Highlights
-
•
We conducted retrospective testing of clinical samples submitted to veterinary diagnostic laboratories for porcine deltacoronavirus.
-
•
Over 2000 samples were tested over three separate years with 3 positive samples detected prior to the PDCoV outbreak detection in February 2014.
-
•
We surveyed 42 early infected swine farms to determine various farm level characteristics, descriptive estimates of biosecurity practices and disease status over time of each operation.
-
•
Clinical signs of PDCoV were reported to be similar to those of PEDv. The average number of animals on each operation exhibiting clinical signs (morbidity) and the average number of case fatalities was greatest for suckling and weaned pigs.
-
•
The survey included questions regarding biosecurity practices for visitors and operation employees; trucks, equipment and drivers; and feed sources. These questions attempted to identify a likely pathway of introduction of PDCoV onto the operations surveyed.
Keywords: Porcine deltacoronavirus, Swine enteric coronaviruses, Case series
Abstract
Porcine deltacoronavirus (PDCoV) was first reported in the United States (US) in February 2014. This was the second novel swine enteric coronavirus detected in the US since May 2013. In this study, we conducted retrospective testing of samples submitted to three veterinary diagnostic laboratories where qualifying biological samples were derived from previously submitted diagnostic case submissions from US commercial swine farms with a clinical history of enteric disease or from cases that had been previously tested for transmissible gastroenteritis virus, PEDV, or rotavirus. Overall, 2286 banked samples were tested from 27 States. Samples were collected in 3 separate years and in 17 different months. Test results revealed 4 positive samples, 3 collected in August 2013 and 1 collected in October 2013. In addition, a case series including 42 operations in 10 States was conducted through administration of a survey. Survey data collected included information on characteristics of swine operations that had experienced PDCoV clinical signs. Special emphasis was placed on obtaining descriptive estimates of biosecurity practices and disease status over time of each operation. Clinical signs of PDCoV were reported to be similar to those of PEDV. The average number of animals on each operation exhibiting clinical signs (morbidity) and the average number of case fatalities was greatest for suckling and weaned pigs. Average operation-level weaned pig morbidity was greatest in the first week of the outbreak while average operation-level suckling pig case fatality was greatest in the second week of the outbreak. The survey included questions regarding biosecurity practices for visitors and operation employees; trucks, equipment and drivers; and feed sources. These questions attempted to identify a likely pathway of introduction of PDCoV onto the operations surveyed.
1. Introduction
In February 2014, porcine deltacornavirus (PDCoV) was first detected in the US (Marthaler et al., 2014). This was the second of two novel enteric coronaviruses to have been detected in the US since May 2013, the first being porcine epidemic diarrhea virus (PEDV). There is very little information available regarding the epidemiology of PDCoV. The virus was first described from a surveillance collection of rectal swabs from pigs in Hong Kong (Woo et al., 2012). Sequencing of PDCoV strains isolated from pigs in the US indicated approximately 99% nucleotide identity with 2 strains of PDCoV from China (Marthaler et al., 2014).
Since June 2013, results of diagnostic samples submitted from pigs and farms suspected to be infected with PEDV were shared through the National Animal Health Laboratory Network (NAHLN). The NAHLN is a cooperative effort between the USDA, Animal and Plant Health Inspection Service (APHIS) and the American Association of Veterinary Laboratory Diagnosticians. It is comprised of laboratories that provide diagnostic services for different diseases, using common testing methods and software platforms to process diagnostic requests and share information. Results of laboratory accessions tested at NAHLN laboratories for PEDV were voluntarily sent weekly to APHIS for compilation and reporting purposes. In April 2014, similar reporting for PDCoV was initiated and eventually combined with the PEDV report. From March 30, 2014 to July 9, 2015, 733 positive laboratory accessions for PDCoV were reported from 17 States (USDA, APHIS, website, 2015). A positive accession does not necessarily represent a single positive herd as multiple accessions may be submitted from a single herd. In June 2014, the USDA published a Federal Order mandating the reporting of PEDV and PDCoV by producers, veterinarians and laboratories. The Federal Order required the inclusion of premises identifier data to allow for more accurate monitoring of disease incidence and spread. From June 2014 to July 9, 2015, 70 premises in 13 States have been confirmed with PDCoV (USDA, APHIS, website, 2015).
Neither PDCoV nor PEDV had been detected in the US prior to May 2013. Identifying the entry pathways of these viruses and characterizing the epidemiology of these pathogens (e.g. PEDV and PDCoV) are of particular interest to farmers, swine veterinarians and government officials. The objectives of the study reported here were to: (a) obtain an estimate of how long PDCoV has been present in US swine through retrospective testing of submissions to veterinary diagnostic laboratories, (b) describe potential factors associated with the introduction of the virus onto pig operations, (c) describe the frequency of clinical signs and, (d) describe the morbidity and case fatality that appears to be associated with PDCoV on commercial swine farms.
