Highlights
-
•
RT-LAMP assays were optimized for diagnosis and subtyping of influenza A viruses.
-
•
RT-LAMP assays were compared with conventional one-step RT-PCR for sensitivity and specificity.
-
•
RT-LAMP assay was found to be more sensitive than conventional RT-PCR.
-
•
Clinical evaluation of RT-LAMP assay was done in comparison with WHO’s rRT-PCR taken as standard.
-
•
RT-LAMP assay can serve as a good alternate for diagnosis and surveillance studies during influenza outbreaks in developing countries.
Abbreviations: IAV, Influenza A viruses; LAMP, loop mediated isothermal amplification; RT, reverse transcription; RT-PCR, reverse transcription polymerase chain reaction; rRT-PCR, real-time RT-PCR; WHO, World Health Organization
Keywords: Influenza A viruses, Molecular detection, Pandemic influenza, RT-LAMP
Abstract
Background
Influenza A viruses (IAVs) have always remain a serious concern for the global economy and public health. A rapid, specific and sensitive detection method is always needed to control the influenza in its early stages by timely intervention of therapy and early clinical management.
Objectives
To develop RT-LAMP assays for detection of influenza A viruses, their further subtyping into seasonal (H1N1, H3N2) and novel pandemic H1N1 viruses and to evaluate clinical applicability of optimized RT-LAMP assays on patients’ samples.
Study design
In this study, we optimized RT-LAMP assay to detect IAVs by using primers against matrix gene and subtyping of IAVs was done by using primers against hemagglutinin gene. Optimized RT-LAMP assays were applied on clinical samples from patients having influenza like illness and results were compared with conventional one-step RT-PCR and real-time RT-PCR.
Results
RT-LAMP assays successfully detected and differentiated IAVs into H1N1, H3N2 and pdm09/H1N1 subtypes. One hundred and sixty seven clinical swab samples from influenza suspected patients were taken and tested with RT-LAMP assay, detecting 30 (17.9%) samples positive for Influenza A virus. Out of 30 samples, 21, 7 and 2 were found positive for pdm09/H1N1, H3N2 and seasonal H1 respectively. Conventional one-step RT-PCR detected a total of 27 (16.2%) samples for influenza A and further subtyping showed 20 and 7 samples positive for pdm09/H1N1 and H3N2 virus respectively whereas none was found positive for seasonal H1N1. RT-LAMP assay demonstrated higher sensitivity (93.8%) than conventional RT-PCR (84.4%) for influenza A viruses detection in clinical samples.
Conclusions
RT-LAMP assay is rapid, sensitive, specific and cost effective method for detection of influenza A viruses than conventional one-step RT-PCR and it can serve as a good alternate for diagnosis and surveillance studies during influenza outbreaks in resource-limited setups of developing countries.
1. Background
Influenza A viruses (IAVs) pose a serious threat to the world and are responsible for severe morbidity and mortality among human population globally. Seasonal outbreaks, epidemics and pandemics have been witnessed for IAVs and have resulted in immense loss to public health and economy (Wright and Neumann, 2013). The devastating potential of Influenza is shown by the facts that annually there are estimated 3–5 million cases of severe illness and 300,000–500,000 deaths around the globe (WHO, 2016). Influenza viruses belong to the family Orthomyxoviridae, characterized by segmented, negative-sense, single stranded RNA genome (Shaw and Palese, 2013). The family contains seven genera viz., Influenzavirus A, Influenzavirus B, Influenzavirus C, Influenzavirus D, Isavirus, Quaranjavirus and Thogotovirus (ICTV, 2016). Out of the four genera of influenza viruses, influenza A and B are the main cause of seasonal epidemics while influenza C causes mild reparatory illness and influenza D affects cattle and not known to cause any infection in humans (Hause et al., 2014, CDC, 2016, Ferguson et al., 2016).
Influenza or “Flu” is a respiratory illness characterized by high grade fever, myalgia, headache, malaise, sore throat, rhinitis and cough (Cox and Subbarao, 1999). Generally influenza is a self limiting disease but may become life-threatening in high risk group individuals including young children, pregnant women, aged people and people with compromised immunity (Cox and Subbarao, 1999, Cox and Subbarao, 2000). Influenza vaccines are updated annually according to the circulating strains and this information is provided by the surveillance system of WHO’s Global Influenza Surveillance and Response System (GISRS) for both the northern and southern hemispheres (WHO, 2012, Webster and Govorkova, 2014). Antivirals treatment proved effective against influenza if given within 24–48 h of the onset of disease (Moscona, 2005). Hence, early and accurate diagnosis of influenza plays a crucial role for timely intervention of therapy, clinical management and thus controlling the spread of disease. Clinical symptoms of influenza and other respiratory infections can be very similar, making it hard to differentiate. Only laboratory diagnosis can provide an accurate influenza diagnosis. These include conventional virus isolation, antigen/antibody detection and molecular methods like reverse transcription polymerase chain reaction (RT-PCR) (Poddar, 2002) and real time RT-PCR (rRT-PCR) (Spackman and Suarez, 2008, Carr et al., 2009, Wang and Taubenberger, 2010). Virus isolation and serological assays are time consuming and take days to weeks to get results making them unsuitable during epidemics and also these methods require highly sophisticated biosafety laboratories. Molecular methods such as RT-PCR and rRT-PCR are although rapid, sensitive and specific but need costly equipments and trained laboratory persons making these assays hard to use in resource-limited laboratories of developing countries.
Loop-mediated isothermal amplification (LAMP) is a specific, efficient and rapid technique that is similar to PCR amplification but the target DNA amplification is under isothermal conditions. This method makes use of a DNA polymerase with strand displacement activity and a set of four specially designed primers that recognize a total of six distinct sequences on the target DNA (Notomi et al., 2000). The method is applicable to amplification of RNA templates when combined with a reverse transcription reaction (RT-LAMP). RT-LAMP assay has been successfully applied for the detection of various RNA viruses including chikungunya (Parida et al., 2007, Lu et al., 2012), dengue (Lu et al., 2012, Parida et al., 2005), Japanese encephalitis (Parida et al., 2006, Toriniwa and Komiya, 2006), severe acute respiratory syndrome (Thai et al., 2004) and West Nile virus (Parida et al., 2004). Influenza viruses and their novel subtypes like H5, H7, H9 and H10, have been effectively detected by RT-LAMP assay (Poon et al., 2005, Ito et al., 2006, Imai et al., 2006, Chen et al., 2008, Zhang et al., 2013, Luo et al., 2015). The specificity of RT-LAMP is extremely high due to use of four primers that recognize six distinct regions on the target DNA. It is faster than PCR because of single temperature is needed for amplification and also cost effective as costly equipments are not required. The amplification results can be viewed directly by using fluorescent dye like SYBR green, reducing the time of the assay.
