Abstract
Substrate ubiquitylation is a reversible process critical to cellular homeostasis that is often dysregulated in many human pathologies including cancer and neurodegeneration. Elucidating the mechanistic details of this pathway could unlock a large store of information useful to the design of diagnostic and therapeutic interventions. Proteomic approaches to the questions at hand have generally utilized mass spectrometry (MS), which has been successful in identifying both ubiquitylation substrates and profiling pan-cellular chain linkages, but is generally unable to connect the two. Interacting partners of the deubiquitylating enzymes (DUBs) have also been reported by MS, although substrates of catalytically competent DUBs generally cannot be. Where they have been used towards the study of ubiquitylation, protein microarrays have usually functioned as platforms for the identification of substrates for specific E3 ubiquitin ligases. Here, we report on the first use of protein microarrays to identify substrates of DUBs, and in so doing demonstrate the first example of microarray proteomics involving multiple (i.e., distinct, sequential and opposing) enzymatic activities. This technique demonstrates the selectivity of DUBs for both substrate and type (mono- versus poly-) of ubiquitylation. This work shows that the vast majority of DUBs are monoubiquitylated in vitro, and are incapable of removing this modification from themselves. This work also underscores the critical role of utilizing both ubiquitin chains and substrates when attempting to characterize DUBs. This article is part of a Special Issue entitled: Ubiquitin Drug Discovery and Diagnostics.
Abbreviations: DUB, deubiquitylase; MS, mass spectrometry; PTM, post-translational modification; ATP, adenosine triphosphate; K48, the forty-eighth amino acid of ubiquitin, a lysine; K63, the sixty-third amino acid of ubiquitin, a lysine; TUBEs, tandem ubiquitin binding entities; c, catalytic (for example, USP2c is the catalytic core domain only of USP2); SDS PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis
Keywords: Ubiquitin, Deubiquitylase, Protein microarray, TUBEs, DUB Chip, Monoubiquitylated
Highlights
►DUB activity depends on both substrate and the type of ubiquitin modification. ►DUBs are monoubiquitylated in vitro. ►Most DUBs do not appear capable of auto-deubiquitylation. ►E3s are hyper-activated for auto-ubiquitylation in the presence of DUBs. ►Microarray proteomics for substrate identification of enzymes that reverse PTMs
1. Introduction
Ubiquitin ligases (E3s) represent the largest family of proteins in humans (~ 620) [1], and largely determine the substrate specificity of ubiquitin transfer to target proteins. De-ubiquitylase enzymes (DUBs), in contrast, remove ubiquitin from proteins to recycle ubiquitin and facilitate proteasomal degradation of substrates, to modify the nature of the linkage (editing), to terminate the signal, and/or to attenuate proteasomal degradation [2]. Ubiquitin itself is found singly on proteins as well as in chains of various linkages, a reference to which of seven lysines of ubiquitin are used to join adjacent moieties within a chain. Much remains to be elucidated concerning the biological meaning of the various linkages, but the current paradigm holds that chains linked through lysine-48 of ubiquitin (K48 chains) signal proteasomal degradation and K63-linked chains are signaling events that modify protein location, interaction, and/or function [3].
Of the two activities regulating substrate ubiquitylation, the E3s have garnered the majority of research interest to date. Despite this fact, it is the DUBs that have attracted the most attention for diagnostic and therapeutic targeting. Reasons for this preference include the simplicity of ubiquitin removal relative to conjugation, a process requiring orchestrated action of three enzymes; the lack of functional redundancy among DUBs relative to the E3s (there are only ~ 90 DUBs [4], [5]); and the success of VelcadeTM (Millennium Pharmaceuticals), a proteasome inhibitor that functionally resembles pan-DUB inhibition. Selective DUB inhibition therefore provides an opportunity for therapeutic efficacy, with decreased likelihood of side effects relative to approaches targeting the whole proteasome.
It is well established that E3s are capable of autoubiquitylation [6], [7], [8], [9] and that many DUBs interact directly with E3s [5]. These DUBs antagonize E3 autoubiquitylation and/or substrate ubiquitylation [10] but little attention has been paid to conversation in the opposite direction, E3 activity towards the DUB. A brief survey of proteomic literature revealed that many DUBs have themselves been putatively identified as being ubiquitylated [11], [12] in addition to a handful that have been definitively demonstrated as such [13], [14], [15], [16], [17], [18], [19]. We therefore took advantage of a DUB protein microarray to investigate the extent of DUB ubiquitylation, and here report that the vast majority of these proteins can be ubiquitylated in vitro. Furthermore, through the use of DUB-specific antibodies and polyubiquitin specific tandem ubiquitin binding entities (TUBEs; [20]), we demonstrate that monoubiquitylation is the predominant form of modification even in vitro among these enzymes. Finally, given the ability of E3s to ubiquitylate themselves, we investigated whether or not the DUBs could remove or prevent their own modification, and found that most could not. Indeed, most were found incapable of completely deubiquitylating other DUBs as well. Interestingly, several different activity profiles were discovered from among the DUBs examined; profiles revealed by the use of actual substrates with isopeptide-linked ubiquitin chains. Particularly in high throughput, this substrate/chain profiling is impossible via MS or alternative biochemical means (e.g., ubiquitin rhodamine or free chains). To our knowledge, this is the first demonstration of microarray proteomics towards the investigation of substrates for enzymes that remove post translational modifications (PTMs); we believe the technique will also prove informative to the study of other reversible PTMs (phosphorylation for example).
2. Materials and methods
2.1. Protein production
Protein production as previously described [21].
2.2. Array production
Array production by GenTel Biosciences, Inc (Madison, WI) as previously described [21].
2.3. Array ubiquitylation
Arrays, stored at − 80 °C were equilibrated to room temperature for 1 h prior to opening package seals to prevent condensation from forming on the array. Arrays were then placed into GenTels SIMplex 16‐Multi Array system (catalog #4-1001). Microarrayed proteins were ubiquitylated by adding 100 μl per subarray of 5 nM E1 (Ube1; LifeSensors #UB101), 100 nM E2 (UBE2D3; LifeSensors #UB201), 20 nM each E3 (as indicated in text; LifeSensors), 0.02 mg/ml ubiquitin (LifeSensors #SI201), and 200 μM ATP (Sigma Aldrich) in array ubiquitylation buffer (50 mM Tris pH 8, 50 mM NaCl, 50 mM MgSO4, 0.1% Tween-20, 1 mM dithiothreitol, 1% bovine serum albumin); reactions were incubated at room temperature for 1 h. Each subarray was washed once with 200 μl phosphate buffered saline with 0.05% Tween-20 (PBST), then three times with phosphate buffered saline (PBS). Arrays were then deubiquitylated or directly visualized, but always kept wet. Lysate-driven ubiquitylation was done by diluting lysate (made as described below) into lysis buffer to the indicated concentrations, and applying directly to arrays for 1 h at room temperature. Arrays were then washed free of lysate (as above) and visualized (Section 2.5).
2.4. Deubiquitylation of pre-ubiquitylated arrays
After washing, arrays were incubated for 1 h with 100 nM of the indicated DUB (LifeSensors, Inc.) in deubiquitylase buffer (20 mM Tris pH 7.5, 150 mM NaCl, 10 mM dithiothreitol, 0.05% CHAPS). Arrays were then washed as described in Section 2.3.
