Graphical abstract

Keywords: Coxsackievirus B3, RT-LAMP, Detection, Optimization
Highlights
-
•
Detection for Coxsackievirus B3 using one-step reverse transcription loop-mediated isothermal amplification (RT-LAMP).
-
•
2- Comparative analysis of CVB3 RT-LAMP assay with RT-PCR.
-
•
3- High sensitivity and selectivity of CVB3 RT-LAMP detection.
Abstract
Coxsackievirus B3 (CVB3) is a member of the genus Enterovirus within the family Picornaviridae and is an important pathogen of viral myocarditis, which accounts for more than 50% viral myocarditis cases. VP1 is major capsid protein that this region has a low homology in both amino acid and nucleotide sequences among Enteroviruses. Therefore we have chosen this region for designed a set of RT-LAMP primers for CVB3 detection. For this the total RNA was extracted from 24-h post infected-HeLa cells with complete cytopathic effect (CPE), and applied to a one-step reverse transcription loop-mediated isothermal amplification reaction (RT-LAMP) using CVB3-specific primers. The optimization of RT-LAMP reaction was carried out with three variables factors including MgSO4 concentration, temperature and time of incubation. Amplification was analyzed by using 2% agarose gel electrophoresis and ethidium bromide and SYBR Green staining. Our results were shown the ladder-like pattern of the VP1 gene amplification. The LAMP reaction mix was optimized and the best result observed at 4 mM MgSO4 and 60 °C for 90 min incubation. RT-LAMP had high sensitivity and specificity for detection of CVB3 infection. This method can be used as a rapid and easy diagnostic test for detection of CVB3 in clinical laboratories.
1. Introduction
The Coxsackieviruses group B consist of six serotypes are members of the genus Enterovirus within the Picornaviridae viral family (Hyypi et al., 1997, Reetoo et al., 2000, Oberste et al., 2004, Knipe and Howley, 2007) that are associated with a wide spectrum of human illness ranging in severity from subclinical infection to rapidly fatal disease, with symptoms as diverse as acute myocarditis, bornholm disease (epidemic pleurodynia), aseptic meningitis, encephalitis, and febrile illness (Reetoo et al., 2000). Coxsackievirus B3 (CVB3) has been recognized as the predominant cause of viral myocarditis in humans, which accounts for more than fifty percent viral myocarditis cases (Shen et al., 2009). The mechanisms of viral myocarditis include direct myocyte injury and subsequent immune-mediated damage of the heart (Yuan et al., 2010). The CVB3 genome is 7400 nucleotides in length and consist a positive single stranded RNA flanked by non-translated regions that encodes 11 proteins within a single open reading frame (Michable et al., 1987, Tracy et al., 1995, Tracy et al., 1997, Dunn et al., 2000). The positive genomic RNA of Enterovirus can be employed directly as an mRNA template for translation of a single polyprotein that processed into 11 mature proteins. Also genomic RNA is employed as a template for viral RNA transcription and synthesizes more copies of genomic RNA by a negative-strand intermediate RNA (Bedard and Semler, 2004, Yuan et al., 2005, Van Ooij et al., 2006, Chang et al., 2010). The viral genome is packaged in a naked capsid approximately 29 nm in diameter (Drescher et al., 2004). The capsid of CVB3, like that of other Enteroviruses, is composed 60 copies each of four viral polypeptides known as VP1 to VP4 (Knowlton et al., 1996, Wu et al., 2013, Jiang et al., 2014). Between all capsid proteins, VP1 is the most external capsid proteins and is the main part in structural of the canyon on the surfaces of Picornavirus, and potentially implicated for pathogenicity of these viruses (De Jesus, 2007, Wu et al., 2013). Moreover, genetic diversity at VP1 is closely associated with the difference in viral serotypes (Wu et al., 2013) and often used as a target for molecular detection of Picornavirus viral family (Yaqing et al., 2012, Nie et al., 2012, Wang et al., 2014, Chen et al., 2014, Ding et al., 2014).
On the other hand, reverse transcription − polymerase chain reaction (RT-PCR) has developed as a common diagnostic method for identification of the infectious agents (Weiss et al., 1991, Dierssen et al., 2008). One advantage of RT-PCR is the usage of this method in comparative studies and diagnosis that could be increase the yield of Enteroviruses detection in tissue culture (Dierssen et al., 2008). But RT-PCR based nucleic acid detection can be time-consuming and requires highly pure RNA extracted which is difficult and impurity in sample can cause effects of inhibitors on PCR sensitivity (Opel et al., 2010, Pan et al., 2011, Ghosh et al., 2015). Loop-mediated isothermal amplification (LAMP) is an alternative isothermal nucleic acid amplification method that has developed by Notomi et al. (2000). Also, reverse transcription LAMP (RT-LAMP) assay which is carried out in a single tube is a simple, high specific, rapid, and cost-effectiveness method (Zhang et al., 2012). Less time-consuming of RT-LAMP than conventional PCR-based methods has been used successfully for rapid detection of pathogenic RNA viruses such as Enteroviruses (Arita et al., 2009, Jaianand et al., 2010, Jaianand et al., 2011, Shi et al., 2011, Nie et al., 2012, Yaqing et al., 2012, Zhang et al., 2012, Zhao et al., 2013, Chen et al., 2014, Ding et al., 2014, Jiang et al., 2014, Zhao et al., 2014, Wang et al., 2014) and less prone to inhibit from impurity of template sample (Caipang et al., 2004, Kaneko et al., 2007, Tomita et al., 2008, Mori and Notomi, 2009, Ghosh et al., 2015). The RT-LAMP assay is very specific in compared to other molecular detection methods, because in this method four primers are used to recognize six specific regions of the target gene for amplification (Zhang et al., 2012).
