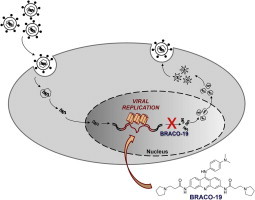
Graphical abstract
Keywords: Antiviral compound, G-rich sequence, G-quadruplex, DNA conformation, DNA secondary structure, Herpes Simplex Virus 1
Highlights
-
•
Large clusters of repeated G-quadruplex folding sequences were identified in the HSV-1 genome.
-
•
BRACO-19, a G-quadruplex ligand, was able to bind and stabilize the HSV-1 G-quadruplexes.
-
•
Stabilization of HSV-1 G-quadruplexes by BRACO-19 inhibited DNA polymerase processing and viral DNA replication.
-
•
BRACO-19 diminished virus production in infected cells.
-
•
HSV-1 G-quadruplex may be regarded as alternative and innovative targets for antiviral therapy.
Abstract
Guanine-rich nucleic acids can fold into G-quadruplexes, secondary structures implicated in important regulatory functions at the genomic level in humans, prokaryotes and viruses. The remarkably high guanine content of the Herpes Simplex Virus-1 (HSV-1) genome prompted us to investigate both the presence of G-quadruplex forming sequences in the viral genome and the possibility to target them with G-quadruplex ligands to obtain anti-HSV-1 effects with a novel mechanism of action. Using biophysical, molecular biology and antiviral assays, we showed that the HSV-1 genome displays multiple clusters of repeated sequences that form very stable G-quadruplexes. These sequences are mainly located in the inverted repeats of the HSV-1 genome. Treatment of HSV-1 infected cells with the G-quadruplex ligand BRACO-19 induced inhibition of virus production. BRACO-19 was able to inhibit Taq polymerase processing at G-quadruplex forming sequences in the HSV-1 genome, and decreased intracellular viral DNA in infected cells. The last step targeted by BRACO-19 was viral DNA replication, while no effect on virus entry in the cells was observed. This work, presents the first evidence of extended G-quadruplex sites in key regions of the HSV-1 genome, indicates the possibility to block viral DNA replication by G-quadruplex-ligand and therefore provides a proof of concept for the use of G-quadruplex ligands as new anti-herpetic therapeutic options.
1. Introduction
G-quadruplexes are nucleic acids secondary structure that may form in G-rich sequences. They are based on the formation of G-quartets, which are stabilized by Hoogsteen hydrogen bonds between guanines (Sen and Gilbert, 1988). G-quartets stack on top of each other to give rise to G-quadruplexes that are further stabilized by the interaction with monovalent cations. DNA strands in G-quadruplex may be oriented in anti-parallel, parallel, or hybrid configuration, and the nucleotide linkers between G-quartet stacks can adopt a multitude of loop structures (Patel et al., 2007, Phan, 2010, Phan et al., 2006, Simonsson, 2001).
G-quadruplexes occur in functionally important regions of the genome (Huppert, 2008, Neidle, 2010): they have been identified in the promoters of a wide range of genes that are important in cell signaling (Balasubramanian et al., 2010, Duquette et al., 2004), suggesting the possibility that G-quadruplexes behave as structural switches of cellular processes, therefore providing a basis for therapeutic intervention (Neidle and Parkinson, 2002, Zhang et al., 2014).
One of the most potent G-quadruplex ligands is BRACO-19 (Fig. 1 a), a 3,6,9-trisubstituted acridine derivative designed to stabilize the quadruplex DNA structures formed in human telomeres (Read et al., 2001). BRACO-19 has been shown to inhibit telomerase activity (Harrison et al., 2003) and to display in vitro and in vivo antitumor activity (Burger et al., 2005, Gowan et al., 2002).
Fig. 1.

The HSV-1 G-quadruplexes and the G-quadruplex ligand. (a) Chemical structure of the G-quadruplex ligand BRACO-19. (b) Schematic representation of the HSV-1 genome. G-quadruplex forming regions and their position within the genome are indicated. The terminal (TRL and TRS) and internal (IRL and IRS) repeats are shown as rectangles. The leading and lagging strands are depicted by a thick and thin line, respectively. G-quadruplex regions indicated above and below the scheme are present in the leading and lagging strand, respectively.
Besides humans, other mammals (Verma et al., 2008), yeast (Hershman et al., 2008) and prokaryotic cells (Beaume et al., 2013, Rawal et al., 2006, Wieland and Hartig, 2009) showed enrichment of putative G-quadruplex forming sequences. In viruses, evidence of significant G-quadruplex regions is increasingly being recognized. In the Epstein–Barr herpes virus (EBV), G-quadruplexes modulate EBV nuclear antigen 1 (EBNA1) activity and translation (Murat et al., 2014); in particular, BRACO-19 inhibited EBNA1-dependent stimulation of viral DNA replication (Norseen et al., 2009). In HIV-1, we have demonstrated a G-quadruplex-mediated transcriptional silencing activity in the SP1 binding region of the long terminal repeat promoter (Perrone et al., 2013a) and in the Nef coding gene (Perrone et al., 2013b). In both cases, G-quadruplex ligands were able to stabilize the G-quadruplex conformations and to exert antiviral activity (Perrone et al., 2014). In the SARS coronavirus, G-quadruplexes targeting by a key viral protein was proposed to control host’s cell response to viral infection (Tan et al., 2009).
The Herpes Simplex Virus type 1 (HSV-1) 152 Kbp genome has 68% guanosines (Gs) and cytosines (Cs) composition, which reaches 84.7% in simple sequence repeats (SSRs), ubiquitously distributed in both coding and non-coding regions with similar relative frequency (Ouyang et al., 2012); in contrast, GC content in the human genome is 41%. The HSV-1 genome is composed of two covalently linked regions of unique sequences, the unique long (UL) and unique short (US) sequences, flanked by large inverted repeated sequences, TRL–IRL and IRS–TRS (Fig. 1b) (Hayward et al., 1975). HSV replication is temporally regulated in rounds of transcription that yield immediate-early, early and late proteins. HSV-1 is a neurotropic virus: after the first lytic infection within mucosal epithelial cells, the virus enters sensory neurons where latency is established. The virus can later reactivate resulting in the generation of new virions that cause recurrent disease (Roizman et al., 2013). HSV-1 most commonly causes vesicular lesions affecting the mucous membranes; however, it can also cause infrequent but serious diseases, such as encephalitis, disseminated neonatal infections and visceral infections in immunocompromised patients (Frangoul et al., 2007, Genereau et al., 1996). The presence of HSV increases sexual transmission of HIV-1 (Freeman et al., 2006).