2.5. Array-signal detection
Array-signal detection using TUBEs was as previously described [22]; FK2 (Millipore) detection was with primary antibody diluted 1:1000 in PBST, and fluorescein-conjugated anti-mouse (Rockland 210–1201) 1:50 in PBST.
2.6. Lysate preparation
HEK293T cells were grown in 10 cm dishes to 80–90% confluency. When stated, cells were treated with 50 μM pan-DUB inhibitor PR-619 (LifeSensors, Inc.) for 1–2 h. Cells were harvested by scraping in ice cold PBS and collected by centrifugation (3000 rpm). Cell pellets were subjected to one freeze–thaw cycle (− 80 °C) and resuspended in cold cell lysis buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 10% glycerol, 1 mM PMSF, protease inhibitor cocktail and 20 μg/ml aprotinin) and vortexed several times. Lysates were centrifuged at 13,000 rpm for 20 min at 4 °C and supernatants collected.
2.7. SDS PAGE gel electrophoresis, protein transfer to nitrocellulose membrane and Western blots
SDS PAGE gel electrophoresis, protein transfer to nitrocellulose membrane and Western blots were all done under standard conditions and procedures except for the following: blots with LifeSensors' biotin-conjugated TUBEs required blocking in 3% bovine serum albumin; blots with LifeSensors' VU-1 anti-ubiquitin were incubated after transfer with 0.5% gluteraldehyde/0.1 M potassium phosphate buffer pH 7.0 for 20 min, and primary incubation was done overnight at 4 °C. All reagents were diluted according to manufacturer's instructions; secondary reagents used were goat anti-rabbit-HRP (Rockland #211-1302), peroxidase conjugated avidin (Rockland #A003-03), and goat anti-mouse HRP (Sigma #A4416). Chicken IgY antibodies to UCHL5 and JOSD1 (LifeSensors, Inc.) were used at 1:1000 dilution in PBST and visualized with horseradish peroxidase-conjugated rabbit anti-chicken IgY (Rockland #803-4302) diluted 1:2000 in PBST.
2.8. DUB activity
DUB activity was assayed using CHOP reporter assay as previously described [22], or by using di-ubiquitin IQF substrates (LifeSensors, Inc.) according to manufacturer's instructions.
2.9. E3 ligase activity
E3 ligase activity was measured using the E3Lite Assay as previously described [23]. Also in modified form using a mixture of E3s (where stated) consisting of Praja1, Murf1, and Carp2; and further modified (where stated) by the addition of DUBs (as listed) to the assay wells for 1 hour incubation at room temperature following the initial ubiquitylation reaction.
2.10. In vitro ubiquitylation/deubiquitylation reactions
Soluble proteins were ubiquitylated by adding 5 nM E1 (Ube1; LifeSensors #UB101), 100 nM E2 (UBE2D3; LifeSensors #UB201), 20 nM each E3 (as indicated in text; LifeSensors), 0.02 mg/ml ubiquitin (LifeSensors #SI201), and 200 μM ATP (Sigma Aldrich) into reactions containing 200 ng substrates (as indicated), all in ubiquitylation buffer (50 mM Tris8, 5 mM MgCl2, 0.1% Tween-20, 1 mM 2-mercaptoethanol). Reactions were incubated at room temperature for 1 h. Where indicated, a second DUB was added to the reaction (concentrations listed in figure), which was allowed to proceed for an additional hour. At the end of reactions, 6 × SDS sample buffer was added, and reactions loaded into PAGE, or stored at − 20 °C until later PAGE analysis.
3. Results
3.1. DUBs are ubiquitylated by various E3s
We tested various E3s for the ability to ubiquitylate a microarray of immobilized DUBs previously developed and described (DUB Chip) [21], [24]. This protein microarray consisted of approximately 40 DUB and ubiquitin-like isopeptidases and enabled us to investigate various conditions under which some of them might be ubiquitylated. Multiple identical arrays were incubated with solution containing E1, E2, ubiquitin, and ATP. Ubiquitylation reactions were initiated by the addition of various E3s into duplicate arrays, individually or in combination (rows as listed in Fig. 1A). All arrays were then thoroughly washed to remove soluble reaction components. Finally, within each experimental condition tested, one of the two identically ubiquitylated arrays was visualized using either an antibody towards ubiquitin (FK2) as the primary detection reagent, or biotinylated tandem ubiquitin binding entities (TUBEs). While the antibody recognizes ubiquitin conjugated to substrate individually (monoubiquitylation) as well as in chains (polyubiquitylation), TUBEs are specific to detection of polyubiquitylation events [20] without preference to specific chain linkage [25]. The absence of signal from reactions lacking E3 demonstrated that observed signals were ubiquitylation dependent (bottom row, Fig. 1A). Each of the E3s shown in Fig. 1A was capable of ubiquitylating a distinct but overlapping subset of immobilized DUBs (rows 1–5 left column) and appeared to work additively when co-incubated (row 6); interestingly, Praja1 was the only E3 tested that exhibited an ability to polyubiquitylate these substrates (rows 1–5, compare left and right columns). On the other hand, all of these E3s were capable of forming polyubiquitin chains in concert with UBE2D3 in the E3Lite autoubiquitylation assay (Fig. 1B). A summary of all recombinant E3/immobilized substrate ubiquitylation events is shown in Fig. 1C. Together, these results show that 1) the vast majority of DUBs were ubiquitylated in vitro, 2) most E3s tested were capable of effecting this modification, 3) the events appeared to be predominantly monoubiquitylation, and 4) the substrates themselves influenced the extent of ubiquitylation (i.e. mono- versus poly-) in an E3 dependent manner.
Fig. 1.
Ubiquitylation of DUB Chip. A. Individual, identical protein microarrays were ubiquitylated with five different E3s (individually as listed), a mixture of three labeled “mix” (Praja1, Carp2, and Murf1), or using reaction mixture lacking an E3 (“No E3”). All substrates were printed in triplicate, and all conditions were tested on two separate (identical) arrays. One of the two was visualized using antibody capable of seeing monoubiquitylation events (left column of arrays labeled “Ab”); the other using polyubiquitin-specific TUBEs (right column of arrays labeled “TUBEs”). B. E3Lite assay (LifeSensors, Inc.) data (signal versus time) showing all E3s used in (A) were capable of forming polyubiquitin chains (either free or E3-conjugated). C. Data from ubiquitylation reactions (A) visually organized by E3 (columns) and substrate (rows). The column labeled “TUBEs” represents the array ubiquitylated with E3 mix and visualized with TUBEs; all other columns represent data from arrays visualized with antibody. The identities of substrates by row are: 1. USP20, 2. USP51c, 3. USP5, 4. Ataxin3, 5. USP7c, 6. AMSH, 7. Otubain1, 8. JOSD1, 9. UCHL3, 10. USP8c, 11. UCHL5, 12. SENP6c, 13. SENP1c, 14. USP21, 15. Otubain2, 16. Ataxin3-like, 17. USP33c, 18. USP15, 19. USP28, 20. sseL, 21. USP18, 22. DEN1, 23. PLPro, 24. SENP2, 25. USP34c, 26. USP14, 27. USP8, 28. hSTAM1, 29. USP2c, 30. USP7, 31. BAP1, 32. JOSD2, 33. YOD1, 34. UCHL1, 35. USP21c, 36. Ubiquitin, 37. Ulp1c, 38. PLP2, 39. USP4.
