Abstract
A novel coronavirus (CoV) has recently been identified as the aetiological agent of severe acute respiratory syndrome (SARS). Nucleocapsid (N) proteins of the Coronaviridae family have no discernable homology, but they share a common nucleolar-cytoplasmic distribution pattern. There are three putative nuclear localization signal (NLS) motifs present in the N. To determine the role of these putative NLSs in the intracellular localization of the SARS–CoV N, we performed a confocal microscopy analysis using rabbit anti-N antisera. In this report, we show that the wild type N was distributed mainly in the cytoplasm. The N-terminal of the N, which contains the NLS1 (aa38–44), was localized to the nucleus. The C-terminus of the N, which contains both NLS2 (aa257–265) and NLS3 (aa369–390) was localized to the cytoplasm and the nucleolus. Results derived from analysis of various deletion mutations show that the region containing amino acids 226–289 is able to mediate nucleolar localization. The deletion of two hydrophobic regions that flanked the NLS3 recovered its activity and localized to the nucleus. Furthermore, deletion of leucine rich region (220-LALLLLDRLNRL) resulted in the accumulation of N to the cytoplasm and nucleolus, and when fusing this peptide to EGFP localization was cytoplasmic, suggesting that the N may act as a shuttle protein. Differences in nuclear/nucleolar localization properties of N from other members of coronavirus family suggest a unique function for N, which may play an important role in the pathogenesis of SARS.
Keywords: SARS–CoV, Nucleocapsid protein, Nuclear localization signal, Nucleolar localization, Cell cycle arrest
1. Introduction
Severe acute respiratory syndrome (SARS) is a life-threatening form of atypical pneumonia that emerged in Guangdong Province, China, resulting in 8098 probable cases and 774 deaths around the world. A number of laboratories worldwide have undertaken the identification of the causative agent (Drosten et al., 2003, Fouchier et al., 2003, Ksiazek et al., 2003, Peiris et al., 2003). The SARS–CoV genome is a single-stranded positive-sense RNA, classified within the order Nidovirales, family Coronaviridae, genus coronavirus of ∼29.7 kb and contains 14 open reading frames. The genome is composed of a stable region encoding functional replicase/transcriptase complex, a variable region representing four coding sequences for viral structural proteins (spike (S), envelope (E), membrane (M) and nucleocapsid (N)), and other putative nonstructural uncharacterized proteins. The gene order of SARS–CoV is similar to that of other coronaviruses; however, phylogenic analyses and sequence comparisons indicate that this virus does not closely resemble any of the previously known human or animal coronaviruses (Marra et al., 2003, Rota et al., 2003, Ruan et al., 2003). Later study suggests that SARS–CoV represents an early split-off from coronavirus group 2 lineage relatively late in coronavirus evalution (Snijder et al., 2003) and this result has found support from other studies using alternative methods (Gorbalenya et al., 2004, Stavrinides and Guttman, 2004).
SARS–CoV N protein is 422 amino acids in length, basic in nature, which suggests it may assist in RNA binding, and can self-associate to form dimers (Surjit et al., 2004b). Recently is has been reported to be phosphoraylated in vitro (Zakhartchouk et al., 2005) although the physiological consequence of such covalent modification is yet to be determined. A recent report showed that phosphorylation of coronavirus infectious bronchitis virus (IBV) N protein determined the recognition of virus RNA (Chen et al., 2005). In addition, serine/arginine rich (SR; aa176–204) motif, which present in SARS–CoV N, may be the site for the location of phosphoserines. Based on the available information, several functions have been postulated for the coronavirus N proteins throughout the virus life cycle (Laude and Masters, 1995), including viral packaging (Davies et al., 1981), viral core formation (He et al., 2004b, Risco et al., 1996) and signal transduction pathway (He et al., 2003) or part of host defense response invoked to counteract viral infection (Chang et al., 2004).
Sub-cellular localization of the viral protein is one of the commonly used approaches to characterize the involvement of the protein in the virus assembly. Although the SARS–CoV N protein harbors a putative nuclear localization signal (NLS) (Marra et al., 2003), the N protein is distributed predominantly in the cytoplasm of SARS–CoV-infected and N gene-transfected cells with a presence of weak fluorescence signals in the nucleus (Chang et al., 2004), therefore, it is not known whether this putative NLS is functional or not. Moreover, Qinfen and coworkers observed a special phenomenon during morphogenesis of SARS–CoV, where virus-like particles appeared in the nucleus of Vero E6 cells 48 h post-infection (Qinfen et al., 2004). On the other hand, an identical nucleolar-cytoplasmic localization pattern is observed for the N proteins of arteriviruses; porcine reproductive and respiratory syndrome virus (PRRSV) and equine arteritis virus (EAV) (Rowland et al., 1999, Tijms et al., 2002) and for N proteins of groups I, II and III coronaviruses; transmissible gastroenteritis virus (TGEV), mouse hepatitis virus (MHV) and IBV, respectively (Hiscox et al., 2001, Wurm et al., 2001). Recently Rowland and Yoo have reported the nucleolar-cytoplasmic shuttling of PRRSV N protein through determining a possible nuclear export signal (NES) (Rowland and Yoo, 2003).
The nucleolus is highly specialized structure that participates in regulation of several host cell processes, including regulation of cell cycle, apoptosis and induction of antiviral responses (Rowland and Yoo, 2003). Targeting a protein to the nucleolus occurs through the diffusion of protein through the nucleoplasm and accumulation in the nucleolus. Localization to the nucleolus is mediated through nucleolar localization signal (NoLS) that is in part an NLS, which functions by interacting with nucleolar components such as nucleolar proteins, rRNA or rDNA (Garcia-Bustos et al., 1991). Since many endogenous proteins that localize in the nucleus have been identified as regulators of the cell cycle (Chen et al., 2002, Garcia-Bustos et al., 1991) this beneficial control over the host cell cycle can either facilitate the early release of progeny virus or help the emerging virus to evade the host immune system for its persistent infection.
In this study, we examined the intracellular localization profile of N protein and a series of deleted mutants in SARS–CoV permissive cell lines and elucidated the possible role of N protein in modulation of the host cell cycle.
2. Materials and methods
2.1. Cells and virus infection
Vero and Vero E6 cells (obtained from China Centre for Type Culture Collection, CCTCC, Wuhan, China) were grown and maintained in Dulbecco's modified Eagle's medium (Gibco-BRL, USA) and minimal essential medium (Gibco) respectively, supplemented with 10% heat-inactivated FBS (Hyclone, Logan, Utah), 100 units/ml penicillin, and 100 μg/ml streptomycin, and incubated at 37 °C in 5% CO2. Vero E6 cells were infected with SARS–CoV WHU strain at the MOI of 0.1, and were seeded after 24 h for both indirect immunofluorescence assay and Western blot analysis or after 48 h for RNA isolation.
2.2. Generation of polyclonal antisera in rabbits
The RNA isolation, RT-PCR, construction of pGEMT-N plasmid contains an authentic copy of the N gene, sequencing (Genbank accession no. AY365036), subcloning of N gene into prokaryotic expression vector, expression and purification of N protein have been described previously (Timani et al., 2004). New Zealand rabbits were kept in conventional conditions and were handled in compliance with Collage of Life Sciences, Wuhan University (Wuhan, China) guidelines for animal care and use. Rabbit was immunized with 0.1–0.2 mg of purified recombinant N protein and was injected subcutaneously at multiple sites on its back. A booster injection was given 2 and 4 weeks later. Blood was drown from the rabbit at 5 weeks following the immunization, the blood was allowed to clot at 4 °C and the antiserum was recovered by centrifugation at 5000 × g for 10 min at 4 °C. A controlled serum was done by injecting normal saline at the same above conditions.
