Abstract
Over 12 years have elapsed since severe acute respiratory syndrome (SARS) triggered the first global alert for coronavirus infections. Virus transmission in humans was quickly halted by public health measures and human infections of SARS coronavirus (SARS-CoV) have not been observed since. However, other coronaviruses still pose a continuous threat to human health, as exemplified by the recent emergence of Middle East respiratory syndrome (MERS) in humans. The work on SARS-CoV widens our knowledge on the epidemiology, pathophysiology and immunology of coronaviruses and may shed light on MERS coronavirus (MERS-CoV). It has been confirmed that T-cell immunity plays an important role in recovery from SARS-CoV infection. Herein, we summarize T-cell immunological studies of SARS-CoV and discuss the potential cross-reactivity of the SARS-CoV-specific immunity against MERS-CoV, which may provide useful recommendations for the development of broad-spectrum vaccines against coronavirus infections.
Keywords: SARS-CoV, MERS-CoV, Vaccine, T-cell, Epitope, Cross-reactivity
Highlights
-
•
T-cell epitopes identified throughout the SARS-CoV proteome may act as candidates for vaccine development.
-
•
Both SARS-CoV and MERS-CoV-recovered donors have had long-lasting memory T-cell immunity.
-
•
The structures of HLA/SARS-CoV-epitopes illuminate the molecular bases of cellular immunogenicity.
-
•
Potential cross-T-cell immune reactivities of SARS-CoV and MERS-CoV benefit vaccine development.
1. Introduction
The successful control of the spread of severe acute respiratory syndrome coronavirus (SARS-CoV) in 2003 facilitated the establishment of an international collaboration system for infectious disease prevention and control (Ksiazek et al., 2003, Rota et al., 2003). During this process, a large number of studies were performed that focused on epidemiology, pathophysiology, immunology, vaccine development and structural studies of SARS-CoV. These studies helped widen knowledge of both SARS-CoV and the whole coronavirus family, as well as promoted methodologies for studying viruses.
Almost ten years after the emergence of SARS, a novel coronavirus termed Middle East respiratory syndrome coronavirus (MERS-CoV) was first reported in the Middle East (Zaki et al., 2012), and this virus was found to be responsible for an outbreak of acute respiratory illness in a public hospital in Zarqa, Jordan during April 2012 (Hijawi et al., 2013). Imported cases have since been reported in France, Germany, Italy, Tunisia, the United States and the United Kingdom (Su et al., 2016, Wong et al., 2015). During 2015, an outbreak of MERS-CoV was reported in South Korea, with 36 deaths in 186 confirmed cases (http://www.who.int/emergencies/mers-cov/en/), in which one patient traveled from South Korea to China, representing the first case of MERS-CoV in China (Su et al., 2015, Wang et al., 2015).
The clinical features of patients infected by MERS-CoV are very similar to SARS, i.e., severe pneumonia (Feng and Gao, 2007, Zaki et al., 2012). SARS-CoV-like viruses have been discovered in insectivorous Rhinolophid bats, and viruses genetically related to MERS-CoV have also been detected in Neoromicia capensis bats from Africa (Corman et al., 2014, Lau et al., 2013, Li et al., 2005b, To et al., 2013). Recently, the bat origins of MERS-CoV were further supported by evidence that bat coronavirus HKU4 also uses the human MERS-CoV receptor CD26 for virus entry (Wang et al., 2014). Additionally, MERS-CoV-like viruses are also widespread in dromedary camels, with sero-epidemiological studies indicating 90% seroprevalence in adult animals (Reusken et al., 2014).
A pivotal role for virus-specific memory T-cells in broad and long-term protection against SARS-CoV infection has been elucidated (Channappanavar et al., 2014, Zhao et al., 2010a). Indeed, the crucial protective role of T-cell immune responses in coronavirus infections has been clearly documented in several animal models, e.g., feline infectious peritonitis virus (FIPV), mouse hepatitis virus (MHV), and avian infectious bronchitis virus (IBV) (Li et al., 2016, Takano et al., 2014, Trujillo et al., 2014). These animal models provide useful references for the study of human infection by SARS-CoV or MERS-CoV. Infection of mice with MHV, a member of the same betacoronavirus group as SARS-CoV and MERS-CoV, was used as an early experimental model to elucidate the role of T-cells in viral clearance and cytotoxicity (Le Prevost et al., 1975) and in T-cell oriented vaccine development (MacNamara et al., 2008, Zust et al., 2007). A primary role for cytotoxic T lymphocytes (CTL) has been demonstrated in protection from MHV and virus clearance (Stohlman et al., 1995).
In this review, we focus on lessons learned from T-cell immunological studies of SARS-CoV, including the immunogenicity of SARS-CoV structural proteins, T-cell epitopes identified, T-cell-oriented vaccine development, and the structural immunology of SARS-CoV based on human leucocyte antigen class I (HLA I)/peptide structures (Hilgenfeld and Peiris, 2013). Based on the knowledge of the T-cell immunity of SARS and recent studies on MERS, the immune correlation and potential T-cell cross-reactivity between SARS-CoV and MERS-CoV are evaluated, which may have implications for vaccine development against human coronavirus infections.
2. Immunogenicity of SARS-CoV and T-cell epitopes
Natural infection with SARS-CoV gives rise to dominant responses against the structural antigens of SARS-CoV in humans and animals (Channappanavar et al., 2014). The spike (S) protein is responsible for both receptor binding and membrane fusion of the virus (Lu et al., 2015), but also acts as a major antigen for both humoral and cellular immunity (Du et al., 2009a). Xu and colleagues detected SARS-CoV-specific T-cell responses from peripheral blood mononuclear cells (PBMCs) of convalescent SARS patients using overlapping peptides covering the whole SARS-CoV proteome (Li et al., 2008). They found that most of the antigenic peptides are located in the structural proteins (especially S protein) rather than non-structural proteins. The biased distribution of T-cell epitopes in the viral proteins may correlate with the different protein synthesis phases during the infection and the diverse abundance of the viral proteins in infected cells or antigen-presenting cells.
The nucleocapsid (N) protein of SARS-CoV can also trigger antibody and T-cell responses in humans, though the neutralizing/protective activities of these antibodies/T-cells still need to be defined (Leung et al., 2004, Lin et al., 2003, Woo et al., 2004). The membrane (M) protein, which is the most abundant protein in the SARS-CoV virion, also acts as a dominant immunogen as revealed by both clustering regions of B-lymphocyte and CTL epitopes (He et al., 2005, Liu et al., 2010b).
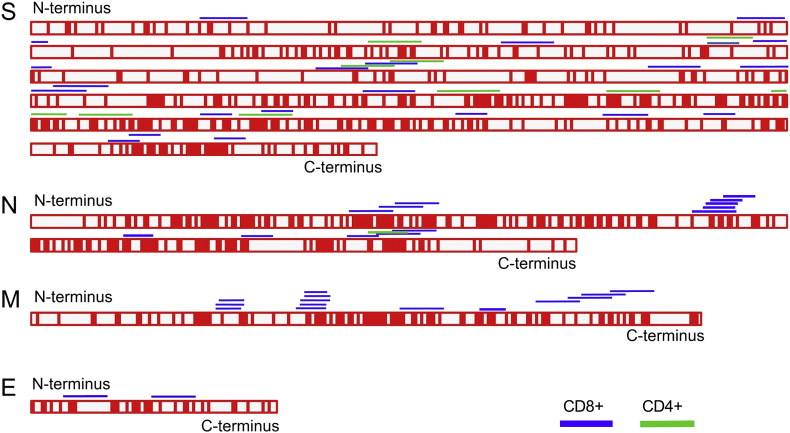
Dozens of T-cell specific epitopes derived from SARS-CoV structural proteins have been identified through diverse strategies in the past years (Table 1 ). We reviewed the reports concerning T-cell specific immunology of SARS-CoV and collected the CD8+ and CD4+ T-cell epitopes defined since the initial SARS outbreak (Fig. 1 ). Among the CD8+ T-cell epitopes (Table 1), most peptides identified thus far are derived from the S protein with HLA-A2 restriction, which is the consequence of the immunodominance of the SARS-CoV S protein and the high coverage of HLA-A2+ populations in different ethnic groups all over the world. HLA-A*1101-, HLA-A*2402-, HLA-B*15-, and HLA-B*4001-restricted T-cell epitopes have also been defined, which can be used to detect T-cell immune responses among populations other than HLA-A2 (Liu et al., 2010b, Oh et al., 2011). Also, several H-2d-, H-2k-, and H-2b-restricted CD4+ T-cell and CD8+ T-cell epitopes have been identified using different mouse strains, which are helpful for SARS-CoV-specific T-cell studies among these animal models and the development of vaccines (Zhao et al., 2007, Zhao et al., 2010a, Zhi et al., 2005). Given that all the CD4+ T cell epitopes identified thus far are derived from the SARS-CoV S protein, further epitope discovery is still needed, especially in other structural proteins.
Table 1.
SARS-CoV-derived T-cell epitopes and their conservation in human coronaviruses. aThe position information is based on the SARS-CoV (strain TJF; GenBank accession no. AY654624) bThe sequences of the corresponding peptides in SARS-CoV, HCoV-OC43 (strain HK04-01; GenBank accession no. JN129834), and HKU1 (strain BJ01-p3; GenBank accession no. KT779555). The variable residues in the peptides compared to MERS-CoV are underlined in bold. cThe references that identified the peptides. dThe references that used the peptides as vaccines or used the peptides to evaluate SARS-related vaccines. eThe positions of HLA-restricted peptides with three or less variable residues between MERS-CoV and any of the three coronaviruses SARS-CoV, HCoV-OC43 and HKU1 are shown in bold.