3.2. Profiling DUBs against multiplexed ubiquitylated substrates
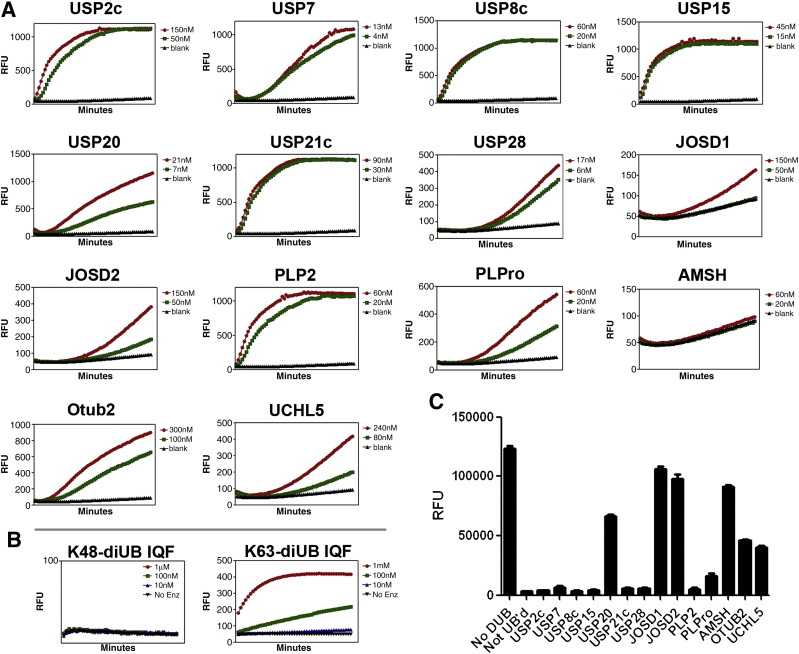
Based on the observation that these E3s together ubiquitylated the vast majority of proteins immobilized on the array, we speculated that this ubiquitylated array might provide a powerful tool to examine the behavior of certain deubiquitylase enzymes against multiple ubiquitylated substrates in parallel. To maximize ubiquitylation events on the array, we utilized a cocktail of E3s (Murf1, Carp2, Praja1). After 1 h, soluble ubiquitylation reaction components were removed from the arrays which were then thoroughly washed before soluble DUBs were applied to the ubiquitylated array (as listed Figs. 2A,C) and incubated for another hour. Washed and dried arrays were then visualized for total ubiquitylation (mono- and poly-; Fig. 2A) or polyubiquitylation events (Fig. 2C), as above. Loss of signal between an array incubated with DUB compared to one mock treated would indicate putative substrates of that DUB. Complete array data for every combination of immobilized substrate and soluble enzyme is presented as Figs. 2B,D. Results indicated that the vast majority of these DUBs were unable to cleave ubiquitin from the immobilized substrates. To verify that the enzymes were enzymatically competent, they were assayed using the CHOP reporter system (LifeSensors, Inc., Fig. 3A). All enzymes tested displayed activity by this assay, save AMSH. Since this DUB is known to prefer K63 linked chains, we incubated it with a K63 linked internally quenched fluorescence based di-ubiquitin substrate (LifeSensors, Inc.). As shown in Fig. 3B, the enzyme displayed robust activity against this substrate, demonstrating that defining activity varies by assay. With this point in mind, we also tested each of the DUBs for their ability to reverse polyubiquitylation signals in the E3Lite assay, which captures and reports autoubiquitylated E3s. As seen in Fig. 3C, utilizing the E3 mix resulted in signal 40-fold higher than the reaction mix lacking an E3. With the possible exceptions of JOSD1, JOSD2, and K63 specific AMSH [26], [27], all DUBs tested demonstrated activity for shortening or removing E3-linked polyubiquitin chains by appreciably reversing the assay signal, including even K48 specific UCHL5 [26]. Importantly, many DUBs were capable of complete signal reversal (USP2c, USP7, USP8c, USP15, USP21c, USP28, PLP2; and 87% reversal for PLPro).
Fig. 2.
Deubiquitylation of DUB Chip. A. Sixteen identical protein microarrays were all ubiquitylated with E3 mix (Praja1, Carp2, and Murf1). After washing away soluble ubiquitylation machinery, individual DUBs (as listed, including mock-treated arrays labeled “No DUB”) were applied to separate arrays, which were then visualized by antibody capable of seeing monoubiquitylation events. Loss of signal indicates putative substrates of the DUB tested. B. Array data from (A) visually organized by DUB tested (columns) and ubiquitylated substrates (rows). The identities of substrates by row are: 1. USP20, 2. USP51c, 3. USP5, 4. Ataxin3, 5. USP7c, 6. AMSH, 7. Otubain1, 8. JOSD1, 9. UCHL3, 10. USP8c, 11. UCHL5, 12. USP21, 13. Otubain2, 14. Ataxin3-like, 15. USP33c, 16. USP15, 17. USP28, 18. sseL, 19. DEN1, 20. PLPro, 21. USP14, 22. USP8, 23. hSTAM1, 24. USP2c, 25. USP7, 26. BAP1, 27. YOD1, 28. UCHL1, 29. USP21c, 30. Ubiquitin, 31. Ulp1c, 32. PLP2, 33. USP4. C. Experiment identical to that described in (A), except that the array was visualized using polyubiquitin-specific TUBEs. D. Array data from (C) visually organized by DUB tested (columns) and ubiquitylated substrates (rows, as above).
Fig. 3.
DUBs examined for behavior against ubiquitylated substrates were active enzymes. A. The fourteen DUBs examined in Fig. 2 all showed activity (signal versus time over 1 h) by CHOP reporter assay (LifeSensors, Inc.), except AMSH. B. AMSH was examined for activity (versus time over 1 h) against different substrates, di-ubiquitin substrates labeled with an internally quenched fluorophore pair. As expected, activity was specific to K63-linkage of the diubiquitin. C. E3Lite assay data (endpoint RFU signal) for triplicate wells either ubiquitylated with E3 mix (Praja1, Carp2, Murf1) and then mock treated with DUB (“No DUB”); mock ubiquitylated using reaction mixture lacking an E3 and then mock treated with DUB (“Not UB'd”); or ubiquitylated with E3 mix and then treated with DUB enzyme (as listed).
Like the ubiquitylated protein microarray, the modified E3Lite assay described above (Fig. 3C) results in immobilization of ubiquitylated proteins (the E3s) presented as potential substrates to DUBs in solution phase. The eight DUBs capable of nearly complete E3Lite signal reversal (listed above) thus demonstrated an ability to function under these parameters, and enabled a more careful look at their behavior towards different substrates, ubiquitylated DUBs.