2.3. Plasmids construction and cell transfection
The pGEMT-N plasmid was used as the template to amplify coding region of N protein with different deletion mutations by PCR. The wild type (1269 nt) and deletion mutant fragments of the N gene were made as shown in (Fig. 3B). Each fragment was digested with BamHI and EcoRI, and ligated into BamHI and EcoRI digested pcDNA3 (Invitrogen), an eukaryotic expression vector, such that transcripts were under the control of the CMV immediate-early promoter. To generate deletion mutant (NΔ220–231) of N protein gene with internal deletion of aa220–231, two PCR fragments were amplified using two pairs of primers. The two fragments were digested with BamHI, XbaI and XbaI, EcoRI restriction enzymes and ligated into pcDNA3. The pCMV-flag-NΔ369–376 recombinant plasmid containing N protein gene with internal deletion of (aa369–376) was constructed using pfu DNA polymerase with Quickchange site directed mutagenesis kit (Stratagene) according to manufacturer's instruction and used as the template to obtain N226–422/Δ369–376 by using the same primers for N226–422 construct.
Fig. 3.
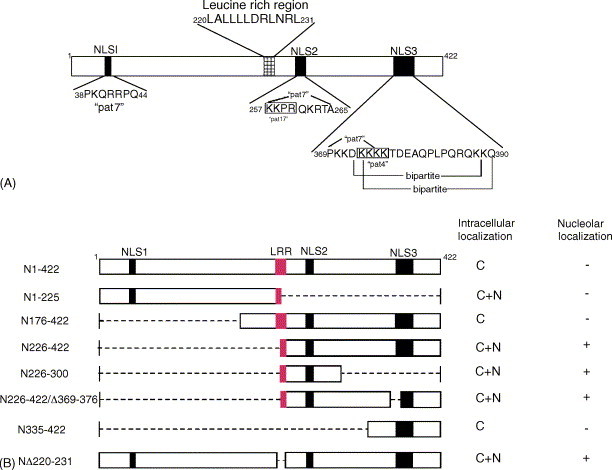
(A) The diagram identifies the location of putative NLSs and LRR domains within SARS–CoV N protein. Amino acid sequence analysis software program, PSORT II (Nakai and Kanehisa, 1992) was used to locate the classical NLS motifs. The numbers identified the amino acid position covered by each domain. The NLS was classified into “pat4”, “pat7” and “bipartite” motifs. For details, see the text. (B) Schematic representation of a summary of different deletion mutants of SARS–CoV N and their respective intracellular localization. Dash lines and numbers indicate the deleted amino acids and amino acids positions, respectively while bars represent translated amino acids. N: nucleus; C: cytoplasm. (+) and (−) designate for presence and absence of protein expression, respectively.
The wild type and mutant fragments of enhanced green fluorescent protein (EGFP)-tagged fusion proteins were made as shown in (Fig. 6). Different parts of the N gene coding sequence were obtained as described above and the PCR fragments were cloned into the BglII and EcoRI sites of pEGFP-N1 vector (Clontech) in-frame with the EGFP. The GNΔ220–231 was obtained as above and cloned into BglII and EcoRI sites of pEGFP-N1. pCMV-flag-NΔ369–376 plasmid was used as the template to generate GN226–422/Δ369–376 by using the same primers for GN226–422 construct. The nucleotide sequence and reading frame of the constructs were verified by restricted digestion and DNA sequencing. The primers sequence used in this study are available from the authors upon request.
Fig. 6.
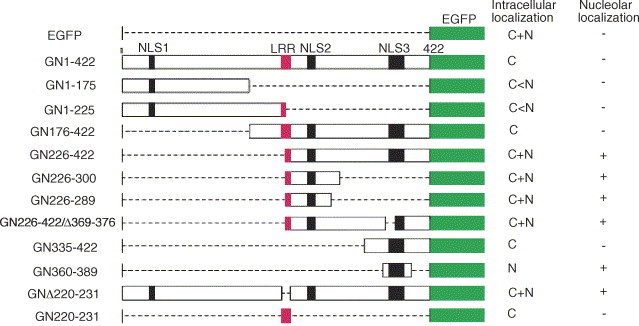
Schematic representation of a summary of wild type EGFP-N fusion protein, different deletion mutants and their respective intracellular localization. The pEGFP-N1 vector was used to express all the constructs. Dash lines and numbers indicate the deleted amino acids and amino acids positions, respectively. The bars represent translated amino acids. N: nucleus; C, cytoplasm. (+) and (−) designate for presence and absence of protein expression, respectively.
Confluent monolayers (60–80%) of Vero and Vero E6 cells grown on 6- or 24-well tissue culture plates (NUNCLON) were transfected with plasmid DNA by using Lipofectamine™ 2000 reagent (Gibco) according to the instructions of the manufacturer.
2.4. Establishments of stable nucleocapsid protein expressing cell line
Vero E6 cells grown on 24-well tissue culture plate were transfected with 0.5 μg of pcDNA-N containing the full length N gene or pcDNA3 as a control and after 48 h, transfected cells were selected with 800 μg/ml of G418 (Life Technologies). Three weeks later, resistant colonies were isolated and screened for N protein expression by RT-PCR and Western blot analysis.
2.5. Western blot analysis
Total proteins were resolved by 10 or 15% SDS-PAGE and transferred onto PVDF membranes (Bio-Rad, Hercules, CA). The residual binding sites on the membranes were blocked by incubation with 5% non-fat dry milk in TBS–T buffer (20 mM Tris–HCl pH 7.6, and 137 mM NaCl containing 0.02% Tween-20) for 2 h. Following the blocking, membranes were subsequently probed with rabbit anti-N polyclonal antibody (1:800 dilution) for 2 h at 4 °C, and then washed three times for 10 min each. The bound antibodies were detected by goat anti-rabbit HRP-conjugated antibody for 1.5 h. After rinsing three times, the specific proteins were visualized by enhanced chemiluminescence reagent (Amersham).
2.6. Indirect immunofluorescence assay and confocal microscopy
Vero or Vero E6 cells grown on coverslips were transfected with 2 μg of plasmid DNA for 6 h. The medium was replaced with maintenance medium for 24 h prior to fixing with ice cold 50% methanol-50% acetone at −20 °C. Coverslips were incubated for 1 h at 37 °C with rabbit anti-N polyclonal antibody (1:200 dilution), and/or mouse monoclonal antibody for B23 protein (Sigma–Aldrich, St. Louis, MO), washed three times with cold PBS, followed by incubation with flourescein isothiocyanate (FITC)-conjugated goat anti-rabbit IgG (Sigma) and/or tetramethyl rhodamine isothiocyanate (TRITC)-conjugated goat anti-mouse IgG (Sigma) for 1 h at 37 °C, respectively, then again washed with cold PBS. Cells were either mounted in mounting media or stained with propidium iodide (PI; Sigma) to visualize nuclear DNA. Fluorescence images were viewed under laser confocal scanning microscope (Leica Laser Technik, Germany) with appropriate filters.