2. Objectives
The objectives of the current study were to (1) optimize RT-LAMP assay for detection of influenza A viruses and their subtypes (H1N1, H3N2 and pdm09/H1N1); (2) determine sensitivity and specificity of RT-LAMP assay; (3) clinical evaluation of RT-LAMP assay and conventional one-step RT-PCR in comparison to WHO recommended rRT-PCR taken as standard.
3. Study design
3.1. Reference strains of influenza A viruses
Reference strains of Influenza A viruses were kindly provided by the department of Microbiology, All India Institute of Medical Sciences (AIIMS), New Delhi, India. These strains include; A/California/7/2009 (H1N1) pdm09-like virus; A/Texas/50/2012 (H3N2)-like virus; and A/Malaysia/2089302/2009(H1N1) virus. These strains were aliquoted and stored at −80 °C until further used.
3.2. RNA extraction from reference strains
Viral RNA was extracted from an aliquot of each reference strain of influenza viruses, using the GeneJET Viral DNA/RNA Purification Kit (Thermo Fisher Scientific, USA) by following the manufacturer’s instructions. RNA was eluted in a final volume of 50 μl in elution buffer (provided with kit) and divided into aliquots of 5 μl each and stored at −80 °C until used. RNA extraction was performed in class II biosafety cabinets.
3.3. RT-LAMP primers
For detection of Influenza A viruses, published primers were taken from matrix (M) gene (Poon et al., 2005). For subtyping of IAVs, primers were selected from hemagglutinin (HA) gene. Published primers were used to detect pandemic influenza H1N1 virus (pdm09/H1N1) (Nakauchi et al., 2011). For seasonal H1N1 and H3N2 virus, HA gene sequence of the respective reference strain was retrieved from GenBank database (GeneBank accession numbers CY118255 and KC892952) and primers were designed by using Primer Explorer version 4 (https://primerexplorer.jp/e/). Four primers consisting of two internal primers (Forward internal primer or FIP and backward internal primer or BIP) and two external primers (F3 and B3) were designed for the study. All primers were chemically synthesized by Sigma-Aldrich (Sigma-Aldrich, Bangalore, India) and were of HPLC grade purity (Table 1 ).
Table 1.
Details of primers used in RT-LAMP assay.
| Primer | Sequence (5′ → 3′) | Genome location | Primer length (bp) | Reference |
|---|---|---|---|---|
| Influenza A/M-gene | ||||
| Inf A-F3 | TGGTGCACTTGCCAGTTG | 375–392 | 18 | Poon et al., 2005 |
| Inf A-B3 | CCAGCCATCTGTTCCATAGC | 593−574 | 20 | |
| Inf A-FIP | TGCTGTGAATCAGCAATCTGTTACAGGA | F1c, 488-467 | 42 | |
| (F1c + F2) | TGGGAACAGTGACC | F2, 410–429 | ||
| Inf A-BIP | AGACAAATGGCTACTACCACCCGTAGT | B1c, 499–519 | 40 | |
| (B1c + B2) | GCTAGCCAGCACC | B2, 567-549 | ||
| A/pdm09H1/HA-gene (pandemic Influenza A) | ||||
| pdmH1-F3 | AGCTAAGAGAGCAATT | 170–185 | 16 | Nakauchi et al., 2011 |
| pdmH1-B3 | TTTCCCTTTATCATTAATGTAGGATTTG | 384−357 | 28 | |
| pdmH1-FIP | ACCTTTGTTCGAGTCATGATTTGGTTCTC | F1c, 264-244 | 51 | |
| (F1c + F2) | AGTGTCATCATTTGAAAGGTTT | F2, 189–213 | ||
| pdmH1-BIP | TAACGGCAGCATGTCCTCATTGTATGAA | B1c, 266–285 | 48 | |
| (B1c + B2) | TTTCCTTTTTTAACTAGCCA | B2, 345-319 | ||
| A/H1/HA-gene (seasonal Influenza A/H1) | ||||
| H1-F3 | CTGTTATGCACATTTACAGCT | 39–59 | 21 | This study |
| H1-B3 | TTCCTAAGATCCACCCGG | 258−241 | 18 | |
| H1-FIP | ACTGTGTCAACAGTGTCGGTTTTGCAGA | F1c, 124-105 | 44 | |
| (F1c + F2) | CACAATATGTATAGGC | F2, 65–86 | ||
| H1-BIP | ACACACTCTGTCAACCTGCTTTTCCAAT | B1c, 147–167 | 41 | |
| (B1c + B2) | TGTAGTGGGGCTA | B2, 225-208 | ||
| A/H3/HA-gene (seasonal Influenza A/H3) | ||||
| H3-F3 | ATAGTAAAACCGGGAGACAT | 754–773 | 20 | This study |
| H3-B3 | TGCTTAACATATCTGGGACA | 980−961 | 20 | |
| H3-FIP | TGGGTGCATCTGATCTCATTATTGATTTT | F1c, 868-844 | 48 | |
| (F1c + F2) | AGGGAATCTAATTGCTCCT | F2, 792–810 | ||
| H3-BIP | GCAAGTCTGAATGCATCACTCCTTTTTG | B1c, 878–899 | 48 | |
| (B1c + B2) | ATCCTGTTTACATTTTGGAA | B2, 949-928 | ||
3.4. RT-LAMP assay
Twenty five microliters of RT-LAMP reaction cocktail was prepared in 0.2 ml tubes using 0.2 μM of each outer primers (F3 and B3); 1.6 μM of each inner primers (FIP and BIP); 1.4 mM of each deoxynucleotides (dNTPs); 20 mM Tris-HCl (pH 8.8); 10 mM (NH4)2SO4; 10 mM KCl; 8 mM MgSO4; 0.8 M betaine; 0.1% Tween 20; 8 U of Bst DNA polymerase (New England Biolabs); 0.5 U of AMV reverse transcriptase (Invitrogen, USA) and 1 μl of SYBR green I (Thermo Fisher Scientific, USA). Two microliters of each reference strain’s RNA was added to their respective tubes (containing strain specific primers) making final volume 25 μl. No template or negative controls were also included in the assay. The reaction tubes were incubated at 63 °C for 1 h either in a water bath or a heating block. The tubes were heated at 80 °C for 5 min to stop the reaction. Amplified products were visualized both by electrophoresis using 2.5% agarose gel and by visual detection of green fluorescence due to added SYBR green I stain (Thermo Fisher Scientific, USA) under ultra violet (UV) light.
