Highlights
-
•
A nanoPCR method was developed for the detection of PEDV.
-
•
The nanoPCR assay was 100-fold more sensitive than a conventional RT-PCR assay.
-
•
The lower detection limit was 2.7 × 10−6 ng/μL of PEDV RNA.
-
•
This test could be applied for clinical diagnosis and field surveillance of PEDV.
Keywords: Porcine epidemic diarrhea virus (PEDV), nanoPCR, RT-PCR, Detection
Abstract
Porcine epidemic diarrhea virus (PEDV) is an important pig pathogen that can cause vomiting, diarrhea, and dehydration, leading to serious damage to the swine industry worldwide. In this study, a nanoparticle-assisted polymerase chain reaction (nanoPCR) assay targeting the N gene of PEDV was developed and the sensitivity and specificity were investigated. Under the optimized conditions for detection of PEDV RNA, the nanoPCR assay was 100-fold more sensitive than a conventional RT-PCR assay. The lower detection limit of the nanoPCR assay was 2.7 × 10−6 ng/μL of PEDV RNA and no cross-reaction was observed with other viruses. This is the first report to demonstrate the application of a nanoPCR assay for the detection of PEDV. The sensitive and specific nanoPCR assay developed in this study can be applied widely in clinical diagnosis and field surveillance of PEDV-infection.
Porcine epidemic diarrhea (PED), a serious and highly contagious swine disease, is characterized clinically by vomiting, diarrhea, and dehydration in suckling piglets (Van Reeth and Pensaert, 1994). Its etiologic agent, porcine epidemic diarrhea virus (PEDV) belongs to the genus Coronavirus of the family Coronaviridae, and is an enveloped, single-stranded RNA virus. The viral disease was discovered in the United Kingdom in 1971 and subsequently reported in many swine-producing countries of Europe and Asia (Puranaveja et al., 2009).
The current methods of detecting PEDV include virus isolation, serology, in situ hybridization, reverse transcription-polymerase chain reaction (RT-PCR), reverse transcription loop-mediated isothermal amplification (RT-LAMP), and TaqMan-based real-time RT-PCR (Hou et al., 2007, Jung and Chae, 2005, Kim and Chae, 2000, Kim et al., 2007, Kusanagi et al., 1992, Kweon et al., 1997, Li et al., 2009, Oh et al., 2005, Rodak et al., 2005, Song et al., 2006). A nanoparticle-assisted polymerase chain reaction (nanoPCR) assay, however, has not been developed for PEDV. Relative to conventional PCR assays, the nanoPCR assay has the potential for increased sensitivity and specificity. In addition, real-time PCR assays require expensive instruments, and LAMP assays are easily contaminated. The nanoPCR is an advanced form of PCR in which solid gold nanometal particles (1–100 nm) form colloidal nanofluids, which increase thermal conductivity. Therefore, PCR assays with nanofluids reach the target temperature more quickly than PCR assays with original liquids, and this reduces the time at non-target temperatures and thereby reduces non-specific amplification and increases specific amplification (Li and Rothberg, 2004, Shen et al., 2009). In addition, the nanoPCR assay does not require specialized instruments beyond standard PCR equipment (Ma et al., 2013). In this study, we developed a highly sensitive and specific nanoPCR method to target the N gene for the rapid detection of PEDV RNA in clinical specimens.
The genome of PEDV consists of at least seven open reading frame (ORF). The N gene is highly conserved. Primers were selected and designed from conserved N gene using the Primer premier 5 software to generate a 477 bp amplicon (Table 1 ). Viral RNA was extracted from 150 μL of supernatant from virus-infected Vero cells or tissue samples using the RNA extraction kit (Qiagen, Inc., USA) following the manufacturer's instruction. The extracts were resuspended in 20 μL of distilled water, aliquoted and stored at −80 °C before amplification was carried out. RNA was used as a template and 10-fold serially diluted in 10 mM Tris-EDTA buffer (pH 8.0) to produce 2.7–2.7 × 10−8 ng/μL. The nanoPCR assay was performed using PEDV-specific primers (Table 1). Briefly, 11 μL RNA extracted from either virus stocks or clinical samples was transcribed into cDNA using 1 μL (10 μM) of reverse primer in a 20 μL reaction volume. The reverse transcription (RT) reaction was performed at 42 °C for 50 min. The cDNA was amplified in a 25 μL reaction mixture containing 1 μL cDNA, 12.5 μL nanoPCR Mixture (GREDBIO, Weihai, China), 0.5 μL Taq DNA polymerase (GREDBIO, Weihai, China), and 10 μM each of forward and reverse primer by following the manufacturer's protocol with the following cycling times and temperatures: 94 °C for 3 min and 30 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 25 s. Five microliters of nanoPCR products were analyzed by agarose gel electrophoresis. The size of PEDV fragments amplified by nanoPCR was 477 bp (Fig. 1 ). Subsequent sequence analysis showed high similarity (100%) between the products obtained with the nanoPCR amplification of the N gene of PEDV (the objective sequences) and the reference sequence of PEDV. The results indicated that the PEDV nanoPCR method is specific.
