in subgenomic mRNA accumulation and about 10-fold reduction of viral titers, while mutation of a conserved Asp residue completely abolished viral RNA replication, both in HCoV-229E and MHV (Ivanov et al., 2004a, Kang et al., 2007). Supplementary studies are needed to better understand the role of this nidovirus-specific enzymatic activity.
5. The SARS-CoV RNA cap structure formation
Although the 5′ ends of SARS-CoV mRNAs have not been characterized yet, they are assumed to carry a cap structure (Fig. 3 ). This assumption is based on the characterization of genomic and subgenomic mRNAs of the coronavirus murine hepatitis virus (MHV) (Lai and Stohlman, 1981, Lai et al., 1982) and the related equine torovirus (van Vliet et al., 2002) belonging also to the Coronaviridae family.
Fig. 3.
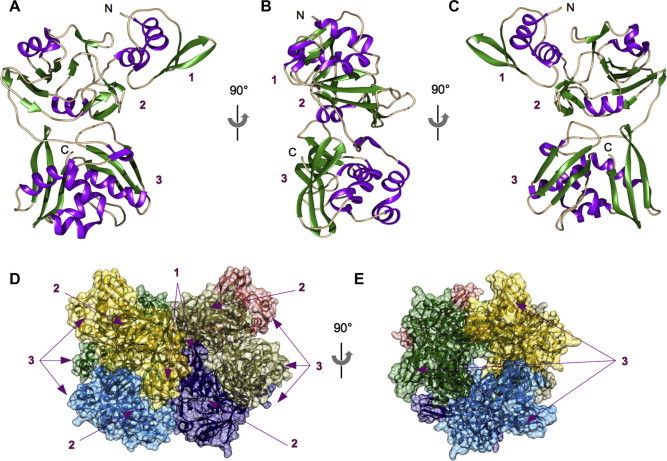
Structure of nsp15, the endoribonuclease from SARS-CoV. (A)–(C) Cartoon representations of nsp15 monomer. The structure consists of three domains: N-terminal domain (1), central domain (2), and C-terminal domain (3). Secondary structures are colored as follows: β-stands are colored in green, α-helix in purple, loops in light brown. (D) and (E) Views of the nsp15 hexamer. (D) Side view of surface representation of the nsp15 hexamer. Each monomer is colored with its own color. Solid arrows indicate the respective subdomains of the monomer. Domains 1 and 2 are involved in the assembly while 3 carries the active site. (E) Surface representation of the hexamer rotated by 90°, presenting a corolla like structure. PDB code: 2H85.
The cap structure promotes initiation of translation, protects mRNAs against cellular 5′–3′-exoribonuclease activities and limits the recognition of viral RNA by the innate immunity guardians (Züst et al., 2011). The synthesis of the cap structure in eukaryotes involves 3 sequential enzymatic activities: (i) an RNA triphosphatase (RTPase) which removes the 5′ γ-phosphate group of the mRNA; (ii) a guanylyltransferase (GTase) which catalyzes the transfer of GMP to the 5′-diphosphate terminus; and (iii) an N7-methyltransferase (MTase) which methylates the cap guanine at the N7-position, thus providing the so-called “cap-0”, 7MeGpppN (for review (Decroly et al., 2012)). Whereas lower eukaryotes, including yeast, employ a cap-0 structure, higher eukaryotes convert cap-0 into cap-1 or cap-2 structures by means of 2′-O-MTases, which methylate the ribose 2′O-position of the first and the second nucleotide of the mRNA, respectively. The methylation on the N7 position of the cap is essential since it prevents the pyrophosphorolytic reversal of the guanylyltransferase reaction, and ensures efficient binding to the eIF4G protein needed for the initiation of RNA translation.
In the 10 years since the SARS-CoV emerged, significant progress has been made on the elucidation of CoV mRNA capping mechanisms. Coronaviruses appear to employ the eukaryotic canonical pathway, described above, for the formation of their cap structure. To date, all known cap-synthesis enzymatic activities are found in the ORF1b and are summarized below.
5.1. The nsp13 RTPase
Nsp13 exhibits RNA 5′-triphosphatase activity in vitro and was proposed to catalyze the first step of the 5′-capping reaction of viral RNAs (Ivanov et al., 2004b), but an actual direct role of nsp13 in RNA capping still awaits experimental evidence.
5.2. The still unknown GTase
The GTase activity is still the last uncharacterized activity of the SARS-CoV capping machinery. It is tempting to speculate that the nsp12 N-terminal-domain could act as the guanylyltransferase. However, to date no functional data are available. Since most GTases were shown to make covalent links with GMP before transfer onto the diphosphate end of RNA, many groups tried to identify the guanylate intermediate. Accordingly, a report showed that no covalent binding of GTP with nsp12 alone or mixed with nsp7, nsp8, nsp10 and nsp13–nsp16 could be detected (Jin et al., 2013). Moreover, nsp12 could not trans-complement yeasts in which GTase activity was deleted (Chen et al., 2009).