In this study, we have developed a one-step RT-LAMP assay for CVB3 detection by designing the specific primers based on the VP1 gene.
2. Materials and methods
2.1. Primer design
RT-LAMP specific primers were designed according to the CVB3 genomic sequence (VP1 gene) in GenBank (Accession No: JX312064) using software PrimerExplorerV4 (Table 1 ). For this, we used multiple sequence alignment by MEGA6 (Molecular Evolutionary Genetics Analysis Version 6.0) software and find the consensus sequences of VP1 from CVB3 for primer design (data was not shown).Each set of primers contained two external primers (F3 and B3), and two internal primers (FIP and BIP). The BIP and FIP included a TTTT spacer between the B1/B2c sequences, and F1c/F2 sequences (Fig. 1 ).
Table 1.
RT-LAMP specific primers.
| Primer | Sequence |
|---|---|
| F3 | GGAGAGTTGCGGATACCGT |
| B3 | ATACCGCTTGGCACCTGA |
| FIP | GTGACGTGTGACCCGTCTCAGTTTTAGGGCCAACCAACTCAGA |
| BIP | TGCAGACACGCCACGTTAAGAATTTTTACACGCATGCTGACCTACA |
Fig. 1.
Locations of primer-binding sequences (CVB3 strain Nancy, GenBank accession no. JX312064).
2.2. Viral culture and RNA extraction
RPMI 1640 medium supplemented with 10% FBS was used for growth and maintenance of HeLa cell cultures. Cells at 90% confluency were infected with CVB3 (Nancy strain) in a medium containing 1% FCS. After 24 h and observed complete cytopathic effect (CPE), the infected-HeLa cells were harvested at 5000 rpm for 5 min. The cell sediment was used for total RNA extraction. For this, after two cycles of freezing and thawing, total RNA was extracted using RNX reagent according to the manufacturers’ instruction. The integrity of the extracted RNA was assessed by resolution on a 1% agarose gel and ethidium bromide staining. RNA samples were stored at −80 °C until amplification using RT- LAMP and RT-PCR detection.
2.3. RT-PCR assay
RT-PCR assay was carried out in a total reaction volume10 μl containing 1 μl of 10X PCR buffer, 1 μM each of primers B3 and F3, 2 mM of MgSO4, 1.5 mM of dNTP, 1 mM of DTT, 5 U of RNase inhibitor, 5 U of cloned AMV reverse transcriptase (Invitrogen), 2.5 U of Taq DNA polymerase, 10 μg of template RNA and RNase free deionized water. The reactions were incubated at 60 °C for 30 min, followed by initial denaturation at 94 °C for 4 min, 35 cycles of 94 °C for 45 s, 60 °C for 45 s and 72 °C for 45 s and a final extension at 72 °C for 5 min. RT-PCR procedure was carried out using an automated thermal cycler (Techgene, Germany) and all PCR products were analyzed by 1% agarose gel electrophoresis.
2.4. Optimized RT-LAMP assay
The RT-LAMP temperature was optimized by incubating the RT-LAMP mixture at 56, 58, 60, 62 and 64 °C for 60 min. The Mg2+ ion concentration was optimized by adding MgSO4 at a final concentration of 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, or 12 mM to RT-LAMP mixture and incubating at 60 °C for 90 min. Also, the time of LAMP reaction was optimized by incubating the RT-LAMP mixture at 30, 60, 90 and 120 min.
2.5. The one step RT- LAMP assay
The CVB3 RT-LAMP assay was carried out in a total 12.5 μl-volume reaction containing 0.8 μM of the inner primers FIP and BIP, 0.1 μM of the outer primers F3 and B3, 1.4 mM dNTPs, 1.25 μl of 10X isothermal amplification buffer, 4 mM of MgSO4, 0.8 M of betaine, 4 U of Bst 2.0 DNA polymerase (New England Biolabs), 5 U of cloned AMV reverse transcriptase (Invitrogen), 1.25 μl of DTT (0.1 M), 0.4 μl of RNasin and 10 μg of target RNA. The mixture was incubated at 60 °C for 90 min and then heated at 80 °C for 10 min to stop of the reaction.