Table 1.
Primers used in the nanoPCR assay for detection of PEDV.
| Type | Sequence (5′–3′) | Positiona |
|---|---|---|
| Forward | ACG GGT GCC ATT ATC CCT CTA T | 26329–26350 |
| Reverse | GAC TGG TTG TTG CCT CTG TTG T | 26784–26805 |
Numbers represents the nucleotide position within the genome of porcine epidemic diarrhea virus (GenBank accession number: KC189944.1).
Fig. 1.
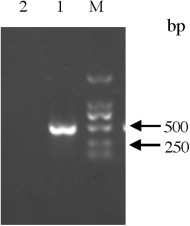
Size of the PEDV product after nanoPCR reaction as determined by gel electrophoresis. DL2000; Lane 1: PEDV; Lane 2: negative control.
To examine the sensitivity of the PEDV nanoPCR, RT-PCR and nanoPCR reaction were conducted using various concentrations of PEDV RNA as template. For this, the PEDV RNA was quantified by NanoDrop 1000 (Thermo Scientific, USA) and was diluted serially 10-fold from 2.7 to 2.7 × 10−8 ng/μL as template. The nanoPCR assay was performed using the optimized reaction parameters. The RT-PCR assay was performed using the same primers and reaction parameters as used for the nanoPCR. Briefly, the conventional RT-PCR assay for the detection of PEDV was performed in a 25-μL reaction mixture containing 1 μL cDNA, 12.5 μL conventional PCR Mixture, 0.5 μL Taq DNA polymerase, and 10 μM each of the forward and the reverse primer. Five microliters of the nanoPCR and the RT-PCR products were analyzed by agarose gel electrophoresis. The detection limit of the nanoPCR was 2.7 × 10−6 ng/μL, whereas that of the RT-PCR was 2.7 × 10−4 copies/μL (Fig. 2 ). Comparisons between the nanoPCR and the RT-PCR amplification indicated that the nanoPCR was 100-fold more sensitive than the RT-PCR assay (Fig. 2A and B).
Fig. 2.
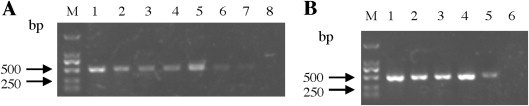
Sensitivity of nanoPCR assay (A) and the RT-PCR assay (B). A serial 10-fold dilution of PEDV RNA was used and analyzed by agarose gel electrophoresis. All experiments were repeated three times and similar results were obtained. M: DL2000; Lane 1: 2.7 ng/μL; Lane 2: 2.7 × 10−1 ng/μL; Lane 3: 2.7 × 10−2 ng/μL; Lane 4: 2.7 × 10−3 ng/μL; Lane 5: 2.7 × 10−4 ng/μL; Lane 6: 2.7 × 10−5 ng/μL; Lane 7: 2.7 × 10−6 ng/μL; Lane 8: 2.7 × 10−7 ng/μL.
The specificity of the nanoPCR assay was evaluated using other animal viruses including transmissible gastroenteritis virus (TGEV), rotavirus (RV), porcine bocavirus (PBoV), and porcine reproductive and respiratory syndrome virus (PRRSV). The nanoPCR assay did not amplified any of these four pig viruses (Fig. 3 ).
Fig. 3.
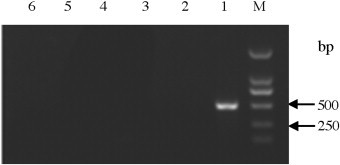
Specificity of the nanoPCR assay. M: DL2000; Lane 1: PEDV; Lane 2: PRRSV; Lane 3: PBoV; Lane 4: TGEV; Lane 5: RV; Lane 6: negative control.