3.5. Conventional one-step RT-PCR
Conventional one-step RT-PCR was performed on RNA samples extracted from the reference strains of IAVs. For detection of IAVs, primer pair was taken from the matrix (M) gene while subtyping primer pairs were taken from hemagglutinin (HA) gene. For diagnosis of pandemic influenza virus, primer pair was selected from the conserved region of HA gene (GeneBank accession number KU933485) using primer blast software (https://www.ncbi.nlm.nih.gov/tools/primer-blast/). Details of primer sets were given in Table 2 . One-step RT-PCR reaction was carried out in final reaction volume of 25 μl by using Verso 1-Step RT-PCR Kit (Thermo Fisher Scientific, USA) by following manufacturer’s instructions. Briefly, reaction mixture was comprised of 12.5 μl of 2X RT-PCR buffers; 1 μl of RT enhancer; 0.2 μM of each forward and reverse primer; 0.5 μl of enzyme mix and 2 μl of extracted RNA. The protocol for optimization of one-step RT-PCR was as follows: 50 °C for 30 min, 95 °C for 15 min, followed by 40 cycles of 95 °C for 30 s, 52 °C–58 °C for 30 s, 72 °C for 45 s and final extension at 72 °C for 10 min. PCR products were separated by electrophoresis using 2% agarose gel and visualized by UV transilluminator.
Table 2.
Details of primers used in conventional one-step RT-PCR assay.
| Primer | Sequence (5′ → 3′) | Primer length (bp) | Amplicon size (bp) | Reference |
|---|---|---|---|---|
| Influenza A/M-gene | ||||
| Inf A-F | AGTCTTCTAACCGAGGTCGAAACG | 24 | 239 | WHO, 2014 |
| Inf A-R | TGGACAAAGCGTCTACGCTGCA | 22 | ||
| Influenza A/H1/HA-gene | ||||
| H1-F | AATTTGCTATGGCTGACGGG | 20 | 183 | Poddar, 2002 |
| H1-R | GCTATAATGTGAAGACACTACAGA | 24 | ||
| Influenza A/H3/HA-gene | ||||
| H3-F | GCGACAGTCCTCATCAGATCCT | 22 | 360 | Poddar, 2002 |
| H3-R | CATTGTTTGGCATAGTCACGTTC | 23 | ||
| Influenza A/pH1/HA-gene | ||||
| pdmH1-F | ACACTAGTAGAGCCGGGAGA | 20 | 261 | This Study |
| pdmH1-R | CAATCCTGTGGCCAGTCTCA | 20 | ||
3.6. Sensitivity and specificity of RT-LAMP
Influenza virus A/California/7/2009 (H1N1) strain with known virus titre as plaque forming units per microliters (PFU/μl) was used to compare sensitivities of RT-LAMP and conventional RT-PCR with WHO recommended Taqman real-time RT-PCR (rRT-PCR). Ten-fold serial dilutions were made from virus strain in separate tubes ranging from 102 to 10−4 PFU per reaction using molecular grade water. RNA was extracted from each dilution and used separately for RT-LAMP, conventional RT-PCR and rRT-PCR. Matrix gene specific primers and 2 μl of RNA from each dilution were used in all assays. Likewise, sensitivities of RT-LAMP, conventional RT-PCR and rRT-PCR were also assessed for subtyping of influenza viruses using HA gene specific primers on H1N1 and H3N2 reference strains. For specificity evaluation, each individual RT-LAMP assay was applied on influenza A viruses (pdm09/H1N1, A/H1N1 and A/H3N2), influenza B virus along with other additional respiratory viruses including respiratory syncytial virus (RSV) and human metapneumovirus (hMPV), and cross-reactivity of the assay was checked.
3.7. Clinical samples
A total of 167 clinical samples were collected from patients with influenza like illness (high grade fever (>38.5 °C), cough, sore throat and myalgia) from Post Graduate Institute of Medical Sciences (PGIMS), Rohtak, Haryana, India from November 2014 to December 2016. One throat and one nasal swab was collected in 2 ml of viral transport medium (Hanks’ balanced salt solution supplemented with 2% BSA, 100 U/ml penicillin and 50 μg/ml streptomycin) from each patient and transported to the laboratory under cold conditions at 4 °C. The samples were processed and RNA was extracted in class II biosafety cabinets.
3.8. Detection and subtyping of influenza A viruses in clinical samples
Clinical samples were evaluated by using optimized RT-LAMP and conventional one-step RT-PCR and detection results were compared with WHO recommended Taqman real-time RT-PCR (rRT-PCR) considering the later as standard. Standardized RT-LAMP and conventional one-step RT-PCR were applied on clinical samples and amplified products were analyzed by gel electrophoresis. Taqman rRT-PCR assay was applied on extracted RNAs from clinical samples for detection of influenza A viruses and further subtyping into H1N1, H3N2 and pdm09/H1N1 subtypes. Taqman rRT-PCR assay was performed in a 25 μl reaction volume using WHO recommended protocol and primers/probes sequences (WHO, 2014).