5.3. The nsp14 N7-MTase
Yeast trans-complementation studies identified the C-terminal domain of SARS-CoV nsp14 as a S-adenosylmethionine (SAM)-dependent N7-guanine methyltransferase having no RNA sequence specificity (Chen et al., 2009). This N7-guanine MTase activity of purified nsp14 was confirmed in vitro on different capped RNAs (Bouvet et al., 2010, Chen et al., 2009) SARS-CoV nsp14 can also use GTP and dGTP as well as cap analogues GpppG, GpppA and, to a lesser extent, m7GpppG as substrates for methylation (Jin et al., 2013). Methylation of GTP by nsp14 triggers inhibition of cellular mRNA translation by m7GTP competing with mRNA caps for binding to eIF4E (Jin et al., 2013).
Amino acids 62–527 of nsp14 are essential for both N7-MTase and ExoN activities, indicating that the two functional domains are structurally linked. Through alanine scanning of SARS-CoV nsp14, residues critical for the N7-MTase activity were identified predominantly in two regions: (i) region ranging from aa 310 to 428, overlapping with the core domain of N7-MTase and (ii) region ranging from aa 73 to 86, located at the N-terminus of the ExoN domain. In contrast, the nsp14 ExoN active site (DEDDh) is not required for nsp14 N7-MTase activity. Furthermore, the sequence motif DxGxPxG/A (where x corresponds to a non-conserved aa) was identified as the key part of the SAM-binding site (Chen et al., 2013). Finally, addition of the nsp10 protein has no significant effect on the nsp14 N7-MTase activity whereas it increases the ExoN activity (Bouvet et al., 2012).
The unique structure and configuration of nsp14 with both ExoN and N7-MTase activities provides an attractive target for drug design.
5.4. The nsp16 2′-O-MTase
The cap formation process at the 5′-end terminates with the addition of a methyl group at the 2′O position of the first nucleotide of the genome, forming a type-1 cap (cap-1) structure (for review Decroly et al., 2012). Since 2003, bioinformatic studies have predicted an AdoMet-dependent mRNA cap (nucleoside-2′-O)-methyltransferase (2′-O-MTase) carried by nsp16 (von Grotthuss et al., 2003) and biochemical studies have next demonstrated that FCoV nsp16 harbors this 2′-O-MTase activity (Decroly et al., 2008). For SARS-CoV nsp16, however, this enzyme activity has been elusive for years. In the case of SARS-CoV nsp16, using yeast two-hybrid system, a strong interaction with nsp10 was detected (Imbert et al., 2008), which was later shown to trigger full nsp16 2′-O-MTase activity (Bouvet et al., 2010). This finding reveals the importance of nsp10, being involved in two distinct regulatory mechanisms (nsp16-2′-O-MTase and nsp14-ExoN). The nsp10-dependent activation of both enzymes may happen simultaneously since nsp10 is translated from ORF1a in excess compared to proteins translated from ORF1b. Nevertheless, an estimation of in vivo protein half-lives is missing to confirm the non-limiting nsp10 level to activate simultaneously nsp14 and nsp16 enzyme activities. Biochemical studies have also shown that the SARS-CoV cap methylation is sequential. The N7-methylation must precede the 2′-O-methylation that is only observed on RNA carrying cap 0 structure (Bouvet et al., 2010). The crystal structure of SARS-CoV nsp16 was solved in complex with nsp10 (Chen et al., 2011, Decroly et al., 2011). Nsp16 fold follows the typical SAM-dependent 2′-O-MTase fold, constituted by a central β-sheet of 7 strands surrounded by 5 α-helices (Fig. 4 , panel A and B), however the topology deviates from the standard SAM-dependent 2′-O-MTases in that nsp16 lacks 2 of 7 α-helices to make it a canonical fold. To compensate this lack of secondary structures nsp10 is needed to stabilize nsp16, whose structure has indeed been solved only in the presence of nsp10. The complex shows in particular how nsp10 rigidifies the SAM binding site and the putative RNA binding groove. Nsp10 contacts and probably holds proper alignment of K46 catalytic residue (first K of the KDKE tetrad) belonging to α-helix Z involved in the 2′-O-methyl reaction. The presence of a magnesium ion is required for methyltransfer, whereas such methylation reactions generally proceed without metal ion. The ion was indeed found to be necessary but for structural reasons. It clamps three distinct secondary structure elements and preserves the general fold of the nsp16 monomer (Fig. 4, panel C, D and E). Crystals soaked into a solution of the inhibitor Sinefungin showed that this SAM analogue targets the SAM binding site and should inhibit nsp16 competitively. Among the few known inhibitors, aurintricarboxylic acid (ATA) was shown to inhibit both SARS-CoV nsp14 and nsp16 MTase activities in vitro (Bouvet et al., 2010). ATA was also shown to be effective in inhibiting SARS-CoV replication (He et al., 2004).
Fig. 4.
