Highlights
-
•
Integrin may act as an alternative receptor for SARS-CoV-2 and could be implicated in its transmission and pathology.
-
•
The spike protein of SARS-CoV-2 acquired a RGD motif known to bind integrins. This motif is absent from other coronaviruses.
-
•
The integrin-binding motif is present at the surface of the spike protein, close to the ACE2 receptor-binding region.
-
•
Integrin binding may be a promising therapeutics target, and should be tested experimentally.
Since December 2019, a novel coronavirus (nCoV) of animal origin started infecting humans, initiating a severe outbreak in China. This virus, named “Severe Acute Respiratory Syndrome-related Coronavirus 2” (SARS-CoV-2), can cause a severe and even fatal respiratory disease, called Coronavirus disease-19 (COVID-19), and lead to acute respiratory distress syndrome (ARDS). The virus is highly contagious and transmission occurs presumably via airborne droplets, and fecal-oral route (Zhang et al., 2020). SARS-CoV-2 belongs to the genus Betacoronavirus of the large family of Coronaviridae (“Betacoronavirus ~ ViralZone,” n.d.). This genus comprises mainly vertebrate respiratory viruses, including HCoV-OC43, which is responsible for 10% of common colds (McIntosh et al., 1970), and SARS, the cause of an epidemic in 2003 with over 8000 infected individuals in 30 countries (Guan et al., 2004). The SARS-CoV-2 genome has now been sequenced: its close similarity to SARS suggests that it has emerged from the same reservoir, namely bats (Zhou et al., 2020).
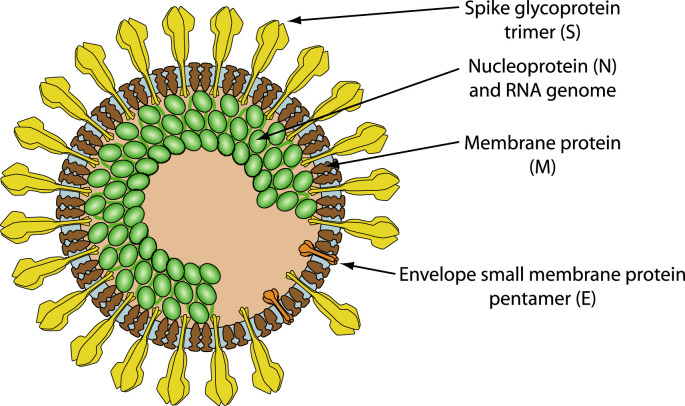
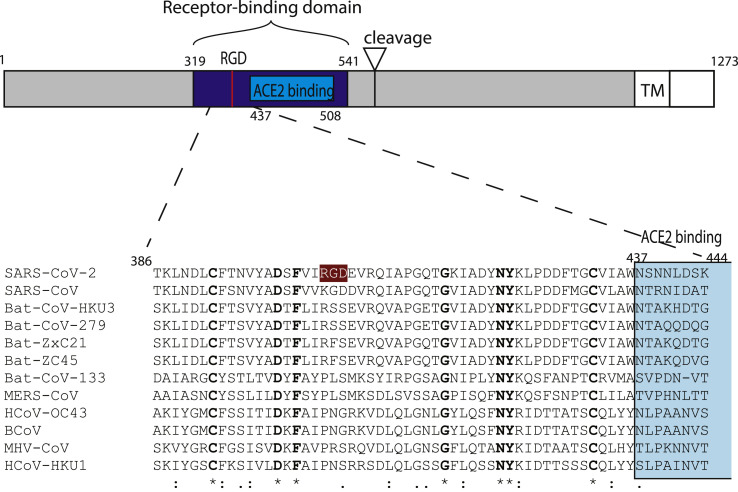
The SARS-CoV-2 spike protein (S) is the main molecule present at the surface of the virion. This large glycoprotein assembles in trimers that form a crown-like structure on the envelope which gives this virus family its name (corona = crown) (Fig. 1 ). The spike protein is a multifunctional protein that contributes to host receptor binding, cell tropism and pathogenesis. It acts by binding host receptors on target cells, inducing endocytosis of virion particle, and then catalyzes the fusion between host and viral membranes, allowing penetration of the virus genome into host cytoplasm. It is also the major target for the host immune system, adding selective pressure to this complex machinery. The angiotensin converting enzyme II (ACE2) is a known cell receptor for SARS in human and bats, and is also used by SARS-CoV-2 (Zhou et al., 2020). We suggest that SARS-CoV-2 may also use integrins as cell receptors in one or more host species, binding to them through a conserved RGD (403–405: Arg-Gly-Asp) motif that is present in the receptor-binding domain of the spike proteins of all SARS-CoV-2 sequences analyzed to date (Fig. 2 ). The motif was identified by a PROSITE scan that included motifs with a high probability of occurrence (PDOC00016) (Sigrist et al., 2013). It is absent from all other coronavirus spike proteins (n = 30) and from all SARS sequences tested (n = 155). The RGD motif is the minimal peptide sequence required for binding proteins of the integrin family, which are commonly used as receptors by many human viruses (Hussein et al., 2015). RGD motif integrin-binding is essential for human metapneumovirus (HMPV) (Chang et al., 2012; Wei et al., 2014), human Adenovirus type 2/5 (Wickham et al., 1994), human cytomegalovirus (HHV-5) (Feire et al., 2004), Kaposi's sarcoma-associated virus (HHV-8) (Hussein et al., 2015), Epstein-Barr virus (HHV-4) (Xiao et al., 2008, p. 2), Rotavirus (RV) (Zárate et al., 2004), and Coxsackievirus A9 (Williams et al., 2004). The conservation of the motif and its localization in the receptor-binding region of the SARS-CoV-2 spike protein suggests that integrins may be alternative receptors for this virus. This could broaden cell tropism and potentially affect viral pathogenicity and transmission.