HSV-1 symptoms are not cured but treated with nucleoside analogues, such as acyclovir (ACV) and its derivatives (Vere Hodge and Field, 2013). However, the virus remains in the body for life, since no cure that eradicates herpes has yet been developed. In addition, emergence of resistance to current anti-herpetic drugs has long created an obstacle for the treatment of HSV-1 (Bacon et al., 2003). Therefore new antiviral approaches able to suppress both lytic and possibly also latent infections are highly required.
Earlier work by Roizman indicated the presence of an alternative conformation of a viral G-rich DNA and RNA sequence around the origin of DNA replication in the HSV-1 genome (McCormick et al., 1992, Roller et al., 1989); however no systematic analysis on the presence of G-quadruplex structures in the HSV-1 nucleic acid has ever been provided.
Here we present the first comprehensive genome analysis and evidence that extended G-rich sequences mainly located in repeats of the HSV-1 genome can fold in stable G-quadruplex structures. We demonstrate that treatment with the G-quadruplex ligand BRACO-19 greatly stabilizes these sequences resulting in decrease of infectious viral particles, reduction of late viral transcripts, inhibition of Taq polymerase processing at the HSV-1 genome, specifically affecting viral DNA replication at G-quadruplex regions.
2. Materials and methods
A detailed description of this section is provided in Supporting information.
3. Results
3.1. Highly conserved putative G-quadruplex forming sequences are present in repetitive regions of the HSV-1 genome
We identified nine regions in the HSV-1 genome with highly repeated putative QGRS (Quadruplex forming G-Rich Sequences) (Table 1 ). Six of them (named gp054a–f) were found in the leading strand of the gp054 gene (Fig. 1b) which encodes UL36, the largest viral protein and essential viral tegument component (McNabb and Courtney, 1992) (Table 1). The gp054 series sequences displayed the G4XTG4XTG4XTG4XT common motif, where X was either a T or a C base, and covered about 220 bps. Four more putative QGRS were clustered at the terminal and internal repeats (both long and short) (Fig. 1b, Table 1): they covered about 900 bps in the leading strand and 700 bps in the lagging strand. The exact role of these regions is as yet unknown and thus these positions were named “un”. All identified sequences were highly conserved among different HSV-1 strains (Table 1). The only exception was the unb sequence, which was identical to gp054b, and was not conserved in the KOS strain. To note that all identified sequences, apart from un3, were characterized by four GGGG-tracts, therefore possessing the ability to fold into 4-tetrad stacked G-quadruplex. This kind of tetraplex is in principle very stable, as denoted by the high G-scores, which indicate the propensity of a sequence to fold into unimolecular G-quadruplex, obtained by these oligonucleotides (Table 1). The un3 sequence displayed four GGG-tracts therefore gaining a lower G-score.
Table 1.
Sequences of putative G-quadruplex forming oligonucleotides in the genome of HSV-1. Sequence repeats within HSV-1 main strains and G-scores are shown.
| Name | Sequence | G-scorea | Strains | Positions |
Repeats and genes |
||
|---|---|---|---|---|---|---|---|
| Leading strand (5′ → 3′) | Lagging strand (5′ → 3′) | Leading strand | Lagging strand | ||||
| gp054a | GGGGTTGGGGCTGGGGTTGGGG | 63 | F | 71527–71620 | – | 4 UL36 | – |
| 17 | 71612–71705 | – | 4 UL36 | – | |||
| H129 | 71561–71654 | – | 4 UL36 | – | |||
| KOS | 71580–71757 | – | 4 UL36 | – | |||
| gp054b | GGGGTTGGGGTTGGGGTTGGGG | 63 | F | 71587–71608 | – | 1 UL36 | – |
| 17 | 71672–71693 | – | 1 UL36 | – | |||
| H129 | 71621–71642 | – | 1 UL36 | – | |||
| KOS | 71616–71637 | – | 1 UL36 | – | |||
| gp054c | GGGGTTGGGGTTGGGGCTGGGG | 63 | F | 71593–71632 | – | 1 UL36 | – |
| 17 | 71678–71717 | – | 1 UL36 | – | |||
| H129 | 71627–71666 | – | 1 UL36 | – | |||
| KOS | 71574–71661 | – | 2 UL36 | – | |||
| gp054d | GGGGCTGGGGCTGGGGCTGGGG | 63 | F | 71623–71722 | – | 4 UL36 | – |
| 17 | 71708–71807 | – | 4 UL36 | – | |||
| H129 | 71657–71756 | – | 4 UL36 | – | |||
| KOS | 71652–71739 | – | 3 UL36 | – | |||
| gp054e | GGGGCTGGGGCTGGGGTTGGGG | 63 | F | 71707–71728 | – | 1 UL36 | – |
| 17 | 71792–71813 | – | 1 UL36 | – | |||
| H129 | 71741–71762 | – | 1 UL36 | – | |||
| KOS | 71562–71745 | – | 2 UL36 | – | |||
| gp054f | GGGGCTGGGGTTGGGGTTGGGG | 63 | F | 71581–71626 | – | 2 UL36 | – |
| 17 | 71666–71711 | – | 2 UL36 | – | |||
| H129 | 71615–71660 | – | 2 UL36 | – | |||
| KOS | 71568–71655 | – | 3 UL36 | – | |||
| unb | GGGGTTGGGGTTGGGGTTGGGG | 63 | F | 4121–4142 | 30075–30096 | 1 TRL | 1 IRL |
| 17 | 4138–4159 | 29987–30008 | 1 TRL | 1 IRL | |||
| H129 | 4114–4135 | 29892–29913 | 1 TRL | 1 IRL | |||
| KOS | – | – | – | – | |||
| un1 | GGGGGAGAGGGGAGAGGGGGGGAGAGGGG | 62 | F | 8966–9147 | 34922–35103 | 5 TRL (b) | 5 IRL (b′) |
| 17 | 9049–9213 | 34898–35062 | 5 TRL (b) | 5 IRL (b′) | |||
| H129 | 9010–9174 | 34804–34968 | 5 TRL (b) | 5 IRL (b′) | |||
| KOS | 8997–9161 | 34768–34932 | 5 TRL (b) | 5 IRL (b′) | |||
| un2 | GGGGGCGAGGGGCGGGAGGGGGCGAGGGG | 62 | F | 150989–151306 | 25157–25474 | 9 TRS (c) | 9 IRS (c′) |
| 17 | 151064–151381 | 25090–25407 | 9 TRS (c) | 9 IRS (c′) | |||
| H129 | 150962–151279 | 25087–25404 | 9 TRS (c) | 9 IRS (c′) | |||
| KOS | 150854–151171 | 25009–25326 | 9 TRS (c) | 9 IRS (c′) | |||
| un3 | GGGAGGAGCGGGGGGAGGAGCGGG | 36 | F | 93–296/151880–152083 | 26047–26250 | 8 TRL, 8 TRS (a) | 8 IR (a′) |
| 17 | 106–309/151929–152132 | 25955–26158 | 8 TRL, 8 TRS (a) | 8 IR (a′) | |||
| H129 | 106–333/151771–151974 | 25896–26111 | 9 TRL, 8 TRS (a) | 9 IR (a′) | |||
| KOS | 118–309/151734–151925 | 25889–26080 | 8 TRL, 8 TRS (a) | 8 IR (a′) | |||
G-score: propensity of a sequence to fold into unimolecular G-quadruplex assigned by the QGRS software.