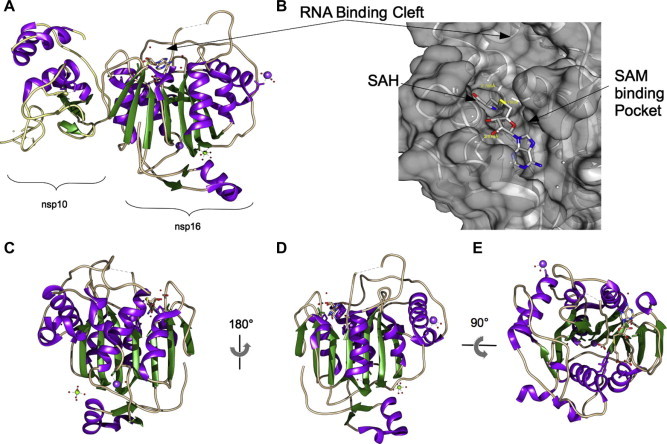
Structure of nsp10/nsp16. (A) Cartoon representation of the nsp10/nsp16 complex with the reaction product SAH and metal ions. Color code is as follow: β-stands are colored in green, α-helix in purple, loops in light brown, water (red) and ions are represented as small spheres (Na2+: pink, Mg2+: green, Zn2+: cream) and the SAH molecule shown in sticks colored following atom type. The nsp16 protein is bound to nsp10 through an interface which does not involve Zn ions present in nsp10. One metal ion is found in nsp16 on the opposite face from the active site to which a SAH molecule is found. (B) Closed caption of the SAM binding site represented in surface and the RNA binding cleft. The surface of the SAM-binding pocket is shown transparent to reveal details of the structural elements forming it. (C)–(E) Ribbon representation of nsp16 structure, in different orientations: (C) and (D) are side views, highlighting the Rossman fold; (E) is a top view of nsp16 showing the dispositions of the helices around the β-sheet. PDB codes: 2XYQ, 2XYV and 2XYR.
6. Conclusion
The picture emerging from recent literature is that nsp7/nsp8/nsp12/nsp13/nsp14 may constitute a minimal viral replisome (Fig. 5 ), partially reconstructed in vitro (Subissi et al., unpublished data). Such assembly is in charge of at least 2 of the 4 capping activities, i.e. RTPase and N7-MTase, as well as unwinding of RNA secondary structures, processive RNA polymerization and most likely mismatch excision. The second step of RNA capping mediated by the guanylyltransferase (GTase) activity is still elusive. It would make sense that it would also carried by the nsp7/nsp8/nsp12/nsp13/nsp14 complex but no data exist to support this hypothesis yet. The nsp10/nsp16 complex is responsible of the last capping step, i.e. methylation of cap-0 to cap-1 through its 2′-O-MTase activity, and it may act during nsp7/nsp8/nsp12/nsp13/nsp14-mediated replication but not necessarily in complex with other nsps.
Fig. 5.
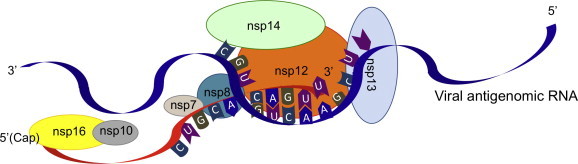
Model of how coronavirus enzymes associate in the replicative complex. A model of the current state of knowledge on coronavirus enzymes and their co-factors is depicted. At the head of the complex nsp13 is in charge of the first capping step and disrupts RNA secondary structures during replication. Nsp12 may be in charge of the GMP transfer to RNA, while the third step of capping is performed by nsp14, bound to nsp12, and also in charge of RNA proofreading during nsp7/nsp8/nsp12-mediated RNA synthesis. Nsp16, in complex with nsp10, performs 2′-O-methylation. Since nsp16 has not been shown to tightly interact with other ORF1b-encoded nsps, nsp10/nsp16 complex may act right after replication has started.
Proofreading could be performed by nsp14 ExoN alone or it could also involve stimulation by nsp10, even though one could speculate that nsp10/nsp14-ExoN complex may be involved in another yet-to-unravel mechanism of RNA processing, e.g., counteracting the innate immune response. Since dsRNA detection by host cell sensors is triggering the cell innate immune response, the nsp10/nsp14-ExoN dsRNA specificity is in agreement with this function. Furthermore, the only other RNA virus-encoded exoribonuclease activity is found in arenaviruses and this nuclease was demonstrated to interfere with innate immunity (Hastie et al., 2011, Martínez-Sobrido et al., 2009). It can be speculated that during replication, nsp14 in the presence of nsp12 corrects RNA nucleotide misincorporation, while in the presence of the nsp14-stimulating factor nsp10, it accomplishes another yet unknown function requiring a greater dsRNA degradation power.
The role of nsp15 in replication is still elusive. This endoribonuclease is also conserved in short genome nidoviruses, in contrast with nsp14-ExoN activity. Since 2003, significant knowledge has been accumulated in coronavirus biology. Nevertheless, many mechanisms used by these viruses remain to be unraveled. Coronaviruses are a constant threat for public health as shown by both the 2003 SARS-CoV pandemics and the current threat of MERS-CoV. Coronaviruses are fascinating due to the originality of their genetic organization, phylogeny, as well as the outstanding diversity of functional proteins encoded in their genome. Within the 16 non structural proteins some are uniquely found in coronaviruses, such as a second non canonical RNA-dependent RNA polymerase (nsp8), the endoribonuclease activity found in nsp15, whose role is still elusive, and the 3′–5′ exonuclease activity found in nsp14 implicated in a novel RNA proofreading mechanism. This ExoN domain is found in middle-sized genome (20 kb) nidoviruses (Mesoniviruses) while it is not conserved in arteriviruses, whose genome is of comparable size to that of other RNA viruses (13–15 kb). Therefore, mesoniviruses were the possible missing link in the evolution between short and long genome nidoviruses. The latter may have evolved up to 32 kb-long genomes with the acquisition of the ExoN proofreading activity critical for coronaviruses to find a viable and optimum mutational rate between error catastrophe and slow evolution. Conversely, short genome nidoviruses might have lost their ExoN proofreading activity and engaged into genome compaction to a level comparable to that of other RNA viruses. In any case, coronaviruses represent an interesting case of integration of multiple enzyme activities into a complex synthesizing and processing viral RNA.
