Abstract
Hepatitis caused by Hepatitis C virus (HCV) is a major cause of chronic liver disease. HCV is transmitted by injection drug use, blood transfusion, hemodialysis, organ transplantation and less frequently sexual intercourse. It has been recognized as a global health problem because of the progression to cirrhosis and hepatocellular carcinoma. Globally, about 170 million people are infected with HCV. Since the discovery of this virus in 1989, the clinical management of chronic hepatitis C infection has undergone a paradigm shift from alpha interferon to direct-acting antiviral (DAA) therapy. However, resistance to many of these antiviral agents has been reported increasingly from all over the globe. This review article focuses on the emerging HCV resistance to DAAs and the relevance of in vitro DAA resistance testing in clinical practice.
Keywords: Directly acting antiviral, Hepatitis C virus, resistance
Introduction
Hepatitis C virus (HCV) is a single-stranded RNA virus belonging to Hepacivirus genus of the Flaviviridae family. It causes hepatitis C which is a major chronic liver disease. Hepatitis C is transmitted by injection drug use, blood transfusion, hemodialysis, organ transplantation and, less frequently, sexual intercourse. It has been recognized as a global health problem because of the progression to cirrhosis and hepatocellular carcinoma. Globally, about 170 million people are infected with HCV. Since the discovery of this virus in 1989, the clinical management of chronic hepatitis C infection has undergone a paradigm shift from alpha interferon to directly acting antiviral (DAA) therapy.[1] However, resistance to many of these antiviral agents has been reported increasingly from all over the globe. The aim of this review article is to focus on the emerging HCV resistance to DAAs and the relevance of in vitro DAA resistance testing in clinical practice.
Structural organization of Hepatitis C virus
HCV is an enveloped virus harboring a 9.6 kb positive-sense single-stranded RNA. The genome carries a long open reading frame (ORF) encoding a polyprotein precursor of 3010 amino acids. Translation of the HCV ORF is directed via a 340-nucleotide-long 5’ nontranslated region (NTR) functioning as an internal ribosome entry site (IRES) which permits the direct binding of ribosomes in close proximity to the start codon of the ORF. The HCV polyprotein is cleaved co- and posttranslationally by cellular and viral proteases into ten different products which are broadly classified as structural [core (C), E1, and E2)] and nonstructural (NS2-5) replicative proteins.[2] Figure 1 shows the structural organization of the HCV genome and the various proteins encoded by it. A summary of various HCV proteins is been depicted in Table 1.
Figure 1.
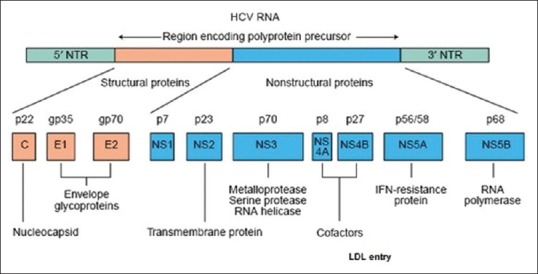
Structural organization of HCV genome[3]
Table 1.
| Proten | Type | Functions |
|---|---|---|
| HCV core protein | Structural | Makes up the viral nucleocapsid. Affects host cell functions like gene transcription, lipid metabolism, apoptosis, and various signaling pathways Directly or indirectly involved in hepatocarcinogenesis and steatosis hepatitis |
| Envelope proteins E1 and E2 | Structural | Play an important role in cell entry. E1 serves as the fusogenic subunit. E2 acts as the receptor binding subunit of the HCV envelope. |
| P7 protein | Structural | P7 has characteristics similar to those of a group of proteins called viroporins. Forms ion channels that play an essential role in virus infection. Essential for virus particle assembly and release of infectious virions in a genotype specific manner |
| NS2 protein | Nonstructural | Essential for completion of the viral replication cycle in vitro and in vivo. |
| NS3 protein | Non-structural | It is a multifunctional protein whose N & C terminals have serine protease & NTPase/helicase activities, respectively |
| NS4A protein | Nonstructural | Acts as a cofactor for NS3 protein. Also required for the phosphorylation of NS5A and can directly interact with NS5A |
| NS4B protein | Nonstructural | Plays an important role in the recruitment of other viral proteins. |
| NS5A protein | Nonstructural | Plays an important role in viral replication and modulation of cell signaling pathways and interferon response. |
| NS5B protein | Nonstructural | Acts as an RNA-dependent RNA polymerase and plays an important role in the synthesis of new RNA genome. |
Natural history of HCV infection
Infection with HCV can present either as acute or chronic hepatitis. Acute hepatitis is usually asymptomatic and rarely leads to hepatic failure. Symptomatic acute HCV infection has a mild clinical course with less than one fourth (<25%) of patients presenting with jaundice. About 60–80% patients with acute infection develop chronic infection.[23] The rate of spontaneous viral clearance in patients with chronic HCV is very low. Approximately, one fifth (20–30%) of patients with chronic HCV infection develop cirrhosis over a period of 10–30 years.[24] 1–4% of cirrhotic patients develop hepatocellular carcinoma per year.[25] There are several factors which may determine the rapidity of disease progression which include: HCV acquisition at an advanced age (>40–55 years), male sex, HIV co-infection, high body mass index, presence of hepatic steatosis, and alcohol consumption. Patients with cirrhosis can decompensate with complications like portal hypertension, ascites, spontaneous bacterial peritonitis, esophageal varices, hepatic encephalopathy and hepatic coma, and hepatorenal syndrome leading to renal failure. Chronic infection can also be associated with extra-hepatic manifestations such as cryoglobulinemia, porphyria cutanea tarda, arthralgia, membranoproliferative glomerulonephritis, Sjogren's syndrome, Raynaud's syndrome, idiopathic thrombocytopenic purpura, and non-Hodgkin's lymphoma.[26]
Problem statement
The global prevalence of HCV infection is estimated at 2–3%.[27,28,29,30] The prevalence of HCV is highest in Africa and the Middle East. China, India, Egypt, Pakistan, and Indonesia account for approximately half of the global HCV-infected subjects.[31,32,33,34] North America, Australia, Japan, and Northern and Western Europe report lower prevalence of HCV infection, with no country showing a rate > 2%. Owing to the paucity of large-scale prevalence studies on hepatitis C in the general population of India, the reported prevalence rates are variable ranging from 0.09–2.02%.[35]