2. Materials and methods
2.1. Retrospective testing
Three veterinary diagnostic laboratories (Iowa State University, Ohio Department of Agriculture, University of Minnesota) tested banked clinical samples for the presence of PDCoV using PDCoV PCR assays (specific assay procedures used in each laboratory). Qualifying biological samples were derived from previously submitted diagnostic case submissions from U.S. commercial swine farms with a clinical history of enteric disease or from cases that had been previously tested for transmissible gastroenteritis virus, PEDV, or rotavirus. Diagnostic samples or extracts from feces, fecal swabs, intestines, or oral fluids from submissions received from April, 2010 to December 1, 2013 were included in the total sample population. Up to 50 cases per month per lab and up to 2 samples per case could be included. Samples from the same case could be pooled in groups of 5. Samples were tested from each month in the date range and from as wide a geographic distribution as possible. Each sample result was to be accompanied by the date of sample collection, State in which the farm submitting samples was located, operation type (sow/boar, nursery, suckling, and grower/finisher) and, the results of any previous diagnostic testing. Data from the three laboratories was combined and the FREQ procedure in SAS used to determine relative frequencies.
2.2. Case series survey
A standardized survey (available from corresponding author), adapted from observational studies conducted to meet similar objectives for PEDV, was used to collect information on characteristics of swine breeding herd operations that had experienced PDCoV clinical signs. Special emphasis was placed on obtaining descriptive estimates of biosecurity practices and disease status over time of each operation. These operations may or may not have also experienced a PEDV outbreak.
Operations included in the study were selected by attending herd veterinarians who were members of the American Association of Swine Veterinarians (AASV). The AASV requested from their membership identification of operations willing to participate in the study. Inclusion criteria for operations were: breeding operations (Farrow to Wean) only, PEDV laboratory confirmed positive herds that subsequently were confirmed positive for PDCoV, or PEDV negative herds (no clinical signs, no laboratory confirmation) that have been confirmed positive for PDCoV, and willingness to complete a survey by either face to face or telephone interview. Attending herd veterinarians identifying operations willing to participate were then required to provide their name and contact information (telephone number and email address), the state where the case farm was located, initial date of PDCoV clinical signs, date of PDCoV laboratory confirmation, and the PEDV status (had the farm previously been confirmed infected with PEDV).
Information from attending herd veterinarians was sent to APHIS field epidemiologists and veterinarians who administered the survey. Training of interviewers was conducted by survey developers prior to survey administration and an interviewer’s manual was provided. In person or telephone interviews with the person most knowledgeable about practices on the operation, usually a herd manager or owner, were conducted from April 28, 2014 to June 30, 2014. The completed questionnaires were sent to APHIS staff at the Center for Epidemiology and Animal Health in Fort Collins, CO.
APHIS staff performed data entry and validation on the entire data set. A data entry interface was used to create an initial dataset from the questionnaires indexed by a farm identification number to denote an observation. Subsequently, each questionnaire was examined. This examination was a preliminary spot check for variable value accuracy using the view table function in SAS. Separate validation and editing code was then executed on the initial dataset to create an edited dataset used in all subsequent estimations. Validation and editing consisted of two main types of checks: valid values and logical checks.
One additional data editing step was done prior to generating descriptive estimates. The questionnaire was unusual in that it asked cross sectional questions at the operation level (e.g., inventory) as well as animal and time level (e.g., number of suckling pigs with clinical signs in the first week of the PDCoV outbreak). There was room in the questionnaire for identification of only two types of ill pigs. When there were more than two types of pigs ill on the operation that observation was split. This resulted in additional rows of data being created using the original farm identification number. Operation level information was replicated in the second row but animal information differed to reflect the third or more ill animal type. This resulted in 56 observations in the final edited dataset rather than the original 42. For subsequent operation level estimations such as average inventory, only one observation was used and identified as operation type. Estimates of morbidity, case fatality, and clinical signs were included and identified as ill pig type. SUDAAN (http://www.rti.org/suddan) was used to complete the statistical estimation due to its ability to estimate standard error automatically.
Due to the large number of estimations and the small variations in the code needed to produce all of them, the SUDAAN code was written in macro form. Three macro types corresponding to procedures were performed: Crosstab, Ratio and Descript. The Crosstab macro estimates frequencies of categorical responses much like the FREQ procedure in SAS. Ratio and Descript estimate proportions were used with selection of macro dependent on the form of the variables summarized (http://www.rti.org/Sudaan/onlinehelp/SUDAAN10/Default.htm).
