Highlights
-
•
Virus-like particle mimetic nanovesicles were prepared using insect cells.
-
•
Surfactant treatment or mechanical extrusion make it possible to display the S protein on VLPs.
-
•
Mechanical extrusion is more effective way to display the S protein on VLPs.
-
•
S protein on the surface of nanovesicles were confirmed by immuno-TEM.
-
•
The purified S protein (SοTM) has the ability to bind a receptor of MERS-CoV.
Keywords: Virus-like particle, Middle East respiratory syndrome, Coronavirus, Silkworm, Vaccine, Bm5 cell
Abstract
Middle East respiratory syndrome coronavirus (MERS-CoV) first emerged in 2012, and over 2000 infections and 800 deaths have been confirmed in 27 countries. However, to date, no commercial vaccine is available. In this study, structural proteins of MERS-CoV were expressed in silkworm larvae and Bm5 cells for the development of vaccine candidates against MERS-CoV and diagnostic methods. The spike (S) protein of MERS-CoV lacking its transmembrane and cytoplasmic domains (SΔTM) was secreted into the hemolymph of silkworm larvae using a bombyxin signal peptide and purified using affinity chromatography. The purified SΔTM forms small nanoparticles as well as the full-length S protein and has the ability to bind human dipeptidyl peptidase 4 (DPP4), which is a receptor of MERS-CoV. These results indicate that bioactive SΔTM was expressed in silkworm larvae. To produce MERS-CoV-like particles (MERS-CoV-LPs), the coexpression of spike proteins was performed in Bm5 cells and envelope (E) and membrane (M) proteins secreted E and M proteins extracellularly, suggesting that MERS-CoV-LPs may be formed. However, this S protein was not displayed on virus-like particles (VLPs) even though E and M proteins were secreted into the culture supernatant. By surfactant treatment and mechanical extrusion using S protein- or three structural protein-expressing Bm5 cells, S protein-displaying nanovesicles with diameters of approximately 100-200 nm were prepared and confirmed by immuno-TEM. The mechanical extrusion method is favorable for obtaining uniform recombinant protein-displaying nanovesicles from cultured cells. The purified SΔTM from silkworm larvae and S protein-displaying nanovesicles from Bm5 cells may lead to the development of nanoparticle-based vaccines against MERS-CoV and the diagnostic detection of MERS-CoV.
1. Introduction
Middle East respiratory syndrome (MERS), which first emerged in Saudi Arabia in 2012, is caused by MERS coronavirus (MERS-CoV). Over 2000 infections have been confirmed in 27 countries, and 800 deaths have occurred (WHO, 2019). MERS-CoV is a single positive-stranded RNA virus and belongs to the group C species of beta-coronavirus (Chan et al., 2012). Bats are the natural reservoir of MERS-CoV, and camels are its intermediate host (Memish et al., 2013; Wang et al., 2014). MERS-CoV may spread to humans through camels and may be transmitted from human to human (Health Protection Agency (HPA) and UK Novel Coronavirus Investigation team, 2013).
MERS-CoV contains four structural proteins, the spike (S), envelope, (E), membrane (M) and nucleocapsid (N) proteins. The S protein, which is a class I fusion protein, is responsible for viral entry into target cells through receptor binding. Dipeptidyl peptidase 4 (DPP4, CD26) is known to be its receptor. During viral infection, S protein is processed into S1 and S2 subunits. The S1 subunit contains the receptor binding domain, and the S2 subunit is required for the membrane fusion of MERS-CoV. Therefore, the S protein is one of the targets for the development of MERS-CoV therapeutics (Du et al., 2017). Some neutralizing antibodies have been developed against the receptor-binding domain of S protein, which blocks the interaction of S protein with DDP4, to inhibit the infection of cells by MERS-CoV (Corti et al., 2015; Jiang et al., 2014). In addition, the S protein and its receptor-binding domain are regarded as promising targets for the development of vaccines against MERS-CoV, even though no vaccine against MERS-CoV is yet commercially available (Ma et al., 2014a, b).
The M protein of severe acute respiratory syndrome (SARS) coronavirus (SARS-CoV) forms enveloped virus-like particles (VLPs) when it is coexpressed with the E or N protein in insect cells and mammalian cells (Mortola and Roy, 2004; Siu et al., 2008). Likewise, VLPs of MERS-CoV (MERS-CoV-LPs) were produced in insect cells by the coexpression of the S, E and M proteins (Wang et al., 2017). The immunization Rhesus macaques with S protein-displaying MERS-CoV-LPs induced the production of receptor-binding domain-specific antibodies and virus-neutralizing antibodies, leading to Th1-mediated immunity. This result indicates that S protein-displaying VLPs are a promising tool for vaccination against MERS-CoV.