| Positiona | MERS-CoV | SARS-CoVb | HCoV-OC43 | HKU1 | MHC restriction | Identificationc | Vaccine evaluationd |
|---|---|---|---|---|---|---|---|
| S411-420 | KQSFSNPTCL | KLPDDFMGCV | RIDTTATSCQ | KIDTTSSSCQ | HLA-A*0201 | (Zhou et al., 2006) | (Zhou et al., 2006) |
| S787-795 | LEPVSISTG | ILPDPLKPT | INFSPVLGC | INFKSLVGC | HLA-A*0201 | (Tsao et al., 2006) | (Tsao et al., 2006) |
| S1042-1050 | LYFMHVGYY | VVFLHVTYV | LYFIHFNYV | LLFMHFSYK | HLA-A2 | ||
| S958-966 | SIGDIIQRL | VLNDILSRL | SLQEILSRL | SLQEILSRL | HLA-A*0201 | (Lv et al., 2009) | (Lv et al., 2009) |
| S978-986e | LINGRLTTL | LITGRLQSL | LINGRLTAL | LINGRLTAL | HLA-A2 | (Wang et al., 2004b) | (Zhou et al., 2006), (Kohyama et al., 2009) |
| S1203-1211 | FIAGLVALA | FIAGLIAIV | ICLAGVAML | ISFSFIIFL | HLA-A2 | ||
| S1167-1175 | SLQQVVKAL | RLNEVAKNL | RLQEAIKVL | LIQESIKSL | HLA-A*0201 | (Wang et al., 2004a) | (Wang et al., 2004a), (Zhou et al., 2006) |
| S1174-1182 | ALNESYIDL | NLNESLIDL | VLNHSYINL | SLNNSYINL | HLA-A*0201 | (Doytchinova and Flower, 2003) | |
| S365-374 | TCSQISPAAI | KCYGVSATKL | TCNNIDAAKI | SCNNFDESKI | H-2d | (Huang et al., 2007) | (Huang et al., 2007), (Du et al., 2008), |
| S436-443 | LKYSYINK | YNYKYRYL | WNKRFGFI | WNRRYGFN | H-2b, H-2b | (Zhi et al., 2005) | (Zhao et al., 2010a) |
| S525-532 | VEYSLYGV | VNFNFNGL | VNYDLYGI | VDYDLYGI | H-2d | ||
| S366-374 | CSQISPAAI | CYGVSATKL | CNNIDAAKI | CNNFDESKI | H-2d | ||
| S884-891 | IFYRLNGV | MAYRFNGI | VQYRINGL | VQYRINGL | H-2Kb | (Poh et al., 2009) | (Poh et al., 2009) |
| S1116-1123 | STNLPPPL | NNTVYDPL | PYVMLNTS | PLVYLNHS | H-2Kb | ||
| N216-224 | GAVGGDLLY | GETALALLL | VTPDMADQI | VKPDMADEI | HLA-B*4001 | (Rivino et al., 2013) | |
| N323-332 | DDHGNPVYFL | MEVTPSGTWL | DEPQKDVYEL | DSPVKDVFEL | HLA-B*4001 | ||
| N223-231 | LYLDLLNRL | LLLDRLNQL | QIASLVLAK | EIANLVLAK | HLA-A*0201 | (Tsao et al., 2006) | (Tsao et al., 2006), (Ohno et al., 2009) |
| N227-235 | LLNRLQALE | RLNQLESKV | LVLAKLGKD | LVLAKLGKE | HLA-A*0201 | ||
| N317-325 | GMSQFKLTH | GMSRIGMEV | FGSKLELAK | FGSKLDLVK | HLA-A*0201 | ||
| N220-228 | GDLLYLDLL | LALLLLDRL | MADQIASLV | MADEIANLV | HLA-A*0201 | (Cheung et al., 2007) | (Cheung et al., 2007) |
| N216-225 | GAVGGDLLYL | GETALALLLL | VTPDMADQIA | VKPDMADEIA | HLA-B*4001 | (Oh et al., 2011) | (Oh et al., 2011) |
| N222-231 | LLYLDLLNRL | LLLLDRLNQL | DQIASLVLAK | DEIANLVLAK | HLA-A*0201 | (Ohno et al., 2009) | (Ohno et al., 2009) |
| N266-275 | TKSFNMVQAF | TKQYNVTQAF | NKQCTVQQCF | NKHCNVQQCF | HLA-B*1525 | (Ng et al., 2016) | |
| N346-354 | NYNKWLELL | QFKDNVILL | GFETIMKVL | GFETIMKVL | HLA-A*2402 | (Liu et al., 2010b) | |
| N362-370 | KTFPKKEKK | KTFPPTEPK | QQQDGMMNM | VNSNQNTDS | HLA-A*1101 | (Blicher et al., 2005) | |
| M60-69 | SMALSIFSAV | TLACFVLAAV | TIILTTFNCV | TITLTIFNCF | HLA-A*0201 | (Liu et al., 2010a) | (Liu et al., 2010a) |
| M88-96 | AMMWISYFV | GLMWLSYFV | IIMWIVYFV | IVIWILYFV | HLA-A*0201 | ||
| M147-155 | HLKMAGMHF | HLRMAGHSL | HLYIQGIKL | HLYIQGVKL | HLA-B*1502 | (Ng et al., 2016) | |
| PP1a3709-3717 | AYLVFVTTL | SMWALVISV | LLMLASLFG | LLFITAFLG | HLA-A*0201 | (Kohyama et al., 2009) | (Kohyama et al., 2009) |
| PP1a1775-1787 | VEHTTPWLL | VQQESSFVM | VRFDVPFLI | TKLNVPFLI | HLA-B*1501 | (Roder et al., 2008) | |
| N353-365 | LLEQNIDAYKTFP | LLNKHIDAYKTFP | VLSENLNAYQQQD | VLEENLNAYVNSN | H-2d,HLA-DR2,DR3 | (Zhao et al., 2010a) | (Zhao et al., 2016) |
Fig. 1.
T-cell antigenic peptides in SARS-CoV structural proteins and the conservation of the corresponding sequences in MERS-CoV. The sequences of the four major structural proteins of SARS-CoV were aligned with that of MERS-CoV. Identical residues are denoted in red and variable residues in white. Peptides with T-cell antigenicity are shown along the protein sequence. Peptides with CD8+ T-cell-specific antigenicity and CD4+ T-cell antigenicity are shown in blue and green, respectively.
Based on an efficient T-cell epitope screening platform, with a combination of bioinformatics, cell biology, mouse models and structural biology (Liu et al., 2011b), several HLA-A2-restricted CD8+ T-cell-specific epitopes derived from the SARS-CoV S and M proteins were identified by our group (Zhou et al., 2006). Interestingly, we also defined a clustering region of CTL epitopes within the transmembrane region of the M protein (Fig. 1) (Liu et al., 2011a), which may correlate with the sequence preference by the peptide-processing related proteins, such as transporter associated with antigen processing (TAP), immunoproteasome and TAP binding protein (tapasin) (Huang et al., 2016). The protein transmembrane regions are usually rich in aliphatic amino acids (e.g. Leu, Ile and Val), which are preferred for the primary anchor residues of HLA-A2 peptides (Sharpe et al., 2010). Further work is needed to elucidate whether the M protein is a good candidate antigen for a prophylactic vaccine inducing both cellular and humoral immunogenicity. We also identified the first HLA-A24-restricted immunodominant T-cell epitope from the SARS-CoV N protein (Liu et al., 2010b). Recently, several MERS-CoV-derived H-2Kd-restricted T-cell epitopes have been identified (Liu et al., 2016). T-cell responses to one of these newly defined peptides (peptide 37-1) have a protective effect against MERS-CoV challenge. The protective efficiency of the peptide is depended on an uncommon interaction of Ile5 in peptide 37-1 with Trp73 of the host MHC I H-2Kd. These T-cell epitopes with different HLA-I-restrictions and also the T-cell epitope screening platform may help to understand the T-cell immunogenicity of different proteins from coronaviruses and provide potential candidates for vaccine development.
3. T-cell-oriented vaccine development for SARS-CoV
Well-defined antigenic peptides act as useful agents in the studies of SARS-specific immunity and immunopathogenesis and, more importantly, as candidates for vaccine development. The major antigens in these T-cell-targeting vaccines are focused on the S or N proteins of SARS-CoV.
3.1. General strategies for T-cell based vaccine development
Due to the low immunogenicity of single peptide vaccinations, many different strategies have been developed to elicit effective T-cell responses and efficient protection by T-cell-based vaccines. DNA vaccines encoding N or S gene segments that cover the immunodominant T-cell epitopes have been developed in mouse models. Cheung et al. produced a potential DNA vaccine candidate expressing an antigenic peptide from the SARS-CoV N protein with a single-chain-trimer (peptide-β2m-MHC I) approach that induces SARS-CoV-specific T-cells with cytotoxicity to N protein-expressing cells (Cheung et al., 2007). Recombinant adeno-associated virus is also an ideal carrier for T-cell immunogens of SARS-CoV, inducing strong mucosal immune responses and protective CTL responses (Du et al., 2008). Mammalian CHO cells-expressing segments of the SARS-CoV S protein also induce potent T-cell immune responses and protection against the virus (Du et al., 2009b, Du et al., 2010). Zhao and colleagues observed that enhanced virus-specific CD8+ T-cells in mice by immunization with SARS-CoV peptide-pulsed dendritic cells also result in earlier virus clearance and increased survival (Zhao et al., 2010a). In a Phase I clinical trial, a single-plasmid DNA vaccine encoding the S protein was well tolerated and induced SARS-CoV-specific CD4+ T-cell responses in all vaccinees, as well as CD8+ T-cell responses in ∼20% of individuals (Martin et al., 2008). MHC II-related antigen presentation in professional antigen-presenting cells may be involved in the dominant CD4+ T-cell response of DNA vaccines, which is required for neutralizing antibodies, though cross-priming to stimulate CD8+ T-cells also occurs (Akbari et al., 1999).
3.2. Adjuvants and their beneficial effects
Different adjuvants have been investigated to increase the efficacy of peptide vaccines. Surface-linked liposomal peptide (Kohyama et al., 2009, Ohno et al., 2009), muramyl dipeptide derivative adjuvant (Chen et al., 2010b), and CpG oligodeoxynucleotide (CpG ODN) (Zhao et al., 2010b, Zhao et al., 2011) can augment peptide-specific T-cell responses, some of the which are protective (Du et al., 2010, Du et al., 2009b, Du et al., 2008, Ohno et al., 2009, Zhao et al., 2010a) and durable (Du et al., 2008, Kohyama et al., 2009, Yang et al., 2009). Virus-like particle (VLP) vaccines containing S protein as the dominant immunogen and adjuvanted with aluminum provided protection against mice challenged with SARS-CoV (Liu et al., 2011c, Lokugamage et al., 2008).
These studies demonstrate that T cells play a crucial role in SARS-CoV clearance and elucidate the avenues for vaccine development against other human coronaviruses, including MERS-CoV (Du et al., 2016, Wang et al., 2016a). However, some SARS-CoV vaccines led to the occurrence of Th2-type immunopathology in spite of antibody and protection against infection with SARS-CoV in mice (Tseng et al., 2012). Therefore, the application of a SARS-CoV vaccine in humans should proceed with caution.
4. Human immune responses against SARS-CoV and memory responses
Since the outbreak of SARS-CoV, the kinetics of the primary and memory immune responses after SARS-CoV infection have been investigated in patients and survivors. The milestones of these studies are presented in Fig. 2 .
Fig. 2.
Milestones of research on humoral and T-cell immunity against SARS-CoV and MERS-CoV. A. Studies that evaluate SARS-CoV-specific memory antibody and cellular responses in SARS patients or recovered donors are summarized along the 12-year time axis after the outbreak of the SARS epidemic. Arrowheads point to the time of sampling. B. Studies on human immunity to MERS-CoV and the cross-reactivity of MERS-CoV and SARS-CoV are listed.