Clinical specimens were collected from different pig farms in the Hebei province. The samples mainly included feces, intestine, and lymph node of diseased pigs. Collecting clinical specimens met the International Guiding Principles for Biomedical Research Involving Animals. Samples were homogenized and centrifuged at 4000 × g for 15 min to obtain a cell-free supernatant. The sample RNAs were extracted as described above. Thirty-five clinical samples were tested by both the nanoPCR and the conventional PCR assays. Eighteen of the thirty-five samples (51%) were positive by the RT-PCR analysis, whereas 23 of 35 samples (66%) were positive by the nanoPCR assay. Eighteen samples (51%) were positive by both methods. Five sample (14%) were positive by nanoPCR, but negative by RT-PCR analysis. The nanoPCR products of the 23 positive sample were subjected subsequently to automated sequencing reactions. The sequence analysis showed high similarity (100%) between the products obtained with the nanoPCR assay and the reference sequence of PEDV. No sample (0%) was positive by RT-PCR and negative by nanoPCR. The results showed that nanoPCR was more sensitive than the conventional RT-PCR assay.
Specific and sensitive diagnostic methods are useful to assess and monitor the transmission of pathogens, which is particularly important for PEDV, considering that its clinical signs are similar to those induced by other animal viruses. The current methods of detecting PEDV include virus isolation, serology, in situ hybridization, RT-PCR, RT-LAMP, and TaqMan-based real-time RT-PCR (Hou et al., 2007, Jung and Chae, 2005, Kim and Chae, 2000, Kim et al., 2007, Kusanagi et al., 1992, Kweon et al., 1997, Li et al., 2009, Oh et al., 2005, Rodak et al., 2005, Song et al., 2006). Virus isolation is one of the conventional methods, but PEDV is not easily adapted to cell lines. Although the addition of trypsin offers a possible solution, but viral titer of PEDV in cell lines remains low (Hofmann and Wyler, 1988). Serology represents the technique used most widely, however, this method has some disadvantages because antibody titers can decrease rapidly after infection (Lager et al., 1997). Conventional RT-PCR is less time-consuming but prone to sample contamination occurring during PCR processing steps, which increases the potential for false-positive results. The nanoPCR is an advanced form of PCR in which solid gold nanometal particles (1–100 nm) form colloidal nanofluids, which increase thermal conductivity. Therefore, PCR assays with nanofluids reach the target temperature more quickly than PCR assays with original liquids, and this reduces the time at non-target temperatures and thereby reduces non-specific amplification and increases specific amplification (Li and Rothberg, 2004, Shen et al., 2009). In addition, the nanoPCR assay does not require specialized instruments beyond standard PCR equipment (Ma et al., 2013). The nanoPCR has been used to detect other pig viruses due to its simplicity and high sensitivity including pseudorabies (Ma et al., 2013), and porcine bocavirus (Ma et al., 2013, Wang et al., 2014)
In this study, a nanoPCR assay targeting the N gene of PEDV was established. The nanoPCR assay is highly sensitive, it can detect 2.7 × 10−6 ng/μL of PEDV RNA, and no cross-reaction with other pig viruses was observed. This is the first report to demonstrate the application of a nanoPCR amplification technique for the detection of PEDV.
Acknowledgement
This work was supported by Colleges and Universities Research Project Foundation on Science & Technology from Hebei Province in China (QN2014220).
References
- Hofmann M., Wyler R. Propagation of the virus of porcine epidemic diarrhea in cell culture. J. Clin. Microbiol. 1988;26:2235–2239. doi: 10.1128/jcm.26.11.2235-2239.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X.L., Yu L.Y., Liu J. Development and evaluation of enzyme-linked immunosorbent assay based on recombinant nucleocapsid protein for detection of porcine epidemic diarrhea (PEDV) antibodies. Vet. Microbiol. 2007;123:86–92. doi: 10.1016/j.vetmic.2007.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K., Chae C. RT-PCR-based dot blot hybridization for the detection and differentiation between porcine epidemic diarrhea virus and transmissible gastroenteritis virus in fecal samples using a non-radioactive digoxigenin cDNA probe. J. Virol. Methods. 2005;123:141–146. doi: 10.1016/j.jviromet.2004.09.012. [DOI] [PubMed] [Google Scholar]