3.2. All putative QGRS fold into highly stable G-quadruplex conformations
Folding of the HSV-1 putative QGRS was determined by circular dichroism (CD) spectroscopy. In physiological concentrations of K+, all tested oligonucleotides displayed CD G-quadruplex signatures that included three different conformations (Vorlickova et al., 2012): hybrid (gp054), antiparallel (un2) and parallel (un1, un3, Fig. 2 a). Usually, G-quadruplex folding occurs in the presence of K+, while in the absence of the monovalent cation oligonucleotides are mostly unfolded. The HSV-1 oligonucleotides, however, displayed peaks characteristic of the G-quadruplex conformation even in the absence of K+ (Fig. S1). Stability of HSV-1 G-quadruplexes in the presence of K+ was assessed by thermal denaturation experiments monitored by CD and UV spectroscopy. Un2 and un3 were the most stable G-quadruplexes with T m above 95 °C, followed by gp054 oligonucleotides. (Fig. 2b, Table 2 ). Because of the very low solubility of un1 in K+, T m values for this sequence were not obtained. In the absence of K+, un3 was the least stable sequence, followed by un1. Un2 and gp054 oligonucleotides showed comparable stability, with the exception of gp054d, which was extremely stable even in the absence of K+ (Table 2).
Fig. 2.

Biophysical and biochemical characterization of HSV-1 G-quadruplex forming sequences. (a) CD spectra and (b) CD thermal unfolding analysis of HSV-1 G-quadruplexes in the presence of K+ 100 mM. In (b) molar ellipticity at the peak wavelength has been plotted against temperature. (c and d) Electrophoresis mobility shift analysis (EMSA) of HSV-1 oligonucleotides folded in the absence (c) or presence (d) of K+ and loaded on native gels lacking or containing K+, respectively. The identity and length of each oligonucleotide are shown above and below the gel image, respectively. M indicate a marker lane loaded with two oligonucleotides with known length and unable to fold into secondary structures.
Table 2.
Stability of HSV-1 G-quadruplex folding sequences measured by UV and CD, and in the presence/absence of K+.
| Sequence | Tm (UV 100 mM K+) | Tm (CD 100 mM K+) | Tm (CD no K+) | ΔTm[100 mM/no K+] (CD) | Tm (CD 100 mM K+, BRACO-19 16 μM) |
|---|---|---|---|---|---|
| gp054a | 87.4 ± 0.4 | 88.1 ± 1.3 | 48.0 ± 1.1 | 40.1 | >95 |
| gp054b | 88.0 ± 0.4 | 87.5 ± 1.1 | 48.4 ± 1.6 | 39.1 | >95 |
| gp054c | 92.3 ± 0.8 | 90.8 ± 0.3 | 51.7 ± 1.3 | 39.1 | >95 |
| gp054d | 91.5 ± 0.1 | 90.7 ± 0.3 | 71.3 ± 2.6 | 19.4 | >95 |
| gp054e | 91.7 ± 0.6 | 87.5 ± 0.5 | 58.0 ± 0.8 | 29.5 | >95 |
| gp054f | 88.1 ± 0.1 | 88.0 ± 0.2 | 51.9 ± 1.4 | 36.1 | >95 |
| un1 | nd | nd | 40.8 ± 9.8 | nd | >95 |
| un2 | >95 | >95 | 50.3 ± 0.9 | >44.7 | >95 |
| un3 | >95 | >95 | 34.7 ± 4.7 | >60.3 | >95 |
The ability of HSV-1 QGRS to fold into G-quadruplex was further assessed by EMSA analysis. Even in the absence of K+, all gp054 sequences run markedly faster than their 22 bp-length control marker, indicating folding in these conditions (Fig. 2c). In particular, gp054d displayed the fastest mobility, confirming its inherent property to fold into G-quadruplex in the absence of K+. Un1 (29-bp) and un2 (28-bp) also migrated faster than the 22-long control marker (see * symbol in Fig. 2c), indicating effective folding. However, a band corresponding to the unfolded un1 was also observable (see § symbol in Fig. 2c). In contrast, the 23 bp-long un3 migrated as a totally unfolded oligonucleotide (compare lane 3 with lane M, Fig. 2c), confirming its lower tendency to fold in the absence of K+ (see lower G-score, Table 1, and lower stability, Table 2). In the presence of K+ (Fig. 2d), all sequences displayed increased migration indicating effective folding. For the gp054 sequences, slower migrating species, ascribable to intermolecular dimeric G-quadruplex forms, were also observed (lanes a–f, Fig. 2d).