3. Results
3.1. Retrospective testing
Overall, 2286 banked samples were tested. Samples were tested from 27 States with total samples per state ranging from 1 to 693. A total of 14.1, 18.1, 5.1 and 22.4% of the samples were obtained from grower/finisher, nursery, suckling and sow/boar operation types respectively. Forty percent of the samples did not identify an operation type. Samples were collected in 3 separate years and in 17 different months (Table 1 ).
Table 1.
Results of PDCoV retrospective PCR testing.
| Month and year sample collected | Number of samples tested | Number samples positive |
|---|---|---|
| April 2010 | 40 | 0 |
| September 2011 | 1 | 0 |
| October 2012 | 9 | 0 |
| November 2012 | 24 | 0 |
| December 2012 | 8 | 0 |
| January 2013 | 88 | 0 |
| February 2013 | 112 | 0 |
| March 2013 | 231 | 0 |
| April 2013 | 273 | 0 |
| May 2013 | 103 | 0 |
| June 2013 | 95 | 0 |
| July 2013 | 102 | 0 |
| August 2013 | 593 | 3 |
| September 2013 | 60 | 0 |
| October 2013 | 78 | 1 |
| November 2013 | 124 | 0 |
| December 2013 | 51 | 0 |
There were 4 positive samples, 3 collected in August 2013 and 1 collected in October 2013. Two of the positive samples were from grower/finisher operation types in Iowa, 1 from a grower/finisher operation type in Minnesota, and 1 from an unknown operation type in Minnesota.
3.2. Case series survey
Survey data was collected on 42 operations in 10 States (Illinois, Indiana, Iowa, Kansas, Michigan, Minnesota, Nebraska, North Carolina, Ohio, and South Dakota) that had laboratory confirmation of PDCoV infection. Although breeding (Farrow to Wean) operations were targeted for survey administration; 2.4, 21.4, 7.1 and 4.8% of operations were identified as Wean to Finish, Farrow to Finish, Farrow to Feeder or other operation type respectively. Breeding operations represented 64.3% of operation types. Other characteristics of the operations completing the survey are included in Table 2 .
Table 2.
Characteristics of operations included in case series survey by operation type.
| Date of clinical sign onset | Farrow to finish |
Farrow to feeder |
Breeding only |
Other |
||||
|---|---|---|---|---|---|---|---|---|
| Pct. | SE | Pct. | SE | Pct. | SE | |||
| 12/1/13 to 12/31/13 | 0.0 | (–) | 0.0 | (–) | 11.2 | (6.1) | 0.0 | (–) |
| 1/1/14 to 1/31/14 | 33.3 | (15.9) | 0.0 | (–) | 33.3 | (15.9) | 0.0 | (–) |
| 2/1/14 to 2/28/14 | 0.0 | (–) | 0.0 | (–) | 29.6 | (8.9) | 0.0 | (–) |
| 3/1/14 to 5/3/14 | 66.7 | (15.9) | 100.0 | (0.0) | 25.9 | (8.5) | 100.0 | (0.0) |
| PEDv occurrence on this operation | Pct. | SE | Pct. | SE | Pct. | SE | Pct. | SE |
| Yes | 77.8 | (14.0) | 66.7 | (27.5) | 70.4 | (8.9) | 50.0 | (35.8) |
| No | 22.2 | (14.0) | 33.3 | (27.5) | 29.6 | (8.9) | 50.0 | (35.8) |
| PEDv occurrence relative to PDCoV | Pct. | SE | Pct. | SE | Pct. | SE | Pct. | SE |
| After | 14.3 | (13.4) | 0.0 | (–) | 21.1 | (9.5) | 0.0 | (–) |
| Before | 71.4 | (17.3) | 100.0 | (0.0) | 63.2 | (11.2) | 100.0 | (0.0) |
| Same day | 14.3 | (13.4) | 0.0 | (–) | 15.7 | (8.5) | 0.0 | (–) |
| PRRS occurrence | Pct. | SE | Pct. | SE | Pct. | SE | Pct. | SE |
| Yes | 14.3 | (13.4) | 0.0 | (–) | 21.1 | (9.5) | 0.0 | (–) |
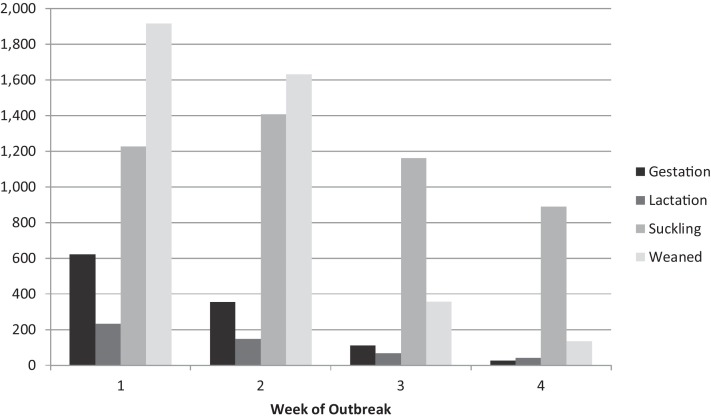
Clinical signs of PDCoV were reported to be similar to those of PEDV. Watery diarrhea occurred in all pig types on nearly 100 percent of operations (Table 3 ). Vomiting, anorexia, depression and fever were also common in all pig types. The average number of animals on each operation exhibiting clinical signs (morbidity) and the average number of case fatalities was greatest for suckling and weaned pigs (Fig. 1, Fig. 2 ). Average operation-level weaned pig morbidity was greatest in the first week of the outbreak while average operation-level suckling pig case fatality was greatest in the second week of the outbreak.