The genetic diversity of the virus further complicates the epidemiology of HCV infection. HCV is classified into 7 genotypes (1–7) which differ by more than 30% sequence diversity and at least 67 subtypes, characterized by about 20% sequence divergence.[36,37] Identification of HCV genotypes and subtypes plays a crucial role in defining the epidemiological patterns and effective treatment. Genotype 1 is distributed worldwide and is responsible for majority of the cases reported in the Americas, Europe, Australia, and Japan. Genotype 1a is distributed widely in Northern Europe and United States. Genotype 1b is commonly found in Europe and Asia. Although, genotype 2 is more prevalent in industrialized countries, it is also found in South America and Asia. Genotype 2a has commonly been isolated from Japan and China. Genotype 2b is widespread in Northern Europe and United States. Genotype 2c is the most common subtype found in Western and Southern Europe, Pakistan, and India. Genotype 3a is prevalent in Europe, United States, Australia, and Southern Asia.[38,39,40] Genotypes 4 and 5 are mainly found in Africa and the Middle East. While genotype 4a is prevalent in Egypt, genotype 4c is highly prevalent in Central Africa. Genotype 5 has mainly been reported from South Africa. Genotype 6 and its numerous subtypes are found mainly in Southeast Asian countries like Thailand, Vietnam, and Myanmar. Genotype 7 has minor clinical relevance and has been recently found in patients from Central Africa and Thailand.[30,40,41,42,43,44,45,46]
Treatment of chronic Hepatitis C—An evolutionary perspective
The treatment of chronic hepatitis C virus infection has been rapidly evolving since early 1990s with interferon-alfa. This injectable drug worked by boosting the immune system instead of specifically attacking the virus. However, the response rates of this drug were low, ranging from 10 to 20%. In 1998, the oral drug ribavirin was added to interferon. The therapeutic response rates increased dramatically, averaging 37 to 43%. In 2002, pegylated interferon-alfa was approved for treatment of chronic hepatitis C. Combined with ribavirin, permanent response rates of 52% were obtained with this drug. In 2011, two protease inhibitors were introduced namely boceprevir and telaprevir.[47] Treatment of chronic HCV with PEG-IFN and ribavirin and, more recently, with PEG-IFN plus ribavirin and first-generation protease inhibitors (Telaprevir and Boceprevir) for HCV-1, have been the standard of care. However, favorable clinical outcomes could not be achieved in majority of the patients.[48,49] Apart from the several side effects which are associated with interferon therapy, there are various contraindications like pregnancy, breast feeding, and allergic reactions. The recent development of a series of new direct antiviral agents (DAAs) that are directed against HCV target proteins used by the virus for its replication and assembly has ushered a new era in the treatment of hepatitis C.[50] The DAAs are molecules that target specific nonstructural proteins of HCV thereby interfering with viral replication and infection. There are four classes of DAAs, which are defined by their mechanism of action and therapeutic targets [Table 2].[51]
Table 2.
| Class of direct antiviral agents | Examples | Mechanism of action | Comments |
|---|---|---|---|
| NS3/4A protease inhibitors | 1st generation: Telaprevir, Boceprevir 2nd generation: Simeprevir Next generation drugs: Glecaprevir, Grazoprevir, Paritaprevir, Voxilaprevir | Bind to the active site of the NS3/4A protease | High potency (varies by HCV genotype) Low barrier to resistance (1a<1b) High potential for drug interactions Can cause rash, anemia, and raised bilirubin Later generation drugs like Glecapravir & Grazoprevir are pangenotypic and are expected to have higher barriers to resistance. |
| NS5A inhibitors | Daclatasvir, Elbasvir, Ledipasvir, Ombitasvir, Pibrentasvir, Velpatasvir | Interact with domain 1 of the NS5A dimer, although the exact mechanism remains to be fully elucidated | Moderate to high potency (consistent across HCV genotypes & subtypes) High barrier to resistance (1a=1b) Low potential for drug interactions; may interact with HIV antiretrovirals (nucleoside reverse transcriptase inhibitors) and ribavirin Can cause mitochondrial toxicity |
| Nucleos(t) ide analog NS5B polymerase inhibitors | Sofosbuvir | These are incorporated into the nascent RNA chain and result in chain termination by compromising the binding of the next incoming nucleotide | Potency varies by HCV genotype Very low barrier to resistance (1a<1b) Variable potential for drug interactions |
| Non-nucleoside NS5B polymerase inhibitors | Dasabuvir | Interact with thumb 1, thumb 2, palm 1 or palm 2 domain of NS5B and inhibit polymerase activity by allosteric mechanisms of action | High potency (against multiple HCV genotypes) Low barrier to resistance (1a<1b) Low to moderate potential for drug interactions |
HCV resistance to DAAs
HCV can develop resistance to DAAs due to the lack of proof-reading activity by RNA dependent RNA polymerase coupled with the high replication capacity of the virus. This leads to the generation of large number of genetically distinct viral variants called “quasispecies”.[55,56] Some quasispecies bear polymorphisms in drug-targeted genes resulting in reduced susceptibility to DAAs.[57] The prevalence of intrinsically resistant variants within a patient's quasispecies is determined by their replicative fitness. Typically, a dominant variant is detectable within the viral quasispecies along with less-fit variants that are present at lower frequencies. The presence of minor populations of resistance-associated variants (RAVs) at the start of treatment may affect the outcomes of the antiviral therapy. Such variants can become dominant as a result of selective pressure exerted by antiviral drugs, subsequently leading to virological breakthrough during treatment or relapse after treatment cessation.[58] Some RAVs such as those conferring resistance to NS5A inhibitors are very fit and persist as the dominant species for months to years after treatment cessation. Understanding drug resistance is important in clinical settings to optimize treatment regimens, increase success rates, and minimize the impact of treatment failure. Table 3 depicts the most common amino acid substitutions detected in DAA non-responding HCV infected patients.