4.1. Humoral immunity to SARS-CoV
IgG against N protein can be detected in the sera as early as 4 days after illness onset, based on immunofluorescence assays and ELISA, with most patients seroconverting by day 14 (Hsueh et al., 2004). IgG and neutralizing antibodies peak at 4 months and then progressively decrease over time (Liu et al., 2006). Eighty-nine percent of the recovered patients have detectable IgG antibodies to SARS-CoV at 24 months post-infection as shown by ELISA, while neutralizing antibodies are detectable in an even higher percentage of donors (Liu et al., 2006). Nevertheless, disease severity may also impact the appearance time and magnitude of the antibody responses. Cameron and colleagues reported that SARS-CoV-infected patients with fatal outcomes display deficient antibody production against the S protein compared to non-severe patients (Cameron et al., 2007). During the long term follow-up of SARS survivors, IgG is only detectable in 2 of 23 recovered donors at 6 years after illness onset (Tang et al., 2011), suggesting diminishing levels of memory B-cells against SARS-CoV.
4.2. T-cell immunity to SARS-CoV
Laboratory investigation of clinical patients demonstrated that SARS-CoV-specific T-cells are important for the recognition and clearance of infected cells, particularly in the lungs of infected individuals (Gu et al., 2005). Although whether the memory T-cell response is sufficient to protect from reinfection requires further investigation, it has been suggested that a more robust CTL response contributes to protection against SARS-CoV in mice (Channappanavar et al., 2014, Chen et al., 2010a, Zhao et al., 2010a). Based on a series of T-cell epitopes identified thus far, as well as the overlapping peptide pools covering full-length antigens of SARS-CoV, durable memory T-cell responses against SARS-CoV have been evaluated in recovered patients (Chen et al., 2005, Fan et al., 2009, Peng et al., 2006, Yang et al., 2006, Yang et al., 2007). Unlike waning serum antibody levels in patients, CTL responses against the S and N proteins can still be detected from the PBMCs of recovered SARS patients 1, 2, 4, 6, and even >10 years post-infection (Da Guan et al., 2015, Ng et al., 2016, Oh et al., 2011, Tang et al., 2011). These findings are expected to have implications for disease surveillance and potential cross-reactivity to other coronaviruses.
4.3. Current knowledge of adaptive immune responses to MERS-CoV
Knowledge of adaptive immunity to MERS-CoV is also based on the most recent clinical data and laboratory investigation of specimens from MERS patients in South Korea (Min et al., 2016, Park et al., 2015), China (Da Guan et al., 2015, Wang et al., 2016b), the United States (Kapoor et al., 2014) and the Kingdom of Saudi Arabia (Corman et al., 2016). Seroconversion for most patients occurs during the second and third week after symptom onset (Corman et al., 2016, Park et al., 2015, Wang et al., 2016b). Similar to SARS patients, weak and delayed antibody responses are associated with more severe disease or fatal outcome in patients infected by MERS-CoV (Corman et al., 2016, Min et al., 2016, Park et al., 2015). However, Corman and colleagues (Corman et al., 2016) discovered that the levels of IgG and neutralizing antibodies are weakly and inversely correlated with viral loads in the lower respiratory tract, suggesting that the presence of antibodies does not lead to the elimination of the virus. For MERS-CoV-specific T-cell responses, PBMCs obtained on day 24 after illness onset show a strong specific T-cell response against the MERS-CoV S protein (Da Guan et al., 2015). Interestingly, persistent and gradual increases of lymphocyte responses after symptom onset in MERS patients with or without mild pneumonia may be required for effective immune responses against MERS-CoV, whereas all of the deceased patients displayed rapid drops in their lymphocyte counts (Min et al., 2016). T-cell immunity-associated cytokines IL-12 and IFN-γ levels were lower in a fatal case than in a patient who survived the infection (Faure et al., 2014).
Seven previously unconfirmed individuals tested positive for MERS-CoV-specific antibodies 13 months after a MERS outbreak in a Jordanian hospital, indicating that the antibodies persisted for at least a year in these asymptomatic cases (Al-Abdallat et al., 2014). However, in a longitudinal study of MERS survivors in Jeddah, Saudi Arabia, only two out of nine survivors remained positive for MERS-specific antibodies when tested 18 months after illness onset (Alshukairi et al., 2016). Furthermore, Arabi and colleagues (Arabi et al., 2016) reported that only 4 (36.7%) of 11 healthcare workers who had a history of laboratory-confirmed MERS-CoV infection had detectable MERS-CoV-antibody levels by ELISA a median of 381 days after infection. These data indicate that MERS-CoV-antibody levels decline more rapidly, compared to SARS-CoV survivors. Thus, it is valuable to investigate the features of T-cell immunity against MERS-CoV in survivors, which may shed light on MERS-CoV vaccine development with long-term protection in the future.
5. The structural immunology of SARS-CoV based on the HLA I/peptide structures
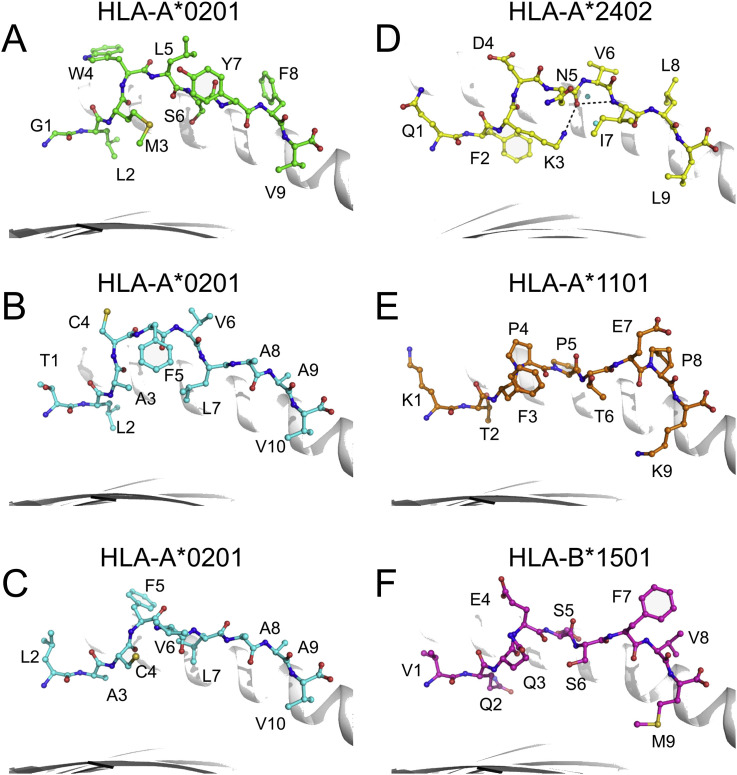
Structural proteome studies of viruses such as SARS-CoV, HIV, and influenza virus have revealed a series of important antibody-recognized epitopes of the major surface antigens and indicate their B-cell-specific antigenic features (Julien et al., 2012, Li et al., 2005a, Zhou et al., 2010). Herein, we summarize the structural characteristics of SARS-CoV-specific T-cell epitopes presented by HLA I molecules and recognized by T-cell receptors, including the structures of three HLA-A*0201, one HLA-A*1101, one HLA-A*2402, and one HLA-B*1501 molecule (Blicher et al., 2005, Liu et al., 2011a, Liu et al., 2010a, Liu et al., 2010b, Roder et al., 2008).
5.1. Structural evidence for T-cell epitope identification
The structural determination of SARS-CoV-derived peptide-HLA I complexes provides corroborative evidence for T-cell epitope identification. Based on the unambiguous electron density of the anchoring residues of the HLA I-bound peptides, we can conclude whether the peptide in question is a typical HLA I-restricted epitope. Similar to other HLA I molecules, the major anchoring residues are in the second (from the N-terminus) and last positions of the peptides in all structures of HLA I complexed with SARS-CoV peptides.
Interestingly, we previously identified two HLA-A2-restricted epitopes, Md3 (TLACFVLAAV) and its C-terminal truncated peptide Md3-C9 (LACFVLAAV) (Liu et al., 2010a, Liu et al., 2011a). Based on the structural determination of the two peptides complexed with HLA-A*0201, we found that they use different residues for the primary anchor at P2 position, though their C-terminal anchors are the same. The Leu2 of peptide Md3 inserts into the B pocket of HLA-A*0201, while the P2 anchor of peptide Md3-C9 is Ala2, which corresponds to Ala3 in Md3. The solvent-exposed conformations of the two peptides are completely distinct, which indicates that these two peptides may function as independent epitopes and correspond to different specific T-cell repertoires (Liu et al., 2014).
5.2. Implications of SARS-CoV epitope-HLA I structure interaction
In the structure of a T-cell epitope from the SARS-CoV N protein complexed with HLA-A*2402, we determined a novel immunodominant peptide presentation strategy (Liu et al., 2010b). In the structure of HLA-A*2402 complexed to peptide N1, the main chain of the central region of N1 exists in a moderately bulged conformation that is stabilized by two intra-chain hydrogen bonds and two water molecules in the peptide binding groove. The featured “A”-shaped conformation of N1 causes the three residues (Asp4, Asn5, and Val6) of the central region to bulge out of the HLA-A*2402 surface for potential T-cell receptor (TCR) docking (Fig. 3 ).
Fig. 3.
Structures of SARS-CoV-derived T-cell peptides presented by HLA I molecules. Conformations of T-cell epitope peptides from SARS-CoV presented by HLA-A*0201 (A-C, PDB codes: 3I6G, 3I6K and 3TO2), HLA-A*2402 (D, PDB code: 3I6L), HLA-A*1101 (E, PDB code: 1X7Q) and HLA-B*1501 (F, PDB code: 3C9N). The peptides are shown as sticks and spheres in different colors. The names and the positions of residues in the peptides are denoted in black letters and numbers. Peptide Md3 (B) and its overlapping peptide Md3-C9 (C) are shown in the similar cyan color, and their residue positions are shown in the corresponding numbers. The peptide N1 (D) forms a bulged conformation that is stabilized by intra-chain hydrogen bonds (dashed black lines) and two water molecules (cyan).
The HLA I-peptide complex structures hold the information necessary for a direct approach to peptide-based vaccine design (Sun et al., 2014). For example, in the structure of the HLA-A*1101-SARS-CoV peptide (Blicher et al., 2005), Thr6 does not make efficient use of the E pocket of the HLA-A*1101 groove, and is thus a potential target for optimization when developing peptide vaccines against SARS-CoV.
The residues in different positions of a peptide play distinct roles in HLA I-binding and TCR docking. Thus, structural studies of HLA I-peptide complexes can have direct visible implications for the T-cell antigenic variability of closely related peptides within different viruses. Based on the structures of T-cell epitopes derived from SARS-CoV, the antigenic variability of MERS-CoV compared to SARS-CoV can be rationally analyzed.