VLPs, which are produced in various expression systems, have been widely developed as vaccines and carriers in drug and gene delivery system (Charlton Hume et al., 2019; Rohovie et al., 2017). In addition to enveloped VLPs, to generate nanovesicles composed of envelope and some functional proteins, the methods to disrupt cells by a surfactant and extrusion through membranes have been also developed (Guo et al., 2018; Mi et al., 2016). Nanovesicles by surfactant treatment or by mechanical extrusion provide the platforms for the vaccination to infectious diseases and drug delivery as well as VLPs and exosomes. Using these methods, a recombinant protein-displaying nanovesicles can be prepared from the cell cultures.
Insect larva and insect cells have been used widely for the production of recombinant proteins including VLPs (Minkner and Park, 2018). Especially, silkworm larvae and pupae are regarded as a favorable host for a large-scale production of recombinant proteins because of its ease to handle, its cost-effectiveness and the capacity of producing proteins (Fuenmayor et al., 2017). These are advantageous to develop the vaccines against infectious diseases.
In this study, the S protein of MERS-CoV was expressed in silkworm larvae as a secretory protein and purified from the hemolymph. In addition, we explored the generation of MERS-CoV-LPs by the coexpression of the S, E and M proteins in silkworms and Bm5 cells and of nanovesicles displaying the S protein by subjecting S protein-expressing cells to surfactant treatment or mechanical extrusion.
2. Materials and methods
2.1. Cell cultivation and silkworms
Bm5 cells were maintained at 27 °C in Sf-900II (Thermo Fisher Scientific K. K., Tokyo, Japan) supplemented with 10% fetal bovine serum (FBS, Thermo Fisher Scientific K. K.) and Antimycotic-Antibiotic (Thermo Fisher Scientific K. K.). In addition, Bm5 cells were also cultivated in non-FBS Sf900II medium. Fourth-instar silkworm larvae were purchased from Ehime Sansyu (Ehime, Japan). Silkworm larvae were reared on an artificial diet, Silkmate 2S (Nosan, Yokohama, Japan) at 25 °C.
2.2. Construction of recombinant baculoviruses
Genes encoding the E and M proteins (GenBank: KF961222.1) were synthesized by Genewiz Japan (Saitama, Japan). A gene encoding the S protein was purchased from Sino Biological (Beijing, China) as a vector (https://www.sinobiological.com/MERS-CoV-NCoV-Novel-coronavirus-Spike-Protein-Codon-Optimized-ORF-mammalian-expression-plasmid-N-Flag-tag-p212233.html). To connect the tag sequences, genes encoding the E and M proteins were amplified using the E and M primer sets (Table 1 ), respectively. The genes encoding the E and M proteins had FLAG tag and PA tag sequences at the 3′-end. The gene encoding the S protein was amplified by PCR using Bx-myc-S-F and the S-R primer set (Table 1). The S protein gene had sequences encoding the bombyxin signal peptide and c-Myc tag instead of that encoding its native signal peptide. To express SΔTM, which does not have its native transmembrane and cytoplasmic domains, the gene encoding SΔTM was amplified by PCR using the bx-myc-S-F and SΔTM-R primer set (Table 1). The gene encoding SΔTM also had the sequence encoding the bombyxin signal peptide and c-Myc tag instead of that encoding its native signal peptide. The gene encoding the E protein was amplified by PCR using the E-F and E-FLAG-R primer set (Table 1). The gene encoding the M protein was amplified by PCR using the M-F and M-PA-R primer set (Table 1). Each amplified gene was inserted into the pFastbac1 vector (Thermo Fisher Scientific K. K.). Each resulting recombinant plasmid was transformed into Escherichia coli BmDH10Bac bacmid (Motohashi et al., 2005), and white colonies were selected. A recombinant BmNPV bacmid (BmNPV/S or BmNPV/SΔTM) containing each gene was extracted from a white colony, and the insertion of each gene into the BmNPV bacmid was checked by PCR using the M13-F and M13-R primer set (Table 1). Each recombinant BmNPV was prepared by the transfection of each constructed BmNPV bacmid into Bm5 cells. For transfection, several micrograms of recombinant BmNPV bacmid was transfected into Bm5 cells with Jet PEI reagent (Polyplus Transfection, New York, NY, USA). After several days, the culture supernatant was collected, followed by titer-up. To express recombinant proteins in Bm5 cells, Bm5 cells were infected with recombinant BmNPVs at an M.O.I. of 1. The titers of recombinant BmNPVs were determined by the protocol described previously (Kato et al., 2009).
Table 1.