Table 3.
Most common amino acid substitutions and genotypes/subtypes in DAA nonresponding HCV infected patients[52,59,60]
| DAAs approved by FDA | Category of DAAs | Most common amino acid substitutions detected in HCV infected patients who failed to achieve SVR | Most common HCV genotype/subtype associated with SVR failure |
|---|---|---|---|
| Boceprevir | NS3/4A protease inhibitor | V36M, T54S, R155K | 1a |
| T54A/S, V55A, A156S, V170A | 1b | ||
| Telaprevir | NS3/4A protease inhibitor | V36M, R155K | 1a |
| V36A, T54A, A156S | 1b | ||
| Simeprevir | NS3/4A protease inhibitor | R155K, D168E/V | 1a |
| Q80R, D168E/V | 1b | ||
| Paritaprevir | NS3/4A protease inhibitor | D168A/V/Y | 1a |
| Y56H, D168V | 1b | ||
| D168V | 4d | ||
| Asunaprevir | NS3/4A protease inhibitor | R155K, D168E | 1a |
| D168E/V/Y | 1b | ||
| Vaniprevir | NS3/4A protease inhibitor | R155K, D168T/V/Y | 1a |
| D168H/T/V | 1b | ||
| NS5A inhibitor | M28T, Q30E/H/R, L31M, H58D, Y93H/N | 1a | |
| L31M/V, Y93H | 1b | ||
| Q30H/S | 4 | ||
| Ledipasvir | NS5A inhibitor | Q30E/R, L31M, Y93C/H/N | 1a |
| Y93H | 1b | ||
| Ombitasvir | NS5A inhibitor | M28V, Q30R | 1a |
| Y93H | 1b | ||
| L28V | 4d | ||
| Sofosbuvir | NS5B nucleotide polymerase inhibitor | S282T | 2 |
| L159F, V321A | 3 | ||
| C316N/H/F | 1b | ||
| Dasabuvir | NS5B non-nucleoside polymerase inhibitor | M414T, S556G | 1a |
| S556G | 1b | ||
| Beclabuvir | NS5B non-nucleoside polymerase inhibitor | A421V, P495L/S | 1a |
Drug resistance testing in clinical practice-An overview
The replication cycle of HCV is error prone and many of these errors either do not have any effect on the progeny viruses or result in progeny viruses that are incompetent to replicate. However, for some newly produced viruses, the transcription errors result in changes in critical coding regions that may change the susceptibility of the virus to one or more anti-viral agents. Subtherapeutic levels of antiviral agents often lead to the emergence of drug-resistant viruses by creating selective pressure for the resistant viruses to emerge as the dominant species. However, viral variants harbouring substitutions associated with resistance to directing-acting antivirals (DAAs) are detectable prior to antiviral therapy in few chronic HCV infected patients and are referred to as baseline resistance-associated substitutions (RASs). These viruses contain substitutions that are designated as treatment-emergent (or treatment-selected) RASs.[61]
Presently, the most clinically significant RASs are in the NS5A position for genotypes 1a and 3.[61] NS5A and NS3 RASs are frequently selected in patients with failure of NS5A or NS3 inhibitor-containing regimens, respectively. NS5B nucleotide RASs are rarely detected (1% of failures) even after exposure to a failing DAA regimen containing a nucleotide inhibitor.[62,63] In addition, any such substitution would likely render the virus replication incompetent. The clinical impact of NS5A RASs is further compounded by their ability to maintain high replication competence in the absence of continued drug pressure. This allows these viral variants to remain as the dominant viral quasispecies for prolonged periods relative to NS3 protease or NS5B nucleotide polymerase inhibitor RASs.[61]
In general, drug-specific RASs need to be present in at least 15% of the viruses of a given patient to reduce the likelihood of achieving SVR.[64] Drug-specific RASs that are found at a lower frequency may not convey sufficient resistance to reduce SVR with currently available DAA regimens. The magnitude of the negative impact of RASs on treatment outcome varies according to treatment regimen and patient factors such as presence of cirrhosis and fold change decrease in potency conferred by the specific RAS(s).[61]
Testing for RASs in clinical practice becomes important when the results would modify treatment management by impacting the duration of therapy and/or inclusion of ribavirin or result in selection of alternative therapy. However, at present the utility of RAS testing varies by both patient characteristics and DAA regimen. Table 4 depicts the regimen-specific recommendations for use of RAS testing in clinical practice.[61]
Table 4.