The G-quadruplex ligand BRACO-19 was next tested by CD analysis for its ability to bind the HSV-1 QGRS in the presence of K+. BRACO-19 converted the gp054 hybrid conformation to an antiparallel-like G-quadruplex (Fig. S2) and further stabilized all HSV-1 oligonucleotides, increasing T m to above 95 °C (Table 2).
Additional evidence of the intrinsic stability of the identified HSV-1 G-quadruplexes within a longer DNA context and on the increased stabilization imparted by BRACO-19 was provided by the Taq polymerase stop assay. All G-quadruplex forming sequences except un3 stopped the polymerase at the first G-rich region encountered by the polymerase even in the absence of K+ (lanes 1, un2, gp054a, gp054d, Fig. 3 ). When K+ was added, strong stops were observed in all G-quadruplex templates (lanes 2, Fig. 3). Upon addition of BRACO-19 the intensity of the initial stop sites dramatically increased (lanes 4, Fig. 3); moreover, an additional intense stop site corresponding to a second G-rich region was observed in the un2 template, possibly indicating the presence of multiple G-quadruplex structures. To note that BRACO-19 stopped the processing of the polymerase to such an extent that the full-length product did not form in all templates, with the exception of un3, which resulted the sequence less susceptible to the activity of the compound (lanes 4, Fig. 3). In contrast, TMPyP2, a porphyrin derivative which shares chemical features with BRACO-19, i.e. a large aromatic surface and cationic moieties, but displays a different G-quadruplex binding mode and markedly lower G-quadruplex binding affinity (Han et al., 2001, Le et al., 2013), used here for comparison, did not induce any stop at G-rich regions and it did not modify the amount of the full-length amplicons (lanes 3, Fig. 3). Moreover, a control sequence devoid of G-tracts did not show any stop site in the presence of K+ and BRACO-19 (lanes 1–4, non-G4 template, Fig. 3), indicating that the observed polymerase inhibition was G-quadruplex dependent and specific.
Fig. 3.
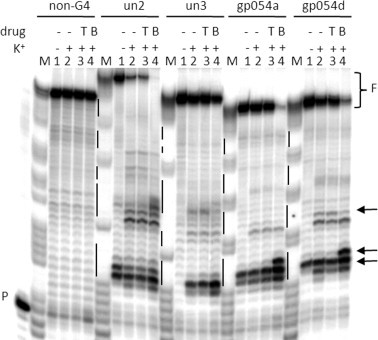
Taq polymerase stop assay on HSV-1 G-quadruplex forming oligonucleotides in the presence of BRACO-19 (B) and TMPyP2 (T). Extended un2, un3, gp054a and gp054d DNA templates (Table S1) in the absence and presence of K+ 10 mM (lanes 1 and 2, respectively) and in the presence of TMPyP2 (T) and BRACO-19 (B) (0.5 μM) (lanes 3 and 4, respectively) were used as templates for the Taq polymerase. A non-G-quadruplex template (non-G4 lanes) was also used as negative control. P indicates primer. F indicates the full-length products. M is a marker lane. Taq pausing regions are indicated by arrows. G tracts are shown on the left of each gel image as vertical lines.
3.3. BRACO-19 displays anti-HSV-1 activity
Given the ability of BRACO-19 to recognize and stabilize HSV-1 G-quadruplexes, we tested the possibility that it displayed antiviral activity. TMPyP2 was used for comparison. Cytotoxicity of both compounds was assessed on Vero cells, a monkey kidney epithelial cell line which sustains HSV-1 infection. Both compounds were non-cytotoxic up to 25 μM, the highest tested dose (CC50 > 25 μM). In HSV-1 infected Vero cells, BRACO-19 demonstrated a statistical significant antiviral effect at 1 μM, which increased up to 70% at 25 μM (EC50 = 8 μM) (Fig. 4 a). In contrast, TMPyP2 displayed EC50 = 25 μM.
Fig. 4.

Antiviral activity of BRACO-19. (a) Plaque assay: infected cells (MOI 1 PFU/cell) were treated with increasing concentrations (0.04 μM–25 μM) of BRACO-19 or TMPyP2; supernatants were collected at 24 h.p.i., the number of plaque forming units determined. (b) mRNA levels of HSV-1 immediate-early, early and late proteins. Infected cells were treated with BRACO-19 or TMPyP2 (5 μM); 7 or 24 h.p.i. total RNA was isolated, retrotranscribed into cDNA and expression of specific genes determined by RT-PCR (Table S2). The mRNAs of the immediate-early US1, ICP4 and UL54, early UL30 DNA polymerase and late proteins UL36 and UL37 were analyzed. RQ are Relative Quantities. In all data sets: n ⩾ 3, mean ± s.d., Student’s t-test, ∗∗P < 0.01.
3.4. BRACO-19 does not inhibit viral entry into cells
Since both BRACO-19 and TMPyP2, as most G-quadruplex binding molecules, are polycationic agents, we first excluded that the observed antiviral activity was due to the interaction of these molecules with the negatively charged heparan sulfate (Campadelli-Fiume et al., 2007, Herold et al., 1991), thus preventing virus attachment to the cells. BRACO-19 or TMPyP2 were added at different times relative to infection until the virus had completed its entry into cells (i.e. 2 h.p.i.) (Sodeik et al., 1997). In this time range we did not observe any difference to the activity obtained by treating the cells 1 h pre-infection (Fig. S3), indicating that neither BRACO-19 nor TMPyP2 act by inhibiting viral entry.
3.5. Stabilization of HSV-1 G-quadruplex forming regions by BRACO-19 impairs viral DNA replication
One cluster of HSV-1 QGRS was embedded in the gp054 gene, which encodes the essential tegument protein UL36; we thus investigated if treatment with BRACO-19 impaired UL36 transcription. Transcript levels of a second tegument protein, UL37, were also assessed as control for a protein whose coding sequence lacks important QGRS clusters. Both viral transcripts were decreased to 30–40% at 24 h.p.i. (Fig. 4b) and this effect was specific for the HSV-1 transcripts (Fig. S4a). The mRNA levels of representative immediate-early and early proteins, whose coding sequences lacked important G-quadruplex forming regions, were also analyzed: BRACO-19 did not affect transcription of these earlier proteins (Fig. 4b). Moreover, TMPyP2 reduced HSV-1 transcript levels to a low extent (100–80%) and it did not differentiate among the different kinetic groups of proteins. The fact that both analyzed late transcripts were affected by BRACO-19 to a similar degree indicates that the observed effect is independent of the presence of QGRS in the UL36 coding sequence; the absence of effect on earlier genes indicates that the compound likely exerts it activity against G-quadruplex-controlled viral mechanisms that mostly influence transcription of late proteins. Since the identified clusters of QGRS were mostly present in non-coding repetitive-element regions, we tested the possibility that QGRS folding induced by BRACO-19 inhibited polymerase processing at the viral DNA level.