4. Results
4.1. Optimization of RT-LAMP and conventional one-step RT-PCR assay
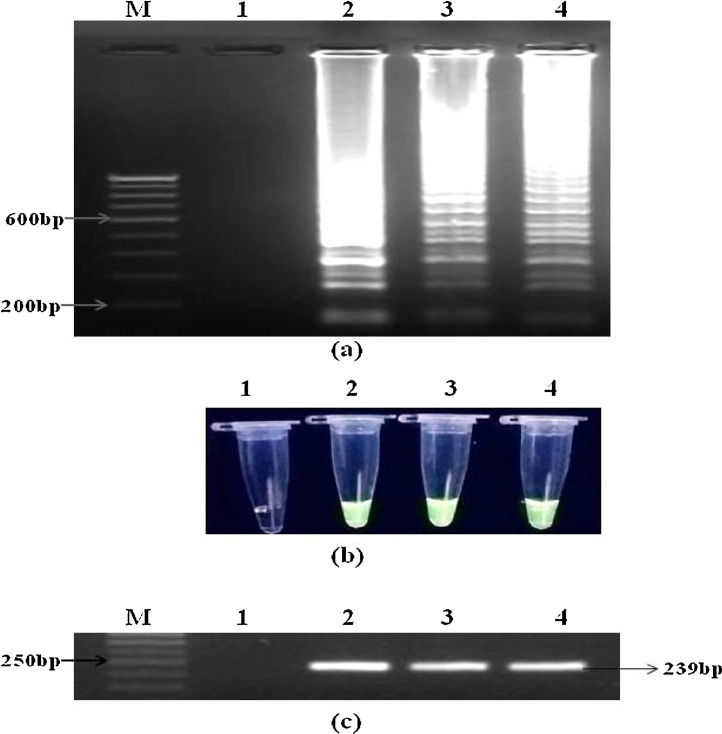
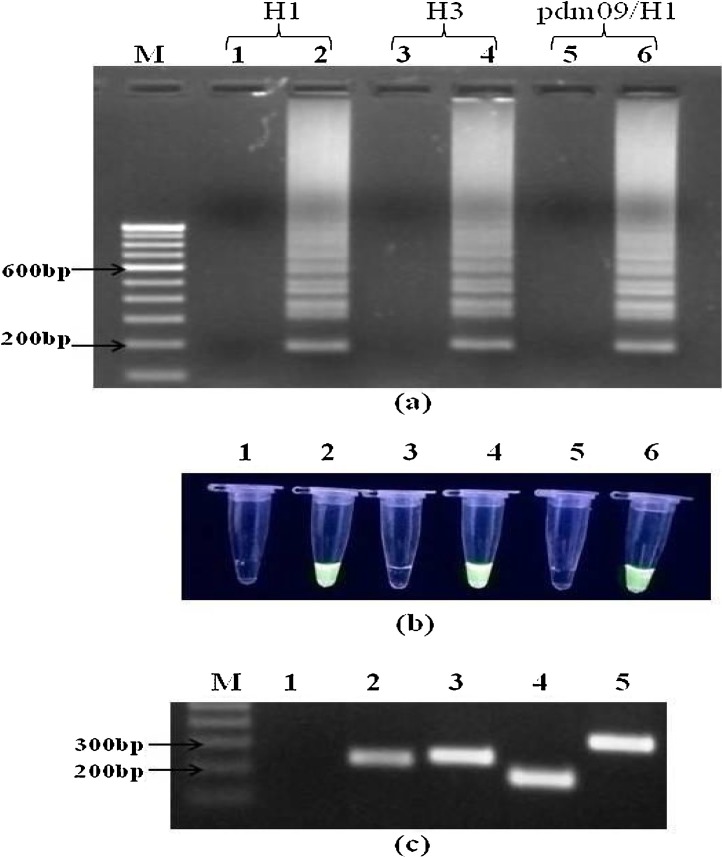
All three reference strains of influenza A virus were detected by matrix gene specific RT-LAMP reaction (Fig. 1 a). On further subtyping, each respective subtype was also detected by HA gene specific RT-LAMP assay (Fig. 2 a). Optimum amplification temperature and incubation time were found to be 63 °C and 1 h respectively. Amplification results were visualized by both gel electrophoresis and green fluorescence detection under UV light (Fig. 1b and 2b). Conventional one-step RT-PCR was able to detect all three influenza A virus strains using matrix gene primers. Optimum annealing temperature and time was found to be 55 °C for 30 s. Amplicons of 239 bp were visualized using 2% agarose gel electrophoresis (Fig. 1 c). For subtyping conventional RT-PCR, annealing at 55 °C for 30 s was found to be optimum for both pdm09/H1N1 and H3N2 subtypes whereas for seasonal H1N1 subtype, annealing temperature of 54 °C for 30 s was optimized. Amplicons of 261 bp, 183 bp and 360 bp were detected for pdm09/H1N1, H1N1 and H3N2 respectively, after 2% agarose gel electrophoresis (Fig. 2c).
Fig. 1.
Detection of influenza A viruses by RT-LAMP and conventional RT-PCR using matrix (M) gene primers. (a) examination of RT-LAMP products in 2.5% agarose gel electrophoresis and direct visualization of green fluorescence in positive amplification reactions in 0.2 ml PCR tubes using SYBR green dye under UV light (b); lane M, 100 bp DNA marker; lane/tube 1, no template control; lane/tube 2, influenza H1N1; lane/tube 3, influenza H3N2; lane/tube 4, influenza pdm09/H1N1. (c) Agarose gel electrophoresis profile of conventional one-step RT-PCR amplified products showing a band of 239 bp. All lanes have similar order as mentioned above except lane M containing 50 bp DNA marker. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 2.
Detection of influenza A viruses subtypes using RT-LAMP and conventional one-step RT-PCR using HA gene primers. (a) agarose (2.5%) gel electrophoresis profile of RT-LAMP assays products (b) visual detection of green fluorescence in 0.2 ml PCR tubes using SYBR green dye under UV light. Lane M, 100 bp DNA marker; lane/tube 2, influenza H1N1; lane/tube 4, influenza H3N2; lane/tube 6, influenza pdm09/H1N1; lane/tube 1, 3, 5 are no template controls for H1N1, H3N2 and pdm09/H1N1 respectively. (c) agarose (2.5%) gel electrophoresis profile of conventional one-step RT-PCR reaction products. Lane M, 100 bp DNA marker; lane 1, no template control; lane 2, influenza A virus detection using M gene primers showing a band of 239 bp; lane 3, influenza A/pdm09/H1N1 showing a band of 261 bp; lane 4, influenza A seasonal H1N1 showing a band of 183 bp; lane 5, influenza A H3N2 showing a band of 360 bp. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4.2. Sensitivity and specificity of RT-LAMP assay
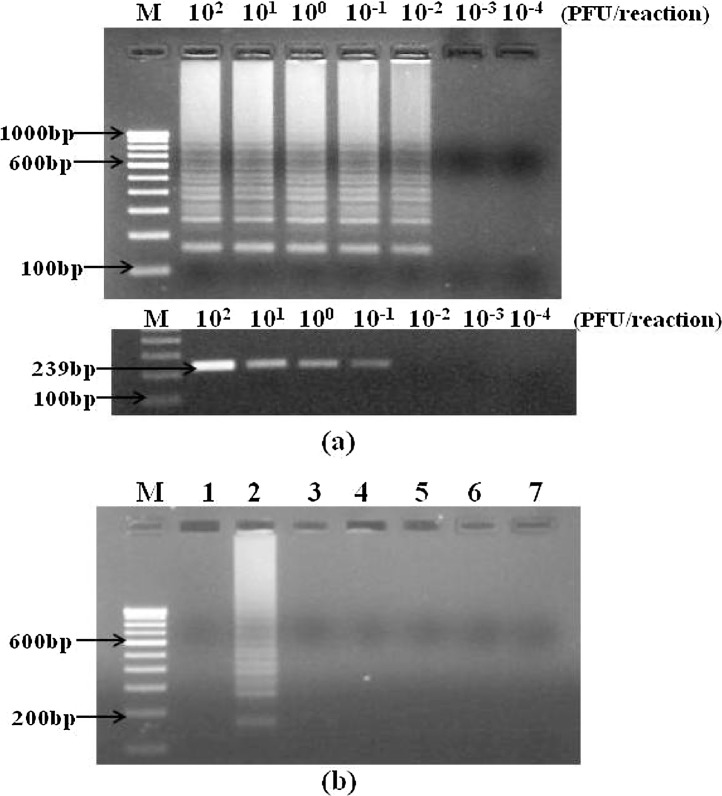
Sensitivity of RT-LAMP assays were assessed for detection of influenza A viruses using M gene primers and for detection of three subtypes i.e. H1N1, H3N2 and pdm09/H1N1, using HA gene primers (Table 3 ). The results were compared with that of conventional one-step RT-PCR and rRT-PCR. The detection limit of M gene specific RT-LAMP was found to be ten times higher (0.01 PFU/reaction) than conventional one-step RT-PCR (0.1 PFU/reaction) (Fig. 3 a) and ten times lower than rRT-PCR (0.001 PFU/reaction) (Table 3 ). The detection limits of HA gene specific RT-LAMP assays for pdm09/H1N1, H3N2 and H1N1 were 0.01, 0.1 and 0.1 PFU per reaction respectively. The detection limits of conventional one-step RT-PCR for pdm09/H1N1, H3N2 and H1N1 were 1, 0.1 and 1 PFU per reaction respectively, making it 