As shown in Fig. 3B,C, the catalytic domains of USP2 and USP8 were each capable of completely deubiquitylating the microarray. The catalytic domain of USP21, on the other hand, appeared more selective in its substrate preference (Figs. 4 , and S1). Although the absence of non-catalytic regulatory regions tempers the ability to draw conclusions, it was nonetheless interesting that USP21c appeared to cut “all or nothing”. That is, wherever USP21c shortened chains it also removed the final monoubiquitin. USP28 displayed little selectivity towards shortening substrate-bound chains (aside from its inability to act on Ataxin3-like), but was incapable of complete substrate deubiquitylation for any of the substrates examined (Figs. 4, S2). A fifth pattern emerged from analysis of USP15 data (Figs. 4, S3); which appeared incapable of completely deubiquitylating any of the substrates examined, yet displayed substrate selectivity for chain shortening. PLP2 (NL63 Coronavirus) identified JOSD1 and UCHL1 as putative substrates (Figs. 4, S4); PLPro (SARS Coronavirus) identified USP4 as a putative substrate and was the only DUB examined that displayed an ability to shorten chains bound to Ataxin3-like (Figs. 4, S5). Notably, there were no DUBs (save catalytic domains) capable of de(mono)ubiquitylating themselves; all DUBs examined were capable of cleaving free chains (Fig. 4, rows 1, TUBEs detection column).
Fig. 4.

Profiling DUB activity requires both chains and substrates. Data from Fig. 2B and D has been arranged to better illustrate activity differences among the DUBs profiled. Five DUB profiles are shown (vertical labels left of the figure). The activity of each of these DUBs is shown against five rows of ubiquitylated substrates (labeled right of the figure for the first profile only). The first four rows of substrates are invariant among all profiles. The variable rows (labeled *) for each of the first three profiled DUBs are intended to show the extent of self-cleavage for these profiled enzymes, and thus are immobilized USP21c, USP28, and USP15, respectively. Within the last two DUB profiles (PLP2 and PLPro) the variable rows are intended to show putative substrates (UCHL1 and USP4, respectively). The boxed column (of triplicate features) shows the effect of treatment of ubiquitylated substrates with each of the DUBs profiled, and is shown in between the maximum possible signal to the left (No DUB) and minimum possible signal to the right (USP8c, a non-specific and highly active DUB). Data is shown from arrays reporting polyubiquitin only (TUBEs detection) and from arrays showing mono- and poly‐ubiquitylation events (Antibody). Complete profiles, arranged identically, are shown in Supplemental Fig. 1, Supplemental Fig. 2, Supplemental Fig. 3, Supplemental Fig. 4, Supplemental Fig. 5.
Profiling DUB activity requires both chains and substrates. Data from Fig. 2B and D has been arranged to better illustrate activity differences among the DUBs profiled. Five DUB profiles are shown (vertical labels left of the figure). The activity of each of these DUBs is shown against five rows of ubiquitylated substrates (labeled right of the figure for the first profile only). The first four rows of substrates are invariant among all profiles. The variable rows (labeled *) for each of the first three profiled DUBs are intended to show the extent of self-cleavage for these profiled enzymes, and thus are immobilized USP21c, USP28, and USP15, respectively. Within the last two DUB profiles (PLP2 and PLPro) the variable rows are intended to show putative substrates (UCHL1 and USP4, respectively). The boxed column (of triplicate features) shows the effect of treatment of ubiquitylated substrates with each of the DUBs profiled, and is shown in between the maximum possible signal to the left (No DUB) and minimum possible signal to the right (USP8c, a non-specific and highly active DUB). Data is shown from arrays reporting polyubiquitin only (TUBEs detection) and from arrays showing mono- and poly‐ubiquitylation events (Antibody). Complete profiles, arranged identically, are shown in Supplementary Figs. 1–5.
3.3. Verification of array-based observations
Because ubiquitylation of array-immobilized DUBs could in theory be a technical artifact of their immobilization (e.g., surface-driven conformational change that exposes an otherwise inaccessible lysine), various DUBs were selected for in vitro ubiquitylation reactions in solution phase with Murf1 as the E3. We used polyubiquitin-specific TUBEs (Fig. 5A) and observed enhanced high molecular weight smearing (characteristic of polyubiquitylation) in reactions containing most DUBs as compared with the no DUB control. The reactions containing USP2c and USP8c were exceptions inasmuch as there was no discernible ubiquitylation in either, likely reflecting the high and indiscriminate deubiquitylase activity of both of these catalytic domains (see Figs. 2A and 3).
Fig. 5.
E3 auto(poly)ubiquitylation and DUB monoubiquitylation in gel. A. Ubiquitylation reactions with Murf1 (shown by array to monoubiquitylate DUB substrates) as the E3 were run in the presence of various DUBs. Reactions were SDS PAGE separated, transferred to nitrocellulose, and probed with TUBEs to observe any high molecular weight polyubiquitylation. Most reactions with DUBs displayed enhanced smearing characteristic of polyubiquitylation, compared to the reaction containing no DUB (first lane). B. Antibody to UCHL5 was used to probe separated reaction components from complete ubiquitylation reactions including Praja1 as the E3 (shown by array to be capable of DUB polyubiquitylation) and UCHL5 as substrate (column labeled (+)). Inclusion of all components except Praja1 (column labeled “-E3”) showed that the observed 8 kD shift (indicated by arrow) above the predominant form of the substrate was ubiquitylation dependent. Similarity of this lane to one containing only UCHL5 protein substrate (column labeled “Pr”) confirms that the antibody was not cross reacting to any ubiquitylation machinery (e.g., ubiquitin or charged E2). C. Identical to Fig. 5B except that blot was visualized with anti-JOSD1 antibody, and JOSD1 was the substrate included in the reactions (or protein alone lane). D. Ubiquitin antibody (VU1) was used to visualize (via immunoblot) soluble components of in vitro ubiquitylation reactions in which E3 was pre-immobilized to the surface of the well (and therefore not present in the gel/blot) and DUB omitted (negative control, lane 1); where all components were soluble including UCHL5 as substrate (positive control, lane 2); and where UCHL5 was included but the E3 (Murf2) was again pre-immobilized to the well surface (lane 3). Arrow indicates expected size of monoubiquitylated UCHL5.
Although confirmatory of DUBs being ubiquitylated per se, polyubiquitylation observed in solution versus monoubiquitylation on the array suggested that chain formation might have been antagonized on the array by substrate immobilization. Alternatively, the observed polyubiquitylation seen in solution might represent enhanced E3 autoubiquitylation activity. To differentiate E3 and DUB ubiquitylation, we used antibodies against UCHL5 and JOSD1 to visualize solution-based in vitro ubiquitylation reactions. UCHL5 has been implicated by MS-based proteomics as being ubiquitylated, but this has not been independently validated [11]. JOSD1 has never been reported to be ubiquitylated. As shown in Fig. 5 both UCHL5 (Fig. 5B) and JOSD1 (Fig. 5C) were exclusively mono-ubiquitylated in vitro. To further distinguish these possibilities (i.e., E3 auto-ubiquitylation and DUB ubiquitylation), Murf2 (shown by array to monoubiquitylate DUBs) was included in ubiquitylation reactions with UCHL5 (shown by array to be ubiquitylated by Murf2). As seen in Fig. 5D (lane 2), and in agreement with Fig. 5A, a high molecular weight smear was observed when all reaction components were soluble. However, prior immobilization of the E3 to the wells of the plate abrogated the appearance of these high molecular weight proteins from the remaining solution-based components and resulted in a single distinct band consistent in size with monoubiquitylated UCHL5 (Fig. 5D lane 3).