Acknowledgements
This work was supported by the French National Research agency, under reference “ANR-08-MIEN-032 and ANR-12-BSV3”, and the Fondation pour la Recherche Médicale (Programme Équipe FRM), and subsequently by the European Union Seventh Framework Programme through the project SILVER (Small Inhibitor Leads Versus Emerging and Negleted RNA viruses, Grant Agreement No. 260644), the Infectiopole Sud, and the Direction Générale pour l’Armement (Contract No. 07co404). LS has a Marie Curie Fellowship through the EUVIRNA project.
References
- Adedeji A.O., Marchand B., Te Velthuis A.J.W., Snijder E.J., Weiss S., Eoff R.L., Singh K., Sarafianos S.G. Mechanism of nucleic acid unwinding by SARS-CoV helicase. PLoS ONE. 2012;7:e36521. doi: 10.1371/journal.pone.0036521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adedeji A.O., Singh K., Calcaterra N.E., DeDiego M.L., Enjuanes L., Weiss S., Sarafianos S.G. Severe acute respiratory syndrome coronavirus replication inhibitor that interferes with the nucleic acid unwinding of the viral helicase. Antimicrob. Agents Chemother. 2012;56:4718–4728. doi: 10.1128/AAC.00957-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn D.-G., Choi J.-K., Taylor D.R., Oh J.-W. Biochemical characterization of a recombinant SARS coronavirus nsp12 RNA-dependent RNA polymerase capable of copying viral RNA templates. Arch. Virol. 2012;157:2095–2104. doi: 10.1007/s00705-012-1404-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnard D.L., Day C.W., Bailey K., Heiner M., Montgomery R., Lauridsen L., Winslow S., Hoopes J., Li J.K.-K., Lee J., Carson D.A., Cottam H.B., Sidwell R.W. Enhancement of the infectivity of SARS-CoV in BALB/c mice by IMP dehydrogenase inhibitors, including ribavirin. Antiviral Res. 2006;71:53–63. doi: 10.1016/j.antiviral.2006.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belshaw R., Gardner A., Rambaut A., Pybus O.G. Pacing a small cage: mutation and RNA viruses. Trends Ecol. Evol. 2008;23:188–193. doi: 10.1016/j.tree.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj K., Guarino L., Kao C.C. The severe acute respiratory syndrome coronavirus Nsp15 protein is an endoribonuclease that prefers manganese as a cofactor. J. Virol. 2004;78:12218–12224. doi: 10.1128/JVI.78.22.12218-12224.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvet M., Debarnot C., Imbert I., Selisko B., Snijder E.J., Canard B., Decroly E. In vitro reconstitution of SARS-coronavirus mRNA cap methylation. PLoS Pathog. 2010;6:e1000863. doi: 10.1371/journal.ppat.1000863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvet M., Imbert I., Subissi L., Gluais L., Canard B., Decroly E. RNA 3′-end mismatch excision by the severe acute respiratory syndrome coronavirus nonstructural protein nsp10/nsp14 exoribonuclease complex. Proc. Natl. Acad. Sci. USA. 2012;109:9372–9377. doi: 10.1073/pnas.1201130109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Cai H., Pan J., Xiang N., Tien P., Ahola T., Guo D. Functional screen reveals SARS coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase. Proc. Natl. Acad. Sci. USA. 2009;106:3484–3489. doi: 10.1073/pnas.0808790106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Su C., Ke M., Jin X., Xu L., Zhang Z., Wu A., Sun Y., Yang Z., Tien P., Ahola T., Liang Y., Liu X., Guo D. Biochemical and structural insights into the mechanisms of SARS coronavirus RNA ribose 2′-O-methylation by nsp16/nsp10 protein complex. PLoS Pathog. 2011;7:e1002294. doi: 10.1371/journal.ppat.1002294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Tao J., Sun Y., Wu A., Su C., Gao G., Cai H., Qiu S., Wu Y., Ahola T., Guo D. Structure-function analysis of severe acute respiratory syndrome coronavirus RNA cap guanine-N7-methyltransferase. J. Virol. 2013;87:6296–6305. doi: 10.1128/JVI.00061-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng A., Zhang W., Xie Y., Jiang W., Arnold E., Sarafianos S.G., Ding J. Expression, purification, and characterization of SARS coronavirus RNA polymerase. Virology. 2005;335:165–176. doi: 10.1016/j.virol.2005.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng V.C.C., Chan J.F.W., To K.K.W., Yuen K.Y. Clinical management and infection control of SARS: lessons learned. Antiviral Res. 2013;100:407–419. doi: 10.1016/j.antiviral.2013.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C.K., Gadthula S., Chen X., Choo H., Olgen S., Barnard D.L., Sidwell R.W. Antiviral activity of nucleoside analogues against SARS-coronavirus (SARS-CoV) Antiviral Chem. Chemother. 2006;17:285–289. doi: 10.1177/095632020601700506. [DOI] [PubMed] [Google Scholar]