Table 3.
Percentage of sites with pig types with clinical signs of PDCoV.
| Clinical Signs | Gestation |
Lactation |
Suckling pigs |
Weaned pigs |
||||
|---|---|---|---|---|---|---|---|---|
| Pct. | SE | Pct. | SE | Pct. | SE | Pct. | SE | |
| Watery diarrhea | 100.0 | (0.0) | 93.5 | (4.5) | 100.0 | (0.0) | 100.0 | (0.0) |
| Vomiting | 57.7 | (9.8) | 61.3 | (8.8) | 70.0 | (8.4) | 75.0 | (21.8) |
| Anorexia | 92.3 | (5.3) | 93.5 | (4.5) | 86.7 | (6.3) | 75.0 | (21.8) |
| Depression | 88.5 | (6.3) | 93.5 | (4.5) | 93.3 | (4.6) | 60.0 | (22.1) |
| Fever | 20.0 | (8.1) | 40.0 | (9.0) | 50.0 | (9.2) | 25.0 | (21.8) |
Fig. 1.
Average number of animals with clinical signs of PDCoV by production type, and by week of the outbreak on the operation.
Fig. 2.
Average number of animals that died of PDCoV by production type, and by week of the outbreak on an operation.
One study objective was to describe potential factors associated with the introduction of the virus onto pig operations. Therefore, a primary focus of the survey was to capture data on management, biosecurity, and biocontainment practices on individual operations. Between 67 and 78% of operations, depending on operation type, had experienced PEDV infection in addition to PDCoV infection. Most operations, independent of operation type, experienced PEDV infection before PDCoV infection. Participants were asked if changes to biosecurity protocols were made in response to either this operation becoming infected with PEDV or concerns about PEDV in general (Table 4, Table 5 ). Specific changes to protocols of personnel entering the pig facilities due to prior PEDV infection included applying disinfectant to the entry mat, requiring hand sanitizer, placing lunches in double bags, requiring use of rubber boots, prohibiting cell phones from entering pig facilities, requiring disposable boots and foot baths, requiring semen deliveries be made off site, and changing down time for employees from 48 to 72 h. Responses for changes to protocols for trucks, equipment and drivers due to prior PEDV infection included biosecurity reviews, delivery and pickup route changes, increased down time for trucks, and requiring trucks to always be washed and disinfected and dried for 24 h and in some cases heated. Responses for changes to feeding practices included changing premix vendors, flushing feed lines after mixing diets that contained porcine products, and eliminating all or some porcine products from the feed (e.g porcine sprayed dried plasma).
Table 4.
Biosecurity changes implemented prior to PDCoV infection because of confirmed PEDv infection by operation type (no wean to finish operation types responses).