As another test of DUB ubiquitylation (and the extent thereof), we made lysate of HEK293T cells and applied it to DUB Chip in various dilutions. Following 1 hour incubation with lysate, arrays were thoroughly washed, then probed for ubiquitin (either FK2 or TUBEs for observing mono- and poly‐events, respectively). As shown in Fig. 6A, the lysate was capable of ubiquitylating immobilized DUBs in a manner consistent with monoubiquitylation. Because the lysate presumably contained various E3s and DUBs, the observed monoubiquitylation might have reflected either deposition of a single ubiquitin or the net effect of DUBs attenuating initially assembled chains. New arrays were therefore treated with HEK293T lysate that had been made in the presence of the pan-DUB inhibitor PR619 (LifeSensors, Inc.) to antagonize DUB activity (e.g., chain shortening). Results were indistinguishable from the untreated lysate (data not shown). This suggested that, consistent with our earlier data (Fig. 1A), DUB monoubiquitylation is a product of single-moiety deposition as opposed to chain shortening. A summary of lysate driven ubiquitylation for each of the proteins on the array is shown as Fig. 6B.
Fig. 6.
HEK293T lysate-driven ubiquitylation of DUB Chip. A. Duplicate protein microarrays were incubated with straight cell lysate (i.e., no inhibitors) at various dilutions, washed, then probed with either antibody (FK2, Millipore; left column) to visualize monoubiquitylation events, or TUBEs (LifeSensors, Inc.) to visualize polyubiquitylation (right column). B. Antibody-visualized data from the above experiment, organized by concentration of lysate (columns) versus substrate (rows, organized and labeled identically to those in Fig. 1C). Note that while most substrates showed dose-dependent ubiquitylation, some were eliminated from analysis by high FK2 background even in the absence of cell lysate (e.g., column 1, rows 10, 29, 35, 36).
For the purposes of verifying array-based deubiquitylation, selected DUBs (the ‘substrates’) were included in complete ubiquitylation reactions for 30 min, at which point a second DUB was added and the reactions were allowed to proceed for an additional hour. Reaction components were then separated by SDS PAGE, transferred to membrane, and probed with antibodies to the DUB ‘substrates’ initially present (JOSD1 and UCHL5) to observe their ubiquitylation status. In agreement with array data, USP2c and USP21c removed ubiquitin from both substrates; PLP2 from only one (ubiquitylated JOSD1); and Ataxin3 showed no activity towards either (Fig. 7 ).
Fig. 7.
Deubiquitylation of soluble ubiquitylated substrates. In vitro ubiquitylation reactions with Praja1 as the E3 were performed in the presence of either JOSD1 or UCHL5 as the intended substrate. After 30 min, a second deubiquitylase enzyme (either USP2c, USP21c, PLP2, or Ataxin3, as indicated) was added to the reactions which were then incubated for another hour. Shown are immunoblots of the separated reaction components probed with primary antibody against the corresponding intended substrate (JOSD1 or UCHL5).
4. Discussion
Of the eighteen DUBs reported to be ubiquitylated in vivo, our array contained twelve: AMSH [13], USP4 [14], USP7 [15], [16], USP15 [17], USP20 [18], Ataxin 3 [11], [12], [19], Bap1 [11], Otub1 [11], UCHL3 [11], UCHL5 [11], USP14 [11], and USP5 [12], all of which were here confirmed as targets of in vitro ubiquitylation. Five of these twelve (Bap1, Otub1, UCHL3, UCHL5, USP14) were initially identified as ubiquitylated in HeLa cells via MS proteomics [11] but never independently confirmed. This study, therefore, represents their first in vitro confirmation as targets of ubiquitylation and suggests putative E3s. Of the 39 substrates examined on the array, the only ubiquitin and ubiquitin-like proteases not ubiquitylated by recombinant E3 mix were SENP1c, SENP6c, and JOSD2. One of these, SENP6c, was ubiquitylated by the lysate, suggesting that our mix lacked an appropriate E3 for this target. UCHL1 was not ubiquitylated by the lysate, despite having been when recombinant E3s were used. This suggests that either the observed in vitro ubiquitylation was artifactual, the lysate used did not contain an appropriate ligase for this substrate, or the lysate contained an UCHL1-specific DUB. Otherwise, the recombinant mix of E3s and the lysate were surprisingly consistent in terms of which substrates were ubiquitylated.
The inability to represent all possible E3/substrate pairings is one explanation for the false negatives that unavoidably accompany any proteomic experiment (MS or array-based). False positives are also inherent to proteomic investigation. In the case of MS-based studies, false positives usually represent indirect effects (for example, proteins identified as hyper-ubiquitylated following DUB mutation/deletion might be substrates for an E3, the inhibitor of which was increasingly degraded by the loss of DUB activity). Array-based proteomics definitively demonstrate direct-effects, but cannot inform as to biological relevance; false positives in this setting generally reflect two proteins capable of interaction that otherwise never do so in cells. However configured, proteomics provides the leads but verification is always required to sort the wheat from the chaff. Cross-complementary strengths and weaknesses make MS and array-based proteomics reciprocal, and suggest a rapid and high-throughput workflow. Namely, performing both types of experiments for the same protein(s) of interest and cross referencing resultant data sets for common hits would identify directly interacting proteins whose interaction occurs in vivo. A slightly modified example of such an approach is the current in vitro verification of in vivo results for Bap1, Otub1, UCHL3, UCHL5, and USP14, previously suggested by MS to be ubiquitylated.
The proteomic alternative to DUB substrate identification (i.e., MS) relies upon a residual di-glycine signature of tryptic ubiquitylated peptides, and is therefore unable to demonstrate chain shortening, the extent of initial ubiquitylation (chain versus mono), or linkage specific activity. Alternative in vitro techniques have profiled the activity of DUBs towards artificial non-isopeptide linked substrates (e.g., UB-Rhodamine) or free chains completely devoid of substrate [27], [28]. Such experiments do not well reflect DUB physiology, since most DUBs do not exist in cells to recognize and cleave free chains (IsoT excepted [29]). The importance of looking at actual substrates with isopeptide-linked chains demonstrated here (Figs. 5, and Supplemental Fig. 1, Supplemental Fig. 2, Supplemental Fig. 3, Supplemental Fig. 4, Supplemental Fig. 5) is intuitively underscored by noting the presence in human (and most eukaryotes) of more DUBs (~ ninety) than unique chain topologies (eight). To our knowledge, this is the first proteomic study of deubiquitylase activity involving true protein substrates that also discriminates between ubiquitin chain shortening versus complete deubiquitylation.
The techniques described here should prove widely informative when applied on a wider proteomic scale (e.g., utilizing a larger, more representative microarray like ProtoArray from Invitrogen, an array containing over 9000 human proteins) and should facilitate investigation into reversal of other PTMs as well (profiling phosphatases, for example). Simple procedural modifications, like utilizing different E2/E3 combinations, or using single-lysine ubiquitin mutants to drive homogeneous chain formation further expand the information potential. Even the current study, which utilized a small array of a single family of enzymes, a single E2 (UBE2D3) reported to form both K11 and K48 linked chains [30], and wild-type ubiquitin, revealed an unexpectedly large breadth of insight into biology of the pathway represented.