2.6. RT-PCR and RT-LAMP assays for artificially infected animals
Eight Balb/c mice, aged 8 weeks, were divided into 2 same groups. The control group was inoculated with medium, while positive group was inoculated with 105 plaque forming units (PFU) of the CVB3 via the intraperitoneal route (IP). After a week, a booster dose was used to inoculate again and 2 weeks later, samples were collected from heart, spleen, feces and blood for RNA extraction and then stored at −80 °C for RT-PCR and RT-LAMP analyses.
2.7. Specificity and sensitivity of RT-PCR and RT-LAMP assays
The specificity of RT-PCR and RT-LAMP assay was determined by CVB3 and some serotypes of genus Enteroviruses include of Coxsackievirus A16, Echovirus and Rhinovirus. Also in order to evaluate the sensitivity of RT-PCR and RT-LAMP assay for detection of CVB3, 7 concentrations of RNA from 10 μg/μl to10 autogram/μl were prepared and used as a template for RT-PCR and RT-LAMP assay. On the other hand, 11-fold serial dilution of CVB3 that has a defined TCID50 titer (per ml), were used for RNA extraction, then, extracted RNA used as template for RT-LAMP assay. Finally, the amplified DNA was analyzed using 2% agarose gel electrophoresis.
2.8. Analysis of RT-LAMP products
To visualize the RT-LAMP products two methods were used: (1) Amplicon was analyzed by running 3 μl of the amplified product on a 2% agarose gel and ethidium bromide staining for 60 min at 60 V in 1X TAE (Tris Acetic acid EDTA) buffer and visualized under a gel documentation system. (2) 4 μl of SYBR Green was added to 10 μl of amplified product and the tubes were then visualized under a UV irradiation source.
3. Results
3.1. Detection of CVB3 by RT-PCR assay
RT-PCR detection was performed using F3/B3 and FIP/BIP primers. Amplification of 234 bp DNA (lane 1) and smear pattern (lane 2) fragments confirmed the CVB3 genome in total RNA extracted from the infected HeLa cells (Fig. 2 ).
Fig. 2.
Analysis of RT-PCR product using 1% agarose gel electrophoresis. Lane 1: RT-PCR product using primers B3/F3, lane 2: RT-PCR product using primers BIP/FIP, lane M: 100 bp DNA leader.
3.2. Detection of CVB3 by RT-LAMP assay
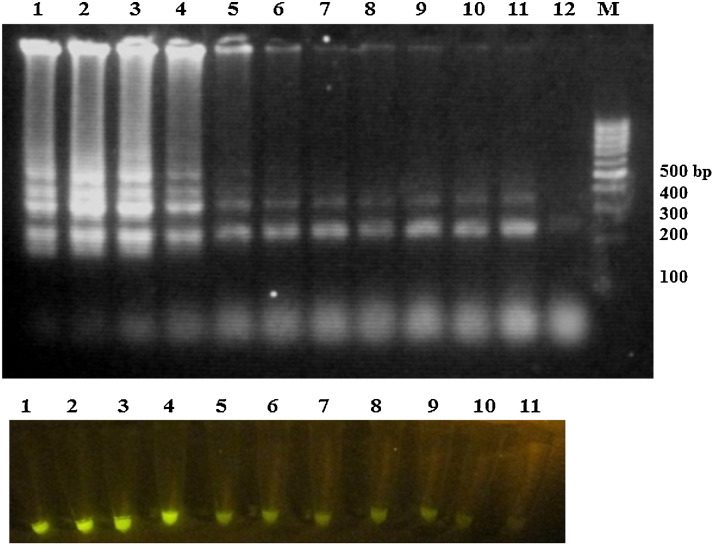
RT-LAMP detection was optimized by three factors: MgSO4 concentrations, temperature and the incubation time of RT-LAMP reaction. Reaction optimization was performed using MgSO4 different concentrations ranging from 2 to 12 mM and the best amplification was achieved using 4 mM MgSO4 (Fig. 3 ).
Fig. 3.
Effect of MgSO4 concentrations on the RT-LAMP reaction: Lane 1: 2 mM, lane 2: 3 mM, lane 3: 4 mM, lane 4: 5 mM, lane 5: 6 mM, lane 6: 7 mM, lane 7: 8 mM, lane 8: 9 mM, lane 9: 10 mM, lane 10: 11 mM and lane 11: 12 mM MgSO4, lane 12: negative control and lane M: 100 bp DNA leader.