2.7. Flow cytometry and colony formation efficiency assays
For cell cycle analysis, ∼60% confluent cells were grown on six-well tissue culture plates. The following day cells were transfected as described above. After 36 h, cells were washed with PBS and removed from the plates by treatment with trypsin-EDTA and re-suspended in 500 μl of ice cold PBS. These cells were fixed with 70% ethanol. Cells were washed twice with PBS after incubation at 4 °C for 30 min or stored at −20 °C until performing the analysis. To remove the double-stranded RNA, cells were further incubated with 1 μl RNase (10 mg/ml) for 30 min at 37 °C, followed by staining with 20 μl PI (1mg/ml) for 15 min. The ∼10,000 cells were analyzed in Coulter Epics XL flow cytometer (Backman Coulter, USA) and the cell cycle analysis was done using the cell quest program by manual sitting regions for G0/G1 and G2/M.
For determination of colony formation efficiency assay, the same number of cells (1 × 102) from stable transfected N protein and neo r as a control were seeded on 35 mm-diameter dishes then cultured for 6–8 days until colonies could be visualized. Colonies were washed with PBS and stained with 0.25% Coomassie Brilliant Blue (R-250) in 50% methanol and 10% acetic acid.
3. Results
3.1. Characterization of anti-N protein antisera
A polyclonal antibody directed against N protein was produced in order to determine its intercellular localization. We generated anti-N rabbit antisera using an Escherichia coli produced fusion protein (SARS–CoV N protein with 6XHis-tag at the N-terminus) as antigen. The antigenicity of the recombinant N protein was confirmed by immunoreactivity with SARS patients sera using ELISA assay, showed high sensitivity and specificity (Timani et al., 2004).
To examine the reactivity and specificity of the N protein antisera, Western blot analysis was performed. The results demonstrated that rabbit antisera notably reacted with N protein from SARS–CoV-infected cells. Intriguingly, a series of shorter bands were also observed (Fig. 1 A, lane 1), most likely they are isomers of N protein due to Caspases degradation. The antisera also strongly reacted to a protein expressed in E. coli cell lysate with an apparent molecular mass of ∼47 kDa (Fig. 1B lane 1). The cell lysate was prepared from Vero E6 constitutively expressing N protein showed a single band with same molecular mass (Fig. 1B lane 2) whereas no protein was detected from sample of cells transfected with an empty vector alone (Fig. 1B lane 3). The pre-immune serum did not react with any specific protein from either E. coli or transfected cell lysates (data not shown). Therefore, we used this polyclonal antiserum for further experiments.
Fig. 1.
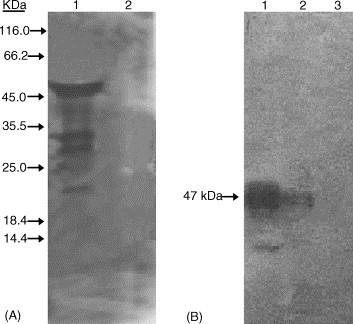
Characterization of the rabbit anti-N polyclonal antibody. (A) Total cell lysates from SARS–CoV-infected Vero E6 (lane 1) and Vero E6 cells only as a control (lane 2) were resolved by 15% SDS-PAGE. (B) Total cell lystaes from E. coli expressing the recombinant N protein after IPTG induction (described elsewhere; (Timani et al., 2004)) (lane 1), N protein constitutively expressing Vero E6 cells (lane 2) and neor control (lane 3) were resolved by 10% SDS-PAGE. Both (A) and (B) were analyzed by Western blot. The transferred PVDF membranes were probed with rabbit anti-N polyclonal antibody. The molecular mass of N protein in the blot was verified as ∼47 kDa. Approximate mobilities of protein markers are indicated on the left of (A).
3.2. Intracellular localization of SARS–CoV N protein in infected and transfected cell lines
To identify the localization of N protein in SARS–CoV-infected Vero E6 cells where also other viral proteins are expressed, cells were fixed at 24 h post-infection and analyzed by indirect immunofluorescence assay using FITC-labeled secondary antibody directed to rabbit anti-N polyclonal antibody. The PI was used to visualize the nuclear DNA and the images were digitally superimposed to depict the distribution of N protein and nuclear DNA. Our results showed that N protein was localized predominantly in the cytoplasm in SARS–CoV-infected cells (Fig. 2 A) whilst in a few cells a weak fluorescence label was also observed in the nucleus (Fig. 2B–D). In a group of infected cells (Fig. 2A), assuming that the amount of fluorescence from the FITC is proportional to the amount of N protein in the infected cells, in some cells a few bright fluorescence granules were found in the cytoplasm while in others a strong fluorescence were observed around the nuclear envelop, more likely the cells are at different infectious stages. No significant fluorescence was observed in pre-immune serum (Fig. 2A, inset panel). A similar observation was found in Vero E6 (Fig. 4A) transfected with plasmid expressing full length N protein.
Fig. 2.
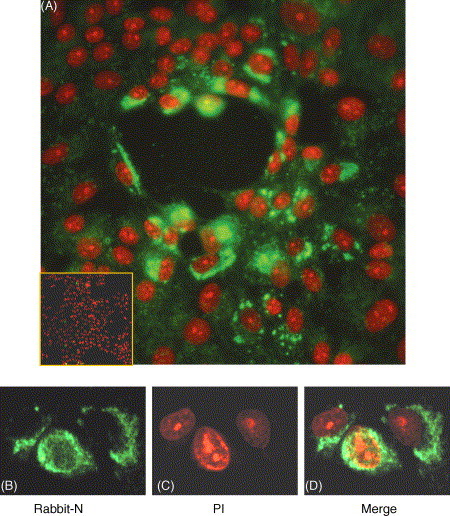
Detection of SARS–CoV N protein by indirect immunofluorescence assay in infected cells. Vero E6 cells were infected with SARS–CoV WHU strain. After 24 h cells were fixed and analyzed by indirect immunofluorescence using rabbit anti-N polyclonal antibody (green) ((A) and (B)) or pre-immune rabbit serum ((A) inset panel), followed by staining with PI (red) ((A) and (C)) to visualize the nuclear DNA. Differentially fluorescing images were gathered separately ((A) and (D)) using confocal microscope. Magnification 20 and 40× for (A) and (B)–(D), respectively.
Fig. 4.
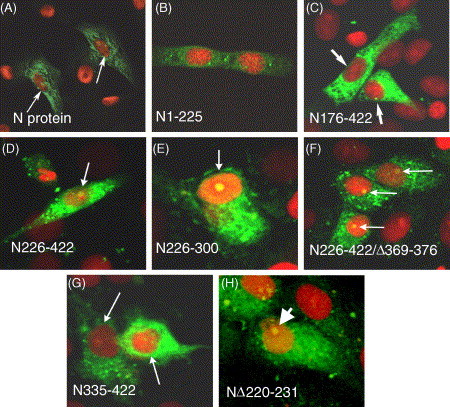
Intracellular localization of various deletion mutants of SARS–CoV N protein in the transfected cells. Vero E6 cells were transfected with different deleted mutants. After 24 h, the cells were fixed and analyzed by indirect immunofluorescence assay using rabbit anti-N polyclonal antibody (green). Additionally, cells were stained with PI to visualize the nuclear DNA (red) and were examined by confocal microscopy. The two colors were then merged and yellow color when it occurs is the region where green and red co-localized. (A)–(H) represents the merged panels of indicated deletion mutants. Arrow points to the cell nucleus or to the yellow region. Magnification 40×.