- De Groot R.J., Baker S.C., Baric R.S., Brown C.S., Drosten C., Enjuanes L., Fouchier R.A.M., Galiano M., Gorbalenya A.E., Memish Z.A., Perlman S., Poon L.L.M., Snijder E.J., Stephens G.M., Woo P.C.Y., Zaki A.M., Zambon M., Ziebuhr J. Middle east respiratory syndrome coronavirus (MERS-CoV): announcement of the coronavirus study group. J. Virol. 2013;87:7790–7792. doi: 10.1128/JVI.01244-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decroly E., Imbert I., Coutard B., Bouvet M., Selisko B., Alvarez K., Gorbalenya A.E., Snijder E.J., Canard B. Coronavirus nonstructural protein 16 is a cap-0 binding enzyme possessing (nucleoside-2′-O)-methyltransferase activity. J. Virol. 2008;82:8071–8084. doi: 10.1128/JVI.00407-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decroly E., Debarnot C., Ferron F., Bouvet M., Coutard B., Imbert I., Gluais L., Papageorgiou N., Sharff A., Bricogne G., Ortiz-Lombardia M., Lescar J., Canard B. Crystal structure and functional analysis of the SARS-coronavirus RNA cap 2′-O-methyltransferase nsp10/nsp16 complex. PLoS Pathog. 2011;7:e1002059. doi: 10.1371/journal.ppat.1002059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decroly E., Ferron F., Lescar J., Canard B. Conventional and unconventional mechanisms for capping viral mRNA. Nat. Rev. Microbiol. 2012;10:51–65. doi: 10.1038/nrmicro2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deming D.J., Graham R.L., Denison M.R., Baric R.S. Processing of open reading frame 1a replicase proteins nsp7 to nsp10 in murine hepatitis virus strain A59 replication. J. Virol. 2007;81:10280–10291. doi: 10.1128/JVI.00017-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson E.F., Sims A.C., Graham R.L., Denison M.R., Baric R.S. Murine hepatitis virus replicase protein nsp10 is a critical regulator of viral RNA synthesis. J. Virol. 2007;81:6356–6368. doi: 10.1128/JVI.02805-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler, J.F., Corman, V.M., Drosten, C., 2013. Antiviral Res. Oct 31;101C:45–56. doi: 10.1016/j.antiviral.2013.10.013. [Epub ahead of print]. PMID: 24184128. [DOI] [PMC free article] [PubMed]
- Eckerle L.D., Lu X., Sperry S.M., Choi L., Denison M.R. High fidelity of murine hepatitis virus replication is decreased in nsp14 exoribonuclease mutants. J. Virol. 2007;81:12135–12144. doi: 10.1128/JVI.01296-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckerle L.D., Becker M.M., Halpin R.A., Li K., Venter E., Lu X., Scherbakova S., Graham R.L., Baric R.S., Stockwell T.B., Spiro D.J., Denison M.R. Infidelity of SARS-CoV Nsp14-exonuclease mutant virus replication is revealed by complete genome sequencing. PLoS Pathog. 2010;6:e1000896. doi: 10.1371/journal.ppat.1000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang S., Chen B., Tay F.P.L., Ng B.S., Liu D.X. An arginine-to-proline mutation in a domain with undefined functions within the helicase protein (Nsp13) is lethal to the coronavirus infectious bronchitis virus in cultured cells. Virology. 2007;358:136–147. doi: 10.1016/j.virol.2006.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E., Koonin E.V. Viral proteins containing the purine NTP-binding sequence pattern. Nucleic Acids Res. 1989;17:8413–8440. doi: 10.1093/nar/17.21.8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E., Pringle F.M., Zeddam J.-L., Luke B.T., Cameron C.E., Kalmakoff J., Hanzlik T.N., Gordon K.H.J., Ward V.K. The palm subdomain-based active site is internally permuted in viral RNA-dependent RNA polymerases of an ancient lineage. J. Mol. Biol. 2002;324:47–62. doi: 10.1016/S0022-2836(02)01033-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham R.L., Becker M.M., Eckerle L.D., Bolles M., Denison M.R., Baric R.S. A live, impaired-fidelity coronavirus vaccine protects in an aged, immunocompromised mouse model of lethal disease. Nat. Med. 2012;18:1820–1826. doi: 10.1038/nm.2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarino L.A., Bhardwaj K., Dong W., Sun J., Holzenburg A., Kao C. Mutational analysis of the SARS virus Nsp15 endoribonuclease: identification of residues affecting hexamer formation. J. Mol. Biol. 2005;353:1106–1117. doi: 10.1016/j.jmb.2005.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie K.M., Kimberlin C.R., Zandonatti M.A., MacRae I.J., Saphire E.O. Structure of the Lassa virus nucleoprotein reveals a dsRNA-specific 3′–5′ exonuclease activity essential for immune suppression. Proc. Natl. Acad. Sci. USA. 2011;108:2396–2401. doi: 10.1073/pnas.1016404108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He R., Adonov A., Traykova-Adonova M., Cao J., Cutts T., Grudesky E., Deschambaul Y., Berry J., Drebot M., Li X. Potent and selective inhibition of SARS coronavirus replication by aurintricarboxylic acid. Biochem. Biophys. Res. Commun. 2004;320:1199–1203. doi: 10.1016/j.bbrc.2004.06.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgenfeld R., Peiris J.M. From SARS to MERS: 10 years of research on highly pathogenic human coronaviruses. Antiviral Res. 2013;100:286–295. doi: 10.1016/j.antiviral.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland J., Spindler K., Horodyski F., Grabau E., Nichol S., VandePol S. Rapid evolution of RNA genomes. Science. 1982;215:1577–1585. doi: 10.1126/science.7041255. [DOI] [PubMed] [Google Scholar]