HSV-1 DNA was extracted from a viral stock and treated with increasing concentrations of BRACO-19 and TMPyP2. Genome regions containing G-rich tracts were amplified by standard PCR using Taq polymerase, which was used as a model DNA polymerase enzyme. A non-G-rich/non-G-quadruplex forming sequence was also amplified as negative control sequence. Amplified DNA corresponding to G-quadruplex regions decreased in a concentration dependent manner in the presence of BRACO-19 (Fig. 5 a, un and gp054, BRACO-19). In contrast, BRACO-19 was not able to disrupt polymerase processing in the non-G-quadruplex region (Fig. 5a, non-G4, BRACO-19) and TMPyP2 had no effect in both G-rich and non-G-rich regions (Fig. 5a, un, gp054, non-G4, TMPyP2). Real-time PCR was employed for quantification purposes. Amplification of the G-quadruplex region was inhibited up to 40% (with respect to the non-treated control, 100%) in the presence of BRACO-19 (4 μM), while no effect was detected in the non-G-quadruplex region (Fig. 5b), confirming lack of activity of the compound against the polymerase enzyme. No activity of TMPyP2 was observed. This data demonstrate that BRACO-19 specifically interacts with G-quadruplex regions in the HSV-1 genome in vitro and inhibits polymerase processing likely due to the steric hindrance caused by multiple tetraplex structures stabilized by the ligand.
Fig. 5.

Inhibition of HSV-1 DNA replication by BRACO-19. (a)-(b) Inhibition of DNA replication in G-quadruplex regions in vitro. (a) Gp054 and un G-quadruplex regions were elongated using the Taq polymerase stop assay, in the presence of increasing concentration of BRACO-19 or TMPyP2 (2-fold dilutions from 16 μM to 0.25 μM). A non-treated sample (lane N) and a sample without DNA (lane C) were used as negative controls; HSV-1 DNA was extracted from HSV-1-infected Vero cells. M indicates the marker lane. (b) Real-time PCR on a gp054 G-quadruplex sequence and a control non-G-quadruplex region was performed in the presence of BRACO-19 or TMPyP2 (2-fold dilutions from 4 μM to 0.25 μM). Quantification of amplified products was obtained by SYBR® green detection. HSV-1 DNA was extracted from HSV-1-infected Vero cells. (c) Quantification of intracellular DNA amounts obtained from infected cells at 3 h and 20 h.p.i., treated with increasing concentration of BRACO-19 (B19) and TMPyP2 (P2), with n ⩾ 3, mean ± s.d., Student’s t-test, ∗∗P < 0.01. RQ are Relative Quantities. (d) Time-of-addition assay of BRACO-19. Vero cells were infected with HSV-1 strain F and BRACO-19 was added at the indicated different time points after infection (X axis). Virus was collected 30 h.p.i. and quantified by plaque assay. The activity of BRACO-19 (B19, 25 μM) was compared with that of the negative control (cnt) and of ACV as reference drugs. The left Y axis refers to BRACO-19 and control data, whereas the right Y axis refers to ACV data. These results are representative of two independent experiments.
We next tested the effect of BRACO-19 on viral DNA amounts in infected cells. HSV-1 infected cells were treated with increasing concentrations of BRACO-19 or TMPyP2 and intracellular viral DNA was extracted and quantified by real-time PCR. DNA extraction was performed at 2 different time points, before (3 h.p.i.) and after (20 h.p.i.) the viral DNA was replicated in cells. BRACO-19 reduced intracellular viral DNA levels to 50% of the untreated control (100%) at the highest concentration at 20 h.p.i. (Fig. 5c). In contrast, at 3 h.p.i. BRACO-19 and TMPyP2 did not affect viral DNA amounts. Moreover, addition of BRACO-19 did not modify cellular DNA amounts (Fig. S4b).
To define the viral step targeted by BRACO-19 in infected cells, a time-of-addition experiment was set up (Daelemans et al., 2011, Pauwels et al., 1990). This assay indicates the last viral step targeted by a compound. ACV was used as reference compound with established mode of action in the time frame of replication events (James and Prichard, 2014). BRACO-19 was added to infected cells every 2 h up to 12 h.p.i. (Fig. 5d). A sharp increase in virus amounts (PFU/ml) was observed between 8 and 10 h.p.i., indicating that BRACO-19 was active at events that occur in this time range. ACV showed a similar increase between 8 and 12 h.p.i. Since ACV acts by inhibiting the viral DNA polymerase during DNA replication, these data confirm the activity of BRACO-19 during viral DNA replication.
4. Discussion and conclusions
In this work we have found that the GC-rich HSV-1 genome presents multiple extended repeated clusters of G-quadruplex forming sequences, covering 1100 bp on the leading strand and 920 bp on the lagging strand (Fig. 1b, Table 1).
A first intriguing aspect is that these sequences display a remarkably high stability: in physiological conditions all but one sequence (un3) displayed T m around 90 °C and were capable of folding into tetraplex even in the absence of K+. In particular, among the gp054 oligonucleotides, gp054d standed out for its improved stability. Interestingly, this is the only sequence that presents one C base in the loop and displays identical CT loops. It has been reported that C can also be involved in G-quartet formation (Lim et al., 2009), therefore C bases may augment the folding stability of this sequence. To our knowledge the HSV-1 G-quadruplex structures reported in this work are the most stable ever reported in an organism.