Fig. 1.
Schematic representation of SARS coronavirus virion (Hulo et al., 2011), based on cryo-electron microscopy (Neuman et al., 2011).
Fig. 2.
Schematic representation of SARS-CoV-2 S-protein with a focus on the receptor-binding domain. The sequences of 12 betacoronavirus were aligned using MAFFT (Katoh et al., 2019). The receptor-binding domain and the ACE2 receptor-binding region are colored in blue and light blue, respectively. The RGD motif of SARS-CoV-2 is highlighted in color. Numbers refer to the SARS-CoV-2 spike protein sequence.
In order to bind integrin, the motif must be present at the surface of the protein. To assess the localization of the RGD motif in the spike protein, we have analyzed its 3D model from SWISS-MODEL (Waterhouse et al., 2018). This model was derived from homology modeling of the SARS-CoV-2 spike glycoprotein sequence from UniProt (P0DTC2; SPIKE_WCPV) with the SARS spike glycoprotein 3D structure (PDB 6ACD) as template (Berman et al., 2000). The RGD motif and other known binding regions were visualized with Jmol, an open-source Java viewer for chemical structures in 3D (http://www.jmol.org/).
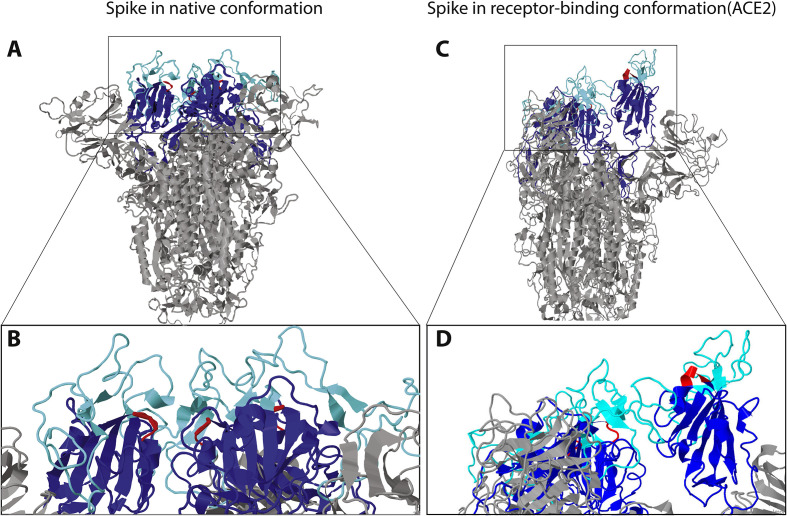
The SARS-CoV-2 spike glycoprotein RGD lies in the receptor-binding domain (amino acids 319 to 541) (Fig. 2) at the border of the subdomain (amino acids 437 to 508) that is specifically involved in the binding to human ACE2 (Li et al., 2005; Xiao et al., 2003). In the unbound state, i.e. the “down” or mushroom-like conformation, the SARS-CoV-2 spike glycoprotein RGD motif defines a small loop (restricted to the motif) between two secondary structures, a β-strand and a α-helix, near the surface of the spike protein (Fig. 3 a and b). Structures of SARS-CoV-2 spike proteins bound to ACE2 receptor motif have been recently made available (Lan et al., 2020, p. 2; Yan et al., 2020). ACE2 binding induces dramatic conformational changes involving one of the receptor-binding domains of the trimer. This adopts a protruding “up” conformation, exposing the ACE2 binding subdomain, as well as the region containing RGD (Fig. 3c and d). However, it is difficult to predict from ACE2 binding how an integrin-binding would look like. It could induce a different conformational change of the receptor-binding domain, exposing the RGD motif to integrin binding.