Regimen-specific recommendations for use of RAS testing in clinical practice[61]
| Recommended | *Rating |
|---|---|
| Elbasvir/grazoprevir NS5A RAS testing is recommended for genotype 1a-infected, treatment-naive or -experienced patients being considered for elbasvir/grazoprevir. If present, a different regimen should be considered. |
I, A |
| Ledipasvir/sofosbuvir NS5A RAS testing can be considered for genotype 1a-infected, treatment-experienced patients without cirrhosis being considered for ledipasvir/sofosbuvir. If clinically importanta resistance is present, a different recommended therapy should be used. NS5A RAS testing can be considered for genotype 1a-infected, treatment-experienced patients with cirrhosis being considered for ledipasvir/sofosbuvir. If clinically importanta resistance is present, a different recommended therapy should be used. |
I, A |
| Sofosbuvir/velpatasvir NS5A RAS testing is recommended for genotype 3-infected, treatment-naive patients with cirrhosis and treatment-experienced patients (with or without cirrhosis) being considered for 12 weeks of sofosbuvir/velpatasvir. If Y93H is present, weight-based ribavirin should be added or sofosbuvir/velpatasvir/voxilaprevir should be used. |
I, A |
| Daclatasvir plus sofosbuvir NS5A RAS testing is recommended for genotype 3-infected, treatment-experienced patients without cirrhosis being considered for 12 weeks of daclatasvir plus sofosbuvir. If Y93H is present, weight-based ribavirin should be added. NS5A RAS testing is recommended for genotype 3-infected, treatment-naive patients with cirrhosis being considered for 24 weeks of daclatasvir plus sofosbuvir. If Y93H is present, treatment should include weight-based ribavirin, or a different recommended therapy used |
I, B |
| NOT RECOMMENDED | |
| Elbasvir/grazoprevir RAS testing is not recommended for any genotype 1b-infected patients being considered for elbasvir/grazoprevir therapy. |
I, A |
| Glecaprevir/pibrentasvir RAS testing is not recommended for patients with genotype 1, 2, 3, 4, 5, or 6 infection being considered for glecaprevir/pibrentasvir for 8, 12, or 16 weeks |
I, A |
| Ledipasvir/sofosbuvir NS5A RAS testing is not recommended for any genotype 1b-infected patients being considered for ledipasvir/sofosbuvir therapy. |
I, A |
| NS5A RAS testing is not recommended for genotype 1a-infected, treatment-naive patients being considered for ledipasvir/sofosbuvir therapy | I, A |
| NS5A RAS testing is not recommended for genotype 1a- or 1b-infected, treatment-naive patients without cirrhosis and with a viral load <6 million IU/mL being considered for an 8-week course of ledipasvir/sofosbuvir therapy. | I, A |
| Paritaprevir/ritonavir/ombitasvir with dasabuvir±weight-based ribavirin, or paritaprevir/ritonavir/ombitasvir + weight-based ribavirin RAS testing is not recommended for genotype 1- or 4-infected, treatment-naive or -experienced patients being considered for therapy with paritaprevir/ritonavir/ombitasvir with dasabuvir±weight-based ribavirin or paritaprevir/ritonavir/ombitasvir + weight-based ribavirin, respectively. |
I, A |
| Sofosbuvir/velpatasvir RAS testing is not recommended for patients with genotype 1, 2, 4, 5, or 6 infection and considered for 12 weeks of sofosbuvir/velpatasvir therapy. |
I, A |
| Sofosbuvir/velpatasvir/voxilaprevir RAS testing is not recommended for patients with genotype 1, 2, 3, 4, 5, or 6 infection and considered for 12 weeks of sofosbuvir/velpatasvir/voxilaprevir therapy. |
I, A |
| Clinically important=greater than 100-fold resistance | |
*I: Evidence and/or general agreement that a given diagnostic evaluation, procedure, or treatment is beneficial, useful, and effective; A: Data derived from multiple randomized clinical trials, meta-analyses, or equivalent; B: Data derived from a single randomized trial, nonrandomized studies, or equivalent
Assays for detecting RASs
The methods to detect RASs in NS3, NS5A, and NS5B include population sequencing (Sanger sequencing) and deep sequencing (Next generation sequencing), which differ in their sensitivity for detecting RASs. For the purposes of clinical care and decisions regarding the choice of appropriate DAA regimen, both methods can be considered equivalent if a ≥15% cutoff point is used for the determination of RASs by NGS. However, the presence of RASs with <15% prevalence should not be considered clinically significant. Phenotypic analysis involves laboratory techniques whereby the degree of drug resistance conferred by an amino acid substitution and the replicative capacity of a particular RAS can be estimated in the presence of a wild-type or consensus strain.[61]
Genotypic analysis
Population-Based Sequencing (Sanger): Population sequencing of the HCV coding region of interest may be performed using reverse transcription polymerase chain reaction (PCR) and standard Sanger sequencing of the bulk PCR product. Although the sensitivity for detection of resistance substitutions is subject to variation, in generally it is 15–25%. As a standard, substitutions are reported as differences compared with a genotype-specific, wild-type strain.
Deep Sequencing Analysis/Next Generation Sequencing (NGS): NGS can increase the sensitivity of detection for minor variants. After sequencing HCV coding regions using PCR, a software algorithm is used to process and align sequencing data via a multistep method to identify the substitutions present at a predetermined level. Although this level, or threshold, can vary, it is often set as low as > 1% for research purposes. To approximate results obtained by population sequencing, NGS thresholds are often set to ≥10%.