HSV-1 G-quadruplexes will likely form in the viral DNA when the duplex is stimulated to unwind, i.e. during replication and transcription events. In eukaryotes G-quadruplex formation at the DNA level has been shown to slow down replication and increase the likelihood of chromosomal breakage, genetic instability (Ribeyre et al., 2009) and genomic rearrangements (Cahoon and Seifert, 2009); moreover, G-quadruplexes accumulation during the S-phase of the cell cycle, the phase at which replication occurs, has been reported (Biffi et al., 2013). DNA processing is allowed by the activity of helicases (Anand et al., 2012), which disentangle the tetraplex structures. HSV-1 probably exploits similar cellular or viral enzymes to unwind its G-quadruplex sequences and allow replication/transcription events to proceed. Moreover, G-quadruplex motifs have been associated with over 90% of DNA replication origins (Besnard et al., 2012).
Here we have used BRACO-19, a tri-substituted acridine characterized by excellent G-quadruplex binding properties, to investigate the effect of HSV-1 G-quadruplex stabilization. We have shown that BRACO-19 stabilized all tested sequences and that it was able to arrest viral DNA processing in vitro. In infected cells, this activity resulted in less viral DNA being synthesized in treated cells, shut down of late, but not immediate-early and early protein mRNAs and overall, in inhibition of viral production.
The activity of BRACO-19 was compared to that of TMPyP2, a molecule that shares the chemical features of a G-quadruplex ligand, but has a much lower binding affinity toward G-quadruplexes (Han et al., 2001). At the highest concentrations, TMPyP2 displayed antiviral activity, which, based on the observation that TMPyP2 was essentially inactive in the polymerase assays and it did not modify the amount of intracellular DNA, likely exploits a non-G-quadruplex related mechanism.
The time-of-addition assay indicated an early time range of activity (8–10 h.p.i.) of BRACO-19, similarly to ACV, which targets viral DNA polymerase. Altogether these data indicate that BRACO-19 inhibits viral DNA replication by stabilizing the extended clusters of G-quadruplex mainly present in non-coding regions of the viral genome.
It has to be noted that (1) the time-of-addition indicates only the last step affected by the antiviral molecule, therefore earlier steps may also be affected by BRACO-19; (2) only large clusters of G-quadruplex sequences have been considered in the present work. Because of the abundance of G nucleotides in the HSV-1 genome, additional single tetraplexes in key regions of the HSV-1 genome may form to promote multiple yet non-identified functions required for the viral biology.
Since its first introduction in the 1980s, ACV has been the antiviral drug of choice for the treatment of HSV-1 infections (Vere Hodge and Field, 2013). However, the emergence of resistance to ACV has created an obstacle for the treatment of HSV-1 (Bacon et al., 2003). Because of the inherent different mechanism of action, G-quadruplex ligands could be envisaged as therapeutic options against HSV-1 strains resistant to current anti-herpetic drugs.
In conclusion, this work provides a proof of concept for the development of selective anti-HSV-1 agents with an innovative mechanism of action.
Transparency declarations
None to declare.
Acknowledgements
This work was supported by the Italian Ministry of University and Research (FIRB-Ideas RBID082ATK_001), by the Bill and Melinda Gates Foundation (GCE Grants OPP1035881 and OPP1097238), by the European Research Council (ERC Consolidator Grant 615879) (S.N.R.); by a Grant from the Canadian Institute of Health Research (L.F.).
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.antiviral.2015.03.016.
Appendix A. Supplementary data
This document contains supplementary information.
References
- Anand R.P., Shah K.A., Niu H., Sung P., Mirkin S.M., Freudenreich C.H. Overcoming natural replication barriers: differential helicase requirements. Nucleic Acids Res. 2012;40:1091–1105. doi: 10.1093/nar/gkr836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacon T.H., Levin M.J., Leary J.J., Sarisky R.T., Sutton D. Herpes simplex virus resistance to acyclovir and penciclovir after two decades of antiviral therapy. Clin. Microbiol. Rev. 2003;16:114–128. doi: 10.1128/CMR.16.1.114-128.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian S., Hurley L.H., Neidle S. Targeting G-quadruplexes in gene promoters: a novel anticancer strategy? Nat. Rev. Drug Discov. 2010;10:261–275. doi: 10.1038/nrd3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaume N., Pathak R., Yadav V.K., Kota S., Misra H.S., Gautam H.K., Chowdhury S. Genome-wide study predicts promoter-G4 DNA motifs regulate selective functions in bacteria: radioresistance of D. radiodurans involves G4 DNA-mediated regulation. Nucleic Acids Res. 2013;41:76–89. doi: 10.1093/nar/gks1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besnard E., Babled A., Lapasset L., Milhavet O., Parrinello H., Dantec C., Marin J.M., Lemaitre J.M. Unraveling cell type-specific and reprogrammable human replication origin signatures associated with G-quadruplex consensus motifs. Nat. Struct. Mol. Biol. 2012;19:837–844. doi: 10.1038/nsmb.2339. [DOI] [PubMed] [Google Scholar]
- Biffi G., Tannahill D., McCafferty J., Balasubramanian S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013;5:182–186. doi: 10.1038/nchem.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger A.M., Dai F., Schultes C.M., Reszka A.P., Moore M.J., Double J.A., Neidle S. The G-quadruplex-interactive molecule BRACO-19 inhibits tumor growth, consistent with telomere targeting and interference with telomerase function. Cancer Res. 2005;65:1489–1496. doi: 10.1158/0008-5472.CAN-04-2910. [DOI] [PubMed] [Google Scholar]