10–100 times less sensitive than RT-LAMP. RT-LAMP demonstrated similar sensitivities as that of rRT-PCR for the detection of pdm09/H1N1 and H3N2 while 10 times less sensitivity for detection of H1N1 subtype (Table 3). Specificity of each RT-LAMP primer set was evaluated against different influenza viruses and other respiratory viruses. All the RT-LAMP primer sets were found to be highly specific for their respective influenza viruses as there was no amplification with other influenza viruses or different respiratory viruses. As shown in Fig. 3b, pdm09/H1N1 RT-LAMP (HA gene) was found to be highly specific for detecting pandemic IAVs as only pdm09/H1N1 reaction showed positive amplification while there was no amplification with other viruses.
Table 3.
Limit of detection of RT-LAMP, conventional one-step RT-PCR and real-time RT-PCR assay using virus titre as plaque forming units (PFU).
| Influenza strain | Target gene | RT-LAMP (PFU/reaction) | Conventional RT-PCR (PFU/reaction) | WHO rRT-PCR (PFU/reaction) |
|---|---|---|---|---|
| A/California/7/2009 (H1N1) pdm09-like virus | M | 0.01 | 0.1 | 0.001 |
| A/California/7/2009 (H1N1) pdm09-like virus | HA | 0.01 | 1 | 0.01 |
| A/Texas/50/2012 (H3N2)-like virus | HA | 0.1 | 0.1 | 0.1 |
| A/Malaysia/2089302/2009 (H1N1)-like virus | HA | 0.1 | 1 | 0.01 |
Fig. 3.
Sensitivity and specificity of RT-LAMP assay. (a) Sensitivity comparison of RT-LAMP assay (upper panel) and conventional RT-PCR (lower panel) for detection of IAVs using M gene primers, was performed using ten-fold serial dilutions of influenza A/California/7/2009(H1N1) strain with concentrations ranging from 100 to 0.0001 PFU/reaction tube. Detection limit of RT-LAMP assay was 0.01 PFU/reaction (upper panel) while conventional RT-PCR (lower panel) showed a detection limit of 0.1 PFU/reaction making RT-LAMP assay ten times more sensitive than conventional RT-PCR. Lane M, 100 bp DNA marker. (b) Influenza pdm09/H1N1 specific RT-LAMP reaction virus using HA gene primers was applied on other respiratory viruses to check the specificity of the reaction. Agarose electrophoresis profile of the RT-LAMP reaction products; lane M, 100 bp DNA maker; lane 1, no template control; lane 2, influenza A pdm09/H1N1; lane 3, influenza A/H1N1; lane 4, influenza A/H3N2; lane 5, influenza B virus; lane 6, respiratory syncytial virus (RSV); lane 7, human metapneumovirus (hMPV).
4.3. Detection and subtyping of influenza A viruses in clinical samples
All clinical samples were analyzed for presence of IAVs and their subtypes (seasonal H1N1, H3N2 and pdm09/H1N1) using WHO recommended Taqman real-time RT-PCR, RT-LAMP assay and conventional RT-PCR assay. Real-time RT-PCR results showed that out of 167 swab samples, 32 (19.2%) were positive for influenza A viruses. Further subtyping of 32 (n = 32) positive samples by real-time RT-PCR showed that 23 (71.9%) samples were positive for pdm09/H1N1, 7 (21.9%) were positive for H3N2 and 2 (6.2%) were positive for H1N1 virus. Matrix gene specific RT-LAMP assay detected 30 (17.9%) samples positive for influenza A viruses which were further sub-typed using HA gene specific RT-LAMP. Results showed that 21 out of 30 (70%) samples were positive for pdm09/H1N1, 7 (23.3%) samples were positive for H3N2, and 2 (6.7%) samples were positive for H1N1 virus. These results showed the comparable sensitivity and specificity of real-time RT-PCR with that of RT-LAMP assay (Table 4 ). Conventional RT-PCR detected 27 (16.2%) samples positive for influenza A viruses out of which 20 (74.1%) were found positive for pdm09/H1N1 and 7 (25.9%) were positive for H3N2 virus where as no sample was detected positive for H1N1 virus. All samples which were found to be negative by real-time RT-PCR also come negative by both RT-LAMP and conventional RT-PCR assay.
Table 4.