Phenotypic analysis
This involves laboratory techniques whereby the degree of drug resistance conferred by an amino acid change as well as the replicative capacity (fitness) of a particular RAS can be estimated in the presence of a wild-type or consensus strain. However, these research techniques are not routinely used for clinical practice. To assess the level of resistance, RASs are typically introduced as point mutations into the backbone of an existing standard HCV genome within an existing cell culture/replicon or an enzyme-based assay. Isolates harboring these RASs are then challenged by appropriate antiviral agents at increasing concentrations and fold changes—based on EC50 or IC50 and EC90 or IC90 values—are determined for inhibition of replication or enzyme activity, respectively, in comparison to wild-type virus. Comparison of replication levels for variants and wild-type constructs in the absence of drug allows for the estimation of fitness.
Directly acting antiviral therapy and primary healthcare
The treatment and monitoring requirements of regimens containing Direct Acting Antivirals (DAAs) are much lower than those containing interferon and ribavirin. Also, DAAs have a much higher efficacy in treating HCV infections. These characteristics mean that initiating treatment and obtaining SVR on completion of treatment in non-specialist environments should be feasible.[65] To achieve the World Health Organization Hepatitis C virus elimination targets, it is essential to increase access to treatment. Hence, DAA treatment can be provided in primary healthcare services in order to improve accessibility and retention in care.[66] Similar SVR rates were demonstrated when services cited in community settings where compared to published studies and real-world clinics in secondary care. However, stronger study designs are needed to confirm the precision of effect size seen in most of these studies.[65]
Currently in India, few states are providing treatment and care for HCV infected patients with DAAs and some healthcare facilities are also providing antiviral treatment for chronic Hepatitis B infection/disease. The National Viral Hepatitis Control Programme (NVHCP) under National Health Mission (NHM), Government of India, aims at smooth transitioning of all state-level programmes to a country-level programme in order to align with the national protocols and guidance on testing and management of different types of viral hepatitis (namely A, B, C, D, and E viruses, respectively). The objectives of NVHCP are as follows[67]:
Enhance community awareness on hepatitis and lay stress on preventive measures among general population, especially high-risk groups and in hotspots.
Provide early diagnosis and management of viral hepatitis at all levels of healthcare.
Develop standard diagnostic and treatment protocols for management of viral hepatitis and its complications.
Strengthen the existing infrastructure facilities, build capacities of existing human resource and raise additional human resources, where required, for providing comprehensive services for management of viral hepatitis and its complications in all districts of the country.
Develop linkages with the existing National programmes towards awareness, prevention, diagnosis, and treatment for viral hepatitis.
Develop a web-based “Viral Hepatitis Information and Management System” to maintain a registry of persons affected with viral hepatitis and its sequelae.
Owing to the recent developments pertaining to the management of chronic HCV infection with DAA therapy at primary healthcare level, it is imperative for physicians to be aware of the emerging resistance to these drugs in order to avoid their misuse.
Conclusion
Management of HCV infection is a challenge for both hepatologists and virologists alike. Emergence of resistance to many currently available anti-HCV drugs is alarming and, therefore, it is imperative to performin vitro RAS testing especially in patients on DAA therapy who fail to attain a sustained viral response. Keeping in mind the emerging concept of management of chronic HCV infections at the primary healthcare level, physicians should be made aware of the importance of emerging resistance to directly acting antiviral drugs to avoid their misuse.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
References
- 1.Modi A, Liang T. Hepatitis C: A clinical review. Oral Dis. 2008;14:10–4. doi: 10.1111/j.1601-0825.2007.01419.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beaulieu PL, Tsantrizos YS. Inhibitors of the HCV NS5B polymerase: New hope for the treatment of hepatitis C infections. Curr Opin Investig Drugs. 2004;5:838–50. [PubMed] [Google Scholar]