- Cahoon L.A., Seifert H.S. An alternative DNA structure is necessary for pilin antigenic variation in Neisseria gonorrhoeae. Science. 2009;325:764–767. doi: 10.1126/science.1175653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campadelli-Fiume G., Amasio M., Avitabile E., Cerretani A., Forghieri C., Gianni T., Menotti L. The multipartite system that mediates entry of herpes simplex virus into the cell. Rev. Med. Virol. 2007;17:313–326. doi: 10.1002/rmv.546. [DOI] [PubMed] [Google Scholar]
- Daelemans D., Pauwels R., De Clercq E., Pannecouque C. A time-of-drug addition approach to target identification of antiviral compounds. Nat. Protoc. 2011;6:925–933. doi: 10.1038/nprot.2011.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duquette M.L., Handa P., Vincent J.A., Taylor A.F., Maizels N. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev. 2004;18:1618–1629. doi: 10.1101/gad.1200804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frangoul H., Wills M., Crossno C., Engel M., Domm J. Acyclovir-resistant herpes simplex virus pneumonia post-unrelated stem cell transplantation: a word of caution. Pediatr. Transplant. 2007;11:942–944. doi: 10.1111/j.1399-3046.2007.00822.x. [DOI] [PubMed] [Google Scholar]
- Freeman E.E., Weiss H.A., Glynn J.R., Cross P.L., Whitworth J.A., Hayes R.J. Herpes simplex virus 2 infection increases HIV acquisition in men and women: systematic review and meta-analysis of longitudinal studies. Aids. 2006;20:73–83. doi: 10.1097/01.aids.0000198081.09337.a7. [DOI] [PubMed] [Google Scholar]
- Genereau T., Lortholary O., Bouchaud O., Lacassin F., Vinceneux P., De Truchis P., Jaccard A., Meynard J.L., Verdon R., Sereni D., Marche C., Coulaud J.P., Guillevin L. Herpes simplex esophagitis in patients with AIDS: report of 34 cases. The Cooperative Study Group on Herpetic Esophagitis in HIV Infection. Clin. Infect. Dis. 1996;22:926–931. doi: 10.1093/clinids/22.6.926. [DOI] [PubMed] [Google Scholar]
- Gowan S.M., Harrison J.R., Patterson L., Valenti M., Read M.A., Neidle S., Kelland L.R. A G-quadruplex-interactive potent small-molecule inhibitor of telomerase exhibiting in vitro and in vivo antitumor activity. Mol. Pharmacol. 2002;61:1154–1162. doi: 10.1124/mol.61.5.1154. [DOI] [PubMed] [Google Scholar]
- Han H., Langley D.R., Rangan A., Hurley L.H. Selective interactions of cationic porphyrins with G-quadruplex structures. J. Am. Chem. Soc. 2001;123:8902–8913. doi: 10.1021/ja002179j. [DOI] [PubMed] [Google Scholar]
- Harrison R.J., Cuesta J., Chessari G., Read M.A., Basra S.K., Reszka A.P., Morrell J., Gowan S.M., Incles C.M., Tanious F.A., Wilson W.D., Kelland L.R., Neidle S. Trisubstituted acridine derivatives as potent and selective telomerase inhibitors. J. Med. Chem. 2003;46:4463–4476. doi: 10.1021/jm0308693. [DOI] [PubMed] [Google Scholar]
- Hayward G.S., Jacob R.J., Wadsworth S.C., Roizman B. Anatomy of herpes simplex virus DNA: evidence for four populations of molecules that differ in the relative orientations of their long and short components. Proc. Natl. Acad. Sci. U.S.A. 1975;72:4243–4247. doi: 10.1073/pnas.72.11.4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold B.C., WuDunn D., Soltys N., Spear P.G. Glycoprotein C of herpes simplex virus type 1 plays a principal role in the adsorption of virus to cells and in infectivity. J. Virol. 1991;65:1090–1098. doi: 10.1128/jvi.65.3.1090-1098.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershman S.G., Chen Q., Lee J.Y., Kozak M.L., Yue P., Wang L.S., Johnson F.B. Genomic distribution and functional analyses of potential G-quadruplex-forming sequences in Saccharomyces cerevisiae. Nucleic Acids Res. 2008;36:144–156. doi: 10.1093/nar/gkm986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppert J.L. Four-stranded nucleic acids: structure, function and targeting of G-quadruplexes. Chem. Soc. Rev. 2008;37:1375–1384. doi: 10.1039/b702491f. [DOI] [PubMed] [Google Scholar]
- James S.H., Prichard M.N. Current and future therapies for herpes simplex virus infections: mechanism of action and drug resistance. Curr. Opin. Virol. 2014;8C:54–61. doi: 10.1016/j.coviro.2014.06.003. [DOI] [PubMed] [Google Scholar]
- Le V.H., Nagesh N., Lewis E.A. Bcl-2 promoter sequence G-quadruplex interactions with three planar and non-planar cationic porphyrins: TMPyP4, TMPyP3, and TMPyP2. PLoS One. 2013;8:e72462. doi: 10.1371/journal.pone.0072462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim K.W., Alberti P., Guedin A., Lacroix L., Riou J.F., Royle N.J., Mergny J.L., Phan A.T. Sequence variant (CTAGGG)n in the human telomere favors a G-quadruplex structure containing a G.C.G.C tetrad. Nucleic Acids Res. 2009;37:6239–6248. doi: 10.1093/nar/gkp630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick L., Roller R.J., Roizman B. Characterization of a herpes simplex virus sequence which binds a cellular protein as either a single-stranded or double-stranded DNA or RNA. J. Virol. 1992;66:3435–3447. doi: 10.1128/jvi.66.6.3435-3447.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNabb D.S., Courtney R.J. Analysis of the UL36 open reading frame encoding the large tegument protein (ICP1/2) of herpes simplex virus type 1. J. Virol. 1992;66:7581–7584. doi: 10.1128/jvi.66.12.7581-7584.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murat P., Zhong J., Lekieffre L., Cowieson N.P., Clancy J.L., Preiss T., Balasubramanian S., Khanna R., Tellam J. G-quadruplexes regulate Epstein–Barr virus-encoded nuclear antigen 1 mRNA translation. Nat. Chem. Biol. 2014;10:358–364. doi: 10.1038/nchembio.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidle S. Human telomeric G-quadruplex: the current status of telomeric G-quadruplexes as therapeutic targets in human cancer. FEBS J. 2010;277:1118–1125. doi: 10.1111/j.1742-4658.2009.07463.x. [DOI] [PubMed] [Google Scholar]