3.3. Subcellular localization of N protein alters upon N- and/or C-terminal deletion mutations
To determine if the N- or C-terminal truncated forms alter the localization pattern of the N protein, a different deletion mutants were constructed (Fig. 3 B) taking into consideration the location of stretches of basic amino acids which are the principle components of NLS and the leucine rich region (LRR; see below). The amino acid sequence analysis of N protein was done by using software program PSORT II (Nakai and Kanehisa, 1992) and three classical putative NLS motifs were identified, labeled NLS1, NLS2 and NLS3 at positions 38–44, 257–265 and 369–390 amino acids, respectively (Fig. 3A). Furthermore, a Kyte–Doolittle hydrophilicity analysis of the protein sequence predicted that NLS motifs are in hydrophilic regions and should be easily accessible. An indirect immunofluorescence assay was used to analyze the intracellular localization of N protein in Vero E6 cells using anti-N polyclonal antibody. All the slides were further stained with PI to visualize the nuclear DNA. The differentially fluorescing images were gathered separately and then digitally superimposed (Fig. 4 A–H). Vero E6 cells transfected with mutant N176–422, which contains NLS1, NLS2 and SR rich motifs were showed a localization pattern similar to the wild type protein (Fig. 4C). Interestingly, mutated protein N226–422 (contains putative NLS2 and NLS3) was localized to the cytoplasm and nucleus accumulated mainly in the nucleolus (bright fluorescence spot inside the nucleus; Fig. 4D). The deletion mutant N226–300 lacking both the NLS1 and NLS3 was also localized to the cytoplasm and the nucleolus (Fig. 4E). To further confirm this, deletion mutant N226–422/Δ369–376 was constructed with internal deletion of the core basic sequence of NLS3 (369-PKKDKKKK) showed a pattern of intranuclear localization similar to N226–422 and N226–300 (Fig. 4F). Furthermore, the deletion mutant N335–422, which contains NLS3, the highest content of basic residues among the three NLS motifs, was unexpectedly present in the cytoplasm only (Fig. 4G). The deletion mutant N1–225 included the NLS1 and SR-rich motifs was localized to the cytoplasm and nucleus but the accumulation in the nucleolus could not be confirmed (Fig. 4B) because the FITC fluorescence was weak even after the experiment was repeated more than once and this could be due to the low antigenicity of the N-terminal of N protein in comparison with C-terminal (personal observation).
To confirm that this bright fluorescent spot was located in the nucleolus rather than being an artifact located in the nucleoplasm, monoclonal antibody specific to nucleolar protein B23 was used. The deletion mutants N226–422 (data not shown), N226–300 (Fig. 5 ) and N226–422/Δ369–376 (data not shown) transfected cells were double labeled with FITC-labeled anti-N polyclonal antibody (Fig. 5A) and TRITC-labeled anti-B23 monoclonal antibody (Fig. 5B); confocal microscopy revealed the co-localization of N and B23 proteins (Fig. 5C) and the nucleoli appeared yellow. These results implied that the region containing amino acids 226–300, which includes putative NLS2 was responsible for the nucleolar localization of the C-terminal N protein.
Fig. 5.
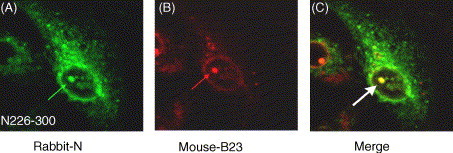
Co-localization of SARS–CoV N deletion mutant N226–300 with nucleolar protein B23 in the nucleolus of transfected cells using indirect immunofluorescence. Vero E6 cells grown on coverslip were transfected with mutant segment N226–300 for 24 h. The cells were fixed then double-labeled with rabbit anti-N polyclonal antibody (green) (A) and mouse monoclonal antibody to the B23 (red) (B). The cells were examined by confocal microscopy. The two colors were then merged and the yellow region in panel (C) is the area where green and red co-localized. The arrow points to the cell nucleolus. Magnification 40×.
3.4. Expression and intracellular localization of N-EGFP fusion proteins
Various deletion mutants of EGFP fusion protein genes were constructed (Fig. 6 ) to study the intracellular localization pattern in another cell line, Vero cells. To eliminate the possibility that positively charged proteins may relocate to the nucleus because of a post-fixation artifact (Lundberg and Johansson, 2002), the coverslips containing cells were overlaid on the slides then directly visualized under confocal microscopy. Our results showed that the wild type EGFP was diffusely localized to the cytoplasm, nucleoplasm and excluded from the nucleolus (Fig. 7 A) which is consistent with passive diffusion. The fusion of N protein to EGFP (GN1–422) altered the distribution of EGFP, resulting in the accumulation of fluorescent protein in the cytoplasm and exclusion from the nucleus (Fig. 7B). The similar observation for the wild type EGFP-N was observed in GN176–422 mutant fragment whereas in few cells a weak fluorescence was also observed in the nucleus (Fig. 7G). The protein products of mutants GN226–422(Fig. 7H and I), GN226–300(Fig. 7J) and GN226–422/Δ369–376(Fig. 7M and N) were accumulated in the cytoplasm and nucleus and had high amounts of fluorescence intensity in the nucleolus. Further deletion from the C-terminal of GN226–300 to generate GN226–289 (Fig. 7K and L) fragment was still localized to the nucleolus. The Nucleoli, when viewed under phase-contrast microscope, are apparent as highly refractive bodies within the nucleus (Fig. 7C, F, I, L, N, Q and T). The GN335–422 fragment was localized in the cytoplasm only (Fig. 7O) which was similar to the localization pattern for N335–422. Interestingly, deletion of 24 amino acids from N-terminal and 33 amino acids from the C-terminal of GN336–422 to generate GN360–389 altered the distribution of EGFP, resulting in accumulation of fluorescent protein in the nucleus and its decrease in the cytoplasm (Fig. 7P and Q). This showed that the peptide region 360–389 contains an active NLS3. Furthermore, GN1–175 and GN1–225 mutant fragments contain NLS1 and NLS1 and SR rich motifs respectively, were localized in the nucleus but excluded from the nucleolus (Fig. 7D and (E and F) respectively) suggesting that N-terminal N protein may also contain an active NLS1 that directs an exogenous protein to the nucleus but not to the nucleolus. These results showed that the region aa226–289 of the N protein contains NoLS and thus its activity is independent from putative NLS1 and NLS3.
Fig. 7.
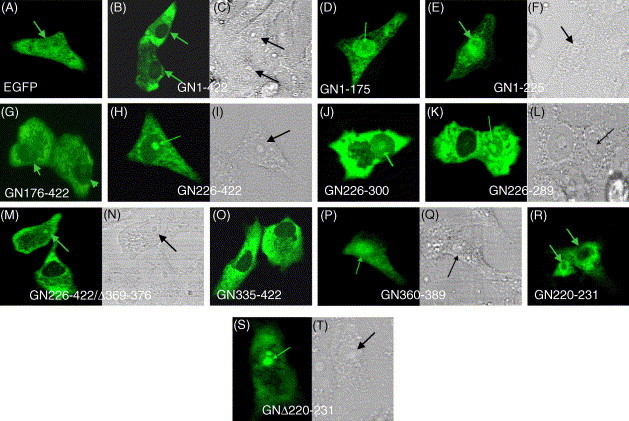
Intracellular localization of EGFP fusion proteins containing segments of N protein deletion mutants. Vero cells were transfected with pEGFP-N1 vector only or with different deletion mutants fused to N-terminal of EGFP. After 24 h cells were analyzed by direct fluorescence using confocal microscopy. The panels represent the distribution of fluorescence signals in the cells transfected with EGFP, EGFP-N and deletion mutants as indicated. ((C), (F), (I), (L), (N), (Q) and (T)) are phase-contrast of the fluorescence images ((B), (E), (H), (K), (M), (P) and (S)) of the same cells, respectively. Arrow points to the cell nucleus or to the nucleolus.