- Ikejiri M., Saijo M., Morikawa S., Fukushi S., Mizutani T., Kurane I., Maruyama T. Synthesis and biological evaluation of nucleoside analogues having 6-chloropurine as anti-SARS-CoV agents. Bioorg. Med. Chem. Lett. 2007;17:2470–2473. doi: 10.1016/j.bmcl.2007.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbert I., Guillemot J.-C., Bourhis J.-M., Bussetta C., Coutard B., Egloff M.-P., Ferron F., Gorbalenya A.E., Canard B. A second, non-canonical RNA-dependent RNA polymerase in SARS coronavirus. EMBO J. 2006;25:4933–4942. doi: 10.1038/sj.emboj.7601368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbert I., Snijder E.J., Dimitrova M., Guillemot J.-C., Lécine P., Canard B. The SARS-coronavirus PLnc domain of nsp3 as a replication/transcription scaffolding protein. Virus Res. 2008;133:136–148. doi: 10.1016/j.virusres.2007.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov K.A., Ziebuhr J. Human coronavirus 229E nonstructural protein 13: characterization of duplex-unwinding, nucleoside triphosphatase, and RNA 5′-triphosphatase activities. J. Virol. 2004;78:7833–7838. doi: 10.1128/JVI.78.14.7833-7838.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov K.A., Hertzig T., Rozanov M., Bayer S., Thiel V., Gorbalenya A.E., Ziebuhr J. Major genetic marker of nidoviruses encodes a replicative endoribonuclease. Proc. Natl. Acad. Sci. USA. 2004;101:12694–12699. doi: 10.1073/pnas.0403127101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov K.A., Thiel V., Dobbe J.C., van der Meer Y., Snijder E.J., Ziebuhr J. Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J. Virol. 2004;78:5619–5632. doi: 10.1128/JVI.78.11.5619-5632.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins G.M., Rambaut A., Pybus O.G., Holmes E.C. Rates of molecular evolution in RNA viruses: a quantitative phylogenetic analysis. J. Mol. Evol. 2002;54:156–165. doi: 10.1007/s00239-001-0064-3. [DOI] [PubMed] [Google Scholar]
- Jin X., Chen Y., Sun Y., Zeng C., Wang Y., Tao J., Wu A., Yu X., Zhang Z., Tian J., Guo D. Characterization of the guanine-N7 methyltransferase activity of coronavirus nsp14 on nucleotide GTP. Virus Res. 2013;176:45–52. doi: 10.1016/j.virusres.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph J.S., Saikatendu K.S., Subramanian V., Neuman B.W., Brooun A., Griffith M., Moy K., Yadav M.K., Velasquez J., Buchmeier M.J., Stevens R.C., Kuhn P. Crystal structure of nonstructural protein 10 from the severe acute respiratory syndrome coronavirus reveals a novel fold with two zinc-binding motifs. J. Virol. 2006;80:7894–7901. doi: 10.1128/JVI.00467-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph J.S., Saikatendu K.S., Subramanian V., Neuman B.W., Buchmeier M.J., Stevens R.C., Kuhn P. Crystal structure of a monomeric form of severe acute respiratory syndrome coronavirus endonuclease nsp15 suggests a role for hexamerization as an allosteric switch. J. Virol. 2007;81:6700–6708. doi: 10.1128/JVI.02817-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H., Bhardwaj K., Li Y., Palaninathan S., Sacchettini J., Guarino L., Leibowitz J.L., Kao C.C. Biochemical and genetic analyses of murine hepatitis virus Nsp15 endoribonuclease. J. Virol. 2007;81:13587–13597. doi: 10.1128/JVI.00547-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keum Y.-S., Jeong Y.-J. Development of chemical inhibitors of the SARS coronavirus: viral helicase as a potential target. Biochem. Pharmacol. 2012;84:1351–1358. doi: 10.1016/j.bcp.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M.M., Stohlman S.A. Comparative analysis of RNA genomes of mouse hepatitis viruses. J. Virol. 1981;38:661–670. doi: 10.1128/jvi.38.2.661-670.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M.M., Patton C.D., Stohlman S.A. Replication of mouse hepatitis virus: negative-stranded RNA and replicative form RNA are of genome length. J. Virol. 1982;44:487–492. doi: 10.1128/jvi.44.2.487-492.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber C., Goeman J.J., Parquet M.D.C., Thi Nga P., Snijder E.J., Morita K., Gorbalenya A.E. The footprint of genome architecture in the largest genome expansion in RNA viruses. PLoS Pathog. 2013;9:e1003500. doi: 10.1371/journal.ppat.1003500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C., Lee J.M., Lee N.-R., Jin B.-S., Jang K.J., Kim D.-E., Jeong Y.-J., Chong Y. Aryl diketoacids (ADK) selectively inhibit duplex DNA-unwinding activity of SARS coronavirus NTPase/helicase. Bioorg. Med. Chem. Lett. 2009;19:1636–1638. doi: 10.1016/j.bmcl.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C., Lee J.M., Lee N.-R., Kim D.-E., Jeong Y.