6. Immune correlation of SARS and MERS
6.1. Potential cross-immune reactivity between SARS-CoV and MERS-CoV
Since MERS-CoV belongs to the genus betacoronavirus with SARS-CoV, it is necessary to investigate whether the immune responses induced by the SARS-CoV have cross-reactivity against MERS-CoV. Chan and colleagues conducted a seroprevalence study on archived sera by using indirect immunofluorescence (IF) screening and confirmatory neutralizing antibody tests from 28 SARS patients in Southern China. Anti-SARS-CoV IF and neutralizing antibodies against SARS-CoV were found in the majority (96.4%) of the SARS patients, as expected. Seventeen (60.7%) SARS patients tested positive for MERS-CoV-specific IgG detected by indirect IF, with titers ranging from 1:20 to 1:320, while seven (25%) had low titers (1:20 or less) of MERS-CoV neutralizing antibody (Chan et al., 2013). However, subsequent studies showed the absence of cross-reactivity between SARS-CoV and MERS-CoV (Agnihothram et al., 2014, Du et al., 2013, Meyer et al., 2014, Reusken et al., 2013). Du and colleagues discovered that a series of SARS-CoV receptor binding domain (RBD)-specific monoclonal antibodies (mAbs) or serum polyclonal antibodies against SARS-CoV S-RBD in vaccinated individuals do not cross-react with or neutralize MERS-CoV (Du et al., 2013).
Based on our sequence analysis (Fig. 1), the amino acid identity of the S protein and RBD of MERS-CoV and SARS-CoV is 29.08% and 18.6%, respectively, which may explain the observed low cross-neutralizing activity. However, the protein sequence identity of the S2 region of the S proteins from MERS-CoV and SARS-CoV, which is responsible for virus fusion with cells, is 40.98%, whereas the identities of the N, M and E proteins are 45.03, 41.63 and 37.80%, respectively. This suggests there may be potential cross-T-cell reactivity between MERS-CoV and SARS-CoV.
Analysis of the known CD8+ T-cell epitopes derived from SARS-CoV showed that 14 of the 21 HLA allele-restricted epitopes are located in the S2 region of the S protein, or N and M proteins, which are more conserved (Fig. 1, Table 1). Furthermore, three peptides from the S2 region of the S protein, two from the N protein, and one from the M protein of MERS-CoV only have two or three variable residues when aligned to the corresponding sequence of SARS-CoV. Among these six peptides, Mn2 (GLMWLSYFV) in the M protein (amino acids 88–96) of SARS-CoV was previously identified as an immunodominant T-cell epitope based on a combination of functional and structural strategies (Liu et al., 2010a). The corresponding peptide (AMMWISYFV) in the M protein of MERS-CoV has three different residues: Gly88Ala, Leu89Met, and Leu92Ile. The structure of peptide Mn2 presented by HLA-A*0201 (Fig. 3A) shows that the P1 and P2 residues, Gly88 and Leu89, are located in the A and B pockets of HLA-A*0201, respectively, as the anchor residues. The corresponding residues Ala88 and Met89 in the MERS-CoV-derived peptide can also act as the anchor residues, which may not influence the conformation of the peptide when binding to HLA-A*0201. The solvent-exposed residue Leu92 in peptide Mn2 of SARS-CoV corresponds to an Ile in MERS-CoV. Considering the similar characteristics of Leu and Ile, this change may only have a limited influence on the TCR binding of the peptide. Thus, the peptides such as Mn2 in SARS-CoV and its parallel peptide in MERS-CoV may have a cross-TCR repertoire, which should be experimentally validated. In a recent study, a relatively conserved CD4+ T cell epitope (Fig. 1) was also recognized in SARS-CoV- and MERS-CoV-infected HLA-DR2 and -DR3 transgenic mice, indicating vaccine strategies targeting conserved epitopes may be broadly applicable with different coronaviruses (Zhao et al., 2016).
The conservation of the corresponding peptides among other human coronaviruses, such as (HCoVs) OC43 and HKU1 is shown in Table 1. Some conserved T-cell epitopes exist among different HCoVs. For an instance, peptide S978-986 from SARS-CoV (LITGRLQSL) is an HLA-A2 restricted CD8+ T-cell epitope. The corresponding peptides among MERS-CoV (LINGRLTTL), HCoV OC43 (LINGRLTTL) and HKU1 (LINGRLTTL) have only one variable residue at position 8. For Mn2 (GLMWLSYFV) of SARS-CoV, the corresponding peptides from HCoV OC43 (IIMWIVYFV) and HKU1 (IVIWILYFV) still have the typical HLA-A2-restricted anchoring residues. These data may also indicate that prior infections with OC43 and HKU1 may also cross-protect against MERS-CoV or SARS-CoV, considering the common existence of cross-reactivity between sequence-related virus epitopes (Zhang et al., 2015), and would be an important topic for experimental investigation.
Da Guan and colleagues (Da Guan et al., 2015) have found that healthcare workers who were infected with SARS-CoV 12 years ago still have detectable responses to SARS-CoV S protein, with a low-level of cross-T-cell responses against MERS-CoV S protein. Interestingly, based on PBMCs obtained from a traveler to Guangdong, China, who acquired this disease during the 2015 MERS outbreak in South Korea, a strong T-cell-specific response against full-length MERS-CoV S protein was detected, but not cross-reactivity against the S1 subunit of the SARS-CoV S protein. This may be due to the low levels of conservation in the S1 subunit, as discussed above, and may also correlate with the low efficacy of stimulating T-cell responses by recombinant proteins.
6.2. Influence of SARS-CoV and MERS-CoV on immunopathogenesis through interaction with their receptors
As a functional SARS-CoV receptor, ACE2 plays a critical role in SARS-CoV-induced lung injury (Kuba et al., 2005). The injection of SARS-CoV S protein into wild-type mice worsens acute lung failure in vivo by both blocking the renin-angiotensin pathway and down-regulation of ACE2 expression. Further, significant pathological features appear in the mice, such as changes in the lung parenchyma and increased lung edema. In contrast, treatment with S protein did not affect the severity of lung failure in Ace2 knockout mice, which indicates that the effect of S protein on acute lung injury is ACE2-specific.
The MERS-CoV receptor is dipeptidyl peptidase 4 (DPP4), also named CD26 (Raj et al., 2013). The interaction of CD26 with its in vivo natural ligand adenosine deaminase (ADA) plays a crucial role in glucose metabolism and likely in T-cell activation, chemotaxis modulation, cell adhesion, and apoptosis (Morimoto and Schlossman, 1998). Our recent structural study of the RBD of MERS-CoV S protein complexed with CD26 indicates potential competitive binding of MERS-CoV S protein to CD26 instead of ADA (Lu et al., 2013), which implies the influence of MERS-CoV infection on the pathogenesis via its receptor CD26, similar to ACE2 in SARS-CoV infection.
Recently, Chu and colleagues demonstrated that MERS-CoV efficiently infects T-cells from the peripheral blood and from human lymphoid organs, including the spleen and tonsils, and induces apoptosis in T-cells (Chu et al., 2016). However, whether the interaction of MERS-CoV S protein and its cellular receptor CD26 is involved in this process requires further exploration. Indeed, the efficacy of antiviral therapies based on mAbs against MERS-CoV (Corti et al., 2015, Li et al., 2015, Tang et al., 2014) may be related to both the direct neutralization of the virions and the blockage of the intervention of functional CD26 by the S protein of the virus (Li et al., 2015).
6.3. Similar immune antagonism strategies of SARS-CoV and MERS-CoV
Pathogenesis studies of SARS-CoV have elucidated diverse strategies used by the virus to evade and block host immune responses, facilitating infection and transmission. A SARS mouse model demonstrates that lethal pneumonia is contributed by dysregulated type I interferon and inflammatory monocyte-macrophage responses (Channappanavar et al., 2016). Recent studies indicate that MERS-CoV has also evolved specific interferon antagonists to counteract the innate immune response (Niemeyer et al., 2013, Yang et al., 2015, Zhou et al., 2014). Serum IFN-α levels were not elevated in a fatal case as compared to a patient who recovered from the infection (Faure et al., 2014).
Similar strategies were developed to evade/inhibit host innate immune functions in both SARS-CoV and MERS-CoV (Vijay and Perlman, 2016). Nsp1 inhibits interferon signaling in SARS-CoV-infected cells by inhibiting phosphorylation of STAT1 (Jauregui et al., 2013) and inhibiting host gene expression by inactivating the translation activity of the ribosomes. Nsp3 interacts with IRF3, inhibiting its phosphorylation, dimerization, and nuclear translocation (Chen et al., 2014). Nsp16 renders viral RNA indistinguishable from host T-cell RNA by effecting 2′-O methylation (Frieman et al., 2010, Menachery et al., 2014). N protein inhibits NFκB signaling and PKR function (Kopecky-Bromberg et al., 2007). The similarity of the immune evasion strategies shared by SARS-CoV and MERS-CoV may indicate common therapeutic targets for drug development.
7. Perspective
Thus far, MERS-CoV vaccines have been shown to provide efficacious protection in animal models, though none of the vaccines developed has been tested in human clinical trials. Major strategies for vaccine development are focused on the elicitation of serum antibodies against the major antigen (S protein) of MERS-CoV (Zhang et al., 2014). Further, passive immunotherapy using convalescent phase human plasma is being considered in MERS patients after its success in animal models (Zhao et al., 2015). However, studies also demonstrate that MERS-CoV S protein-derived vaccines induce specific CD8+ T-cell and virus-neutralizing antibodies, which could contribute to complete protection against MERS-CoV in animal models (Lan et al., 2014, Volz et al., 2015). Based on the investigations of immune memory against SARS-CoV in follow-up studies of recovered patients (discussed above), T-cell responses can provide robust long-term memory and possess a considerable potential for cross-reactivity with heterotypic coronaviruses. Thus, vaccines combining both cellular and humoral responses should be considered for coronavirus prevention. The similarities between the immunopathogenesis of SARS-CoV and MERS-CoV through interaction with cell receptors and blockage of host innate immune responses also point to potential therapeutic targets in patients infected with other pathogenic coronaviruses.
Financial supports
This work was supported by grants from the China Mega-Project on Infectious Disease Prevention (Grants 2016ZX10004222-003, 2014ZX10004001-006 and 2013ZX10004608-002), the National 973 Project of the China Ministry of Science and Technology (Grant 2015CB910500 and 2011CB504700), and the National Natural Science Foundation of China (Grants 81401312, 31390432 and 81373141). The funders had no role in the study design, data collection and interpretation, or the decision to submit the work for publication. G.F.G. is a leading principal investigator of the National Natural Science Foundation of China Innovative Research Group (Grant 81321063).
Conflict of interest
No conflicts of interest.
Acknowledgements
We are grateful to Ms. Xiaoying Liang for her kind and professional assistance with the preparation of the figures.
Contributor Information
William J. Liu, Email: liujun@ivdc.chinacdc.cn.
George F. Gao, Email: gaofu@chinacdc.cn.