3.5. The leucine rich region may act as nuclear export signal
Why the wild type N and its mutant fragment N176–422 localize in the cytoplasm, although the presence of aa226–289 peptide region responsible for the nucleolar localization in its C-terminal? To address this question, we performed amino acid sequence analysis of the of N protein and showed that it contains LRR (220-LALLLLDRLNRL-231; Fig. 3A) which satisfies the consensus requirements for the classical NES; X-R(2–4)-X-R2-X-R-X, where X is leucine, isoleucine or valine and R is any amino acid (Henderson and Eleftheriou, 2000, Hope, 1999). Furthermore, Kyte–Doolittle hydrophilicity analysis showed that this region has highest hydrophobicity. To determine if the N protein LLR sequence is functional, different deletion mutants were constructed, NΔ220–231 and GNΔ220–231. These mutants contain the full length N protein with an internal deletion of LRR (Fig. 3, Fig. 6), cloned to pcDNA3 and pEGFP-N1, respectively. In addition, the peptide aa220–231 was placed at the C-terminal of EGFP to produce GN220–231. The Vero cells transfected with mutant plasmids were examined by indirect immnuofluorescence using rabbit anti-N polyclonal antibody or direct fluorescence for EGFP fusion mutant fragments. The NΔ220–231 localized in the cytoplasm and nucleus mainly in the nucleolus similar to the localization pattern of N226–300 (Fig. 4H). The localization pattern for GNΔ220–231 was similar to GN226–289, i.e. the localization resulted in accumulation of fluorescent protein in the cytoplasm and nucleus with high fluorescence intensity in the nucleolus (Fig. 7S and T). To determine if this LRR could export a heterogous protein from the nucleus, a construct GN220–231 was used. This construct was localized predominantly in the cytoplasm (Fig. 7R) whilst in some cells, fluorescent granules were surrounding the nuclear envelope with very low fluorescence label in the nucleus (data not shown). These observations suggest that LRR may act as an NES and this also might explain the cytoplasmic localization of mutant proteins N176–422 (Fig. 4C) and GN176–422 (Fig. 7G). Another point to note is ∼20% of transfected cells only showed nucleolar localization of C-terminal N protein; this may be due to other unknown factors favoring the protein to the cytoplasm.
The GN226–422 and N226–422 mutant fragments contain the half part of the LRR (220-LALLLL) at its N-terminal and the GN1–225 contains the other half (226-DRLNRL) at its C-terminal (Fig. 3, Fig. 6), shown in red box), were localized to the cytoplasm and nucleus, thus the full peptide sequence of LRR is required for export of protein out from the nucleus.
3.6. Stable cell line constitutively expressing N protein affects cell cycle
To study if the constitutively expressing N protein could affect the cell cycle, Vero E6 cells were stably transfected with N protein expression vector and the expression level of N protein and mRNA were detected by Western blot (Fig. 1 lane 2) and RT-PCR (data not shown), respectively. Then we determined the colony formation efficiency of N-expressing Vero E6 cells. The same numbers of cells were seeded onto culture dishes and the colonies were visualized as described under “Section 2”. The result shown in (Fig. 8 ) indicated that colony formation efficiency was greatly reduced in N-expressing Vero E6 cells compared with G418-resistent control cells. Likewise, we sought to determine the effect of N protein expression on cell cycle distribution by flow cytometric analysis. Our results showed a prolonged G0/G1 phase with a shortened S phase compared with control cells (Table 1 ). The experiment was repeated three times and the results were reproducible. These results indicated that Vero E6 cells constitutively expressing N protein exhibited cell growth retardation most likely through G0/G1 arrest.
Fig. 8.
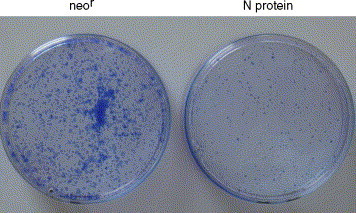
SARS–CoV N expression inhibits the colony formation efficiency. Vero E6 cells of constitutively expressing N protein and neor control cells (1 × 102) were seeded on 35 mm culture dishes and cultured until colonies were visually seen. Colonies were stained as described under “Section 2”. The assay was carried out in triplicate and the results were reproducible.
Table 1.
Effect of SARS–CoV nucleocapsid protein expression on cell cycle
| Neor (%) | N proteina (%) | |
|---|---|---|
| G0/G1 | 67.9 ± 0.5 | 74.0 ± 0.4 |
| S | 18.0 ± 1.0 | 12.7 ± 0.6 |
| G2/M | 14.1 ± 1.1 | 13.3 ± 0.7 |
N protein-expressing Vero E6 cell line in a constitutive system was analyzed by flow cytometry. The values represent the mean ± SD of triplicate experiments of total cell population in each phase of cell cycle.
4. Discussion
Molecules can enter the nucleus by passive diffusion or active transport mechanisms, depending on their size (Macara, 2001). Small molecules up to size of 50–60 kDa or less than 10 nm in diameter can diffuse passively through NPC, but most of proteins are transported by energy driven transport mechanisms (Richardson et al., 1988). Active transport of proteins is mediated by NLS. These sequences are recognized by proteins of importin super-family (importin α and β) that mediate the transport across the nuclear envelope using RanGTP (Macara, 2001).
NLS are generally classified as one of the three types (Hicks and Raikhel, 1995, Nakai and Kanehisa, 1992). The “pat4” motif consists of continuous stretch of four basic amino acids (lysine or arginine) or three basic amino acids associated with histidine or proline. The “pat7” sequence starts with proline and is followed by a three residues segment containing three basic residues out of four (Garcia-Bustos et al., 1991). The third type of NLS, known as “bipartite” motif, consists of 2 basic amino acids, a 10 amino acids spacer, and a 5 amino acids segment containing at least three basic residues. Using these criteria, three NLS motifs were predicted to be present in the N protein, for the positions of NLS and their respective NLS types, see (Fig. 3).
Our study showed that region containing aa226–289 overlapping the NLS2 was capable of reproducing the same nucleolar-cytoplasmic localization pattern as the C-terminal N protein (aa226–422) and inactivation or complete removal of NLS3 has no effect on the C-terminal N nuclear translocation. Interestingly, the C-terminal N (aa209–422) was mapped as the region, which participated in the formation of the N–N protein interaction (Surjit et al., 2004b). Due to the high concentration of basics residues, this region could involved in the interaction with RNA, DNA or nucleolar proteins such as B23 and nucleolin which contain stretches of acidic residues. The minimal peptide sequence required for nucleolar localization was not elucidated in this study. However, the NoLS associated with most viral proteins is usually no longer than 20 amino acids and possesses at least nine basic residues, including one continuous stretch of four basic amino acids or two stretches of three basic amino acids (Garcia-Bustos et al., 1991, Kubota et al., 1989). A comparison with known viral NoLS sequences shows little sequence homology except for the incorporation of an NLS2 which contains “pat4” and “pat7” sequence motifs furthermore, this region (aa226–289) contains 14 basic residues.