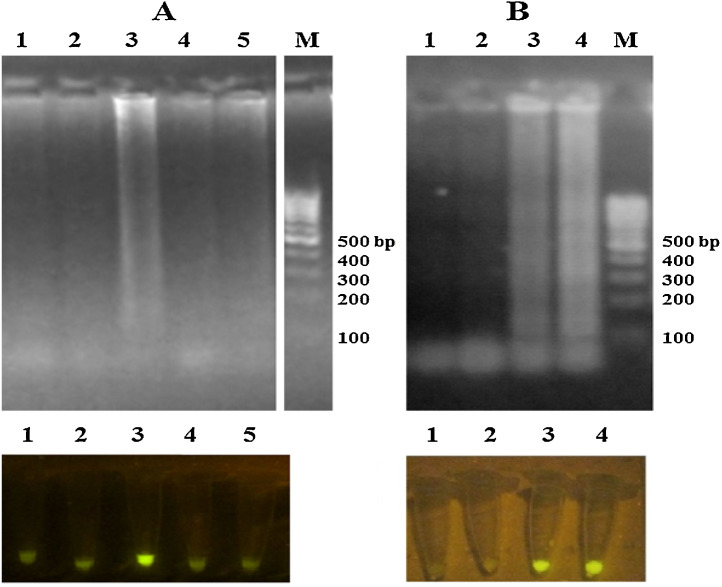
On the other hand, the effect of temperature on the RT-LAMP reaction was determined in 56, 58, 60, 62 and 64 °C. The data shown that 60 °C was better than other temperatures for amplification (Fig. 4 -A). Also, in order to determination of the optimum incubation time for RT-LAMP reaction, RT-LAMP amplification was performed in 30, 60, 90 and120 min. The results showed that RT-LAMP amplification is completed after 120 min (Fig. 4-B).
Fig. 4.
A) Effect of temperature on the RT-LAMP reaction: Lane 1: 56 °C, lane 2: 58 °C, lane 3: 60 °C, lane 4: 62 °C, lane 5: 64 °C and lane M: 100 bp DNA marker. B) Effect of incubation time on the RT-LAMP amplification: Lane 1: 30 min, lane 2: 60 min, lane 3: 90 min, lane 4: 120 min and lane M: 100 bp DNA leader.
3.3. Specificity of RT-PCR and RT-LAMP assays
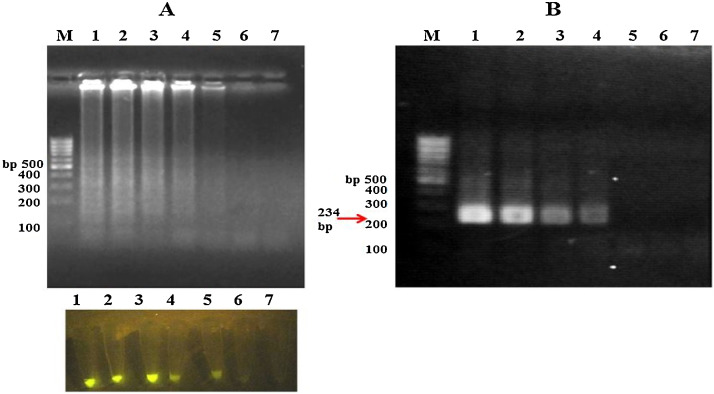
According to use of four specific LAMP primers, the selectivity of RT-PCR and RT-LAMP assays shown that these techniques were highly specific for CVB3 detection. In conclusion RT-PCR and RT-LAMP results were negative for Coxsackievirus A16, Echovirus and Rhinovirus (Fig. 5 ).
Fig. 5.
The specificity of RT-LAMP (A) and RT-PCR (B) was determined using some serotypes of genus Enteroviruses. Lane 1: Coxsackievirus A16, lane 2: Echovirus, lane 3: Rhinovirus, Lane 4: Coxsackievirus B3 (positive control), lane 5: without template (negative control) and lane M: 100 bp DNA leader.
3.4. Sensitivity of RT-PCR and RT-LAMP assays
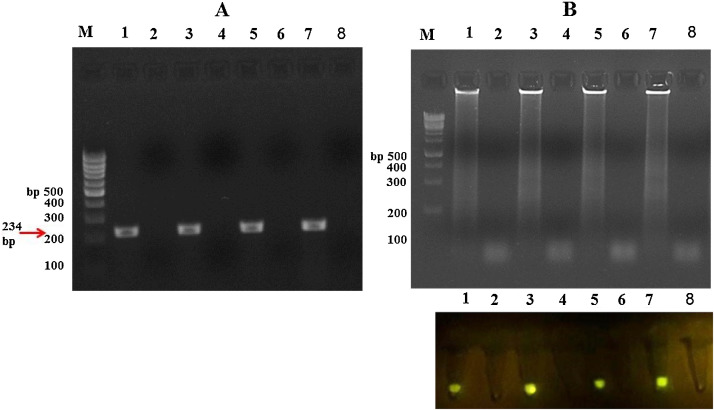
In order to evaluate the sensitivity of RT-PCR and RT-LAMP detections of CVB3, we used 7 different concentrations of total RNA from 10 μg to 10 ag. The data shown that the sensitivity of RT-PCR and RT- LAMP were 10 pg and 100 fg, respectively. Therefore, the sensitivity of RT-LAMP assay was 100 fold higher than RT-PCR assay (Fig. 6 ). Also, sensitivity of RT-LAMP determined about 10 −2 TCID50/ml (Fig. 7 ).
Fig. 6.