References
- Agnihothram S., Gopal R., Yount B.L., Jr., Donaldson E.F., Menachery V.D., Graham R.L., Scobey T.D., Gralinski L.E., Denison M.R., Zambon M., Baric R.S. Evaluation of serologic and antigenic relationships between Middle Eastern respiratory syndrome coronavirus and other coronaviruses to develop vaccine platforms for the rapid response to emerging coronaviruses. J. Infect. Dis. 2014;209:995–1006. doi: 10.1093/infdis/jit609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbari O., Panjwani N., Garcia S., Tascon R., Lowrie D., Stockinger B. DNA vaccination: transfection and activation of dendritic cells as key events for immunity. J. Exp. Med. 1999;189:169–178. doi: 10.1084/jem.189.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Abdallat M.M., Payne D.C., Alqasrawi S., Rha B., Tohme R.A., Abedi G.R., Al Nsour M., Iblan I., Jarour N., Farag N.H., Haddadin A., Al-Sanouri T., Tamin A., Harcourt J.L., Kuhar D.T., Swerdlow D.L., Erdman D.D., Pallansch M.A., Haynes L.M., Gerber S.I., Jordan M.-C.I.T. Hospital-associated outbreak of Middle East respiratory syndrome coronavirus: a serologic, epidemiologic, and clinical description. Clin. Infect. Dis. 2014;59:1225–1233. doi: 10.1093/cid/ciu359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alshukairi A.N., Khalid I., Ahmed W.A., Dada A.M., Bayumi D.T., Malic L.S., Althawadi S., Ignacio K., Alsalmi H.S., Al-Abdely H.M., Wali G.Y., Qushmaq I.A., Alraddadi B.M., Perlman S. Antibody response and disease severity in healthcare worker MERS survivors. Emerg. Infect. Dis. 2016;22(6) doi: 10.3201/eid2206.160010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabi Y.M., Hajeer A.H., Luke T., Raviprakash K., Balkhy H., Johani S., Al-Dawood A., Al-Qahtani S., Al-Omari A., Al-Hameed F., Hayden F.G., Fowler R., Bouchama A., Shindo N., Al-Khairy K., Carson G., Taha Y., Sadat M., Alahmadi M. Feasibility of using convalescent plasma immunotherapy for MERS-CoV infection, Saudi Arabia. Emerg. Infect. Dis. 2016;22:1554–1561. doi: 10.3201/eid2209.151164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blicher T., Kastrup J.S., Buus S., Gajhede M. High-resolution structure of HLA-A*1101 in complex with SARS nucleocapsid peptide. Acta Crystallogr. D. Biol. Crystallogr. 2005;61:1031–1040. doi: 10.1107/S0907444905013090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron M.J., Ran L., Xu L., Danesh A., Bermejo-Martin J.F., Cameron C.M., Muller M.P., Gold W.L., Richardson S.E., Poutanen S.M., Willey B.M., DeVries M.E., Fang Y., Seneviratne C., Bosinger S.E., Persad D., Wilkinson P., Greller L.D., Somogyi R., Humar A., Keshavjee S., Louie M., Loeb M.B., Brunton J., McGeer A.J., Canadian S.R.N., Kelvin D.J. Interferon-mediated immunopathological events are associated with atypical innate and adaptive immune responses in patients with severe acute respiratory syndrome. J. Virol. 2007;81:8692–8706. doi: 10.1128/JVI.00527-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K.H., Chan J.F., Tse H., Chen H., Lau C.C., Cai J.P., Tsang A.K., Xiao X., To K.K., Lau S.K., Woo P.C., Zheng B.J., Wang M., Yuen K.Y. Cross-reactive antibodies in convalescent SARS patients' sera against the emerging novel human coronavirus EMC (2012) by both immunofluorescent and neutralizing antibody tests. J. Infect. Dis. 2013;67:130–140. doi: 10.1016/j.jinf.2013.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channappanavar R., Fehr A.R., Vijay R., Mack M., Zhao J., Meyerholz D.K., Perlman S. Dysregulated type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-infected mice. Cell Host Microbe. 2016;19:181–193. doi: 10.1016/j.chom.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channappanavar R., Fett C., Zhao J., Meyerholz D.K., Perlman S. Virus-specific memory CD8 T cells provide substantial protection from lethal severe acute respiratory syndrome coronavirus infection. J. Virol. 2014;88:11034–11044. doi: 10.1128/JVI.01505-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Hou J., Jiang X., Ma S., Meng M., Wang B., Zhang M., Zhang M., Tang X., Zhang F., Wan T., Li N., Yu Y., Hu H., Yang R., He W., Wang X., Cao X. Response of memory CD8+ T cells to severe acute respiratory syndrome (SARS) coronavirus in recovered SARS patients and healthy individuals. J. Immunol. 2005;175:591–598. doi: 10.4049/jimmunol.175.1.591. [DOI] [PubMed] [Google Scholar]
- Chen J., Lau Y.F., Lamirande E.W., Paddock C.D., Bartlett J.H., Zaki S.R., Subbarao K. Cellular immune responses to severe acute respiratory syndrome coronavirus (SARS-CoV) infection in senescent BALB/c mice: CD4+ T cells are important in control of SARS-CoV infection. J. Virol. 2010;84:1289–1301. doi: 10.1128/JVI.01281-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Yang X., Zheng Y., Yang Y., Xing Y., Chen Z. SARS coronavirus papain-like protease inhibits the type I interferon signaling pathway through interaction with the STING-TRAF3-TBK1 complex. Protein Cell. 2014;5:369–381. doi: 10.1007/s13238-014-0026-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.Z., Liu G., Senju S., Wang Q., Irie A., Haruta M., Matsui M., Yasui F., Kohara M., Nishimura Y. Identification of SARS-COV spike protein-derived and HLA-A2-restricted human CTL epitopes by using a new muramyl dipeptidederivative adjuvant. Int. J. Immunopathol. Pharmacol. 2010;23:165–177. doi: 10.1177/039463201002300115. [DOI] [PubMed] [Google Scholar]
- Cheung Y.K., Cheng S.C., Sin F.W., Chan K.T., Xie Y. Induction of T-cell response by a DNA vaccine encoding a novel HLA-A*0201 severe acute respiratory syndrome coronavirus epitope. Vaccine. 2007;25:6070–6077. doi: 10.1016/j.vaccine.2007.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H., Zhou J., Wong B.H., Li C., Chan J.F., Cheng Z.S., Yang D., Wang D., Lee A.C., Li C., Yeung M.L., Cai J.P., Chan I.H., Ho W.K., To K.K., Zheng B.J., Yao Y., Qin C., Yuen K.Y. Middle East respiratory syndrome coronavirus efficiently infects human primary T Lymphocytes and activates the extrinsic and intrinsic apoptosis pathways. J. Infect. Dis. 2016;213:904–914. doi: 10.1093/infdis/jiv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Albarrak A.M., Omrani A.S., Albarrak M.M., Farah M.E., Almasri M., Muth D., Sieberg A., Meyer B., Assiri A.M., Binger T., Steinhagen K., Lattwein E., Al-Tawfiq J., Muller M.A., Drosten C., Memish Z.A. Viral shedding and antibody response in 37 patients with Middle East respiratory syndrome coronavirus infection. Clin. Infect. Dis. 2016;62:477–483. doi: 10.1093/cid/civ951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Ithete N.L., Richards L.R., Schoeman M.C., Preiser W., Drosten C., Drexler J.F. Rooting the phylogenetic tree of Middle East respiratory syndrome coronavirus by characterization of a conspecific virus from an African bat. J. Virol. 2014;88:11297–11303. doi: 10.1128/JVI.01498-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti D., Zhao J., Pedotti M., Simonelli L., Agnihothram S., Fett C., Fernandez-Rodriguez B., Foglierini M., Agatic G., Vanzetta F., Gopal R., Langrish C.J., Barrett N.A., Sallusto F., Baric R.S., Varani L., Zambon M., Perlman S., Lanzavecchia A. Prophylactic and postexposure efficacy of a potent human monoclonal antibody against MERS coronavirus. Proc. Natl. Acad. Sci. U. S. A. 2015;112:10473–10478. doi: 10.1073/pnas.1510199112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Guan W., Mok C.K., Chen Z.L., Feng L.Q., Li Z.T., Huang J.C., Ke C.W., Deng X., Ling Y., Wu S.G., Niu X.F., Perera R.A., Da Xu Y., Zhao J., Zhang L.Q., Li Y.M., Chen R.C., Peiris M., Chen L., Zhong N.S. Characteristics of traveler with Middle East respiratory syndrome, China, 2015. Emerg. Infect. Dis. 2015;21:2278–2280. doi: 10.3201/eid2112.151232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doytchinova I., Flower D. The HLA-A2-supermotif: a QSAR definition. Organ. Biomol. Chem. 2003;1:2648–2654. doi: 10.1039/b300707c. [DOI] [PubMed] [Google Scholar]
- Du L., He Y., Zhou Y., Liu S., Zheng B.J., Jiang S. The spike protein of SARS-CoV–a target for vaccine and therapeutic development. Nat. Rev. Microbiol. 2009;7:226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., Ma C., Jiang S. Antibodies induced by receptor-binding domain in spike protein of SARS-CoV do not cross-neutralize the novel human coronavirus hCoV-EMC. J. Infect. 2013;67:348–350. doi: 10.1016/j.jinf.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., Tai W., Zhou Y., Jiang S. Vaccines for the prevention against the threat of MERS-CoV. Expert Rev. Vac. 2016;15:1123–1134. doi: 10.1586/14760584.2016.1167603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., Zhao G., Chan C.C., Li L., He Y., Zhou Y., Zheng B.J., Jiang S. A 219-mer CHO-expressing receptor-binding domain of SARS-CoV S protein induces potent immune responses and protective immunity. Viral Immunol. 2010;23:211–219. doi: 10.1089/vim.2009.0090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., Zhao G., Li L., He Y., Zhou Y., Zheng B.J., Jiang S. Antigenicity and immunogenicity of SARS-CoV S protein receptor-binding domain stably expressed in CHO cells. Biochem. Biophys. Res. Commun. 2009;384:486–490. doi: 10.1016/j.bbrc.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., Zhao G., Lin Y., Sui H., Chan C., Ma S., He Y., Jiang S., Wu C., Yuen K.Y., Jin D.Y., Zhou Y., Zheng B.J. Intranasal vaccination of recombinant adeno-associated virus encoding receptor-binding domain of severe acute respiratory syndrome coronavirus (SARS-CoV) spike protein induces strong mucosal immune responses and provides long-term protection against SARS-CoV infection. J. Immunol. 2008;180:948–956. doi: 10.4049/jimmunol.180.2.948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y.Y., Huang Z.T., Li L., Wu M.H., Yu T., Koup R.A., Bailer R.T., Wu C.Y. Characterization of SARS-CoV-specific memory T cells from recovered individuals 4 years after infection. Arch. Virol. 