| Site types | Percent sites |
|||||||
|---|---|---|---|---|---|---|---|---|
| Farrow to finish |
Farrow to feeder |
Breeding only |
Other |
|||||
| Pct. | SE | Pct. | SE | Pct. | SE | Pct. | SE | |
| Change to visitor biosecurity protocols | ||||||||
| Yes | 42.9 | (18.9) | 50.0 | (35.8) | 16.7 | (8.9) | 100.0 | (0.0) |
| No | 57.1 | (18.9) | 50.0 | (35.8) | 83.3 | (8.9) | 0.0 | (–) |
| Don't know | 0.0 | (–) | 0.0 | (–) | 0.0 | (–) | 0.0 | (–) |
| Change to personnel entering the hog and pig facilities protocols | ||||||||
| Yes | 28.6 | (17.3) | 50.0 | (35.8) | 31.6 | (10.8) | 100.0 | (0.0) |
| No | 71.4 | (17.3) | 50.0 | (35.8) | 68.4 | (10.8) | 0.0 | (–) |
| Don't know | 0.0 | (–) | 0.0 | (–) | 0.0 | (–) | 0.0 | (–) |
| Change to protocols for trucks, equipment, or drivers | ||||||||
| Yes | 28.6 | (17.3) | 0.0 | (–) | 31.6 | (10.8) | 0.0 | (–) |
| No | 71.4 | (17.3) | 100.0 | (0.0) | 63.2 | (11.2) | 100.0 | (0.0) |
| Don't know | 0.0 | (–) | 0.0 | (–) | 5.2 | (5.2) | 0.0 | (–) |
| Change to feed processing | ||||||||
| Yes | 28.6 | (17.3) | 50.0 | (35.8) | 15.8 | (8.5) | 0.0 | (–) |
| No | 71.4 | (17.3) | 50.0 | (35.8) | 78.9 | (9.3) | 100.0 | (0.0) |
| Don't know | 0.0 | (–) | 0.0 | (–) | 5.3 | (5.2) | 0.0 | (–) |
| Change to feed ingredients | ||||||||
| Yes | 85.7 | (13.4) | 100.0 | (0.0) | 15.8 | (8.5) | 0.0 | (–) |
| No | 14.3 | (13.4) | 0.0 | (–) | 78.9 | (9.5) | 100.0 | (0.0) |
| Don't know | 0.0 | (–) | 0.0 | (–) | 5.3 | (5.2) | 0.0 | (–) |
Table 5.
Biosecurity changes implemented prior to PDCoV infection because of general concerns about PEDv. Percent of sites by type.
| Site types | Percent sites |
|||||||
|---|---|---|---|---|---|---|---|---|
| Farrow to Finish |
Farrow to Feeder |
Breeding only |
Other |
|||||
| Pct. | SE | Pct. | SE | Pct. | SE | Pct. | SE | |
| Change to visitor biosecurity protocols | ||||||||
| Yes | 55.5 | (16.8) | 100.0 | (0.0) | 37.0 | (9.4) | 100.0 | (0.0) |
| No | 44.4 | (16.8) | 0.0 | (–) | 62.9 | (9.4) | 0.0 | (–) |
| Don’t know | 0.0 | (–) | 0.0 | (–) | 0.0 | (–) | 0.0 | (–) |
| Change to personnel entering the hog and pig facilities protocols | ||||||||
| Yes | 11.1 | (10.6) | 0.0 | (–) | 51.9 | (9.7) | 100.0 | 0.0 |
| No | 88.9 | (10.6) | 100.0 | 0.0 | 44.4 | (9.7) | 0.0 | (–) |
| Don’t know | 0.0 | (–) | 0.0 | (–) | 3.7 | (3.7) | 0.0 | (–) |
| Change to protocols for trucks, equipment, or drivers | ||||||||
| Yes | 77.8 | (14.0) | 0.0 | (–) | 63 | (9.4) | 50.0 | (35.8) |
| No | 22.2 | (14.0) | 100.0 | 0.0 | 37.0 | (9.4) | 50.0 | (35.8) |
| Don’t know | 0.0 | (–) | 0.0 | (–) | 0.0 | (–) | 0.0 | (–) |
| Change to feed processing | ||||||||
| Yes | 11.1 | (10.6) | 33.3 | (27.5) | 3.8 | (3.8) | 0.0 | (–) |
| No | 88.9 | (10.6) | 66.7 | (27.5) | 96.2 | (3.8) | 100.0 | (0.0) |
| Don’t know | 0.0 | (–) | 0.0 | (–) | 0.0 | (–) | 0.0 | (–) |
| Change to feed ingredients | ||||||||
| Yes | 22.2 | (14.0) | 33.3 | (27.5) | 51.9 | (9.7) | 0.0 | (–) |
| No | 77.8 | (14.0) | 66.7 | (27.5) | 48.1 | (9.7) | 100 | 0.0 |
| Don’t know | 0.0 | (–) | 0.0 | (–) | 0.0 | (–) | 0.0 | (–) |
The survey included questions regarding biosecurity practices for visitors and operation employees; trucks, equipment and drivers; and feed sources. These questions attempted to identify a likely pathway of introduction of PDCoV onto the operations surveyed. Participants were asked if in the 10 days before the onset of PDCoV if someone from a foreign country visited the operation and if someone from this operation, who works with pigs (including their veterinarian or their partners), returned from a foreign country. For all operation types, 100% of operations responded that no visitors from foreign countries visited the site and 88.9% and 11.1% responded that no one from their operation who works with pigs visited a foreign country or they did not know if they had visited a foreign country respectively. In the 10 days before the onset of clinical signs of PDCoV, 22.2, 33.3 and 59.3% of Farrow to Finish, Farrow to Feeder and breeding operations had non-employees enter the pig facilities. For sites that had any visitors during the 10 days before the onset of PDCoV, Table 6 shows the percentage of sites by number of visits of each type of visitor.