Sensitivity of RT-LAMP (A) and RT-PCR (B) assays for CVB3 detection. 7 different concentrations of total RNA were prepared and used as template. Lane 1: 10 μg, lane 2: 100 ng, lane 3: 1 ng, lane 4: 10 pg, lane 5: 100 fg, lane 6: 1 fg and lane 7: 10 ag of total RNA, and lane M: 100 bp DNA leader.
Fig. 7.
Sensitivity of RT-LAMP according to TCID50. 11 different TCID50/ml from CVB3 were used for RNA extraction and RT-LAMP assay. Lane 1: 106, lane 2: 105, lane 3:10 4, lane 4: 103, lane 5: 102, lane 6: 101, lane 7: 1, lane 8: 10−1, lane 9: 10−2, lane 10: 10−3 and lane 11: 10−4 TCID50/ml, lane M: 100 bp DNA leader.
3.5. RT-LAMP assay for artificially infected animals
The RT-PCR and RT-LAMP detections of CVB3 were evaluated using Balb/C mice inoculated with 105 PFU of CVB3. Samples from heart, spleen, feces and blood were collected two- weeks post-infection. According to Fig. 8 , amplification in infected samples confirmed presence of CVB3 RNA.
Fig. 8.
RT-PCR (A) and RT-LAMP (B) detections for heart of infected mice (lane 1), un-infected mice (lane 2), spleen of infected mice (lane 3), un-infected mice (lane 4), feces of infected mice (lane 5), un-infected mice (lane 6), and blood of infected mice (lane 7), un-infected mice (lane 8).
4. Discussion
In this study, The CVB3 one-step RT-LAMP assay was developed and optimized at 60 °C for 90 min to amplify and detection of VP1 gene. We demonstrated in the normal and infected animal samples, RT-LAMP could be used for infected tissue detection. The identification of enteroviral-specific RNA sequences in myocardial patients is very important. CVB3 as a membrane from Enteroviruse is a major etiological agent in human myocarditis and idiopathic dilated cardiomyopathy (Shen et al., 2009). Several serological methods were used for Enteroviruses detection (Gao et al., 2016), but because of the disadvantages of these methods, the use of molecular techniques is more important. The LAMP method is an isothermal amplification technique for molecular detection of target DNA that was developed by Notomi et al. (2000). Advantages of LAMP method are amplification takes place at one temperature so it does not require thermal cycler, and also in this assay four primers used that recognizes 6 specific regions on the target DNA, therefore amplification is more specific. The RT-LAMP assay has been developed for simple and fast detection of various RNA viruses such as pandemic influenza A H1N1 Virus (Ma et al., 2010), coronavirus (Shirato et al., 2014), Ebola Virus (Kurosaki et al., 2016) and some of other viruses. In recent years, several publications have been reported using the LAMP method for detection of bacterial pathogens or fungal contaminants (Niessen et al., 2013).
Different diagnostic methods such as conventional RT-PCR, real-time RT-PCR and NASBA, have been used for detection of CVB3 (Weiss et al., 1991, Dierssen et al., 2008, Saeedinia et al., 2008). The sensitivity of the conventional RT-PCR has been established at approximately 1 to100 PFU of virus per gram of tissue (Shen et al., 2009), the sensitivity of NASBA similar to RT-PCR (Saeedinia et al., 2008), and the sensitivity of the real-time PCR assay have been reported 100 copies per reaction for CVB3 (Dierssen et al., 2008). Most researchers that have developed RT-LAMP method for detection of Enteroviruses have shown that sensitivity of the RT-LAMP was approximately 10 copies per reaction that was 10-fold higher sensitive than RT-PCR (Jaianand et al., 2010, Yaqing et al., 2012, Zhao et al., 2014).The RT-LAMP assay developed in this work and comparative analysis of RT-LAMP and RT-PCR sensitivity suggested that RT-LAMP was 100-fold higher sensitive than RT-PCR. Also due to the designing four primers that recognize six specific areas in target sequence, RT-LAMP assay has a high specificity, therefore cross- reactivity with other Enterovirus were not observed.
5. Conclusion
The CVB3 RT-LAMP detection developed in our study showed high sensitivity and selectivity in compared with RT-PCR. Also, the one-step CVB3 RT-LAMP assay for the first time evaluated in this work and could be used in clinical samples.
Acknowledgments
We would like to thank the research council of Malek-Ashtar university for the financial support of this investigation.