Fig. 3.
A) and B) Model of SARS-CoV-2 structure provided by SWISSMODEL and visualized with Jmol. A) The mushroom-like fold of the model is the classical one in absence of ligand binding. Ligand binding causes a drastic conformational change leading to the protrusion of one of the trimeric binding domains, further exposing the RGD-loop. The receptor-binding domain has been colored in blue, with a focus in light blue on the part binding the receptor ACE2. The RGD motif is colored in red. B) Enlarged view of the receptor-binding domain. The region demonstrated to bind ACE2, as well as the RGD-loops, are located at the surface of the domain even in the absence of ligand. Same colors as in A. C) and D) Model of SARS-CoV-2 structure in the conformational state of ACE2-binding provided by SWISSMODEL and visualized with Jmol. C) The receptor-binding domain of the trimer is in the “up” conformation exposing the RGD motif. Same color as in Fig. 2; ACE2 is not represented. D) Close-up view of the receptor-binding domain, highlighting the location of the RGD motif at the very surface of the domain. Same colors as in A.
Integrins are heterodimeric cell surface receptors that play roles in cell adhesion, cell migration, and cell signaling processes (Hynes, 2002). Viral proteins with RGD motifs promote infection by binding integrin heterodimers such as αVβ1, αVβ3, αVβ5, αVβ6, αVβ8, α5β1, α8β1 and αIIbβ3 (Hussein et al., 2015), activating transducing pathways involving phosphatidylinositol-3 kinase (PI–3K) or mitogen-activate protein kinase (MAPK), which promote virus entry and infection of the host cell. Adenoviruses of serotypes 2 and 5 possess such proteins, as does human metapneumovirus (Chang et al., 2012; Wei et al., 2014). This virus is the closest to SARS-CoV-2 in terms of organ tropism and symptoms, although it mainly affects children. Interestingly, the metapneumovirus protein F's RGD motif adopts a fold similar to that of SARS-CoV-2, with a small loop inserted between two secondary structures (in this case two β-sheets) (Cox et al., 2012). Given these apparent similarities, it is plausible that SARS-CoV-2 acquired integrin-binding to promote virus entry into host cells, but experimental proof of this is required. Binding to integrin may play a supplemental role to ACE2 binding, like facilitating endocytosis by signaling through the integrin. Alternatively, the virus could infect different target cells by binding to ACE2 or to integrins.
There are currently few antiviral molecules that are effective against SARS-CoV-2 (Wang et al., 2020) Agents that block integrin binding may provide a promising avenue of research. Known blockers of integrin binding include the antibody natalizumab (a α4β1/β7 integrin antagonist) for the treatment of multiple sclerosis/Crohn's disease, the small molecule tirofiban (an αIIbβ3 inhibitor) for the treatment of acute coronary syndrome, as well as inhibitors of αV RGD-binding integrin (Hatley et al., 2018). Anti-integrin pharmacotherapy is certainly possible, although no treatment is yet available that prevents virus-integrin binding, suggesting that further research is necessary to neutralize these pathogens.
Acknowledgements
This work was supported by the Swiss Federal Government through the State Secretariat for Education, Research and Innovation SERI.
References
- Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betacoronavirus ViralZone page. https://viralzone.expasy.org/764?outline=all_by_species [WWW document], n.d. accessed 2.20.20.