The deletion mutants N335–422 and its EGFP fusion segment; GN335–422 region including NLS3 and containing one “pat4”, two “pat7” and two “bipartite” motifs, were unexpectedly localized to the cytoplasm and excluded from the nucleus. However, truncations of the peptide from the N- and C-terminal to produce peptide GN360–389 was localized to the nucleus and small amounts of fluorescence remained in the cytoplasm. Sequence analysis of the C-terminal N revealed the presence of two hydrophobic regions, 351-VILL and 392-VTLL that flanked the NLS3 at N- and C-terminal, respectively. In other words, the activity of NLS3 was recovered by removing the flanked hydrophobic regions, suggesting that these regions could have hidden the NLS3 from exposing to the surface of the protein. Furthermore, the N-terminal fragments GN1–176 and GN1–225 that were localized to nucleus were excluded from the nucleolus and weak fluorescence was observed in the cytoplasm.
The results also revealed that LRR (220-LALLLLDRLNRL) may act as NES. Since, few cells showed a weak expression of wild type N protein in the nucleus (Fig. 2D), possible reasons could be that LRR exhibits a strong export signal favoring the N to the cytoplasm (for review, see reference (Rowland and Yoo, 2003)). Furthermore, the location of LRR is just close to the N-terminal of NLS2, which might have affected its function; these findings suggest that N may act as shuttle protein. Moreover, additional experiments are needed to study the molecular mechanism of SARS–CoV N protein LRR. The PRRSV coronavirus and EVA arterivirus N proteins utilize the CRM1-mediated nuclear export pathway. Inactivation of this pathway with leptomycin B (LMB) resulted in nuclear retention of N, demonstrating that protein shuttles between cytoplasm and nucleus before playing role in virus assembly (Rowland and Yoo, 2003, Tijms et al., 2002). When GN1–422 expressing cells were treated with 20 ng/ml of LMB for 4 h or for prolong period of time (overnight) or even at higher concentration (100 ng/ml) for 2 h, the localization of N protein was unaltered, i.e. predominantly cytoplasmic (data not shown), possible reasons could be that N protein exported via exportin-independent pathway or LRR could act as masking signal rather than NES.
Why does the SARS–CoV N protein exhibits different and complex localization pattern is unknown and needs to be addressed in future. Recent reports demonstrated that SARS–CoV induced apoptosis in virus-infected cells (Mizutani et al., 2004a, Mizutani et al., 2004b, Yan et al., 2004), whilst Surjit et al. showed that SARS–CoV N induced apoptosis in COS-1 cells under stressed condition and activated Caspase-3 and -7 (Surjit et al., 2004a). The appearance of shorter form of N protein late in the infection has been observed with different strains of TGEV (Eleouet et al., 2000) and another report demonstrated the cleavage of recombinant N of SARS–CoV by Caspase-3 but not Caspase-6 in vitro and the pattern of peptide fragments distribution were similar to that in infected cells (Ying et al., 2004) which is consistent with our result (Fig. 1A, lane 1). Taken together, we hypothesized that at a late stage of infection, the activation of Caspases during the apoptosis of the infected cells by SARS–CoV leads to the cleavage of N protein at particular sites. Some of the cleavage products may contain an active NLS leading to its translocation to the nucleus and that may disrupt or usurp nuclear function whereas self-association of some N molecules (He et al., 2004a) may protect N from cleavage by Caspases and remain in the cytoplasm to participate in the virus assembly.
In our present study, further investigation of the effect of N protein on the cell growth was carried out. Vero E6 cells constitutively expressing N protein exhibited a pronounced retardation of cell growth by reduced colony formation efficiency concomitantly with a shortened S phase and a prolonged G0/G1 phase of the cell cycle. Many viral proteins that have nuclear/nucleolar localization properties were reported to disrupt cell cycle, for instance, TGEV and IBV N proteins cause aberrant cell division by disrupting cytokinesis and this inhibits or delays cell growth (Chen et al., 2002, Wurm et al., 2001) and in HeLa cells, HIV-I Vpr which is a nucleolar-cytoplasmic protein causes growth retardation and G2 arrest (Nishizawa et al., 1999). While in HCV NS5A, N-terminally deleted form but not the full length localizes to the nucleus and the protein causes growth inhibition and cell cycle perturbations through G2 arrest (Arima et al., 2001, Song et al., 2000). Recently Chen and Makino demonstrated that MHV coronavirus infection in asynchronously growing cells led to the inhibition of host DNA synthesis and accumulation of cells in the G0/G1 phase and they subsequently attributed to a replicase protein (Chen and Makino, 2004, Chen et al., 2004). Future studies need to be designed to elucidate the role of N protein in regulation of cell cycle, e.g. interaction with nucleolar proteins and activation or suppression cell cycle regulators. Thus, the localization to the nucleus/nucleolus implies that N protein may involve in the regulating of cell cycle possibly by delaying the cell growth to promote intracellular conditions for virus assembly and sequestering ribosomes for translation of viral proteins or by participating in the modulation of nucleolar functions, a strategy to optimize virus replication [see (Hiscox, 2003) for a review]. The identification of different nuclear and nucleolar localization properties of N protein from other members of Coronaviridae family suggests a unique function for N protein and will enable us to investigate the role of N protein in the pathogenesis of SARS and that N protein might have contributed in some way to the severity of the recent out break.
Acknowledgments
We thank the SARS virus research group of Wuhan University, College of Life Sciences (Wuhan, China). The authors are grateful to Dr. Stephen J. Polyak (Department of Laboratory Medicine, University of Washington, Seattle) for valuable suggestions. We thank Wei Peng for assistance with confocal microscopy. Dr. Songya Lu for assistance with the flow cytometry. Ms. Zhenghui Wu for techniqual support. Dr. Timani is supported by Chinese Scholarship Council nominated by Ministry of Higher Studies, Lebanon.