- Kim O., Chae C. In situ hybridization for the detection and localization of porcine epidemic diarrhea virus in the intestinal tissues from naturally infected piglets. Vet. Pathol. 2000;37:62–67. doi: 10.1354/vp.37-1-62. [DOI] [PubMed] [Google Scholar]
- Kim S.H., Kim I.J., Pyo H.M., Tark D.S., Song J.Y., Hyun B.H. Multiplex real-time RT-PCR for the simultaneous detection and quantification of transmissible gastroenteritis virus and porcine epidemic diarrhea virus. J. Virol. Methods. 2007;146:172–177. doi: 10.1016/j.jviromet.2007.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusanagi K., Kuwahara H., Katoh T., Nunoya T., Ishikawa Y., Samejima T., Tajima M. Isolation and serial propagation of porcine epidemic diarrhea virus in cell cultures and partial characterization of the isolate. J. Vet. Med. Sci. 1992;54:313–318. doi: 10.1292/jvms.54.313. [DOI] [PubMed] [Google Scholar]
- Kweon C.H., Lee J.G., Han M.G., Kang Y.B. Rapid diagnosis of porcine epidemic diarrhea virus infection by polymerase chain reaction. J. Vet. Med. Sci. 1997;59:231–232. doi: 10.1292/jvms.59.231. [DOI] [PubMed] [Google Scholar]
- Lager K.M., Mengeling W.L., Brockmeier S.L. Duration of homologous porcine reproductive and respiratory syndrome virus immunity in pregnant swine. Vet. Microbiol. 1997;58:127–133. doi: 10.1016/s0378-1135(97)00159-4. [DOI] [PubMed] [Google Scholar]
- Li H., Rothberg L. Colorimetric detection of DNA sequences based on electrostatic interactions with unmodified gold nanoparticles. Proc. Natl. Acad. Sci. U. S. A. 2004;101:14036–14039. doi: 10.1073/pnas.0406115101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Zhou Q.F., Xue C.Y., Ma J.Y., Zhu D.Z., Cao Y.C. Rapid detection of porcine reproductive and respiratory syndrome virus by reverse transcription loop-mediated isothermal amplification assay. J. Virol. Methods. 2009;155:55–60. doi: 10.1016/j.jviromet.2008.09.012. [DOI] [PubMed] [Google Scholar]
- Ma X., Cui Y., Qiu Z., Zhang B., Cui S. A nanoparticle-assisted PCR assay to improve the sensitivity for rapid detection and differentiation of wild-type pseudorabies virus and gene-deleted vaccine strains. J. Virol. Methods. 2013;193:374–378. doi: 10.1016/j.jviromet.2013.07.018. [DOI] [PubMed] [Google Scholar]
- Oh J.S., Song D.S., Yang J.S., Song J.Y., Moon H.J., Kim T.Y., Park B.K. Comparison of an enzyme-linked immunosorbent assay with serum neutralization test for serodiagnosis of porcine epidemic diarrhea virus infection. J. Vet. Sci. 2005;6:349–352. [PubMed] [Google Scholar]
- Puranaveja S., Poolperm P., Lertwatcharasarakul P., Kesdaengsakonwut S., Boonsoongnern A., Urairong K., Kitikoon P., Choojai P., Kedkovid R., Teankum K., Thanawongnuwech R. Chinese-like strain of porcine epidemic diarrhea virus, Thailand. Emerg. Infect. Dis. 2009;15:1112–1115. doi: 10.3201/eid1507.081256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodak L., Valicek L., Smid B., Nevorankova Z. An ELISA optimized for porcine epidemic diarrhoea virus detection in faeces. Vet. Microbiol. 2005;105:9–17. doi: 10.1016/j.vetmic.2004.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C., Yang W., Ji Q., Maki H., Dong A., Zhang Z. NanoPCR observation: different levels of DNA replication fidelity in nanoparticle-enhanced polymerase chain reactions. Nanotechnology. 2009;20:455103. doi: 10.1088/0957-4484/20/45/455103. [DOI] [PubMed] [Google Scholar]
- Song D.S., Kang B.K., Oh J.S., Ha G.W., Yang J.S., Moon H.J., Jang Y.S., Park B.K. Multiplex reverse transcription-PCR for rapid differential detection of porcine epidemic diarrhea virus, transmissible gastroenteritis virus, and porcine group A rotavirus. J. Vet. Diagn. Invest. 2006;18:278–281. doi: 10.1177/104063870601800309. [DOI] [PubMed] [Google Scholar]
- Van Reeth K., Pensaert M. Prevalence of infections with enzootic respiratory and enteric viruses in feeder pigs entering fattening herds. Vet. Rec. 1994;135:594–597. [PubMed] [Google Scholar]
- Wang X., Bai A., Zhang J., Kong M., Cui Y., Ma X., Ai X., Tang Q., Cui S. A new nanoPCR molecular assay for detection of porcine bocavirus. J. Virol. Methods. 2014;202:106–111. doi: 10.1016/j.jviromet.2014.02.029. [DOI] [PubMed] [Google Scholar]