2009;154:1093–1099. doi: 10.1007/s00705-009-0409-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure E., Poissy J., Goffard A., Fournier C., Kipnis E., Titecat M., Bortolotti P., Martinez L., Dubucquoi S., Dessein R., Gosset P., Mathieu D., Guery B. Distinct immune response in two MERS-CoV-infected patients: can we go from bench to bedside? PloS One. 2014;9:e88716. doi: 10.1371/journal.pone.0088716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., Gao G.F. Towards our understanding of SARS-CoV, an emerging and devastating but quickly conquered virus. Comp. Immunol. Microbiol. Infect. Dis. 2007;30:309–327. doi: 10.1016/j.cimid.2007.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieman M.B., Chen J., Morrison T.E., Whitmore A., Funkhouser W., Ward J.M., Lamirande E.W., Roberts A., Heise M., Subbarao K., Baric R.S. SARS-CoV pathogenesis is regulated by a STAT1 dependent but a type I, II and III interferon receptor independent mechanism. PLoS Pathog. 2010;6:e1000849. doi: 10.1371/journal.ppat.1000849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J., Gong E., Zhang B., Zheng J., Gao Z., Zhong Y., Zou W., Zhan J., Wang S., Xie Z., Zhuang H., Wu B., Zhong H., Shao H., Fang W., Gao D., Pei F., Li X., He Z., Xu D., Shi X., Anderson V.M., Leong A.S. Multiple organ infection and the pathogenesis of SARS. J. Exp. Med. 2005;202:415–424. doi: 10.1084/jem.20050828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Zhou Y., Siddiqui P., Niu J., Jiang S. Identification of immunodominant epitopes on the membrane protein of the severe acute respiratory syndrome-associated coronavirus. J. Clin. Microbiol. 2005;43:3718–3726. doi: 10.1128/JCM.43.8.3718-3726.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijawi B., Abdallat M., Sayaydeh A., Alqasrawi S., Haddadin A., Jaarour N., Alsheikh S., Alsanouri T. Novel coronavirus infections in Jordan, April 2012: epidemiological findings from a retrospective investigation. East Mediterr Health J. 2013;19(Suppl. 1):S12–S18. [PubMed] [Google Scholar]
- Hilgenfeld R., Peiris M. From SARS to MERS: 10 years of research on highly pathogenic human coronaviruses. Antivir. Res. 2013;100:286–295. doi: 10.1016/j.antiviral.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsueh P.R., Huang L.M., Chen P.J., Kao C.L., Yang P.C. Chronological evolution of IgM, IgA, IgG and neutralisation antibodies after infection with SARS-associated coronavirus. Clin. Microbiol. Infect. 2004;10:1062–1066. doi: 10.1111/j.1469-0691.2004.01009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Cao Y., Du J., Bu X., Ma R., Wu C. Priming with SARS CoV S DNA and boosting with SARS CoV S epitopes specific for CD4+ and CD8+ T cells promote cellular immune responses. Vaccine. 2007;25:6981–6991. doi: 10.1016/j.vaccine.2007.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M., Zhang W., Guo J., Wei X., Phiwpan K., Zhang J., Zhou X. Improved transgenic mouse model for studying HLA class I antigen presentation. Sci. Rep. 2016;6:33612. doi: 10.1038/srep33612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jauregui A.R., Savalia D., Lowry V.K., Farrell C.M., Wathelet M.G. Identification of residues of SARS-CoV nsp1 that differentially affect inhibition of gene expression and antiviral signaling. PloS One. 2013;8:e62416. doi: 10.1371/journal.pone.0062416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien J.-P., Lee P.S., Wilson I.A. Structural insights into key sites of vulnerability on HIV-1 Env and influenza HA. Immunol. Rev. 2012;250:180–198. doi: 10.1111/imr.12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor M., Pringle K., Kumar A., Dearth S., Liu L., Lovchik J., Perez O., Pontones P., Richards S., Yeadon-Fagbohun J., Breakwell L., Chea N., Cohen N.J., Schneider E., Erdman D., Haynes L., Pallansch M., Tao Y., Tong S., Gerber S., Swerdlow D., Feikin D.R. Clinical and laboratory findings of the first imported case of Middle East respiratory syndrome coronavirus to the United States. Clin. Infect. Dis. 2014;59:1511–1518. doi: 10.1093/cid/ciu635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohyama S., Ohno S., Suda T., Taneichi M., Yokoyama S., Mori M., Kobayashi A., Hayashi H., Uchida T., Matsui M. Efficient induction of cytotoxic T lymphocytes specific for severe acute respiratory syndrome (SARS)-associated coronavirus by immunization with surface-linked liposomal peptides derived from a non-structural polyprotein 1a. Antivir. Res. 2009;84:168–177. doi: 10.1016/j.antiviral.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopecky-Bromberg S.A., Martinez-Sobrido L., Frieman M., Baric R.A., Palese P. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J. Virol. 2007;81:548–557. doi: 10.1128/JVI.01782-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A.E., Humphrey C.D., Shieh W.J., Guarner J., Paddock C.D., Rota P., Fields B., DeRisi J., Yang J.Y., Cox N., Hughes J.M., LeDuc J.W., Bellini W.J., Anderson L.J., Group S.W. A novel coronavirus associated with severe acute respiratory syndrome. New Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- Kuba K., Imai Y., Rao S., Gao H., Guo F., Guan B., Huan Y., Yang P., Zhang Y., Deng W., Bao L., Zhang B., Liu G., Wang Z., Chappell M., Liu Y., Zheng D., Leibbrandt A., Wada T., Slutsky A.S., Liu D., Qin C., Jiang C., Penninger J.M. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat. Med. 2005;11:875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J., Deng Y., Chen H., Lu G., Wang W., Guo X., Lu Z., Gao G.F., Tan W. Tailoring subunit vaccine immunity with adjuvant combinations and delivery routes using the Middle East respiratory coronavirus (MERS-CoV) receptor-binding domain as an antigen. PloS One. 2014;9:e112602. doi: 10.1371/journal.pone.0112602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.K., Li K.S., Tsang A.K., Lam C.S., Ahmed S., Chen H., Chan K.H., Woo P.C., Yuen K.Y. Genetic characterization of Betacoronavirus lineage C viruses in bats reveals marked sequence divergence in the spike protein of pipistrellus bat coronavirus HKU5 in Japanese pipistrelle: implications for the origin of the novel Middle East respiratory syndrome coronavirus. J. Virol. 2013;87:8638–8650. doi: 10.1128/JVI.01055-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Prevost C., Levy-Leblond E., Virelizier J.L., Dupuy J.M. Immunopathology of mouse hepatitis virus type 3 infection. Role of humoral and cell-mediated immunity in resistance mechanisms. J. Immunol. 1975;114:221–225. [PubMed] [Google Scholar]
- Leung D.T., Tam F.C., Ma C.H., Chan P.K., Cheung J.L., Niu H., Tam J.S., Lim P.L. Antibody response of patients with severe acute respiratory syndrome (SARS) targets the viral nucleocapsid. J. Infect. Dis. 2004;190:379–386. doi: 10.1086/422040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C.K., Wu H., Yan H., Ma S., Wang L., Zhang M., Tang X., Temperton N.J., Weiss R.A., Brenchley J.M., Douek D.C., Mongkolsapaya J., Tran B.H., Lin C.L., Screaton G.R., Hou J.L., McMichael A.J., Xu X.N. T cell responses to whole SARS coronavirus in humans. J. Immunol. 2008;181:5490–5500. doi: 10.4049/jimmunol.181.8.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Li W., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- Li H., Wang Y., Han Z., Wang Y., Liang S., Jiang L., Hu Y., Kong X., Liu S. Recombinant duck enteritis viruses expressing major structural proteins of the infectious bronchitis virus provide protection against infectious bronchitis in chickens. Antivir. Res. 2016;130:19–26. doi: 10.1016/j.antiviral.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H., Wang H., Crameri G., Hu Z., Zhang H., Zhang J., McEachern J., Field H., Daszak P., Eaton B.T., Zhang S., Wang L.F. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- Li Y., Wan Y., Liu P., Zhao J., Lu G., Qi J., Wang Q., Lu X., Wu Y., Liu W., Zhang B., Yuen K.Y., Perlman S., Gao G.F., Yan J. A humanized neutralizing antibody against MERS-CoV targeting the receptor-binding domain of the spike protein. Cell Res. 2015;25:1237–1249. doi: 10.1038/cr.2015.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., Shen X., Yang R.F., Li Y.X., Ji Y.Y., He Y.Y., Shi M.D., Lu W., Shi T.L., Wang J., Wang H.X., Jiang H.L., Shen J.H., Xie Y.H., Wang Y., Pei G., Shen B.F., Wu J.R., Sun B. Identification of an epitope of SARS-coronavirus nucleocapsid protein. Cell Res. 2003;13:141–145. doi: 10.1038/sj.cr.7290158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Qi J., Gao F., Yan J., Gao G.F. Functional and structural definition of a clustering region of HLA-A2-restricted cytotoxic T lymphocyte epitopes. Sci. Tech. Rev. China. 2011;29:8. [Google Scholar]
- Liu J., Sun Y., Qi J., Chu F., Wu H., Gao F., Li T., Yan J., Gao G.F. The membrane protein of severe acute respiratory syndrome coronavirus acts as a dominant immunogen revealed by a clustering region of novel functionally and structurally defined cytotoxic T-lymphocyte epitopes. J. Infect. Dis. 2010;202:1171–1180. doi: 10.1086/656315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Wu P., Gao F., Qi J., Kawana-Tachikawa A., Xie J., Vavricka C.J., Iwamoto A., Li T., Gao G.F. Novel immunodominant peptide presentation strategy: a featured HLA-A*2402-restricted cytotoxic T-lymphocyte epitope stabilized by intrachain hydrogen bonds from severe acute respiratory syndrome coronavirus nucleocapsid protein. J. Virol. 2010;84:11849–11857. doi: 10.1128/JVI.01464-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Zhang S., Tan S., Zheng B., Gao G.F. Revival of the identification of cytotoxic T-lymphocyte epitopes for immunological diagnosis, therapy and vaccine development. Exp. Biol. Med. 2011;236:253–267. doi: 10.1258/ebm.2010.010278. [DOI] [PubMed] [Google Scholar]
- Liu P., Liu D., Yang X., Gao J., Chen Y., Xiao X., Liu F., Zou J., Wu J., Ma J., Zhao F., Zhou X., Gao G.F., Zhu B. Characterization of human alphabetaTCR repertoire and discovery of D-D fusion in TCRbeta chains. Protein Cell. 2014;5:603–615. doi: 10.1007/s13238-014-0060-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Fontanet A., Zhang P.H., Zhan L., Xin Z.T., Baril L., Tang F., Lv H., Cao W.C. Two-year prospective study of the humoral immune response of patients with severe acute respiratory syndrome. J. Infect. Dis. 2006;193:792–795. doi: 10.1086/500469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W.J., Lan J., Liu K., Deng Y., Yao Y., Wu S., Chen H., Bao L., Zhang H., Zhao M., Wang Q., Han L., Chai Y., Qi J., Zhao J., Meng S., Qin C., Gao G.F., Tan W. Protective T-cell responses featured by concordant recognition of MERS-CoV-derived CD8+ T-cell epitopes and host MHC. J. Immunol. 2016 doi: 10.4049/jimmunol.1601542. (in press) [DOI] [PubMed] [Google Scholar]