Identification of influenza A viruses and subtypes (H1N1, H3N2 and pdm09/H1N1) in clinical samples from Haryana, India, using RT-LAMP and conventional one-step RT-PCR, confirmed by WHO recommended rRT-PCR. Sensitivity and specificity were calculated by taking rRT-PCR as standard. The numbers in parentheses indicate the real-time RT-PCR results. ND: not determined.
| Detection and subtyping of influenza A viruses |
||||||
|---|---|---|---|---|---|---|
| Influenza A virus | Influenza A/H1N1 | Influenza A/H3N2 | Influenza A/pdm09/H1N1 | Negative | Total | |
| RT-LAMP assay | 30 (32) | 2 (2) | 7 (7) | 21 (23) | 137 (135) | 167 |
| Conventional RT-PCR | 27 (32) | 0 (2) | 7 (7) | 20 (23) | 140 (135) | 167 |
| Sensitivity/specificity of RT-LAMP assay (%) | 93.8/100 | ND | 100/100 | 91.3/100 | ||
| Sensitivity/specificity of conventional RT-PCR (%) | 84.4/100 | ND | 100/100 | 87/100 | ||
5. Discussion
Influenza surveillance has importance not only in diagnosis of circulating strains of the virus but also to upgrade the available vaccine on annual basis to provide protection against influenza. Control of novel influenza A viruses mainly depends of early and accurate identification of virus strain followed by early intervention of antiviral therapy. So, a rapid, sensitive, specific and cost effective method of detection and subtyping of influenza A viruses is always seems fruitful in this scenario. Virus isolation using egg embryo culture or cell culture is considered as ‘gold standard’ for influenza virus detection but the whole process is labour intensive and lengthy taking 3–7 days for obtaining results (Ellis and Zambon, 2002). Serological methods are also not very reliable for influenza surveillance as these are time consuming and require analysis of serum samples from both acute phase and recovering phase. Rapid detection kit based assays are although fast but are not very consistent, have low sensitivity and may produce false results (Wang and Taubenberger, 2010, Ellis and Zambon, 2002, Kim and Poudel, 2013). Molecular detection methods have emerged as very useful tools in comparison to time consuming conventional virus isolation and serological methods. Molecular methods based on conventional RT-PCR and real-time RT-PCR have been proved useful in diagnosis and surveillance of influenza viruses, but most of these assays require highly sophisticated instruments and are not feasible to conduct in resource-limited settings of developing countries.
Therefore, present study was focused on RT-LAMP assay for detection of influenza A viruses and further subtyping into seasonal influenza (H1 and H3) and novel pandemic influenza A virus (pdm09/H1N1). RT-LAMP assay has been successfully applied to detect influenza A viruses like swine flu (H1N1), pdmH1N1, avian influenza H5, H7, H9 and H10 subtypes (Poon et al., 2005, Ito et al., 2006, Imai et al., 2006, Chen et al., 2008, Luo et al., 2015, Kubo et al., 2010, Parida et al., 2011, Bao et al., 2012). In this study, we used matrix gene based primers for detection of influenza A viruses (Poon et al., 2005) and hemagglutinin gene based primers for their further subtyping. Matrix gene based RT-LAMP assay was able to amplify all three reference strains of influenza A virus and hemagglutinin gene based RT-LAMP assay successfully detected the respective subtypes. No cross-reactivity was observed for other related viruses and influenza subtypes making the reaction highly specific.
Sensitivity of a detection assay is an important deciding factor when it comes to applicability of the assay. In this study we have compared RT-LAMP with conventional RT-PCR and Taqman real-time RT-PCR. In our study, detection limit of Taqman rRT-PCR was found to be ten times more than RT-LAMP for influenza A and its subtype H1N1 whereas for detection of pdm09/H1N1 and H3N2 both assays showed similar values. Sensitivity comparison of RT-LAMP with conventional RT-PCR showed that RT-LAMP was 10–100 times more sensitive than conventional RT-PCR. Previous studies have also established that RT-LAMP is more sensitive than conventional RT-PCR and has comparable sensitivity with real-time RT-PCR for detection of influenza viruses (Poon et al., 2005, Kubo et al., 2010, Parida et al., 2011, Bao et al., 2012, Sharma and Kaushik, 2016).
In the present study, a total of 167 clinical samples were analyzed for the presence of IAVs and their further subtyping was done. We have used RT-LAMP assay, conventional RT-PCR and rRT-PCR assay in parallel reactions for diagnosis of influenza in clinical samples. Real-time RT-PCR reaction was performed using WHO recommended primers (WHO, 2014) and was used as standard to compare RT-LAMP and conventional RT-PCR assay. Real-time RT-PCR detected 32 samples positive for influenza A viruses, which on further subtyping showed 2, 7 and 23 samples for influenza H1N1, H3N2 and pdm09/H1N1 respectively. Sensitivity of conventional RT-PCR for detection of influenza A viruses and subtype pdm09/H1N1 was 84.4% (27/32) and 86.9% (20/23) respectively. Whereas RT-LAMP assay showed sensitivity of 93.8% (30/32) for influenza A viruses and 91.3% (21/23) for subtype pdm09/H1N1 detection, indicating higher sensitivity than conventional RT-PCR. Both these assays detected H3 subtype with equal sensitivity of 100% (7/7). For seasonal H1 subtype, sensitivity and specificity could not be determined due to small number of positive samples. The results showed that RT-LAMP assay can be applied successfully for the diagnostic and surveillance studies of influenza viruses.
Detection time is a crucial factor of any diagnostic assay. RT-LAMP assays were completed in 1 h and results were shown after 30 min of agarose gel electrophoresis. So RT-LAMP assay gave efficient results within one and a half hours of sample processing (RNA extraction) making this assay fast when compared with conventional RT-PCR and real time RT-PCR which gave results within 2–3 h (Gu et al., 2010). Previous studies have reported that by using two accessory loop primers the amplification time can be reduced from 60 min to 30–40 min (Luo et al., 2015, Kubo et al., 2010, Gu et al., 2010, Imai et al., 2007, Peng et al., 2011). For the sake of simplicity and making our reaction more cost effective we stuck to the basic protocol of RT-LAMP assay and use only four primers for amplification which didn’t compromise with sensitivity of the assay. RT-LAMP assay can be modified to detect samples in real-time by using turbidity measurement by turbidimeter as turbidity of reaction increases due to release of pyrophosphates during amplification. Moreover, addition of a florescent dye like calcein or SYBR green in the reaction can be used for direct visualization of the amplification products under UV light thus eliminating the need of gel electrophoresis making the reaction more rapid and less prone to cross-contaminations (Luo et al., 2015, Kubo et al., 2010, Bao et al., 2012, Gu et al., 2010, Peng et al., 2011) .We used SYBR green I dye (Sigma-Aldrich, USA) for direct visualization under UV light and both visual detection and agarose gel electrophoresis produced similar results. This showed that RT-LAMP assay can be applied as a point-of-care test during influenza outbreaks due to its simplicity, high sensitivity and less turn-around time.