References
- Arita M., Ling H., Yan D., Nishimura Y., Yoshida H., Wakita T., Shimizu H. Development of a reverse transcription-loop-mediated isothermal amplification (RT-LAMP) system for a highly sensitive detection of enterovirus in the stool samples of acute flaccid paralysis cases. BMC Infect. Dis. 2009;9(1) doi: 10.1186/1471-2334-9-208. (p.1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedard K.M., Semler B.L. Regulation of Picornavirus gene expression. Microbes Infect. 2004;6(7):702–713. doi: 10.1016/j.micinf.2004.03.001. [DOI] [PubMed] [Google Scholar]
- Caipang C.M., Haraguchi I., Ohira T., Hirono I., Aoki T. Rapid detection of a fish iridovirus using loop-mediated isothermal amplification (LAMP) J. Virol. Methods. 2004;121:155–161. doi: 10.1016/j.jviromet.2004.06.011. [DOI] [PubMed] [Google Scholar]
- Chang G.H., Lin L., Luo Y.J., Cai L.J., Wu X.Y., Xu H.M., Zhu Q.Y. Sequence analysis of six enterovirus 71 strains with different virulences in humans. Virus Res. 2010;151(1):66–73. doi: 10.1016/j.virusres.2010.04.001. [DOI] [PubMed] [Google Scholar]
- Chen Q., Hu Z., Zhang Q., Yu M. Development and evaluation of a real-time method of simultaneous amplification and testing of enterovirus 71 incorporating a RNA internal control system. J. Virol. Methods. 2014;196:139–144. doi: 10.1016/j.jviromet.2013.11.003. [DOI] [PubMed] [Google Scholar]
- De Jesus N.H. Epidemics to eradication: the modern history of poliomyelitis. Virol. J. 2007;4(1):1. doi: 10.1186/1743-422X-4-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierssen U., Rehren F., Henke-Gendo C., Harste G., Heim A. Rapid routine detection of enterovirus RNA in cerebrospinal fluid by a one-step real-time RT-PCR assay. J. Clin. Virol. 2008;42(1):58–64. doi: 10.1016/j.jcv.2007.11.016. [DOI] [PubMed] [Google Scholar]
- Ding X., Nie K., Shi L., Zhang Y., Guan L., Zhang D., Qi S., Ma X. Improved detection limit in rapid detection of human enterovirus 71 and coxsackievirus A16 by a novel reverse transcription–isothermal multiple-self-matching-initiated amplification assay. J. Clin. Microbiol. 2014;52(6):1862–1870. doi: 10.1128/JCM.03298-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drescher K.M., Kono K., Bopegamage S., Carson S.D., Tracy S. Coxsackievirus B3 infection and type 1 diabetes development in NOD mice: insulitis determines susceptibility of pancreatic islets to virus infection. Virology. 2004;329:381–394. doi: 10.1016/j.virol.2004.06.049. [DOI] [PubMed] [Google Scholar]
- Dunn J.J., Chapman N.M., Tracy S., Romero J. Genomic determinants of cardiovirulence in coxsackievirus B3 clinical isolates: localization to the 5′ nontranslated region. J. Virol. 2000;74(10):4787–4794. doi: 10.1128/jvi.74.10.4787-4794.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C., Ding Y., Zhou P., Feng J., Qian B., Lin Z., Wang L., Wang J., Zhao C., Li X., Cao M. Serological detection and analysis of anti-VP1 responses against various enteroviruses (EV)(EV-A, EV-B and EV-C) in Chinese individuals. Sci. Rep. 2016;6:21979. doi: 10.1038/srep21979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh R., Nagavardhini A., Sengupta A., Sharma M. Development of Loop-Mediated Isothermal Amplification (LAMP) assay for rapid detection of Fusarium oxysporum f. sp. ciceris-wilt pathogen of chickpea. BMC Res. Notes. 2015;8(1) doi: 10.1186/s13104-015-0997-z. (p.1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyypi T., Hovi T., Knowles N.J., Stanway G. Classification of enteroviruses based on molecular and biological properties. J. Gen. Virol. 1997;78(1):1–11. doi: 10.1099/0022-1317-78-1-1. [DOI] [PubMed] [Google Scholar]
- Jaianand K., Gunasekaran P., Rajkumar M., Sheriff A.K. Development of a new method for diagnosis of Coxsackie B5 viruses by Reverse transcription Loop-mediated isothermal amplification. Indian J. Med. Microbiol. 2010;1 doi: 10.4103/0255-0857.81780. (p.03) [DOI] [PubMed] [Google Scholar]
- Jaianand K., Saravanan N., Gunasekaran P., Sheriff A.K. Development of a new method for diagnosis of Group B Coxsackie genome by reverse transcription loop-mediated isothermal amplification. Indian J. Med. Microbiol. 2011;29(2) doi: 10.4103/0255-0857.81780. (p.110) [DOI] [PubMed] [Google Scholar]