Table 6.
For sites that had any visitors during the 10 days before the date of onset of PDCoV, percentage of sites by number of visits of each type of visitor.
| Type of visitor | Percent sites |
||||||
|---|---|---|---|---|---|---|---|
| Number of visits |
|||||||
| 1–5 |
6–10 |
11–15 |
Total | ||||
| Pct. | SE | Pct. | SE | Pct. | SE | ||
| Feed truck | 57.5 | (7.9) | 35.0 | (7.6) | 7.5 | (4.2) | 100.0 |
| Electric company | 100.0 | (0.0) | 0.0 | (–) | 0.0 | (–) | 100.0 |
| Gas/propane company | 100.0 | (0.0) | 0.0 | (–) | 0.0 | (–) | 100.0 |
| Diesel or fuel oil delivery | 100.0 | (0.0) | 0.0 | (–) | 0.0 | (–) | 100.0 |
| Veterinarian | 100.0 | (0.0) | 0.0 | (–) | 0.0 | (–) | 100.0 |
| Company service person | 77.8 | (9.9) | 22.2 | (9.9) | 0.0 | (–) | 100.0 |
| Vaccination crew | 100.0 | (0.0) | 0.0 | (–) | 0.0 | (–) | 100.0 |
| Pig truck bringing pigs of any age onto farm | 92.9 | (7.0) | 7.1 | (7.0) | 0.0 | (–) | 100.0 |
| Pig truck removing pigs of any age from farm | 78.4 | (6.8) | 18.9 | (6.5) | 2.7 | (2.7) | 100.0 |
| Trash pickup | 100.0 | (0.0) | 0.0 | (–) | 0.0 | (–) | 100.0 |
| Building contractor | 50.0 | (35.8) | 50.0 | (35.8) | 0.0 | (–) | 100.0 |
| Equipment sales/installation | 100.0 | (0.0) | 0.0 | (–) | 0.0 | (–) | 100.0 |
| Cattle/sheep (or other nonswine livestock or poultry) hauler | 100.0 | (0.0) | 0.0 | (–) | 0.0 | (–) | 100.0 |
| Other swine producer | 0.0 | (–) | 0.0 | (–) | 0.0 | (–) | 100.0 |
| Friends or family | 0.0 | (–) | 0.0 | (–) | 0.0 | (–) | 100.0 |
| Feed/nutrition consultant | 100.0 | (0.0) | 0.0 | (–) | 0.0 | (–) | 100.0 |
| Professional exterminator | 100.0 | (0.0) | 0.0 | (–) | 0.0 | (–) | 100.0 |
| Renderer | 66.7 | (27.5) | 33.3 | (27.5) | 0.0 | (–) | 100.0 |
| Other visitors | 81.8 | (11.8) | 18.2 | (11.8) | 0.0 | (–) | 100.0 |
Cleaning and disinfection protocols for trucks either picking up or delivering pigs were similar across all operation types. The percentage of sites requiring that the animal area inside of the truck be cleaned and then disinfected prior to picking up pigs were 88.9, 100.0 and 88.9 for Farrow to Finish, Farrow to Feeder and breeding operations respectively. These same percentages were observed for sites requiring the animal area outside of the truck be cleaned prior to picking up pigs. However, a lower percentage of Farrow to Feeder (33.3) and breeding operations (46.2) required the animal area outside of the truck to be disinfected prior to picking up pigs.
Sites with ill gestating sows and gilts were almost evenly split in thirds as to the nature of feed purchased in 10 days before the date of onset of PDCoV. Between 27% and 35% purchased either complete feed or components of feed that were then mixed off-site by company or non-company personnel. Nearly 29% of sites with ill gestating sows and gilts that had feed purchased and delivered in the 10 days before the date of onset of PDCoV sourced feed ingredients in the gestation ration from outside the US. Slightly over 30% of sites with feed delivered in the 10 day period received feed within 1–6 days of the outbreak. Sixty percent of sites with ill suckling pigs received spray dried plasma in the 10 days before the date of onset of PDCoV while 40% received blood products or pork fat.
4. Discussion
One objective of this study was to estimate the initial point in time of PDCoV introduction into the US through retrospective testing of submissions to veterinary diagnostic laboratories. Samples testing PDCoV PCR positive in August and October of 2013 indicates that PDCoV was present at least 3–6 months prior to the reports of clinical outbreaks on farms. It has been suggested that the PDCoV, similar to other enteric coronaviruses in swine, does not survive well in warmer temperatures. The earliest detections in August and October may not have propagated or spread rapidly across the US due to warm summer and fall temperatures.