- 3.Ashfaq UA, Javed T, Rehman S, Nawaz Z, Riazuddin S. An overview of HCV molecular biology, replication and immune responses. Virol J. 2011;8:161. doi: 10.1186/1743-422X-8-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bukh J, Purcell RH, Miller RH. Sequence analysis of the core gene of 14 hepatitis C virus genotypes. Proc Natl Acad Sci USA. 1994;91:8239–43. doi: 10.1073/pnas.91.17.8239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Santolini E, Migliaccio G, La Monica N. Biosynthesis and biochemical properties of the hepatitis C virus core protein. J Virol. 1994;68:3631–41. doi: 10.1128/jvi.68.6.3631-3641.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hope RG, Murphy DJ, McLauchlan J. The domains required to direct core proteins of hepatitis C virus and GB virus-B to lipid droplets share common features with plant oleosin proteins. J Biol Chem. 2002;277:4261–70. doi: 10.1074/jbc.M108798200. [DOI] [PubMed] [Google Scholar]
- 7.Lerat H, Honda M, Beard MR, Loesch K, Sun J, Yang Y, et al. Steatosis and liver cancer in transgenic mice expressing the structural and nonstructural proteins of hepatitis C virus. Gastroenterology. 2002;122:352–65. doi: 10.1053/gast.2002.31001. [DOI] [PubMed] [Google Scholar]
- 8.Tellinghuisen TL, Rice CM. Interaction between hepatitis C virus proteins and host cell factors. Curr Opin Microbiol. 2002;5:419–27. doi: 10.1016/s1369-5274(02)00341-7. [DOI] [PubMed] [Google Scholar]
- 9.Drummer HE, Maerz A, Poumbourios P. Cell surface expression of functional hepatitis C virus E1 and E2 glycoproteins. FEBS Lett. 2003;546:385–90. doi: 10.1016/s0014-5793(03)00635-5. [DOI] [PubMed] [Google Scholar]
- 10.Goffard A, Callens N, Bartosch B, Wychowski C, Cosset FL, Montpellier C, et al. Role of N-linked glycans in the functions of hepatitis C virus envelope glycoproteins. J Virol. 2005;79:8400–9. doi: 10.1128/JVI.79.13.8400-8409.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Griffin SD, Beales LP, Clarke DS, Worsfold O, Evans SD, Jaeger J, et al. The p7 protein of hepatitis C virus forms an ion channel that is blocked by the antiviral drug, Amantadine. FEBS Lett. 2003;535:34–8. doi: 10.1016/s0014-5793(02)03851-6. [DOI] [PubMed] [Google Scholar]
- 12.Steinmann E, Penin F, Kallis S, Patel AH, Bartenschlager R, Pietschmann T. Hepatitis C virus p7 protein is crucial for assembly and release of infectious virions. PLoS Pathog. 2007;3:e103. doi: 10.1371/journal.ppat.0030103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khromykh AA, Westaway EG. Subgenomic replicons of the flavivirus Kunjin; 1997. pp. 1497–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pietschmann T, Kaul A, Koutsoudakis G, Shavinskaya A, Kallis S, Steinmann E, et al. Construction and characterization of infectious intragenotypic and intergenotypic hepatitis C virus chimeras. Proc Natl Acad Sci USA. 2006;103:7408–13. doi: 10.1073/pnas.0504877103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gallinari P, Brennan D, Nardi C, Brunetti M, Tomei L, Steinkuhler C, et al. Multiple enzymatic activities associated with recombinant NS3 protein of hepatitis C virus. J Virol. 1998;72:6758–69. doi: 10.1128/jvi.72.8.6758-6769.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wolk B, Sansonno D, Krausslich HG, Dammacco F, Rice CM, Blum HE, et al. Subcellular localization, stability, and trans-cleavage competence of the hepatitis C virus NS3-NS4A complex expressed in tetracycline-regulated cell lines. J Virol. 2000;74:2293–304. doi: 10.1128/jvi.74.5.2293-2304.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Asabe SI, Tanji Y, Satoh S, Kaneko T, Kimura K, Shimotohno K. The N-terminal region of hepatitis C virus-encoded NS5A is important for NS4A-dependent phosphorylation. J Virol. 1997;71:790–6. doi: 10.1128/jvi.71.1.790-796.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin C, Wu JW, Hsiao K, Su MS. The hepatitis C virus NS4A protein: Interactions with the NS4B and NS5A proteins. J Virol. 1997;71:6465–71. doi: 10.1128/jvi.71.9.6465-6471.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hugle T, Fehrmann F, Bieck E, Kohara M, Krausslich HG, Rice CM, et al. The hepatitis C virus nonstructural protein 4B is an integral endoplasmic reticulum membrane protein. Virology. 2001;284:70–81. doi: 10.1006/viro.2001.0873. [DOI] [PubMed] [Google Scholar]
- 20.Macdonald A, Crowder K, Street A, McCormick C, Harris M. The hepatitis C virus NS5A protein binds to members of the Src family of tyrosine kinases and regulates kinase activity. J Gen Virol. 2004;85:721–9. doi: 10.1099/vir.0.19691-0. [DOI] [PubMed] [Google Scholar]
- 21.Reed KE, Xu J, Rice CM. Phosphorylation of the hepatitis C virus NS5A protein in vitro and in vivo: Properties of the NS5A-associated kinase. J Virol. 1997;71:7187–97. doi: 10.1128/jvi.71.10.7187-7197.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Behrens SE, Tomei L, De Francesco R. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. Embo J. 1996;15:12–2. [PMC free article] [PubMed] [Google Scholar]
- 23.Hoofnagle JH. Course and outcome of hepatitis C. Hepatology. 2002;36(5 Suppl 1):S21–9. doi: 10.1053/jhep.2002.36227. [DOI] [PubMed] [Google Scholar]