References
- Arima N., Kao C.Y., Licht T., Padmanabhan R., Sasaguri Y. Modulation of cell growth by the hepatitis C virus nonstructural protein NS5A. J. Biol. Chem. 2001;276(16):12675–12684. doi: 10.1074/jbc.M008329200. [DOI] [PubMed] [Google Scholar]
- Chang M.S., Lu Y.T., Ho S.T., Wu C.C., Wei T.Y., Chen C.J., Hsu Y.T., Chu P.C., Chen C.H., Chu J.M., Jan Y.L., Hung C.C., Fan C.C., Yang Y.C. Antibody detection of SARS–CoV spike and nucleocapsid protein. Biochem. Biophys. Res. Commun. 2004;314(4):931–936. doi: 10.1016/j.bbrc.2003.12.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.J., Makino S. Murine coronavirus replication induces cell cycle arrest in G0/G1 phase. J. Virol. 2004;78(11):5658–5669. doi: 10.1128/JVI.78.11.5658-5669.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.J., Sugiyama K., Kubo H., Huang C., Makino S. Murine coronavirus nonstructural protein p28 arrests cell cycle in G0/G1 phase. J. Virol. 2004;78(19):10410–10419. doi: 10.1128/JVI.78.19.10410-10419.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Gill A., Dove B.K., Emmett S.R., Kemp C.F., Ritchie M.A., Dee M., Hiscox J.A. Mass spectroscopic characterization of the coronavirus infectious bronchitis virus nucleoprotein and elucidation of the role of phosphorylation in RNA binding by using surface plasmon resonance. J. Virol. 2005;79(2):1164–1179. doi: 10.1128/JVI.79.2.1164-1179.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Wurm T., Britton P., Brooks G., Hiscox J.A. Interaction of the coronavirus nucleoprotein with nucleolar antigens and the host cell. J. Virol. 2002;76(10):5233–5250. doi: 10.1128/JVI.76.10.5233-5250.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies H.A., Dourmashkin R.R., Macnaughton M.R. Ribonucleoprotein of avian infectious bronchitis virus. J. Gen. Virol. 1981;53(Pt 1):67–74. doi: 10.1099/0022-1317-53-1-67. [DOI] [PubMed] [Google Scholar]
- Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., Berger A., Burguiere A.M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.C., Muller S., Rickerts V., Sturmer M., Vieth S., Klenk H.D., Osterhaus A.D., Schmitz H., Doerr H.W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348(20):1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- Eleouet J.F., Slee E.A., Saurini F., Castagne N., Poncet D., Garrido C., Solary E., Martin S.J. The viral nucleocapsid protein of transmissible gastroenteritis coronavirus (TGEV) is cleaved by caspase-6 and -7 during TGEV-induced apoptosis. J. Virol. 2000;74(9):3975–3983. doi: 10.1128/jvi.74.9.3975-3983.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouchier R.A., Kuiken T., Schutten M., van Amerongen G., van Doornum G.J., van den Hoogen B.G., Peiris M., Lim W., Stohr K., Osterhaus A.D. Aetiology: Koch's postulates fulfilled for SARS virus. Nature. 2003;423(6937):240. doi: 10.1038/423240a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Bustos J., Heitman J., Hall M.N. Nuclear protein localization. Biochim. Biophys. Acta. 1991;1071(1):83–101. doi: 10.1016/0304-4157(91)90013-m. [DOI] [PubMed] [Google Scholar]
- Gorbalenya A.E., Snijder E.J., Spaan W.J. Severe acute respiratory syndrome coronavirus phylogeny: toward consensus. J. Virol. 2004;78(15):7863–7876. doi: 10.1128/JVI.78.15.7863-7866.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He R., Dobie F., Ballantine M., Leeson A., Li Y., Bastien N., Cutts T., Andonov A., Cao J., Booth T.F., Plummer F.A., Tyler S., Baker L., Li X. Analysis of multimerization of the SARS coronavirus nucleocapsid protein. Biochem. Biophys. Res. Commun. 2004;316(2):476–483. doi: 10.1016/j.bbrc.2004.02.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He R., Leeson A., Andonov A., Li Y., Bastien N., Cao J., Osiowy C., Dobie F., Cutts T., Ballantine M., Li X. Activation of AP-1 signal transduction pathway by SARS coronavirus nucleocapsid protein. Biochem. Biophys. Res. Commun. 2003;311(4):870–876. doi: 10.1016/j.bbrc.2003.10.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He R., Leeson A., Ballantine M., Andonov A., Baker L., Dobie F., Li Y., Bastien N., Feldmann H., Strocher U., Theriault S., Cutts T., Cao J., Booth T.F., Plummer F.A., Tyler S., Li X. Characterization of protein-protein interactions between the nucleocapsid protein and membrane protein of the SARS coronavirus. Virus Res. 2004;105(2):121–125. doi: 10.1016/j.virusres.2004.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson B.R., Eleftheriou A. A comparison of the activity, sequence specificity, and CRM1-dependence of different nuclear export signals. Exp. Cell Res. 2000;256(1):213–224. doi: 10.1006/excr.2000.4825. [DOI] [PubMed] [Google Scholar]
- Hicks G.R., Raikhel N.V. Protein import into the nucleus: an integrated view. Annu. Rev. Cell Dev. Biol. 1995;11:155–188. doi: 10.1146/annurev.cb.11.110195.001103. [DOI] [PubMed] [Google Scholar]
- Hiscox J.A. The interaction of animal cytoplasmic RNA viruses with the nucleus to facilitate replication. Virus Res. 2003;95(1–2):13–22. doi: 10.1016/S0168-1702(03)00160-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiscox J.A., Wurm T., Wilson L., Britton P., Cavanagh D., Brooks G. The coronavirus infectious bronchitis virus nucleoprotein localizes to the nucleolus. J. Virol. 2001;75(1):506–512. doi: 10.1128/JVI.75.1.506-512.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope T.J. The ins and outs of HIV Rev. Arch. Biochem. Biophys. 1999;365(2):186–191. doi: 10.1006/abbi.1999.1207. [DOI] [PubMed] [Google Scholar]
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A.E., Humphrey C.D., Shieh W.J., Guarner J., Paddock C.D., Rota P., Fields B., DeRisi J., Yang J.Y., Cox N., Hughes J.M., LeDuc J.W., Bellini W.J., Anderson L.J. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348(20):1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- Kubota S., Siomi H., Satoh T., Endo S., Maki M., Hatanaka M. Functional similarity of HIV-I rev and HTLV-I rex proteins: identification of a new nucleolar-targeting signal in rev protein. Biochem. Biophys. Res. Commun. 1989;162(3):963–970. doi: 10.1016/0006-291x(89)90767-5. [DOI] [PubMed] [Google Scholar]
- Laude H., Masters P.S. The coronavirus nucleocapsid protein. In: Siddell I.S.G., editor. The Coronaviridae. Plenum Press Inc.; New York: 1995. pp. 141–163. [Google Scholar]
- Lundberg M., Johansson M. Positively charged DNA-binding proteins cause apparent cell membrane translocation. Biochem. Biophys. Res. Commun. 2002;291(2):367–371. doi: 10.1006/bbrc.2002.6450. [DOI] [PubMed] [Google Scholar]