The data for PDCoV after December 2013 demonstrate an exponential epidemiology curve (Fig. 3 ), suggesting a point-source origin followed by highly contagious propagated spread (Smith, 1995). Others have speculated that the virus has been silently circulating prior to and in-between these times and remained undetected by clinical observations because it is less virulent resulting in milder signs than the PEDV. Thachil et al. (2015) described a new ELISA test for PDCoV and following retrospective serological testing of 395 banked samples submitted for gastrointestinal disease with this ELISA suggested that PDCoV has been present in the US since 2010. The results from this serologic testing are based on an estimated test specificity of 95%, based on a negative population cohort of 30 animals. The confidence interval was not provided in this report.
Fig. 3.
Number of PDCoV positive herds or submissions August 2013 through May 2014.
We investigated reported clinical signs, morbidity and case fatality associated with PDCoV in the participating herds, particularly for suckling age pigs. Comparisons to other morbidity and case fatality data are made with published estimates of PEDV morbidity and case fatality since those for PDCoV are lacking. The first published mortality figures associated with PEDV were reported to be 90–95% mortality in suckling pigs which is numerically higher than our highest value of 44.2% (Stevenson et al., 2013). Similar mortality was found by others (Schlueter et al., 2015, Dufrense and Robbins, 2014). Results from farms in the study reported here suggest that case fatality for PDCoV is lower in pre-weaned pigs than found with PEDV.
In addition to clinical signs of PDCoV, we also attempted to describe factors associated with the introduction of the virus to the sites. Prior to this we asked about changes made in biosecurity brought about by PEDV. Initially, it appeared, based on responses, that the threat of PEDV can prompt more changes than its actual occurrence in certain types of sites. For example, breeding operations that actually had previously experienced PEDV outbreak seemed less likely to make changes to protocols surrounding visitors, personnel, trucks or equipment than those that were simply worried about PEDV in general.
A substantial amount of guidance has been established for biosecurity protocols concerning general visitors, farm personnel, deliveries/pick-ups and feed since the entrance of PEDV into the U.S. in 2013 (www.pork.org/pork-checkoff-research/PEDV/). There has not been a separate effort to update this research for PDCoV. Most sites had multiple deliveries of feed, company service person visits, and renderer entry. Most also had pigs picked up for some reason such as permanent removal to an abattoir. Lowe et al. (2014) described how this stream is a concern since the prevalence of PEDV in vehicles leaving a harvest facility appeared to increase as the prevalence of contaminated trucks came into the facility. It is fortunate that sites in the study changed truck protocol to include trailer heating based on the work of Thomas et al. (2013). Other researchers suggest that the transportation of dead animals (e.g., rendering) inherently has a high risk for transmission of PEDV and probably PDCoV (Henry, 2014).
There has been a focus on feed as a source for transmission of PEDV since an early investigation (Nugent, 2015). In a survival analysis done on incidence data from North Carolina, having a feed truck enter a premise increased the probability of PEDV infection 44%. In addition to the vehicle or the driver, feed itself is a known risk for PEDV and most likely for PDCoV (Dee et al., 2014, Yeske, 2014). Although more than half of sites with ill suckling pigs received spray dried plasma prior to infection, there appeared to be no common source and the risk of this material likely depends on how it is manufactured (Pujols and Segalés, 2014, Sampedro et al., 2015).
The study reported here, provides useful information on the potential time of introduction of PDCoV into the U.S. The large number of samples and extended coverage of those samples in time and geography provide confidence in the estimated date the virus entered the U.S. This paper also provides the first description of clinical signs, morbidity and mortality across multiple operations (location and operation type) and also describes the processes and procedures in place on sites involved in initial PDCoV outbreaks. It is limited by its small size and by the nature of recent coronaviruses, which appear to enter swine operations by varied routes. It is therefore difficult to directly ascertain specific introductory pathways through the results of this survey.
References
- Dee S., Clement T., Schelkopf A., Nerem J., Knudsen D., Christopher J., Nelson E. An evaluation of contaminated complete feed as a vehicle for porcine epidemic diarrhea virus infection of naïve pigs following consumption via natural feeding behavior: proof of concept. BMC Vet. Res. 2014;10:176. doi: 10.1186/s12917-014-0176-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufrense L., Robbins R. Field experience with porcine epidemic diarrhea. Proceedings of the American Association of Swine Veterinarians 45th Annual Meeting; March 1–4 2014, Dallas, Texas, USA; 2014. pp. 613–616. [Google Scholar]
- Henry, S., 2014. Porcine Epidemic Diarrhea Virus (PEDV), Delta Corona (Sdcv) and PED Variant (Pdcov). Presentation. http://anrs.oregonstate.edu/system/files/u3084/PED%20Madras%20Oregon%202014.pdf (accessed 10.07.15.).