- 24.Thomas DL, See LB. Natural history of hepatitis C. Clin Liver Dis. 2005;9:383–98. doi: 10.1016/j.cld.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 25.Fattovich G, Stroffolini T, Zagni I, Donato F. Hepatocellular carcinoma in cirrhosis: Incidence and risk factors. Gastroenterology. 2004;127(5 Suppl 1):S35–50. doi: 10.1053/j.gastro.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 26.Liang TJ, Rehermann B, See LB, Hoofnagle JH. Pathogenesis, natural history, treatment, and prevention of hepatitis C. Ann Intern Med. 2000;132:296–305. doi: 10.7326/0003-4819-132-4-200002150-00008. [DOI] [PubMed] [Google Scholar]
- 27.Lavanchy D. The global burden of hepatitis C. Liver Int. 2009;29(Suppl 1):74–81. doi: 10.1111/j.1478-3231.2008.01934.x. [DOI] [PubMed] [Google Scholar]
- 28.Global Burden Of Hepatitis C Working Group. Global burden of disease (GBD) for hepatitis C. J Clin Pharmacol. 2004;44:20–9. doi: 10.1177/0091270003258669. [DOI] [PubMed] [Google Scholar]
- 29.Lavanchy D. Evolving epidemiology of hepatitis C virus. Clin Microbiol Infect. 2011;17:107–15. doi: 10.1111/j.1469-0691.2010.03432.x. [DOI] [PubMed] [Google Scholar]
- 30.Hepatitis C--global prevalence (update) Wkly Epidemiol Rec. 1999;74:425–7. [PubMed] [Google Scholar]
- 31.Nerrienet E, Pouillot R, Lachenal G, Njouom R, Mfoupouendoun J, Bilong C, et al. Hepatitis C virus infection in cameroon: A cohort-effect. J Med Virol. 2005;76:208–14. doi: 10.1002/jmv.20343. [DOI] [PubMed] [Google Scholar]
- 32.Guerra J, Garenne M, Mohamed MK, Fontanet A. HCV burden of infection in Egypt: Results from a nationwide survey. J Viral Hepat. 2012;19:560–7. doi: 10.1111/j.1365-2893.2011.01576.x. [DOI] [PubMed] [Google Scholar]
- 33.Mukhopadhyaya A. Hepatitis C in India. J Biosci. 2008;33:465–73. doi: 10.1007/s12038-008-0065-0. [DOI] [PubMed] [Google Scholar]
- 34.Simmonds P, Bukh J, Combet C, Deléage G, Enomoto N, Feinstone S, et al. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology. 2005;42:962–73. doi: 10.1002/hep.20819. [DOI] [PubMed] [Google Scholar]
- 35.Gottwein JM, Bukh J. Cutting the gordian knot-development and biological relevance of hepatitis C virus cell culture systems. Adv Virus Res. 2008;71:51–133. doi: 10.1016/S0065-3527(08)00002-X. [DOI] [PubMed] [Google Scholar]
- 36.Smith DB, Bukh J, Kuiken C, Muerhoff AS, Rice CM, Stapleton JT, et al. Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: Updated criteria and genotype assignment web resource. Hepatology. 2014;59:318–27. doi: 10.1002/hep.26744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chlabicz S, Flisiak R, Kowalczuk O, Grzeszczuk A, Pytel-Krolczuk B, Prokopowicz D, et al. Changing HCV genotypes distribution in Poland--relation to source and time of infection. J Clin Virol. 2008;42:156–9. doi: 10.1016/j.jcv.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 38.Tallo T, Norder H, Tefanova V, Krispin T, Schmidt J, Ilmoja M, et al. Genetic characterization of hepatitis C virus strains in Estonia: Fluctuations in the predominating subtype with time. J Med Virol. 2007;79:374–82. doi: 10.1002/jmv.20828. [DOI] [PubMed] [Google Scholar]
- 39.Katsoulidou A, Sypsa V, Tassopoulos NC, Boletis J, Karafoulidou A, Ketikoglou I, et al. Molecular epidemiology of hepatitis C virus (HCV) in Greece: Temporal trends in HCV genotype-specific incidence and molecular characterization of genotype 4 isolates. J Viral Hepat. 2006;13:19–27. doi: 10.1111/j.1365-2893.2005.00649.x. [DOI] [PubMed] [Google Scholar]
- 40.Sievert W, Altraif I, Razavi HA, Abdo A, Ahmed EA, Alomair A, et al. Asystematic review of hepatitis C virus epidemiology in Asia, Australia and Egypt. Liver Int. 2011;31(Suppl 2):61–80. doi: 10.1111/j.1478-3231.2011.02540.x. [DOI] [PubMed] [Google Scholar]
- 41.Chao DT, Abe K, Nguyen MH. Systematic review: Epidemiology of hepatitis C genotype 6 and its management. Aliment Pharmacol Ther. 2011;34:286–96. doi: 10.1111/j.1365-2036.2011.04714.x. [DOI] [PubMed] [Google Scholar]
- 42.Nguyen MH, Keeffe EB. Prevalence and treatment of hepatitis C virus genotypes 4, 5, and 6. Clin Gastroenterol Hepatol. 2005;3:S97–101. doi: 10.1016/s1542-3565(05)00711-1. [DOI] [PubMed] [Google Scholar]
- 43.Mauss S, Berger F, Vogel M, Pfeiffer-Vornkahl H, Alshuth U, Rockstroh JK, et al. Treatment results of chronic hepatitis C genotype 5 and 6 infections in Germany. Z Gastroenterol. 2012;50:441–4. doi: 10.1055/s-0031-1282072. [DOI] [PubMed] [Google Scholar]
- 44.Bostan N, Mahmood T. An overview about hepatitis C: A devastating virus. Crit Rev Microbiol. 2010;36:91–133. doi: 10.3109/10408410903357455. [DOI] [PubMed] [Google Scholar]
- 45.Bunchorntavakul C, Chavalitdhamrong D, Tanwandee T. Hepatitis C genotype 6: A concise review and response-guided therapy proposal. World J Hepatol. 2013;5:496–504. doi: 10.4254/wjh.v5.i9.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cornberg M, Razavi HA, Alberti A, Bernasconi E, Buti M, Cooper C, et al. Asystematic review of hepatitis C virus epidemiology in Europe, Canada and Israel. Liver Int. 2011;31(Suppl 2):30–60. doi: 10.1111/j.1478-3231.2011.02539.x. [DOI] [PubMed] [Google Scholar]
- 47.Porter L. Hepatitis C: The Evolution of Treatment [Internet]. Lucinda Porter, RN header image. 2015. cited 2018 Jun 25. Available from: http://www.lucindaporterrn.com/hepatitis-c-the-evolution-of-treatment/