The present study demonstrated, most obviously, that the majority of DUBs can be ubiquitylated, and that the predominant form of this modification is monoubiquitin. We showed that most E3s tested are capable of effecting this modification for a distinct but overlapping set of substrates. Because most E3s transferred only a single ubiquitin (Figs. 1A, 5B–D, 6A, and 7) despite being shown capable of auto-polyubiquitylation, we know that there was only one acceptor lysine per DUB; this highlights the degree of specificity maintained in vitro considering that, for example, JOSD1 has 15 lysines and UCHL5 has 25. Because we identified an E3 capable of DUB polyubiquitylation (Praja 1, Fig. 1A), we know that even in vitro, the substrate is capable of influencing the extent to which the modification occurs in an E3-dependent manner. The fact that cell lysates also monoubiquitylated these substrates suggests that the mono-events observed here have biological relevance. We demonstrated that E3s appear hyperactivated for autoubiquitylation in the presence of most DUBs (Fig. 5). This effect could be mediated by conformational change occurring after deposition of ubiquitin to substrate, or through simple interaction with the DUB. Because USP33c and DEN1 were not ubiquitylated in vitro by Murf1 (Fig. 1C column 3, rows 17 and 22, respectively) but mediated this E3-hyperactivation under similar conditions (Fig. 5A), we have limited evidence to support the latter model. Because ubiquitin itself was present as a substrate on the array, we learned that most E3s here tested do not generate free chains in vitro, but Praja1 does (Fig. 1A compare TUBEs to FK2 detection). We demonstrated that most DUBs do not auto-deubiquitylate; nor, in fact, do most act to completely deubiquitylate any other DUBs. By visualizing both poly- and mono-ubiquitin on substrates we were able to observe several characteristic activity behaviors from among the DUBs; behaviors not apparent in studies with free chains considering that all DUBs tested cleaved free chains (loss of signal in Fig. 4 rows 1, TUBEs detection, or Supplemental Fig. 1, Supplemental Fig. 2, Supplemental Fig. 3, Supplemental Fig. 4, Supplemental Fig. 5 rows 30 TUBEs detection). Namely, we have demonstrated that some DUBs cleave chains promiscuously (USP28); some cleave chains with substrate specificity (USP15); some are selective by substrate and cleave “all or nothing” (USP21c) consistent with a model in which the DUB removes chains in toto but only from certain substrates; and finally, some DUBs promiscuously cleave chains, but show selectivity in the removal of the final mono-UB (PLPro and PLP2). Interestingly, some of these profiles have been proposed based on structural prediction of differential interaction among DUB, substrate, and ubiquitin [31].
Two obvious and unexplored questions arising from the current study are 1) what is the biological significance of DUB monoubiquitylation, and 2) is there physiological significance to the substrates identified in vitro for the Coronaviral DUBs (e.g. promoting infection of host)? The most well-studied consequence to DUB ubiquitylation is the hyperactivation of Ataxin3 activity [19]. Preliminary experiments aimed at exploring this possibility for other selected enzymes suggested that this was not, in fact, a functional consequence shared by other DUBs (data not shown). Nothing has yet been attempted to address the second question, but it is worth noting that a ubiquitylated membrane receptor has been reported to be substrate of USP4 (itself the putative substrate of PLPro) [32]; and manipulation of host receptors seems a likely target for invading microorganisms that require endocytosis (and lysosomal avoidance) for infection.
The ability of substrates to dictate the extent of their own ubiquitylation and selectively interact with DUBs points to a model of simple structural recognition, in agreement with recent discoveries [33], [34]. While regulatory cofactors, non-functional inhibitory interactions, spatio-temporal regulation, carefully balanced opposing enzymatic activities, and all other manners of cellular complexity will undoubtedly impact physiology, this system nonetheless appears particularly amenable to reductionist study. Regardless of which substrates or ligases are used, the current work shows both technical feasibility and biological advantage of examining chains and substrates together when characterizing DUB activity.
The following are the supplementary data related to this article.
Complete DUB activity profile of USP21c against all ubiquitylated substrates. Rows are substrates (labeled as in Fig. 1C); columns are labeled according to which DUB was incubated with these substrates where “No DUB” shows maximal ubiquitylation and USP8c shows minimal ubiquitylation possible following DUB treatment. This profile was labeled “all or nothing” based on the observation that USP21c could not cut chains from all substrates (see primarily TUBEs detection rows 3, 7, 11, 23); but where it did cut chains, it also removed the substrate-proximal mono-ubiquitin as well (see primarily antibody detection rows 1, 6, 10, 12).
Complete DUB activity profile of USP28 against all ubiquitylated substrates. (See Supplemental Fig. 1 for explanation of rows and columns.) This profile showed no specificity for chain cleavage (see TUBEs detection, all rows) with the exception of an inability to cleave chains from Ataxin3-like, row 14. However, it was unable to remove the substrate-proximal monoubiquitin from any substrates (see antibody detection, all rows).
Complete DUB activity profile of USP28 against all ubiquitylated substrates. (See Supplemental Fig. 1 for explanation of rows and columns.) This profile showed no specificity for chain cleavage (see TUBEs detection, all rows) with the exception of an inability to cleave chains from Ataxin3-like, row 14. However, it was unable to remove the substrate-proximal monoubiquitin from any substrates (see antibody detection, all rows).
Complete DUB activity profile of USP15 against all ubiquitylated substrates. (See Supplemental Fig. 1 for explanation of rows and columns.) USP15 did not appear capable of removing the substrate-proximal monoubiquitin from any substrates (see antibody detection, all rows). However, it displayed substrate specificity for chains (TUBEs detection, compare the diminution of signal in rows 2, 7, 12, 23 to the undiminished signals of rows 3, 4, 17).
Complete DUB activity profile of USP15 against all ubiquitylated substrates. (See Supplemental Fig. 1 for explanation of rows and columns.) USP15 did not appear capable of removing the substrate-proximal monoubiquitin from any substrates (see antibody detection, all rows). However, it displayed substrate specificity for chains (TUBEs detection, compare the diminution of signal in rows 2, 7, 12, 23 to the undiminished signals of rows 3, 4, 17).
Complete DUB activity profile of PLP2 against all ubiquitylated substrates. (See Supplemental Fig. 1 for explanation of rows and columns.) This profile showed no specificity for chain cleavage (see TUBEs detection, all rows) with the exception of an inability to cleave chains from Ataxin3-like, row 14. UCHL1 (row 28) and maybe JOSD1 (row 8) were identified as putative substrates (see antibody detection).
Complete DUB activity profile of PLP2 against all ubiquitylated substrates. (See Supplemental Fig. 1 for explanation of rows and columns.) This profile showed no specificity for chain cleavage (see TUBEs detection, all rows) with the exception of an inability to cleave chains from Ataxin3-like, row 14. UCHL1 (row 28) and maybe JOSD1 (row 8) were identified as putative substrates (see antibody detection).
Complete DUB activity profile of PLPro against all ubiquitylated substrates. (See Supplemental Fig. 1 for explanation of rows and columns.) This profile showed no specificity for chain cleavage (see TUBEs detection, all rows) and represents the only non-core domain DUB to cleave chains from Ataxin3-like, row 14. USP4 (row 33, antibody detection) was identified as a putative substrate.