- Neidle S., Parkinson G. Telomere maintenance as a target for anticancer drug discovery. Nat. Rev. Drug Discov. 2002;1:383–393. doi: 10.1038/nrd793. [DOI] [PubMed] [Google Scholar]
- Norseen J., Johnson F.B., Lieberman P.M. Role for G-quadruplex RNA binding by Epstein–Barr virus nuclear antigen 1 in DNA replication and metaphase chromosome attachment. J. Virol. 2009;83:10336–10346. doi: 10.1128/JVI.00747-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang Q., Zhao X., Feng H., Tian Y., Li D., Li M., Tan Z. High GC content of simple sequence repeats in Herpes simplex virus type 1 genome. Gene. 2012;499:37–40. doi: 10.1016/j.gene.2012.02.049. [DOI] [PubMed] [Google Scholar]
- Patel D.J., Phan A.T., Kuryavyi V. Human telomere, oncogenic promoter and 5′-UTR G-quadruplexes: diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 2007;35:7429–7455. doi: 10.1093/nar/gkm711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels R., Andries K., Desmyter J., Schols D., Kukla M.J., Breslin H.J., Raeymaeckers A., Van Gelder J., Woestenborghs R., Heykants J. Potent and selective inhibition of HIV-1 replication in vitro by a novel series of TIBO derivatives. Nature. 1990;343:470–474. doi: 10.1038/343470a0. [DOI] [PubMed] [Google Scholar]
- Perrone R., Nadai M., Frasson I., Poe J.A., Butovskaya E., Smithgall T.E., Palumbo M., Palu G., Richter S.N. A dynamic G-quadruplex region regulates the HIV-1 long terminal repeat promoter. J. Med. Chem. 2013;56:6521–6530. doi: 10.1021/jm400914r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrone R., Nadai M., Poe J.A., Frasson I., Palumbo M., Palu G., Smithgall T.E., Richter S.N. Formation of a unique cluster of G-quadruplex structures in the HIV-1 Nef coding region: implications for antiviral activity. PLoS One. 2013;8:e73121. doi: 10.1371/journal.pone.0073121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrone R., Butovskaya E., Daelemans D., Palu G., Pannecouque C., Richter S.N. Anti-HIV-1 activity of the G-quadruplex ligand BRACO-19. J. Antimicrob. Chemother. 2014 doi: 10.1093/jac/dku280. [DOI] [PubMed] [Google Scholar]
- Phan A.T. Human telomeric G-quadruplex: structures of DNA and RNA sequences. FEBS J. 2010;277:1107–1117. doi: 10.1111/j.1742-4658.2009.07464.x. [DOI] [PubMed] [Google Scholar]
- Phan A.T., Kuryavyi V., Patel D.J. DNA architecture: from G to Z. Curr. Opin. Struct. Biol. 2006;16:288–298. doi: 10.1016/j.sbi.2006.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawal P., Kummarasetti V.B., Ravindran J., Kumar N., Halder K., Sharma R., Mukerji M., Das S.K., Chowdhury S. Genome-wide prediction of G4 DNA as regulatory motifs: role in Escherichia coli global regulation. Genome Res. 2006;16:644–655. doi: 10.1101/gr.4508806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read M., Harrison R.J., Romagnoli B., Tanious F.A., Gowan S.H., Reszka A.P., Wilson W.D., Kelland L.R., Neidle S. Structure-based design of selective and potent G quadruplex-mediated telomerase inhibitors. Proc. Natl. Acad. Sci. U.S.A. 2001;98:4844–4849. doi: 10.1073/pnas.081560598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeyre C., Lopes J., Boule J.B., Piazza A., Guedin A., Zakian V.A., Mergny J.L., Nicolas A. The yeast Pif1 helicase prevents genomic instability caused by G-quadruplex-forming CEB1 sequences in vivo. PLoS Genet. 2009;5:e1000475. doi: 10.1371/journal.pgen.1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roizman B., Knipe D.M., Whitley R.J. sixth ed. Lippincott Williams & Wilkins; Philadelphia, PA, USA: 2013. Herpes Simplex Viruses. [Google Scholar]
- Roller R.J., McCormick A.L., Roizman B. Cellular proteins specifically bind single- and double-stranded DNA and RNA from the initiation site of a transcript that crosses the origin of DNA replication of herpes simplex virus 1. Proc. Natl. Acad. Sci. U.S.A. 1989;86:6518–6522. doi: 10.1073/pnas.86.17.6518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen D., Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988;334:364–366. doi: 10.1038/334364a0. [DOI] [PubMed] [Google Scholar]
- Simonsson T. G-quadruplex DNA structures–variations on a theme. Biol. Chem. 2001;382:621–628. doi: 10.1515/BC.2001.073. [DOI] [PubMed] [Google Scholar]
- Sodeik B., Ebersold M.W., Helenius A. Microtubule-mediated transport of incoming herpes simplex virus 1 capsids to the nucleus. J. Cell Biol. 1997;136:1007–1021. doi: 10.1083/jcb.136.5.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J., Vonrhein C., Smart O.S., Bricogne G., Bollati M., Kusov Y., Hansen G., Mesters J.R., Schmidt C.L., Hilgenfeld R. The SARS-unique domain (SUD) of SARS coronavirus contains two macrodomains that bind G-quadruplexes. PLoS Pathog. 2009;5:e1000428. doi: 10.1371/journal.ppat.1000428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vere Hodge R.A., Field H.J. Antiviral agents for herpes simplex virus. Adv. Pharmacol. 2013;67:1–38. doi: 10.1016/B978-0-12-405880-4.00001-9. [DOI] [PubMed] [Google Scholar]
- Verma A., Halder K., Halder R., Yadav V.K., Rawal P., Thakur R.K., Mohd F., Sharma A., Chowdhury S. Genome-wide computational and expression analyses reveal G-quadruplex DNA motifs as conserved cis-regulatory elements in human and related species. J. Med. Chem. 2008;51:5641–5649. doi: 10.1021/jm800448a. [DOI] [PubMed] [Google Scholar]
- Vorlickova M., Kejnovska I., Sagi J., Renciuk D., Bednarova K., Motlova J., Kypr J. Circular dichroism and guanine quadruplexes. Methods. 2012;57:64–75. doi: 10.1016/j.ymeth.2012.03.011. [DOI] [PubMed] [Google Scholar]
- Wieland M., Hartig J.S. Investigation of mRNA quadruplex formation in Escherichia coli. Nat. Protoc. 2009;4:1632–1640. doi: 10.1038/nprot.2009.111. [DOI] [PubMed] [Google Scholar]
- Zhang S., Wu Y., Zhang W. G-quadruplex structures and their interaction diversity with ligands. ChemMedChem. 2014;9:899–911. doi: 10.1002/cmdc.201300566. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This document contains supplementary information.