- 48.Ghany MG, Nelson DR, Strader DB, David LT, Leonard BS. An update on treatment of genotype 1 chronic hepatitis C virus infection: 2011 practice guidelines by the American Association for the Study of Liver Diseases. Hepatology. 2011;54:1433–44. doi: 10.1002/hep.24641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jacobson IM, Pawlotsky JM, Afdhal NH, Dusheiko GM, Forns X, Jensen DM, et al. A practical guide for the use of Boceprevir and Telaprevir for the treatment of hepatitis C. J Viral Hepatitis. 2012;19(Suppl 2):1–26. doi: 10.1111/j.1365-2893.2012.01590.x. [DOI] [PubMed] [Google Scholar]
- 50.Manns M, Cornberg M. Sofosbuvir: The final nail in the coffin for hepatitis C. Lancet Infectious Diseases. 2013;13:378–9. doi: 10.1016/S1473-3099(13)70074-4. [DOI] [PubMed] [Google Scholar]
- 51.Poordad F, Dieterich D. Treating hepatitis C: Current standard of care and emerging direct-acting antiviral agents. J Viral Hepat. 2012;19:449. doi: 10.1111/j.1365-2893.2012.01617.x. [DOI] [PubMed] [Google Scholar]
- 52.Lontok E, Harrington P, Howe A, Kieffer T, Lennerstrand J, Lenz O, et al. Hepatitis C virus drug resistance-associated substitutions: State of the art summary. Hepatology. 2015;62:1623–32. doi: 10.1002/hep.27934. [DOI] [PubMed] [Google Scholar]
- 53.Schaefer EA, Chung RT. Anti-hepatitis C virus drugs in development. Gastroenterology. 2012;142:1340. doi: 10.1053/j.gastro.2012.02.015. [DOI] [PubMed] [Google Scholar]
- 54.Pockros PJ. [Last accessed on 2018 Jun 25];Direct-acting antivirals for the treatment of hepatitis C virus infection. UpToDate. [Google Scholar]
- 55.Pawlotsky JM, Germanidis G, Frainais PO, Bouvier M, Soulier A, Pellerin M, et al. Evolution of the hepatitis C virus second envelope protein hypervariable region in chronically infected patients receiving α interferon therapy. J Virol. 1999;73:6490–9. doi: 10.1128/jvi.73.8.6490-6499.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Neumann AU, Lam NP, Dahari H, Gretch DR, Wiley TE, Layden TJ, et al. Hepatitis C viral dynamicsin vivo and the antiviral efficacy of interferon-α therapy. Science. 1998;282:103–7. doi: 10.1126/science.282.5386.103. [DOI] [PubMed] [Google Scholar]
- 57.Pawlotsky JM. Hepatitis: HCV variability, the immune system and resistance to antiviral drugs. Nat Rev Gastroenterol Hepatol. 2009;6:383–5. doi: 10.1038/nrgastro.2009.102. [DOI] [PubMed] [Google Scholar]
- 58.Pawlotsky JM. Treatment failure and resistance with direct-acting antiviral drugs against hepatitis C virus. Hepatology. 2011;53:1742–51. doi: 10.1002/hep.24262. [DOI] [PubMed] [Google Scholar]
- 59.Vermehren J, Sarrazin C. The role of resistance in HCV treatment. Best Pract Res Clin Gastroenterol. 2012;26:487–503. doi: 10.1016/j.bpg.2012.09.011. [DOI] [PubMed] [Google Scholar]
- 60.Kati W, Koev G, Irvin M, Beyer J, Liu Y, Krishnan P, et al. In vitro activity and resistance profile of dasabuvir, a nonnucleoside hepatitis C virus polymerase inhibitor. Antimicrob Agents Chemother. 2015;59:1505–11. doi: 10.1128/AAC.04619-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hcvguidelines.org. (2018). HCV Resistance Primer | HCV Guidance. [online] [Last accessed 2018 Jun 29]. Available from: https://www.hcvguidelines.org/evaluate/resistance .
- 62.Svarovskaia ES, Dvory-Sobol H, Parkin N, Hebner C, Gontcharova V, Martin R, et al. Infrequent development of resistance in genotype 1-6 hepatitis C virus-infected subjects treated with sofosbuvir in phase 2 and 3 clinical trials. Clin Infect Dis. 2014;59:1666–74. doi: 10.1093/cid/ciu697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wyles D, Mangia A, Cheng W, Shafran S, Schwabe C, Ouyang W, et al. Long-term persistence of HCV NS5A resistance associated substitutions after treatment with the HCV NS5A inhibitor, ledipasvir, without sofosbuvir. Antivir Ther. 2018;23:229–38. doi: 10.3851/IMP3181. [DOI] [PubMed] [Google Scholar]
- 64.Pawlotsky JM. Hepatitis C virus resistance to direct-acting antiviral drugs in interferon-free regimens. Gastroenterology. 2016;151:70–86. doi: 10.1053/j.gastro.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 65.Radley A, Robinson E, Aspinall EJ, Angus K, Tan L, Dillon JS. A systematic review and meta-analysis of community and primary-care-based hepatitis C testing and treatment services that employ direct acting antiviral drug treatments? BMC Health Serv Res. 2019;19:765. doi: 10.1186/s12913-019-4635-7. doi: 10.1186/s12913-019-4635-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wade AJ, Doyle JS, Gane E, Stedman C, Draper B, Iser D, et al. Community-based provision of direct-acting antiviral therapy for hepatitis C: Study protocol and challenges of a randomized controlled trial. Trials. 2018;19:383. doi: 10.1186/s13063-018-2768-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.[Internet]. Mohfw.gov.in. 2019 [cited 2019 Nov 20] Available from: https://mohfw.gov.in/sites/default/files/National%20Action%20Plan_Lowress_Reference%20file.pdf .