- Lowe, J., Gauger, P., Harmon, K., Zhang, J., Connor, J., Yeske, P., Loula, T., Levis, I., Dufresne, L., 2014. Role of transportation in spread of porcine epidemic diarrhea virus infection, US [dispatch]. Emerging Infectious Diseases [Internet]. May. http://wwwnc.cdc.gov/eid/article/20/5/13-1628_article (accessed 12.07.15.). [DOI] [PMC free article] [PubMed]
- Marthaler, D., Bruner, L., Collins, J., Rossow, K., 2014. Third strain of porcine epidemic diarrhea virus, US [letter]. Emerging Infectious Diseases [Internet]. December. http://wwwnc.cdc.gov/eid/article/20/12/14-0908_article#r7 (accessed 12.07.15.). [DOI] [PMC free article] [PubMed]
- Nugent R. State of the knowledge: the relationships between PEDV/PDCoV transmission and feed. Proceedings of the American Association of Swine Veterinarians 46th Annual Meeting; February 28–March 3, 2015, Orlando, Florida, USA; 2015. [Google Scholar]
- Pujols J., Segalés J. Survivability of porcine epidemic diarrhea virus (PEDV) in bovine plasma submitted to spray drying processing and held at different time by temperature storage conditions. Vet. Microbiol. 2014;174:427–432. doi: 10.1016/j.vetmic.2014.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampedro F., Bueno I., Bergeron J., Urriola P., Davies P. Risk assessment of feed ingredients of porcine origin as vehicles for transmission of porcine epidemic diarrhea virus (PEDV). roceedings of the American Association of Swine Veterinarians 46th Annual Meeting; February 28–March 3, 2015, Orlando, Florida, USA; 2015. pp. 63–64. [Google Scholar]
- Schlueter E., Johnson C., Thompson N. Managing the initial break. Proceedings of the American Association of Swine Veterinarians 46th Annual Meeting; February 28–March 3, 2015, Orlando, Florida, USA; 2015. pp. 411–416. [Google Scholar]
- Smith R. 2nd ed. CRC Press, Inc.; 1995. Veterinary Clinical Epidemiology: A Problem Oriented Approach. [Google Scholar]
- Stevenson G., Hoang H., Schwartz K., Burrough E., Sun D., Madson D., Cooper V., Pillatzki A., Gauger P., Schmitt B., Koster L., Killian M., Yoon K. Emergence of Porcine epidemic diarrhea virus in the US: clinical signs, lesions, and viral genomic sequences. J. Vet. Diag. Invest. 2013;25:649–654. doi: 10.1177/1040638713501675. [DOI] [PubMed] [Google Scholar]
- Thachil A., Gerber P., Xiao C., Huang Y., Opriessnig T., Halbur P. A porcine deltacoronavirus serological survey using an indirect PDCoV anti-IgG ELISA confirms that PDCoV infection in US pigs is low and has been present since. Proceedings of the American Association of Swine Veterinarians 46th Annual Meeting; February 28–March 3, 2015, Orlando, Florida, USA; 2010. p. 65. [Google Scholar]
- Thomas P., Karriker L., Ramirez A., Zhang J., Ellingson J., Holtkamp D. Evaluation of time and temperature sufficient to inactivate porcine epidemic diarrhea virus in swine feces on metal surfaces. Proceedings of the 23rd IPVS Congress; June 8–11, 2014, Cancun, Mexico; 2013. p. 249. [Google Scholar]
- USDA, 2015. Swine Enteric Coronavirus Disease (SECD) Situation Report-July 9, 2015. http://www.aphis.usda.gov/animal_health/animal_dis_spec/swine/downloads/secd_sit_rep_07_09_15.pdf. (accessed 10.07.15.).
- Woo P., Lau S., Lam C., Lau C., Tsang A., Lau J., Bai R., Teng J., Tsang C., Wang M., Zheng B., Chan K., Yuen K. Discovery of seven novel mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeske, P. 2014. Evidence of Feed Transmitting PEDV. Presentation: Leman Preconference Session. 2014 Allen D. Leman Swine Conference, September 13–16, 2014, St. Paul RiverCentre, St. Paul, Minnesota. https://docs.google.com/a/umn.edu/file/d/0BzGsnfsQ28heSTZMTFBqWG5rZlE/edit?pli=1. (accessed 10.07.15.).