- Macara I.G. Transport into and out of the nucleus. Microbiol. Mol. Biol. Rev. 2001;65(4):570–594. doi: 10.1128/MMBR.65.4.570-594.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S., Khattra J., Asano J.K., Barber S.A., Chan S.Y., Cloutier A., Coughlin S.M., Freeman D., Girn N., Griffith O.L., Leach S.R., Mayo M., McDonald H., Montgomery S.B., Pandoh P.K., Petrescu A.S., Robertson A.G., Schein J.E., Siddiqui A., Smailus D.E., Stott J.M., Yang G.S., Plummer F., Andonov A., Artsob H., Bastien N., Bernard K., Booth T.F., Bowness D., Czub M., Drebot M., Fernando L., Flick R., Garbutt M., Gray M., Grolla A., Jones S., Feldmann H., Meyers A., Kabani A., Li Y., Normand S., Stroher U., Tipples G.A., Tyler S., Vogrig R., Ward D., Watson B., Brunham R.C., Krajden M., Petric M., Skowronski D.M., Upton C., Roper R.L. The Genome sequence of the SARS-associated coronavirus. Science. 2003;300(5624):1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- Mizutani T., Fukushi S., Saijo M., Kurane I., Morikawa S. Importance of Akt signaling pathway for apoptosis in SARS–CoV-infected Vero E6 cells. Virology. 2004;327(2):169–174. doi: 10.1016/j.virol.2004.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani T., Fukushi S., Saijo M., Kurane I., Morikawa S. Phosphorylation of p38 MAPK and its downstream targets in SARS coronavirus-infected cells. Biochem. Biophys. Res. Commun. 2004;319(4):1228–1234. doi: 10.1016/j.bbrc.2004.05.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai K., Kanehisa M. A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics. 1992;14(4):897–911. doi: 10.1016/S0888-7543(05)80111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa M., Myojin T., Nishino Y., Nakai Y., Kamata M., Aida Y. A carboxy-terminally truncated form of the Vpr protein of human immunodeficiency virus type 1 retards cell proliferation independently of G(2) arrest of the cell cycle. Virology. 1999;263(2):313–322. doi: 10.1006/viro.1999.9905. [DOI] [PubMed] [Google Scholar]
- Peiris J.S., Lai S.T., Poon L.L., Guan Y., Yam L.Y., Lim W., Nicholls J., Yee W.K., Yan W.W., Cheung M.T., Cheng V.C., Chan K.H., Tsang D.N., Yung R.W., Ng T.K., Yuen K.Y. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361(9366):1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qinfen Z., Jinming C., Xiaojun H., Huanying Z., Jicheng H., Ling F., Kunpeng L., Jingqiang Z. The life cycle of SARS coronavirus in Vero E6 cells. J. Med. Virol. 2004;73(3):332–337. doi: 10.1002/jmv.20095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson W.D., Mills A.D., Dilworth S.M., Laskey R.A., Dingwall C. Nuclear protein migration involves two steps: rapid binding at the nuclear envelope followed by slower translocation through nuclear pores. Cell. 1988;52(5):655–664. doi: 10.1016/0092-8674(88)90403-5. [DOI] [PubMed] [Google Scholar]
- Risco C., Anton I.M., Enjuanes L., Carrascosa J.L. The transmissible gastroenteritis coronavirus contains a spherical core shell consisting of M and N proteins. J. Virol. 1996;70(7):4773–4777. doi: 10.1128/jvi.70.7.4773-4777.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.H., Tong S., Tamin A., Lowe L., Frace M., DeRisi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., Gunther S., Osterhaus A.D., Drosten C., Pallansch M.A., Anderson L.J., Bellini W.J. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300(5624):1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- Rowland R.R., Kervin R., Kuckleburg C., Sperlich A., Benfield D.A. The localization of porcine reproductive and respiratory syndrome virus nucleocapsid protein to the nucleolus of infected cells and identification of a potential nucleolar localization signal sequence. Virus Res. 1999;64(1):1–12. doi: 10.1016/s0168-1702(99)00048-9. [DOI] [PubMed] [Google Scholar]
- Rowland R.R., Yoo D. Nucleolar-cytoplasmic shuttling of PRRSV nucleocapsid protein: a simple case of molecular mimicry or the complex regulation by nuclear import, nucleolar localization and nuclear export signal sequences. Virus Res. 2003;95(1–2):23–33. doi: 10.1016/S0168-1702(03)00161-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Y.J., Wei C.L., Ee A.L., Vega V.B., Thoreau H., Su S.T., Chia J.M., Ng P., Chiu K.P., Lim L., Zhang T., Peng C.K., Lin E.O., Lee N.M., Yee S.L., Ng L.F., Chee R.E., Stanton L.W., Long P.M., Liu E.T. Comparative full-length genome sequence analysis of 14 SARS coronavirus isolates and common mutations associated with putative origins of infection. Lancet. 2003;361(9371):1779–1785. doi: 10.1016/S0140-6736(03)13414-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Bredenbeek P.J., Dobbe J.C., Thiel V., Ziebuhr J., Poon L.L., Guan Y., Rozanov M., Spaan W.J., Gorbalenya A.E. Unique and conserved features of genome and proteome of SARS–coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 2003;331(5):991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J., Nagano-Fujii M., Wang F., Florese R., Fujita T., Ishido S., Hotta H. Nuclear localization and intramolecular cleavage of N-terminally deleted NS5A protein of hepatitis C virus. Virus Res. 2000;69(2):109–117. doi: 10.1016/s0168-1702(00)00206-9. [DOI] [PubMed] [Google Scholar]
- Stavrinides J., Guttman D.S. Mosaic evolution of the severe acute respiratory syndrome coronavirus. J. Virol. 2004;78(1):76–82. doi: 10.1128/JVI.78.1.76-82.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surjit M., Liu B., Jameel S., Chow V.T., Lal S.K. The SARS coronavirus nucleocapsid protein induces actin reorganization and apoptosis in COS-1 cells in the absence of growth factors. Biochem. J. 2004;383(Pt 1):13–18. doi: 10.1042/BJ20040984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surjit M., Liu B., Kumar P., Chow V.T., Lal S.K. The nucleocapsid protein of the SARS coronavirus is capable of self-association through a C-terminal 209 amino acid interaction domain. Biochem. Biophys. Res. Commun. 2004;317(4):1030–1036. doi: 10.1016/j.bbrc.2004.03.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijms M.A., van der Meer Y., Snijder E.J. Nuclear localization of non-structural protein 1 and nucleocapsid protein of equine arteritis virus. J. Gen. Virol. 2002;83(Pt 4):795–800. doi: 10.1099/0022-1317-83-4-795. [DOI] [PubMed] [Google Scholar]
- Timani K.A., Ye L., Zhu Y., Wu Z., Gong Z. Cloning, sequencing, expression, and purification of SARS-associated coronavirus nucleocapsid protein for serodiagnosis of SARS. J. Clin. Virol. 2004;30(4):309–312. doi: 10.1016/j.jcv.2004.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurm T., Chen H., Hodgson T., Britton P., Brooks G., Hiscox J.A. Localization to the nucleolus is a common feature of coronavirus nucleoproteins, and the protein may disrupt host cell division. J. Virol. 2001;75(19):9345–9356. doi: 10.1128/JVI.75.19.9345-9356.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H., Xiao G., Zhang J., Hu Y., Yuan F., Cole D.K., Zheng C., Gao G.F. SARS coronavirus induces apoptosis in Vero E6 cells. J. Med. Virol. 2004;73(3):323–331. doi: 10.1002/jmv.20094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying W., Hao Y., Zhang Y., Peng W., Qin E., Cai Y., Wei K., Wang J., Chang G., Sun W., Dai S., Li X., Zhu Y., Li J., Wu S., Guo L., Dai J., Wan P., Chen T., Du C., Li D., Wan J., Kuai X., Li W., Shi R., Wei H., Cao C., Yu M., Liu H., Dong F., Wang D., Zhang X., Qian X., Zhu Q., He F. Proteomic analysis on structural proteins of Severe Acute Respiratory Syndrome coronavirus. Proteomics. 2004;4(2):492–504. doi: 10.1002/pmic.200300676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakhartchouk A.N., Viswanathan S., Mahony J.B., Gauldie J., Babiuk L.A. Severe acute respiratory syndrome coronavirus nucleocapsid protein expressed by an adenovirus vector is phosphorylated and immunogenic in mice. J. Gen. Virol. 2005;86(Pt 1):211–215. doi: 10.1099/vir.0.80530-0. [DOI] [PubMed] [Google Scholar]