- Jiang P., Liu Y., Ma H.C., Paul A.V., Wimmer E. Picornavirus morphogenesis. Microbiol. Mol. Biol. Rev. 2014;78(3):418–437. doi: 10.1128/MMBR.00012-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko H., Kawana T., Fukushima E., Suzutani T. Tolerance of loop-mediated isothermal amplification to a culture medium and biological substances. J. Biochem. Biophys. Methods. 2007;70(3):499–501. doi: 10.1016/j.jbbm.2006.08.008. [DOI] [PubMed] [Google Scholar]
- Knipe D.M., Howley P.M. Picornaviridae, the Viruses and Their Replication. Lippincott Williams & Wilkins; 2007. Fields virology; pp. 795–838. [Google Scholar]
- Knowlton K.U., Jeon E.S., Berkley N., Wessely R., Huber S. A mutation in the puff region of VP2 attenuates the myocarditic phenotype of an infectious cDNA of the Woodruff variant of coxsackievirus B3. J. Virol. 1996;70(11):7811–7818. doi: 10.1128/jvi.70.11.7811-7818.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosaki Y., Magassouba N.F., Oloniniyi O.K., Cherif M.S., Sakabe S., Takada A., Hirayama K., Yasuda J. Development and evaluation of reverse transcription-Loop-Mediated isothermal amplification (RT-LAMP) assay coupled with a portable device for rapid diagnosis of ebola virus disease in Guinea. PLoS Negl. Trop. Dis. 2016;10(2) doi: 10.1371/journal.pntd.0004472. (p.e0004472) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X.J., Shu Y.L., Nie K., Qin M., Wang D.Y., Gao R.B., Wang M., Wen L.Y., Han F., Zhou S.M., Zhao X. Visual detection of pandemic influenza A H1N1 Virus 2009 by reverse-transcription loop-mediated isothermal amplification with hydroxynaphthol blue dye. J. Virol. Methods. 2010;167(2):214–217. doi: 10.1016/j.jviromet.2010.03.027. [DOI] [PubMed] [Google Scholar]
- Michable A., Lindberg P., Stalhandske K., Petterson U. Genome of Coxsackievirus B3. Virology. 1987;156(1):50–63. doi: 10.1016/0042-6822(87)90435-1. [DOI] [PubMed] [Google Scholar]
- Mori Y., Notomi T. Loop-mediated isothermal amplification (LAMP): a rapid, accurate, and cost-effective diagnostic method for infectious diseases. J. Infect. Chemother. 2009;15(2):62–69. doi: 10.1007/s10156-009-0669-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie K., Qi S.X., Zhang Y., Luo L., Xie Y., Yang M.J., Zhang Y., Li J., Shen H., Li Q., Ma X.J. Evaluation of a direct reverse transcription loop-mediated isothermal amplification method without RNA extraction for the detection of human enterovirus 71 subgenotype C4 in nasopharyngeal swab specimens. PLoS One. 2012;7(12) doi: 10.1371/journal.pone.0052486. (p.e52486) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niessen L., Luo J., Denschlag C., Vogel R.F. The application of loop-mediated isothermal amplification (LAMP) in food testing for bacterial pathogens and fungal contaminants. Food Microbiol. 2013;36(2):191–206. doi: 10.1016/j.fm.2013.04.017. [DOI] [PubMed] [Google Scholar]
- Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28(12):e63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberste M.S., Peñaranda S., Pallansch M.A. RNA recombination plays a major role in genomic change during circulation of coxsackie B viruses. J. Virol. 2004;78(6):2948–2955. doi: 10.1128/JVI.78.6.2948-2955.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opel K.L., Chung D., McCord B.R. A study of PCR inhibition mechanisms using real time PCR. J. Forensic Sci. 2010;55(1):25–33. doi: 10.1111/j.1556-4029.2009.01245.x. [DOI] [PubMed] [Google Scholar]
- Pan L., Zhang L., Wang G., Liu Q., Yu Y., Wang S., Yu H., He J. Rapid, simple, and sensitive detection of Anaplasma phagocytophilum by loop-mediated isothermal amplification of the msp2 gene. J. Clin. Microbiol. 2011;49(12):4117–4120. doi: 10.1128/JCM.01085-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reetoo K.N., Osman S.A., Illavia S.J., Cameron-Wilson C.L., Banatvala J.E., Muir P. Quantitative analysis of viral RNA kinetics in coxsackievirus B3-induced murine myocarditis: biphasic pattern of clearance following acute infection, with persistence of residual viral RNA throughout and beyond the inflammatory phase of disease. J. Gen. Virol. 2000;81(11):2755–2762. doi: 10.1099/0022-1317-81-11-2755. [DOI] [PubMed] [Google Scholar]
- Saeedinia A., Shamsara M., Zeinoddini M., Sadeghi V., Maghsoudi N. Evaluation of nucleic acid sequence based amplification (NASBA) and Reverse Transcription Polymerase Chain Reaction for detection of coxsackievirus B3 in cell culture and animal tissue samples. Iran. J. Biotechnol. 2008;6(4):222–228. [Google Scholar]