- Liu Y.V., Massare M.J., Barnard D.L., Kort T., Nathan M., Wang L., Smith G. Chimeric severe acute respiratory syndrome coronavirus (SARS-CoV) S glycoprotein and influenza matrix 1 efficiently form virus-like particles (VLPs) that protect mice against challenge with SARS-CoV. Vaccine. 2011;29:6606–6613. doi: 10.1016/j.vaccine.2011.06.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokugamage K.G., Yoshikawa-Iwata N., Ito N., Watts D.M., Wyde P.R., Wang N., Newman P., Kent Tseng C.T., Peters C.J., Makino S. Chimeric coronavirus-like particles carrying severe acute respiratory syndrome coronavirus (SCoV) S protein protect mice against challenge with SCoV. Vaccine. 2008;26:797–808. doi: 10.1016/j.vaccine.2007.11.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G., Hu Y., Wang Q., Qi J., Gao F., Li Y., Zhang Y., Zhang W., Yuan Y., Bao J., Zhang B., Shi Y., Yan J., Gao G.F. Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26. Nature. 2013;500:227–231. doi: 10.1038/nature12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G., Wang Q., Gao G.F. Bat-to-human: spike features determining 'host jump' of coronaviruses SARS-CoV, MERS-CoV, and beyond. Trends Microbiol. 2015;23:468–478. doi: 10.1016/j.tim.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv Y., Ruan Z., Wang L., Ni B., Wu Y. Identification of a novel conserved HLA-A*0201-restricted epitope from the spike protein of SARS-CoV. BMC Immunol. 2009;10:61. doi: 10.1186/1471-2172-10-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacNamara K.C., Bender S.J., Chua M.M., Watson R., Weiss S.R. Priming of CD8+ T cells during central nervous system infection with a murine coronavirus is strain dependent. J. Virol. 2008;82:6150–6160. doi: 10.1128/JVI.00106-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin J.E., Louder M.K., Holman L.A., Gordon I.J., Enama M.E., Larkin B.D., Andrews C.A., Vogel L., Koup R.A., Roederer M., Bailer R.T., Gomez P.L., Nason M., Mascola J.R., Nabel G.J., Graham B.S., Team V.R.C.S. A SARS DNA vaccine induces neutralizing antibody and cellular immune responses in healthy adults in a Phase I clinical trial. Vaccine. 2008;26:6338–6343. doi: 10.1016/j.vaccine.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., Yount B.L., Jr., Josset L., Gralinski L.E., Scobey T., Agnihothram S., Katze M.G., Baric R.S. Attenuation and restoration of severe acute respiratory syndrome coronavirus mutant lacking 2'-o-methyltransferase activity. J. Virol. 2014;88:4251–4264. doi: 10.1128/JVI.03571-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer B., Drosten C., Muller M.A. Serological assays for emerging coronaviruses: challenges and pitfalls. Virus Res. 2014;194:175–183. doi: 10.1016/j.virusres.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min C.K., Cheon S., Ha N.Y., Sohn K.M., Kim Y., Aigerim A., Shin H.M., Choi J.Y., Inn K.S., Kim J.H., Moon J.Y., Choi M.S., Cho N.H., Kim Y.S. Comparative and kinetic analysis of viral shedding and immunological responses in MERS patients representing a broad spectrum of disease severity. Sci. Rep. 2016;6:25359. doi: 10.1038/srep25359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto C., Schlossman S.F. The structure and function of CD26 in the T-cell immune response. Immunol. Rev. 1998;161:55–70. doi: 10.1111/j.1600-065x.1998.tb01571.x. [DOI] [PubMed] [Google Scholar]
- Ng O.W., Chia A., Tan A.T., Jadi R.S., Leong H.N., Bertoletti A., Tan Y.J. Memory T cell responses targeting the SARS coronavirus persist up to 11 years post-infection. Vaccine. 2016;34:2008–2014. doi: 10.1016/j.vaccine.2016.02.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemeyer D., Zillinger T., Muth D., Zielecki F., Horvath G., Suliman T., Barchet W., Weber F., Drosten C., Muller M.A. Middle East respiratory syndrome coronavirus accessory protein 4a is a type I interferon antagonist. J. Virol. 2013;87:12489–12495. doi: 10.1128/JVI.01845-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H.L., Chia A., Chang C.X., Leong H.N., Ling K.L., Grotenbreg G.M., Gehring A.J., Tan Y.J., Bertoletti A. Engineering T cells specific for a dominant severe acute respiratory syndrome coronavirus CD8 T cell epitope. J. Virol. 2011;85:10464–10471. doi: 10.1128/JVI.05039-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S., Kohyama S., Taneichi M., Moriya O., Hayashi H., Oda H., Mori M., Kobayashi A., Akatsuka T., Uchida T., Matsui M. Synthetic peptides coupled to the surface of liposomes effectively induce SARS coronavirus-specific cytotoxic T lymphocytes and viral clearance in HLA-A*0201 transgenic mice. Vaccine. 2009;27:3912–3920. doi: 10.1016/j.vaccine.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park W.B., Perera R.A., Choe P.G., Lau E.H., Choi S.J., Chun J.Y., Oh H.S., Song K.H., Bang J.H., Kim E.S., Kim H.B., Park S.W., Kim N.J., Man Poon L.L., Peiris M., Oh M.D. Kinetics of serologic responses to MERS coronavirus infection in humans, South Korea. Emerg. Infect. Dis. 2015;21:2186–2189. doi: 10.3201/eid2112.151421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H., Yang L.T., Wang L.Y., Li J., Huang J., Lu Z.Q., Koup R.A., Bailer R.T., Wu C.Y. Long-lived memory T lymphocyte responses against SARS coronavirus nucleocapsid protein in SARS-recovered patients. Virology. 2006;351:466–475. doi: 10.1016/j.virol.2006.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poh W.P., Narasaraju T., Pereira N.A., Zhong F., Phoon M.C., Macary P.A., Wong S.H., Lu J., Koh D.R., Chow V.T. Characterization of cytotoxic T-lymphocyte epitopes and immune responses to SARS coronavirus spike DNA vaccine expressing the RGD-integrin-binding motif. J. Med. Virol. 2009;81:1131–1139. doi: 10.1002/jmv.21571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj V.S., Mou H., Smits S.L., Dekkers D.H., Muller M.A., Dijkman R., Muth D., Demmers J.A., Zaki A., Fouchier R.A., Thiel V., Drosten C., Rottier P.J., Osterhaus A.D., Bosch B.J., Haagmans B.L. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reusken C., Mou H., Godeke G.J., van der Hoek L., Meyer B., Muller M.A., Haagmans B., de Sousa R., Schuurman N., Dittmer U., Rottier P., Osterhaus A., Drosten C., Bosch B.J., Koopmans M. Specific serology for emerging human coronaviruses by protein microarray. Euro Surveil. 2013;18:20441. doi: 10.2807/1560-7917.es2013.18.14.20441. [DOI] [PubMed] [Google Scholar]
- Reusken C.B., Messadi L., Feyisa A., Ularamu H., Godeke G.J., Danmarwa A., Dawo F., Jemli M., Melaku S., Shamaki D., Woma Y., Wungak Y., Gebremedhin E.Z., Zutt I., Bosch B.J., Haagmans B.L., Koopmans M.P. Geographic distribution of MERS coronavirus among dromedary camels, Africa. Emerg. Infect. Dis. 2014;20:1370–1374. doi: 10.3201/eid2008.140590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivino L., Tan A.T., Chia A., Kumaran E.A., Grotenbreg G.M., MacAry P.A., Bertoletti A. Defining CD8+ T cell determinants during human viral infection in populations of Asian ethnicity. J. Immunol. 2013;191:4010–4019. doi: 10.4049/jimmunol.1301507. [DOI] [PubMed] [Google Scholar]
- Roder G., Kristensen O., Kastrup J.S., Buus S., Gajhede M. Structure of a SARS coronavirus-derived peptide bound to the human major histocompatibility complex class I molecule HLA-B*1501. Acta Crystallogr. Sect. F. Struct. Biol. Cryst. Commun. 2008;64:459–462. doi: 10.1107/S1744309108012396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.H., Tong S., Tamin A., Lowe L., Frace M., DeRisi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., Gunther S., Osterhaus A.D., Drosten C., Pallansch M.A., Anderson L.J., Bellini W.J. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- Sharpe H.J., Stevens T.J., Munro S. A comprehensive comparison of transmembrane domains reveals organelle-specific properties. Cell. 2010;142:158–169. doi: 10.1016/j.cell.2010.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stohlman S.A., Bergmann C.C., van der Veen R.C., Hinton D.R. Mouse hepatitis virus-specific cytotoxic T lymphocytes protect from lethal infection without eliminating virus from the central nervous system. J. Virol. 1995;69:684–694. doi: 10.1128/jvi.69.2.684-694.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S., Wong G., Liu Y., Gao G.F., Li S., Bi Y. MERS in South Korea and China: a potential outbreak threat? Lancet. 2015;385:2349–2350. doi: 10.1016/S0140-6736(15)60859-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S., Wong G., Shi W., Liu J., Lai A.C., Zhou J., Liu W., Bi Y., Gao G.F. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trend Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Zhang Y., Zhao B., Deng M., Liu J., Li X., Hou J., Gui M., Zhang S., Li X., Gao G.F., Meng S. A new unconventional HLA-A2-restricted epitope from HBV core protein elicits antiviral cytotoxic T lymphocytes. Protein Cell. 2014;5:317–327. doi: 10.1007/s13238-014-0041-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano T., Tomizawa K., Morioka H., Doki T., Hohdatsu T. Evaluation of protective efficacy of the synthetic peptide vaccine containing the T-helper 1 epitope with CpG oligodeoxynucleotide against feline infectious peritonitis virus infection in cats. Antivir. Ther. 2014;19:645–650. doi: 10.3851/IMP2735. [DOI] [PubMed] [Google Scholar]
- Tang F., Quan Y., Xin Z.T., Wrammert J., Ma M.J., Lv H., Wang T.B., Yang H., Richardus J.H., Liu W., Cao W.C. Lack of peripheral memory B cell responses in recovered patients with severe acute respiratory syndrome: a six-year follow-up study. J. Immunol. 2011;186:7264–7268. doi: 10.4049/jimmunol.0903490. [DOI] [PubMed] [Google Scholar]