In conclusion, this study showed that RT-LAMP assay can be used for detection and surveillance studies during the time of influenza outbreaks as a good alternative for conventional RT-PCR and real-time RT-PCR. RT-LAMP assay is highly cost effective and easy to perform as sophisticated instruments like a thermocycler and fluorescent data analyzing computers (as in real-time RT-PCR) are not needed and the whole reaction can be done in a controlled temperature water bath or dry heating block. The results of the study clearly showed that RT-LAMP assay can be applied successfully to the clinical samples and the results were compatible with that of the WHO’s recommended real-time RT-PCR. The RT-LAMP assay is a simple, rapid, specific, sensitive and cost effective method for detection and subtyping of influenza A viruses and may prove useful in the resource-poor laboratories of the developing countries during influenza outbreaks.
Contributions
VS has designed the study, performed the experiments, analyses and writing, DC participated in designing the study, SK designed the study, performed analyses and writing.
Funding
This work was funded by a major research project grant (40–108/2011) of University Grants Commission (UGC), New Delhi, India.
Ethical clearance
This study was approved by the Human Ethical Committee (letter no. PHY/13/362), Maharshi Dayanand University, Rohtak, Haryana, India and detailed informed consent was taken from each patient before taking swab samples.
Competing interests
None declared.
Acknowledgements
Authors are thankful to Council of Scientific & Industrial Research, New Delhi, India for providing financial support in form of Senior Research Fellowship to Vikrant Sharma, to Department of Microbiology, All India Institute of Medical Sciences (AIIMS), New Delhi, India for providing reference strains of influenza A viruses and to Post Graduate Institute of Medical Sciences (PGIMS), Rohtak, Haryana, India for providing clinical samples for the study.
Contributor Information
Vikrant Sharma, Email: viky.mdu@gmail.com.
Dhruva Chaudhry, Email: drchaudhrydhruva@gmail.com.
Samander Kaushik, Email: samanderkaushik@gmail.com.
References
- Bao H., Wang X., Zhao Y., Sun X., Li Y., Xiong Y. Development of a reverse transcription loop-mediated isothermal amplification method for the rapid detection of avian influenza virus subtype H7. J. Virol. Methods. 2012;179:33–37. doi: 10.1016/j.jviromet.2011.08.023. [DOI] [PubMed] [Google Scholar]
- CDC . 2016. Types of Influenza Viruses.https://www.cdc.gov/flu/about/viruses/types.htm [Google Scholar]
- Carr M.J., Gunson R., Maclean A., Coughlan S., Fitzgerald M., Scully M. Development of a real-time RT-PCR for the detection of swine-lineage influenza A (H1N1) virus infections. J. Clin. Virol. 2009;45:196–199. doi: 10.1016/j.jcv.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.T., Zhang J., Sun D.H., Ma L.N., Liu X.T., Cai X.P. Development of reverse transcription loop-mediated isothermal amplification for rapid detection of H9 avian influenza virus. J. Virol. Methods. 2008;151:200–203. doi: 10.1016/j.jviromet.2008.05.009. [DOI] [PubMed] [Google Scholar]
- Cox N.J., Subbarao K. Influenza. Lancet. 1999;354:1277–1282. doi: 10.1016/S0140-6736(99)01241-6. [DOI] [PubMed] [Google Scholar]
- Cox N.J., Subbarao K. Global epidemiology of influenza: past and present. Annu. Rev. Med. 2000;51:407–421. doi: 10.1146/annurev.med.51.1.407. [DOI] [PubMed] [Google Scholar]
- Ellis J.S., Zambon M.C. Molecular diagnosis of influenza. Rev. Med. Virol. 2002;12:375–389. doi: 10.1002/rmv.370. [DOI] [PubMed] [Google Scholar]
- Ferguson A.K., Olivier S., Genova W.B., Epperson D.R., Smith L. Pathogenesis of influenza D virus in cattle. J. Virol. 2016;90:5636–5642. doi: 10.1128/JVI.03122-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H., Qi X., Li X., Jiang H., Wang Y., Liu F. Rapid and specific detection of H3 swine influenza virus using reverse transcription loop-mediated isothermal amplification method. J. Appl. Microbiol. 2010;108:1145–1154. doi: 10.1111/j.1365-2672.2009.04520.x. [DOI] [PubMed] [Google Scholar]
- Hause B.M., Collin E.A., Liu R., Huang B., Sheng Z., Lu W. Characterization of a novel influenza virus in cattle and swine: proposal for a new genus in the Orthomyxoviridae family. mBio. 2014;5 doi: 10.1128/mBio.00031-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ICTV . 2016. Virus Taxonomy 2016.Release, https://talk.ictvonline.org/taxonomy/ [Google Scholar]
- Imai M., Ninomiya A., Minekawa H., Notomi T., Ishizaki T., Tashiro M. Development of H5-RT-LAMP (loop-mediated isothermal amplification) system for rapid diagnosis of H5 avian influenza virus infection. Vaccine. 2006;24:6679–6682. doi: 10.1016/j.vaccine.2006.05.046. [DOI] [PubMed] [Google Scholar]
- Imai M., Ninomiya A., Minekawa H., Notomi T., Ishizaki T., Tu P.V. Rapid diagnosis of H5N1 avian influenza virus infection by newly developed influenza H5 hemagglutinin gene-specific loop-mediated isothermal amplification method. J. Virol. Methods. 2007;141:173–180. doi: 10.1016/j.jviromet.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Ito M., Watanabe M., Nakagawa N., Ihara T., Okuno Y. Rapid detection and typing of influenza A and B by loop-mediated isothermal amplification: comparison with immunochromatography and virus isolation. J. Virol. Methods. 2006;135:272–275. doi: 10.1016/j.jviromet.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Kim D.K., Poudel B. Tools to detect influenza virus. Yonsei Med. J. 2013;54:560–566. doi: 10.3349/ymj.2013.54.3.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo T., Agoh M., Mai L.Q., Fukushima K., Nishimura H., Yamaguchi A. Development of a reverse transcription-loop-mediated isothermal amplification assay for detection of pandemic (H1N1) 2009 virus as a novel molecular method for diagnosis of pandemic influenza in resource-limited settings. J. Clin. Microbiol. 2010;48:728–735. doi: 10.1128/JCM.01481-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Li X., Mo Z., Jin F., Wang B., Zhao H. Rapid identification of chikungunya and dengue virus by a real-time reverse transcription-loop-mediated isothermal amplification method. Am. J. Trop. Med. Hyg. 2012;87:947–953. doi: 10.4269/ajtmh.2012.11-0721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S., Xie Z., Xie L., Liu J., Xie Z., Deng X. Reverse-transcription, loop-mediated isothermal amplification assay for the sensitive and rapid detection of H10 subtype avian influenza viruses. Virol. J. 2015;12:145–151. doi: 10.1186/s12985-015-0378-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscona A. Neuraminidase inhibitors for influenza. N. Engl. J. Med. 2005;353:1363–1373. doi: 10.1056/NEJMra050740. [DOI] [PubMed] [Google Scholar]