- Chang A., Masante C., Buchholz U.J., Dutch R.E. Human metapneumovirus (HMPV) binding and infection are mediated by interactions between the HMPV fusion protein and heparan sulfate. J. Virol. 2012;86:3230–3243. doi: 10.1128/JVI.06706-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox R.G., Livesay S.B., Johnson M., Ohi M.D., Williams J.V. The human metapneumovirus fusion protein mediates entry via an interaction with RGD-binding integrins. J. Virol. 2012;86:12148–12160. doi: 10.1128/JVI.01133-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feire A.L., Koss H., Compton T. Cellular integrins function as entry receptors for human cytomegalovirus via a highly conserved disintegrin-like domain. Proc. Natl. Acad. Sci. U.S.A. 2004;101:15470–15475. doi: 10.1073/pnas.0406821101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y., Peiris J.S.M., Zheng B., Poon L.L.M., Chan K.H., Zeng F.Y., Chan C.W.M., Chan M.N., Chen J.D., Chow K.Y.C., Hon C.C., Hui K.H., Li J., Li V.Y.Y., Wang Y., Leung S.W., Yuen K.Y., Leung F.C. Molecular epidemiology of the novel coronavirus that causes severe acute respiratory syndrome. Lancet Lond. Engl. 2004;363:99–104. doi: 10.1016/s0140-6736(03)15259-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatley R.J.D., Macdonald S.J.F., Slack R.J., Le J., Ludbrook S.B., Lukey P.T. An αv-RGD integrin inhibitor toolbox: drug discovery insight, challenges and opportunities. Angew. Chem., Int. Ed. Engl. 2018;57:3298–3321. doi: 10.1002/anie.201707948. [DOI] [PubMed] [Google Scholar]
- Hulo C., de Castro E., Masson P., Bougueleret L., Bairoch A., Xenarios I., Le Mercier P. ViralZone: a knowledge resource to understand virus diversity. Nucleic Acids Res. 2011;39:D576–D582. doi: 10.1093/nar/gkq901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussein H.A.M., Walker L.R., Abdel-Raouf U.M., Desouky S.A., Montasser A.K.M., Akula S.M. Beyond RGD: virus interactions with integrins. Arch. Virol. 2015;160:2669–2681. doi: 10.1007/s00705-015-2579-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes R.O. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- Katoh K., Rozewicki J., Yamada K.D. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Briefings Bioinf. 2019;20:1160–1166. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Crystal structure of the 2019-nCoV spike receptor-binding domain bound with the ACE2 receptor. 2020. bioRxiv 2020.02.19.956235. [DOI] [PubMed]
- Li F., Li W., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- McIntosh K., Kapikian A.Z., Turner H.C., Hartley J.W., Parrott R.H., Chanock R.M. Seroepidemiologic studies of coronavirus infection in adults and children. Am. J. Epidemiol. 1970;91:585–592. doi: 10.1093/oxfordjournals.aje.a121171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman B.W., Kiss G., Kunding A.H., Bhella D., Baksh M.F., Connelly S., Droese B., Klaus J.P., Makino S., Sawicki S.G., Siddell S.G., Stamou D.G., Wilson I.A., Kuhn P., Buchmeier M.J. A structural analysis of M protein in coronavirus assembly and morphology. J. Struct. Biol. 2011;174:11–22. doi: 10.1016/j.jsb.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist C.J.A., de Castro E., Cerutti L., Cuche B.A., Hulo N., Bridge A., Bougueleret L., Xenarios I. New and continuing developments at PROSITE. Nucleic Acids Res. 2013;41:D344–D347. doi: 10.1093/nar/gks1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020 doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F., T., de Beer T., AP, Rempfer C., Bordoli L., Lepore R., Schwede T. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46(W1):296–303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y., Zhang Y., Cai H., Mirza A.M., Iorio R.M., Peeples M.E., Niewiesk S., Li J. Roles of the putative integrin-binding motif of the human metapneumovirus fusion (f) protein in cell-cell fusion, viral infectivity, and pathogenesis. J. Virol. 2014;88:4338–4352. doi: 10.1128/JVI.03491-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham T.J., Filardo E.J., Cheresh D.A., Nemerow G.R. Integrin alpha v beta 5 selectively promotes adenovirus mediated cell membrane permeabilization. J. Cell Biol. 1994;127:257–264. doi: 10.1083/jcb.127.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams C.H., Kajander T., Hyypiä T., Jackson T., Sheppard D., Stanway G. Integrin alpha v beta 6 is an RGD-dependent receptor for coxsackievirus A9. J. Virol. 2004;78:6967–6973. doi: 10.1128/JVI.78.13.6967-6973.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J., Palefsky J.M., Herrera R., Berline J., Tugizov S.M. The Epstein-Barr virus BMRF-2 protein facilitates virus attachment to oral epithelial cells. Virology. 2008;370:430–442. doi: 10.1016/j.virol.2007.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X., Chakraborti S., Dimitrov A.S., Gramatikoff K., Dimitrov D.S. The SARS-CoV S glycoprotein: expression and functional characterization. Biochem. Biophys. Res. Commun. 2003;312:1159–1164. doi: 10.1016/j.bbrc.2003.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R., Zhang Y., Guo Y., Xia L., Zhou Q. Structural basis for the recognition of the 2019-nCoV by human ACE2. 2020. bioRxiv 2020.02.19.956946. [DOI] [PMC free article] [PubMed]
- Zárate S., Romero P., Espinosa R., Arias C.F., López S. VP7 mediates the interaction of rotaviruses with integrin alphavbeta3 through a novel integrin-binding site. J. Virol. 2004;78:10839–10847. doi: 10.1128/JVI.78.20.10839-10847.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Du R.-H., Li B., Zheng X.-S., Yang X.-L., Hu B., Wang Y.-Y., Xiao G.-F., Yan B., Shi Z.-L., Zhou P. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg. Microb. Infect. 2020;9:386–389. doi: 10.1080/22221751.2020.1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020 doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]