- Shen Y., Kan Q.C., Xu W., Chu Y.W., Xiong S.D. Coxsackievirus B3 infection induced viral myocarditis by regulating the expression pattern of chemokines in cardiac myocytes. Iran J. Allergy Asthma Immunol. 2009;8(1) (p.1) [PubMed] [Google Scholar]
- Shi W., Li K., Ji Y., Jiang Q., Shi M., Mi Z. Development and evaluation of reverse transcription-loop-mediated isothermal amplification assay for rapid detection of enterovirus 71. BMC Infect. Dis. 2011;11(1) doi: 10.1186/1471-2334-11-197. (p.1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirato K., Yano T., Senba S., Akachi S., Kobayashi T., Nishinaka T., Notomi T., Matsuyama S. Detection of Middle East respiratory syndrome coronavirus using reverse transcription loop-mediated isothermal amplification (RT-LAMP) Virol. J. 2014;11(1) doi: 10.1186/1743-422X-11-139. (p.1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita N., Mori Y., Kanda H., Notomi T. Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat. Protoc. 2008;3(5):877–882. doi: 10.1038/nprot.2008.57. [DOI] [PubMed] [Google Scholar]
- Tracy S., Tu Z., Chapman N., Hufnagel G. Genetics of coxsackievirus B3 cardiovirulence. Eur. Heart J. 1995;16(Suppl. O):15–17. doi: 10.1093/eurheartj/16.suppl_o.15. [DOI] [PubMed] [Google Scholar]
- Tracy S., Chapman N., Mahy B. The Coxsackie B viruses. Curr. Top. Microbiol. Immunol. 1997;223:1–303. [Google Scholar]
- Van Ooij M.J., Vogt D.A., Paul A., Castro C., Kuijpers J., van Kuppeveld F.J., Cameron C.E., Wimmer E., Andino R., Melchers W.J. Structural and functional characterization of the coxsackievirus B3 CRE (2C): role of CRE (2C) in negative-and positive-strand RNA synthesis. J. Gen. Virol. 2006;87(1):103–113. doi: 10.1099/vir.0.81297-0. [DOI] [PubMed] [Google Scholar]
- Wang D., Wang X., Geng Y., An C. Detection of Enterovirus 71 gene from clinical specimens by reverse-transcription loop-mediated isothermal amplification. Indian J. Med. Microbiol. 2014;32(2) doi: 10.4103/0255-0857.129779. (p.124) [DOI] [PubMed] [Google Scholar]
- Weiss L.M., Movahed L.A., Billingham M.E., Cleary M.L. Detection of Coxsackievirus B3 RNA in myocardial tissues by the polymerase chain reaction. Am. J. Pathol. 1991;138(2) (p.497) [PMC free article] [PubMed] [Google Scholar]
- Wu J.S., Zhao N., Pan H., Wang C.M., Wu B., Zhang H.M., He H.X., Liu D., Amer S., Liu S.L. Patterns of polymorphism and divergence in the VP1 gene of enterovirus 71 circulating in the Asia-Pacific region between 1994 and 2013. J. Virol. Methods. 2013;193(2):713–728. doi: 10.1016/j.jviromet.2013.07.051. [DOI] [PubMed] [Google Scholar]
- Yaqing H., Wenping Z., Zhiyi Y., Xionghu W., Shouyi Y., Hong Y., Yingchun D., Guifang H. Detection of human Enterovirus 71 reverse transcription loop-mediated isothermal amplification (RT-LAMP) Lett. Appl. Microbiol. 2012;54(3):233–239. doi: 10.1111/j.1472-765X.2011.03198.x. [DOI] [PubMed] [Google Scholar]
- Yuan J., Yu M., Lin Q.W., Cao A.L., Yu X., Dong J.H., Wang J.P., Zhang J.H., Wang M., Guo H.P., Cheng X., Liao Y.H. Th17Cells contribute to viral replication in coxsackievirus B3-induced acute viral myocarditis. J. Immunol. 2005;185:4004–4010. doi: 10.4049/jimmunol.1001718. [DOI] [PubMed] [Google Scholar]
- Yuan J., Yu M., Lin Q.W., Cao A.L., Yu X., Dong J.H., Wang J.P., Zhang J.H., Wang M., Guo H.P., Cheng X. Th17 cells contribute to viral replication in coxsackievirus B3-induced acute viral myocarditis. J. Immunol. 2010;185(7):4004–4010. doi: 10.4049/jimmunol.1001718. [DOI] [PubMed] [Google Scholar]
- Zhang H., Nie K., Liu Y., Luo L., Huang W., Zhou S., Yang M., Chen Y., Luo J., Gao L., Ma X. Evaluation of reverse transcription loop-mediated isothermal amplification assays for rapid detection of human enterovirus 71 and coxsackievirus A16 in clinical samples. Adv. Infect. Dis. 2012;2(4):110. [Google Scholar]
- Zhao G.P., Yin G.Y., Zhao H., Li G.L., Meng B., Hu X., Li Y.X., Wang Y.Z., Zhao H.B. Two reverse-transcription-loop-mediated isothermal amplification systems for highly sensitive detection of enterovirus 71 and coxsackievirus A16. Afr. J. Microbiol. Res. 2013;7(35):4443–4450. [Google Scholar]
- Zhao H.B., Yin G.Y., Zhao G.P., Huang A.H., Wang J.H., Yang S.F., Gao H.S., Kang W.J. Development of loop-mediated isothermal amplification (LAMP) for universal detection of enteroviruses. Indian J. Microbiol. 2014;54(1):80–86. doi: 10.1007/s12088-013-0399-7. [DOI] [PMC free article] [PubMed] [Google Scholar]