- Nakauchi M., Yoshikawa T., Nakai H., Sugata K., Yoshikawa A., Asano Y. Evaluation of reverse transcription loop-mediated isothermal amplification assays for rapid diagnosis of pandemic influenza A/H1N1 2009 virus. J. Med. Virol. 2011;83:10–15. doi: 10.1002/jmv.21934. [DOI] [PubMed] [Google Scholar]
- Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:e63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parida M.M., Posadas G., Inoue S., Hasebe F., Morita K. Real-time reverse transcription loop-mediated isothermal amplification for rapid detection of West Nile virus. J. Clin. Microbiol. 2004;42:257–263. doi: 10.1128/JCM.42.1.257-263.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parida M.M., Horioke K., Ishida H., Dash P.K., Saxena P., Jana A.M. Rapid detection and differentiation of dengue virus serotypes by a real-time reverse transcription-loop-mediated isothermal amplification assay. J. Clin. Microbiol. 2005;43:2895–2903. doi: 10.1128/JCM.43.6.2895-2903.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parida M.M., Santhosh S.R., Dash P.K., Tripathi N.K., Saxena P., Ambuj S. Development and evaluation of reverse transcription-loop-mediated isothermal amplification assay for rapid and real-time detection of Japanese encephalitis virus. J. Clin. Microbiol. 2006;44:4172–4178. doi: 10.1128/JCM.01487-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parida M.M., Santhosh S.R., Dash P.K., Tripathi N.K., Lakshmi V., Mamidi N. Rapid and real-time detection of chikungunya virus by reverse transcription loop-mediated isothermal amplification assay. J. Clin. Microbiol. 2007;45:351–357. doi: 10.1128/JCM.01734-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parida M.M., Shukla J., Sharma S., Santhosh S.R., Ravi V., Mani R. Development and evaluation of reverse transcription loop-mediated isothermal amplification assay for rapid and real-time detection of the swine-origin influenza A H1N1 virus. J. Mol. Diagn. 2011;13:100–107. doi: 10.1016/j.jmoldx.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y., Xie Z., Liu J., Pang Y., Deng X., Xie Z. Visual detection of H3 subtype avian influenza viruses by reverse transcription loop-mediated isothermal amplification assay. Virol. J. 2011;8:337–346. doi: 10.1186/1743-422X-8-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poddar R. Evaluation of a single step multiplex RT-PCR for influenza virus type and subtype detection in respiratory samples. J. Clin. Lab. Anal. 2002;16:163–166. doi: 10.1002/jcla.10036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon L.L., Leung C.S., Chan K.H., Lee J.H., Yuen K.Y., Guan Y. Detection of human influenza A viruses by loop-mediated isothermal amplification. J. Clin. Microbiol. 2005;43:427–430. doi: 10.1128/JCM.43.1.427-430.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma V., Kaushik S. Comparative analysis of molecular methods for detection of influenza viruses. Br. Microbiol. Res. J. 2016;17:1–10. [Google Scholar]
- Shaw M.L., Palese P. Orthomyxoviridae. In: Knipe D.M., Howley P.M., editors. Fields Virology. 6th Ed. Lippincott Williams & Wilkins Philadelphia; 2013. pp. 1151–1185. [Google Scholar]
- Spackman E., Suarez D.L. Type A influenza virus detection and quantitation by real-time RT-PCR. Methods Mol. Bio. 2008;436:19–26. doi: 10.1007/978-1-59745-279-3_4. [DOI] [PubMed] [Google Scholar]
- Thai H.T.C., Le M.Q., Vuong C.D., Parida M.M., Minekawa H., Notomi T. Development and evaluation of a novel loop-mediated isothermal amplification method for rapid detection of severe acute respiratory syndrome coronavirus. J. Clin. Microbiol. 2004;42:1956–1961. doi: 10.1128/JCM.42.5.1956-1961.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toriniwa H., Komiya T. Rapid detection and quantification of Japanese encephalitis virus by real-time reverse transcription loop-mediated isothermal amplification. Microbiol. Immunol. 2006;50:379–387. doi: 10.1111/j.1348-0421.2006.tb03804.x. [DOI] [PubMed] [Google Scholar]
- WHO . 2012. Weekly Epidemiological Record (WER), Vaccines Against Influenza, WHO Position Paper, November 2012.http://www.who.int/wer/2012/wer8747/en/ [Google Scholar]
- WHO . 2014. WHO Information for Molecular Diagnosis of Influenza Virus–Update. March 2014.http://www.who.int/influenza/gisrs_laboratory/molecular_diagnosis/en/ [Google Scholar]
- WHO . 2016. Influenza (Seasonal) Fact Sheet November 2016.http://www.who.int/mediacentre/factsheets/fs211/en/ [Google Scholar]
- Wang R., Taubenberger J.K. Methods for molecular surveillance of influenza. Expert Rev. Anti. Infect. Ther. 2010;8:517–527. doi: 10.1586/eri.10.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster R.G., Govorkova E.A. Continuing challenges in influenza. Ann. N. Y. Acad. Sci. 2014;1323:115–139. doi: 10.1111/nyas.12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright G., Neumann Y. Orthomyxoviruses. In: Knipe D.M., Howley P.M., editors. Fields Virology. 6th Ed. Lippincott Williams & Wilkins Philadelphia; 2013. pp. 1186–1243. [Google Scholar]
- Zhang J., Feng Y., Hu D., Lv H., Zhu J., Cao M. Rapid and sensitive detection of H7N9 avian influenza virus by use of reverse transcription-loop-mediated isothermal amplification. J. Clin. Microbiol. 2013;51:3760–3764. doi: 10.1128/JCM.01907-13. [DOI] [PMC free article] [PubMed] [Google Scholar]