Complete DUB activity profile of PLPro against all ubiquitylated substrates. (See Supplemental Fig. 1 for explanation of rows and columns.) This profile showed no specificity for chain cleavage (see TUBEs detection, all rows) and represents the only non-core domain DUB to cleave chains from Ataxin3-like, row 14. USP4 (row 33, antibody detection) was identified as a putative substrate.
Acknowledgements
Sincere gratitude to Steven J. Orcutt and Mark J. Engleka for helpful discussion and critically reviewing this manuscript; to David Kommander for helpful discussion; to K.G. Suresh Kumar for providing HEK293T cell lysate; and to Michael J. Eddins for providing E3Lite data (Fig. 1B). This work was supported by NIH SBIR 5 R44 RR024807-03.
Footnotes
This article is part of a Special Issue entitled: Ubiquitin Drug Discovery and Diagnostics.
References
- 1.Li W., Bengtson M.H., Ulbrich A., Matsuda A., Reddy V.A., Orth A., Chanda S.K., Batalov S., Joazeiro C.A. Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle's dynamics and signaling. PLoS One. 2008;3:e1487. doi: 10.1371/journal.pone.0001487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reyes-Turcu F.E., Ventii K.H., Wilkinson K.D. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu. Rev. Biochem. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kerscher O., Felberbaum R., Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 4.Nijman S.M., Luna-Vargas M.P., Velds A., Brummelkamp T.R., Dirac A.M., Sixma T.K., Bernards R. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 5.Sowa M.E., Bennett E.J., Gygi S.P., Harper J.W. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Skowyra D., Koepp D.M., Kamura T., Conrad M.N., Conaway R.C., Conaway J.W., Elledge S.J., Harper J.W. Reconstitution of G1 cyclin ubiquitination with complexes containing SCFGrr1 and Rbx1. Science. 1999;284:662–665. doi: 10.1126/science.284.5414.662. [DOI] [PubMed] [Google Scholar]
- 7.Suzuki H., Chiba T., Kobayashi M., Takeuchi M., Furuichi K., Tanaka K. In vivo and in vitro recruitment of an IkappaBalpha-ubiquitin ligase to IkappaBalpha phosphorylated by IKK, leading to ubiquitination. Biochem. Biophys. Res. Commun. 1999;256:121–126. doi: 10.1006/bbrc.1999.0296. [DOI] [PubMed] [Google Scholar]
- 8.Yang Y.L., Li X.M. The IAP family: endogenous caspase inhibitors with multiple biological activities. Cell Res. 2000;10:169–177. doi: 10.1038/sj.cr.7290046. [DOI] [PubMed] [Google Scholar]
- 9.Mallery D.L., Vandenberg C.J., Hiom K. Activation of the E3 ligase function of the BRCA1/BARD1 complex by polyubiquitin chains. EMBO J. 2002;21:6755–6762. doi: 10.1093/emboj/cdf691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marfany G., Denuc A. To ubiquitinate or deubiquitinate: it all depends on the partners. Biochem. Trans. 2008;36:833–838. doi: 10.1042/BST0360833. [DOI] [PubMed] [Google Scholar]
- 11.Meierhofer D., Wang X., Huang L., Kaiser P. Quantitative analysis of global ubiquitination in HeLa cells by mass spectrometry. J. Proteome Res. 2008;7:4566–4576. doi: 10.1021/pr800468j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andrews P.S., Schneider S., Yang E., Michaels M., Chen H., Tang J., Emkey R. Identification of substrates of SMURF1 ubiquitin ligase activity utilizing protein microarrays. Assay Drug Dev. Technol. 2010;8:471–487. doi: 10.1089/adt.2009.0264. [DOI] [PubMed] [Google Scholar]
- 13.Li H., Seth A. An RNF11: Smurf2 complex mediates ubiquitination of the AMSH protein. Oncogene. 2004;23:1801–1808. doi: 10.1038/sj.onc.1207319. [DOI] [PubMed] [Google Scholar]
- 14.Wada K., Kamitani T. UnpEL/Usp4 is ubiquitinated by Ro52 and deubiquitinated by itself. Biochem. Biophys. Res. Commun. 2006;342:253–258. doi: 10.1016/j.bbrc.2006.01.144. [DOI] [PubMed] [Google Scholar]
- 15.Boutell C., Canning M., Orr A., Everett R.D. Reciprocal activities between herpes simplex virus type 1 regulatory protein ICP0, a ubiquitin E3 ligase, and ubiquitin-specific protease USP7. J. Virol. 2005;79:12342–12354. doi: 10.1128/JVI.79.19.12342-12354.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fernandez-Montalvan A., Bouwmeester T., Joberty G., Mader R., Mahnke M., Pierrat B., Schlaeppi J.M., Worpenberg S., Gerhartz B. Biochemical characterization of USP7 reveals post-translational modification sites and structural requirements for substrate processing and subcellular localization. FEBS J. 2007;274:4256–4270. doi: 10.1111/j.1742-4658.2007.05952.x. [DOI] [PubMed] [Google Scholar]
- 17.B.K.J. Hetfeld, A. Helfrich, B. Kapelari, H. Scheel, K. Hofmann, A. Guterman, M. Glickman, R. Schade, P.M. Kloetzel, W. Dubiel, The zinc finger of the CSN-associated deubiquitinating enzyme USP15 is essential to rescue the E3 ligase Rbx1, Current Biology 15 (2005) 1217–1221. [DOI] [PubMed]
- 18.Li Z., Wang D., Na X., Schoen S.R., Messing E.M., Wu G. Identification of a deubiquitinating enzyme subfamily as substrates of the von Hippel–Lindau tumor suppressor. Biochem. Biophys. Res. Commun. 2002;294:700–709. doi: 10.1016/S0006-291X(02)00534-X. [DOI] [PubMed] [Google Scholar]
- 19.Todi S.V., Winborn B.J., Scaglione K.M., Blount J.R., Travis S.M., Paulson H.L. Ubiquitination directly enhances activity of the deubiquitinating enzyme ataxin-3. EMBO J. 2009;28:372–382. doi: 10.1038/emboj.2008.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hjerpe R., Aillet F., Lopitz-Otsoa F., Lang V., England P., Rodriques M.S. Efficient protection and isolation of ubiquitylated proteins using tandem ubiquitin-binding entities. EMBO Rep. 2009;10:1250–1258. doi: 10.1038/embor.2009.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loch C.M., Cuccherini C.L., Leach C.A., Strickler J.E. Deubiquitylase, DeSUMOylase, and DeISGylase activity microarrays for assay of substrate preference and functional modifiers. Mol. Cell. Proteomics. 2011;10 doi: 10.1074/mcp.M110.002402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nicholson B., Leach C.A., Goldenberg S.J., Francis D.M., Kodrasov M.P., Tian X., Shanks J., Sterner D.E., Bernal A., Mattern M.R., Wilkinson K.D., Butt T.R. Characterization of ubiquitin and ubiquitin-like protein isopeptidase activities. J. Protein Sci. 2008;17:1035–1043. doi: 10.1110/ps.083450408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marblestone J.G., Kumar K.G.S., Eddins M.J., Leach C.A., Sterner D.E., Mattern M., Nicholson B. Novel approach for characterizing ubiquitin E3 ligase function. J. Biomol. Screen. 2010;15:1220–1228. doi: 10.1177/1087057110380456. [DOI] [PubMed] [Google Scholar]