-J., Chong Y. Investigation of the pharmacophore space of severe acute respiratory syndrome coronavirus (SARS-CoV) NTPase/helicase by dihydroxychromone derivatives. Bioorg. Med. Chem. Lett. 2009;19:4538–4541. doi: 10.1016/j.bmcl.2009.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N.-R., Kwon H.-M., Park K., Oh S., Jeong Y.-J., Kim D.-E. Cooperative translocation enhances the unwinding of duplex DNA by SARS coronavirus helicase nsP13. Nucleic Acids Res. 2010;38:7626–7636. doi: 10.1093/nar/gkq647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Receptor recognition and cross-species infection of SARS coronavirus. Antiviral Res. 2013;100:246–254. doi: 10.1016/j.antiviral.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H., Wang H., Crameri G., Hu Z., Zhang H., Zhang J., McEachern J., Field H., Daszak P., Eaton B.T., Zhang S., Wang L.-F. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- Martínez-Sobrido L., Emonet S., Giannakas P., Cubitt B., García-Sastre A., de la Torre J.C. Identification of amino acid residues critical for the anti-interferon activity of the nucleoprotein of the prototypic arenavirus lymphocytic choriomeningitis virus. J. Virol. 2009;83:11330–11340. doi: 10.1128/JVI.00763-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthes N., Mesters J.R., Coutard B., Canard B., Snijder E.J., Moll R., Hilgenfeld R. The non-structural protein Nsp10 of mouse hepatitis virus binds zinc ions and nucleic acids. FEBS Lett. 2006;580:4143–4149. doi: 10.1016/j.febslet.2006.06.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minskaia E., Hertzig T., Gorbalenya A.E., Campanacci V., Cambillau C., Canard B., Ziebuhr J. Discovery of an RNA virus 3′–5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc. Natl. Acad. Sci. USA. 2006;103:5108–5113. doi: 10.1073/pnas.0508200103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nga P.T., del Parquet M., Lauber C., Parida M., Nabeshima T., Yu F., Thuy N.T., Inoue S., Ito T., Okamoto K., Ichinose A., Snijder E.J., Morita K., Gorbalenya A.E. Discovery of the first insect nidovirus, a missing evolutionary link in the emergence of the largest RNA virus genomes. PLoS Pathog. 2011;7:e1002215. doi: 10.1371/journal.ppat.1002215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyrc K., Bosch B.J., Berkhout B., Jebbink M.F., Dijkman R., Rottier P., van der Hoek L. Inhibition of human coronavirus NL63 infection at early stages of the replication cycle. Antimicrob. Agents Chemother. 2006;50:2000–2008. doi: 10.1128/AAC.01598-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricagno S., Coutard B., Grisel S., Brémond N., Dalle K., Tocque F., Campanacci V., Lichière J., Lantez V., Debarnot C., Cambillau C., Canard B., Egloff M.P. Crystallization and preliminary X-ray diffraction analysis of Nsp15 from SARS coronavirus. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2006;62:409–411. doi: 10.1107/S1744309106009407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricagno S., Egloff M.-P., Ulferts R., Coutard B., Nurizzo D., Campanacci V., Cambillau C., Ziebuhr J., Canard B. Crystal structure and mechanistic determinants of SARS coronavirus nonstructural protein 15 define an endoribonuclease family. Proc. Natl. Acad. Sci. USA. 2006;103:11892–11897. doi: 10.1073/pnas.0601708103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki S.G., Sawicki D.L., Younker D., Meyer Y., Thiel V., Stokes H., Siddell S.G. Functional and genetic analysis of coronavirus replicase-transcriptase proteins. PLoS Pathog. 2005;1:e39. doi: 10.1371/journal.ppat.0010039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seybert A., Posthuma C.C., van Dinten L.C., Snijder E.J., Gorbalenya A.E., Ziebuhr J. A complex zinc finger controls the enzymatic activities of nidovirus helicases. J. Virol. 2005;79:696–704. doi: 10.1128/JVI.79.2.696-704.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddell S., Sawicki D., Meyer Y., Thiel V., Sawicki S. Identification of the mutations responsible for the phenotype of three MHV RNA-negative ts mutants. Adv. Exp. Med. Biol. 2001;494:453–458. doi: 10.1007/978-1-4615-1325-4_66. [DOI] [PubMed] [Google Scholar]