- Tang X.C., Agnihothram S.S., Jiao Y., Stanhope J., Graham R.L., Peterson E.C., Avnir Y., Tallarico A.S., Sheehan J., Zhu Q., Baric R.S., Marasco W.A. Identification of human neutralizing antibodies against MERS-CoV and their role in virus adaptive evolution. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E2018–E2026. doi: 10.1073/pnas.1402074111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To K.K., Hung I.F., Chan J.F., Yuen K.Y. From SARS coronavirus to novel animal and human coronaviruses. J. Thorac. Dis. 2013;5:S103–S108. doi: 10.3978/j.issn.2072-1439.2013.06.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo J.A., Gras S., Twist K.A., Croft N.P., Channappanavar R., Rossjohn J., Purcell A.W., Perlman S. Structural and functional correlates of enhanced antiviral immunity generated by heteroclitic CD8 T cell epitopes. J. Immunol. 2014;192:5245–5256. doi: 10.4049/jimmunol.1400111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao Y.P., Lin J.Y., Jan J.T., Leng C.H., Chu C.C., Yang Y.C., Chen S.L. HLA-A*0201 T-cell epitopes in severe acute respiratory syndrome (SARS) coronavirus nucleocapsid and spike proteins. Biochem. Biophys. Res. Commun. 2006;344:63–71. doi: 10.1016/j.bbrc.2006.03.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng C.T., Sbrana E., Iwata-Yoshikawa N., Newman P.C., Garron T., Atmar R.L., Peters C.J., Couch R.B. Immunization with SARS coronavirus vaccines leads to pulmonary immunopathology on challenge with the SARS virus. PloS One. 2012;7:e35421. doi: 10.1371/journal.pone.0035421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijay R., Perlman S. Middle East respiratory syndrome and severe acute respiratory syndrome. Curr. Opin. Virol. 2016;16:70–76. doi: 10.1016/j.coviro.2016.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volz A., Kupke A., Song F., Jany S., Fux R., Shams-Eldin H., Schmidt J., Becker C., Eickmann M., Becker S., Sutter G. Protective efficacy of recombinant modified vaccinia virus ankara delivering Middle East respiratory syndrome coronavirus spike glycoprotein. J. Virol. 2015;89:8651–8656. doi: 10.1128/JVI.00614-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B.M., Chen H.B., Jiang X.D., Zhang M.H., Wan T., Li N., Zhou X.Y., Wu Y.F., Yang F., Yu Y.Z., Wang X.N., Yang R.F., Cao X.T. Identification of an HLA-A*0201-restricted CD8(+) T-cell epitope SSp-1 of SARS-CoV spike protein. Blood. 2004;104:200–206. doi: 10.1182/blood-2003-11-4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Qi J., Yuan Y., Xuan Y., Han P., Wan Y., Ji W., Li Y., Wu Y., Wang J., Iwamoto A., Woo P.C., Yuen K.Y., Yan J., Lu G., Gao G.F. Bat origins of MERS-CoV supported by bat coronavirus HKU4 usage of human receptor CD26. Cell Host Microbe. 2014;16:328–337. doi: 10.1016/j.chom.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Wong G., Lu G., Yan J., Gao G.F. MERS-CoV spike protein: targets for vaccines and therapeutics. Antivir. Res. 2016;133:165–177. doi: 10.1016/j.antiviral.2016.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W.L., Wang H.J., Deng Y., Song T., Lan J.M., Wu G.Z., Ke C.W., Tan W.J. Serological study of an imported case of Middle East respiratory syndrome and his close contacts in China, 2015. Biomed. Environ. Sci. 2016;29:219–223. doi: 10.3967/bes2016.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Liu D., Shi W., Lu R., Wang W., Zhao Y., Deng Y., Zhou W., Ren H., Wu J., Wang Y., Wu G., Gao G.F., Tan W. Origin and possible genetic recombination of the Middle East respiratory syndrome coronavirus from the first imported case in China: phylogenetics and coalescence analysis. mBio. 2015;6 doi: 10.1128/mBio.01280-15. e01280–01215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.D., Sin W.Y., Xu G.B., Yang H.H., Wong T.Y., Pang X.W., He X.Y., Zhang H.G., Ng J.N., Cheng C.S., Yu J., Meng L., Yang R.F., Lai S.T., Guo Z.H., Xie Y., Chen W.F. T-cell epitopes in severe acute respiratory syndrome (SARS) coronavirus spike protein elicit a specific T-cell immune response in patients who recover from SARS. J. Virol. 2004;78:5612–5618. doi: 10.1128/JVI.78.11.5612-5618.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong G., Liu W., Liu Y., Zhou B., Bi Y., Gao G.F. MERS, SARS, and Ebola: the role of super-spreaders in infectious disease. Cell Host Microbe. 2015;18:398–401. doi: 10.1016/j.chom.2015.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Wong B.H., Tsoi H.W., Fung A.M., Chan K.H., Tam V.K., Peiris J.S., Yuen K.Y. Detection of specific antibodies to severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein for serodiagnosis of SARS coronavirus pneumonia. J. Clin. Microbiol. 2004;42:2306–2309. doi: 10.1128/JCM.42.5.2306-2309.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K., Sun K., Srinivasan K.N., Salmon J., Marques E.T., Xu J., August J.T. Immune responses to T-cell epitopes of SARS CoV-N protein are enhanced by N immunization with a chimera of lysosome-associated membrane protein. Gene Ther. 2009;16:1353–1362. doi: 10.1038/gt.2009.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Peng H., Zhu Z., Li G., Huang Z., Zhao Z., Koup R.A., Bailer R.T., Wu C. Persistent memory CD4+ and CD8+ T-cell responses in recovered severe acute respiratory syndrome (SARS) patients to SARS coronavirus M antigen. J. Gen. Virol. 2007;88:2740–2748. doi: 10.1099/vir.0.82839-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L.T., Peng H., Zhu Z.L., Li G., Huang Z.T., Zhao Z.X., Koup R.A., Bailer R.T., Wu C.Y. Long-lived effector/central memory T-cell responses to severe acute respiratory syndrome coronavirus (SARS-CoV) S antigen in recovered SARS patients. Clin. Immunol. 2006;120:171–178. doi: 10.1016/j.clim.2006.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Ye F., Zhu N., Wang W., Deng Y., Zhao Z., Tan W. Middle East respiratory syndrome coronavirus ORF4b protein inhibits type I interferon production through both cytoplasmic and nuclear targets. Sci. Rep. 2015;5:17554. doi: 10.1038/srep17554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. New Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- Zhang N., Jiang S., Du L. Current advancements and potential strategies in the development of MERS-CoV vaccines. Expert Rev. Vac. 2014;13:761–774. doi: 10.1586/14760584.2014.912134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Bakshi R.K., Suneetha P.V., Fytili P., Antunes D.A., Vieira G.F., Jacobs R., Klade C.S., Manns M.P., Kraft A.R., Wedemeyer H., Schlaphoff V., Cornberg M. Frequency, private specificity, and cross-reactivity of preexisting hepatitis C virus (HCV)-Specific CD8+ T cells in HCV-seronegative individuals: implications for vaccine responses. J. Virol. 2015;89:8304–8317. doi: 10.1128/JVI.00539-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Huang Q., Wang W., Zhang Y., Lv P., Gao X.M. Identification and characterization of dominant helper T-cell epitopes in the nucleocapsid protein of severe acute respiratory syndrome coronavirus. J. Virol. 2007;81:6079–6088. doi: 10.1128/JVI.02568-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Perera R.A., Kayali G., Meyerholz D., Perlman S., Peiris M. Passive immunotherapy with dromedary immune serum in an experimental animal model for Middle East respiratory syndrome coronavirus infection. J. Virol. 2015;89:6117–6120. doi: 10.1128/JVI.00446-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Zhao J., Mangalam A.K., Channappanavar R., Fett C., Meyerholz D.K., Agnihothram S., Baric R.S., David C.S., Perlman S. Airway memory CD4(+) T cells mediate protective immunity against emerging respiratory coronaviruses. Immunity. 2016;44:1379–1391. doi: 10.1016/j.immuni.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Zhao J., Perlman S. T cell responses are required for protection from clinical disease and for virus clearance in severe acute respiratory syndrome coronavirus-infected mice. J. Virol. 2010;84:9318–9325. doi: 10.1128/JVI.01049-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K., Wang H., Wu C. The immune responses of HLA-A*0201 restricted SARS-CoV S peptide-specific CD8(+) T cells are augmented in varying degrees by CpG ODN, PolyI: C and R848. Vaccine. 2011;29:6670–6678. doi: 10.1016/j.vaccine.2011.06.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K., Yang B., Xu Y., Wu C. CD8+ T cell response in HLA-A*0201 transgenic mice is elicited by epitopes from SARS-CoV S protein. Vaccine. 2010;28:6666–6674. doi: 10.1016/j.vaccine.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhi Y., Kobinger G.P., Jordan H., Suchma K., Weiss S.R., Shen H., Schumer G., Gao G., Boyer J.L., Crystal R.G., Wilson J.M. Identification of murine CD8 T cell epitopes in codon-optimized SARS-associated coronavirus spike protein. Virology. 2005;335:34–45. doi: 10.1016/j.virol.2005.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Chu H., Li C., Wong B.H., Cheng Z.S., Poon V.K., Sun T., Lau C.C., Wong K.K., Chan J.Y., Chan J.F., To K.K., Chan K.H., Zheng B.J., Yuen K.Y. Active replication of Middle East respiratory syndrome coronavirus and aberrant induction of inflammatory cytokines and chemokines in human macrophages: implications for pathogenesis. J. Infect. Dis. 2014;209:1331–1342. doi: 10.1093/infdis/jit504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M., Xu D., Li X., Li H., Shan M., Tang J., Wang M., Wang F.S., Zhu X., Tao H., He W., Tien P., Gao G.F. Screening and identification of severe acute respiratory syndrome-associated coronavirus-specific CTL epitopes. J. Immunol. 2006;177:2138–2145. doi: 10.4049/jimmunol.177.4.2138. [DOI] [PubMed] [Google Scholar]
- Zhou T., Georgiev I., Wu X., Yang Z.Y., Dai K., Finzi A., Kwon Y.D., Scheid J.F., Shi W., Xu L., Yang Y., Zhu J., Nussenzweig M.C., Sodroski J., Shapiro L., Nabel G.J., Mascola J.R., Kwong P.D. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science. 2010;329:811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zust R., Cervantes-Barragan L., Kuri T., Blakqori G., Weber F., Ludewig B., Thiel V. Coronavirus non-structural protein 1 is a major pathogenicity factor: implications for the rational design of coronavirus vaccines. PLoS Pathog. 2007;3:e109. doi: 10.1371/journal.ppat.0030109. [DOI] [PMC free article] [PubMed] [Google Scholar]