- 24.Balut C.M., Loch C.M., Devor D.C. Role of ubiquitylation and USP8-dependent deubiquitylation in the endocytosis and lysosomal targeting of plasma membrane KCa3.1. FASEB J. 2011;25:3938–3948. doi: 10.1096/fj.11-187005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Altun M., Kramer H.B., Willems L.I., McDermott J.L., Leach C.A., Goldenberg S.J., Kumar K.G., Konietzny R., Fischer R., Kogan E., Mackeen M.M., McGouran J., Khoronenkova S.V., Parsons J.L., Dianov G.L., Nicholson B., Kessler B.M. Activity-based chemical proteomics accelerates inhibitor development for deubiquitylating enzymes. Chem. Biol. 2011;18:1401–1412. doi: 10.1016/j.chembiol.2011.08.018. [DOI] [PubMed] [Google Scholar]
- 26.Yao T., Song L., Xu W., DeMartino G.N., Florens L., Swanson S.K., Washburn M.P., Conaway R.C., Conaway J.W., Cohen R.E. Proteasome recruitment and activation of the Uch37 deubiquitinating enzyme by Adrm1. Nat. Cell Biol. 2006;8:994–1002. doi: 10.1038/ncb1460. [DOI] [PubMed] [Google Scholar]
- 27.Komander D., Reyes-Turcu F.E., Licchesi J.D.F., Odenwaelder P., Wilkinson K.D. Molecular discrimination of structurally equivalent Lys 63-linked and linear polyubiquitin chains. EMBO Rep. 2009;10:466–473. doi: 10.1038/embor.2009.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Licchesi J.D., Mieszczanek J., Mevissen T.E., Rutherford T.J., Akutsu M., Virdee S., El Oualid F., Chin J.W., Ovaa H., Bienz M., Komander D. An ankyrin-repeat ubiquitin-binding domain determines TRABID's specificity for atypical ubiquitin chains. Nat. Struct. Mol. Biol. 2012;19:62–71. doi: 10.1038/nsmb.2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilkinson K.D., Tashayev V.L., O'Connor L.B., Larsen C.N., Kasperek E., Pickart C.M. Metabolism of the polyubiquitin degradation signal: structure, mechanism, and role of isopeptidase T. Biochemistry. 1995;34:14535–14546. doi: 10.1021/bi00044a032. [DOI] [PubMed] [Google Scholar]
- 30.David Y., Ziv T., Admon A., Navon A. The E2 ubiquitin-conjugating enzymes direct polyubiquitination to preferred lysines. J. Biol. Chem. 2010;285:8595–8604. doi: 10.1074/jbc.M109.089003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reyes-Turcu F.E., Wilkinson K.D. Polyubiquitin binding and disassembly by deubiquitinating enzymes. Chem. Rev. 2009;109:1495–1508. doi: 10.1021/cr800470j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zezula J., Freissmuth M. The A(2A)-adenosine receptor: a GPCR with unique features? Br. J. Pharmacol. 2008;153(Suppl. 1):S184–S190. doi: 10.1038/sj.bjp.0707674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.David Y., Ternette N., Edelmann M.J., Ziv T., Gayer B., Sertchook R., Dadon Y., Kessler B.M., Navon A. E3 ligases determine ubiquitination site and conjugate type by enforcing specificity on E2 enzymes. J. Biol. Chem. 2011;286:44104–44115. doi: 10.1074/jbc.M111.234559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schaefer J.B., Morgan D.O. Protein-linked ubiquitin chain structure restricts activity of deubiquitinating enzymes. J. Biol. Chem. 2011;286:45186–45196. doi: 10.1074/jbc.M111.310094. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Complete DUB activity profile of USP21c against all ubiquitylated substrates. Rows are substrates (labeled as in Fig. 1C); columns are labeled according to which DUB was incubated with these substrates where “No DUB” shows maximal ubiquitylation and USP8c shows minimal ubiquitylation possible following DUB treatment. This profile was labeled “all or nothing” based on the observation that USP21c could not cut chains from all substrates (see primarily TUBEs detection rows 3, 7, 11, 23); but where it did cut chains, it also removed the substrate-proximal mono-ubiquitin as well (see primarily antibody detection rows 1, 6, 10, 12).
Complete DUB activity profile of USP28 against all ubiquitylated substrates. (See Supplemental Fig. 1 for explanation of rows and columns.) This profile showed no specificity for chain cleavage (see TUBEs detection, all rows) with the exception of an inability to cleave chains from Ataxin3-like, row 14. However, it was unable to remove the substrate-proximal monoubiquitin from any substrates (see antibody detection, all rows).
Complete DUB activity profile of USP28 against all ubiquitylated substrates. (See Supplemental Fig. 1 for explanation of rows and columns.) This profile showed no specificity for chain cleavage (see TUBEs detection, all rows) with the exception of an inability to cleave chains from Ataxin3-like, row 14. However, it was unable to remove the substrate-proximal monoubiquitin from any substrates (see antibody detection, all rows).
Complete DUB activity profile of USP15 against all ubiquitylated substrates. (See Supplemental Fig. 1 for explanation of rows and columns.) USP15 did not appear capable of removing the substrate-proximal monoubiquitin from any substrates (see antibody detection, all rows). However, it displayed substrate specificity for chains (TUBEs detection, compare the diminution of signal in rows 2, 7, 12, 23 to the undiminished signals of rows 3, 4, 17).
Complete DUB activity profile of USP15 against all ubiquitylated substrates. (See Supplemental Fig. 1 for explanation of rows and columns.) USP15 did not appear capable of removing the substrate-proximal monoubiquitin from any substrates (see antibody detection, all rows). However, it displayed substrate specificity for chains (TUBEs detection, compare the diminution of signal in rows 2, 7, 12, 23 to the undiminished signals of rows 3, 4, 17).
Complete DUB activity profile of PLP2 against all ubiquitylated substrates. (See Supplemental Fig. 1 for explanation of rows and columns.) This profile showed no specificity for chain cleavage (see TUBEs detection, all rows) with the exception of an inability to cleave chains from Ataxin3-like, row 14. UCHL1 (row 28) and maybe JOSD1 (row 8) were identified as putative substrates (see antibody detection).
Complete DUB activity profile of PLP2 against all ubiquitylated substrates. (See Supplemental Fig. 1 for explanation of rows and columns.) This profile showed no specificity for chain cleavage (see TUBEs detection, all rows) with the exception of an inability to cleave chains from Ataxin3-like, row 14. UCHL1 (row 28) and maybe JOSD1 (row 8) were identified as putative substrates (see antibody detection).
Complete DUB activity profile of PLPro against all ubiquitylated substrates. (See Supplemental Fig. 1 for explanation of rows and columns.) This profile showed no specificity for chain cleavage (see TUBEs detection, all rows) and represents the only non-core domain DUB to cleave chains from Ataxin3-like, row 14. USP4 (row 33, antibody detection) was identified as a putative substrate.
Complete DUB activity profile of PLPro against all ubiquitylated substrates. (See Supplemental Fig. 1 for explanation of rows and columns.) This profile showed no specificity for chain cleavage (see TUBEs detection, all rows) and represents the only non-core domain DUB to cleave chains from Ataxin3-like, row 14. USP4 (row 33, antibody detection) was identified as a putative substrate.