- Smith E.C., Blanc H., Vignuzzi M., Denison M.R. Coronaviruses lacking exoribonuclease activity are susceptible to lethal mutagenesis: evidence for proofreading and potential therapeutics. PLoS Pathog. 2013;9:e1003565. doi: 10.1371/journal.ppat.1003565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Bredenbeek P.J., Dobbe J.C., Thiel V., Ziebuhr J., Poon L.L.M., Guan Y., Rozanov M., Spaan W.J.M., Gorbalenya A.E. Unique and conserved features of genome and proteome of SARS-Coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 2003;331:991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su D., Lou Z., Sun F., Zhai Y., Yang H., Zhang R., Joachimiak A., Zhang X.C., Bartlam M., Rao Z. Dodecamer structure of severe acute respiratory syndrome coronavirus nonstructural protein nsp10. J. Virol. 2006;80:7902–7908. doi: 10.1128/JVI.00483-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner J.A., Watt R.M., Chai Y.-B., Lu L.-Y., Lin M.C., Peiris J.S.M., Poon L.L.M., Kung H.-F., Huang J.-D. The severe acute respiratory syndrome (SARS) coronavirus NTPase/helicase belongs to a distinct class of 5′–3′ viral helicases. J. Biol. Chem. 2003;278:39578–39582. doi: 10.1074/jbc.C300328200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner J.A., Zheng B.-J., Zhou J., Watt R.M., Jiang J.-Q., Wong K.-L., Lin Y.-P., Lu L.-Y., He M.-L., Kung H.-F., Kesel A.J., Huang J.-D. The adamantane-derived bananins are potent inhibitors of the helicase activities and replication of SARS coronavirus. Chem. Biol. 2005;12:303–311. doi: 10.1016/j.chembiol.2005.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Te Velthuis A.J.W., Arnold J.J., Cameron C.E., van den Worm S.H.E., Snijder E.J. The RNA polymerase activity of SARS-coronavirus nsp12 is primer dependent. Nucleic Acids Res. 2010;38:203–214. doi: 10.1093/nar/gkp904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Te Velthuis A.J.W., van den Worm S.H.E., Snijder E.J. The SARS-coronavirus nsp7 + nsp8 complex is a unique multimeric RNA polymerase capable of both de novo initiation and primer extension. Nucleic Acids Res. 2012;40:1737–1747. doi: 10.1093/nar/gkr893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Vliet A.L.W., Smits S.L., Rottier P.J.M., de Groot R.J. Discontinuous and non-discontinuous subgenomic RNA transcription in a nidovirus. EMBO J. 2002;21:6571–6580. doi: 10.1093/emboj/cdf635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Brunn A., Teepe C., Simpson J.C., Pepperkok R., Friedel C.C., Zimmer R., Roberts R., Baric R., Haas J. Analysis of intraviral protein–protein interactions of the SARS coronavirus ORFeome. PLoS ONE. 2007;2:e459. doi: 10.1371/journal.pone.0000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Grotthuss M., Wyrwicz L.S., Rychlewski L. MRNA cap-1 methyltransferase in the SARS genome. Cell. 2003;113:701–702. doi: 10.1016/S0092-8674(03)00424-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.F., Eaton B.T. Bats, civets and the emergence of SARS. Curr. Top. Microbiol. Immunol. 2007;315:325–344. doi: 10.1007/978-3-540-70962-6_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Huang J.-D., Wong K.-L., Wang P.-G., Zhang H.-J., Tanner J.A., Spiga O., Bernini A., Zheng B.-J., Niccolai N. On the mechanisms of bananin activity against severe acute respiratory syndrome coronavirus. FEBS J. 2011;278:383–389. doi: 10.1111/j.1742-4658.2010.07961.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y., Ma Q., Restle T., Shang W., Svergun D.I., Ponnusamy R., Sczakiel G., Hilgenfeld R. Nonstructural proteins 7 and 8 of feline coronavirus form a 2:1 heterotrimer that exhibits primer-independent RNA polymerase activity. J. Virol. 2012;86:4444–4454. doi: 10.1128/JVI.06635-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Zhai Y., Sun F., Lou Z., Su D., Xu Y., Zhang R., Joachimiak A., Zhang X.C., Bartlam M., Rao Z. New antiviral target revealed by the hexameric structure of mouse hepatitis virus nonstructural protein nsp15. J. Virol. 2006;80:7909–7917. doi: 10.1128/JVI.00525-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N., Tanner J.A., Wang Z., Huang J.-D., Zheng B.-J., Zhu N., Sun H. Inhibition of SARS coronavirus helicase by bismuth complexes. Chem. Commun. Cambridge Engl. 2007:4413–4415. doi: 10.1039/b709515e. [DOI] [PubMed] [Google Scholar]
- Yang N., Tanner J.A., Zheng B.-J., Watt R.M., He M.-L., Lu L.-Y., Jiang J.-Q., Shum K.-T., Lin Y.-P., Wong K.-L., Lin M.C.M., Kung H.-F., Sun H., Huang J.-D. Bismuth complexes inhibit the SARS coronavirus. Angew. Chem. Int. Ed. Engl. 2007;46:6464–6468. doi: 10.1002/anie.200701021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M.-S., Lee J., Lee J.M., Kim Y., Chin Y.-W., Jee J.-G., Keum Y.-S., Jeong Y.-J. Identification of myricetin and scutellarein as novel chemical inhibitors of the SARS coronavirus helicase, nsP13. Bioorg. Med. Chem. Lett. 2012;22:4049–4054. doi: 10.1016/j.bmcl.2012.04.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D.M.E., Fouchier R.A.M. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- Zuo Y., Deutscher M.P. Exoribonuclease superfamilies: structural analysis and phylogenetic distribution. Nucleic Acids Res. 2001;29:1017–1026. doi: 10.1093/nar/29.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Züst R., Cervantes-Barragan L., Habjan M., Maier R., Neuman B.W., Ziebuhr J., Szretter K.J., Baker S.C., Barchet W., Diamond M.S., Siddell S.G., Ludewig B., Thiel V. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat. Immunol. 2011;12:137–143. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]