Primers used.
| Name | 5′ 3′ |
|---|---|
| Bx-myc-S-F | CGCGAATCCATGAAGATACTCCTTGCTATTGCATTAATGTTGTCAACAGTAATGTGGGTGTCAACAGAACAAAAACTCATCTCAGAAGAGGATCTGTATGTGGATGTGGGACCTGAC |
| S-R | GCGGAATTCTTAATGTACGTGCACCTTGTGG |
| SΔTM-R | GCGGAATTCCCACTTGTTGTAGTAGGTGTAGTTGC |
| E-F | GCGAATTCATGTTACCCTTTGTCCAAGA |
| E-FLAG-R | GCAAGCTTTTACTTGTCATCGTCATCCTTGTAGTCAACCCACTCGTCAGGTGGTA |
| M-F | GCGAATTCATGTCTAATATGACGCAACTCACTG |
| M-R | GCAAGCTTCTACACCACATCATCTTCGGCACCTGGCATGGCAACGCCAGCTCGAAGCAATGCAA |
| M13-F | GTTTTCCCAGTCACGAC |
| M13-R | CAGGAAACAGCTATGAC |
| α-1-F | AACGCTCTATGGTCTAAAGATTTACTCCGGAATATTAATAG |
| β-1-R | AAACGTGCAATAGTATCCAGTTTTAGATTTCACTTATCTGG |
| β-2-F | AAACTGGATACTATTGCACGTTTACTCCGGAATATTAATAG |
| γ-2-R | AAACATCAGGCATCATTAGGTTTTAGATTTCACTTATCTGG |
| γ-3-F | AAACCTAATGATGCCTGATGTTTACTCCGGAATATTAATAG |
| ω-3-R | AAACTAAGCTATGTGAACCGTTTTAGATTTCACTTATCTGG |
| ω-pFB-F | AAACACTGACATTGACTTGGTTTCCCGGTCCGAAGCGCGCG |
| α-pFB-R | AAATCTTTAGACCATAGAGCGTTCTATTAATATTCCGGAGT |
To coexpress S, E and M proteins, a recombinant BmNPV/S/E/M bacmid containing these gene expression cassettes was constructed. The gene expression cassettes were amplified by PCR using the α-β primer set (α-1-F and β-1-R, Table 1), β-γ primer set (β-2-F and γ-2-R, Table 1) and γ-ω primer set (γ-3-F and ω-3-R, Table 1), respectively (Weissmann et al., 2016). pFastbac 1, in which the polyhedrin promoter and the multicloning site were deleted, was amplified by PCR using the ω-pFB-F and α-pFB-R primer set (Table 1). These 4 PCR fragments were assembled simultaneously by the Gibson assembly method (Gibson, 2011). Using the constructed vector containing 3 gene expression cassettes, a recombinant BmNPV/S/E/M bacmid was constructed.
2.3. Preparation of protein extracts and purification of SΔTM from hemolymph
The culture supernatant was separated from Bm5 cells by centrifugation, and the collected Bm5 cells were suspended in phosphate-buffered saline (PBS, pH 7.4). Each sample was mixed with 2 × sample buffer containing 2-mercaptoethanol (Nacalai Tesque, Kyoto, Japan), followed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
A recombinant BmNPV solution was injected into silkworm larvae to express each structural protein. To purify SΔTM from the hemolymph, the collected hemolymph was centrifuged to remove hemocytes and insoluble materials, and the supernatant was dialyzed with PBS overnight. After dialysis, SΔTM was purified by anti c-Myc antibody beads (10D11) (FUJIFILM Wako pure chemical). Elution was performed with 0.1-M glycine-HCl (pH 3.5), and the elution fractions were immediately neutralized with 1.5-M Tris-HCl (pH 7.4).
The protein concentration in each sample was determined using a Pierce BCA Protein Assay Kit (Thermo Fisher Scientific).
2.4. SDS-PAGE and western blot
Proteins were separated by SDS-PAGE using 10% or 12% polyacrylamide gel. The gels were then subjected to western blotting. Proteins were transferred onto a polyvinylidene fluoride (PVDF) membrane using the Mini Trans-Blot Electrophoretic Transfer Cell (Bio-Rad, Hercules, CA, USA). The blocking step was carried out in 5% skim milk in Tris-buffered saline containing 0.1% Tween 20 (TBST, pH 7.6), followed by the incubation of the membrane with each primary antibody, namely, mouse anti c-Myc monoclonal antibody (FUJIFILM Wako Pure Chemical, Osaka, Japan), anti-DDDDK-tag monoclonal antibody (Medical & Biological Laboratories, Nagoya, Japan) or rat anti-PA tag monoclonal antibody (FUJIFILM Wako Pure Chemical). Each primary antibody was diluted 1,000-fold before use. After washing with TBS-T, the membrane was incubated with each secondary antibody, namely, 10,000-fold-diluted sheep HRP-linked IgG (GE Healthcare Japan, Tokyo, Japan) or goat anti-rat IgG-HRP (Santa Cruz Biotechnology, Dallas, USA). Detection based on the HRP reaction was carried out using Immobilon Western Chemiluminescent HRP Substrate (Merck Millipore Japan, Tokyo, Japan). Protein bands were detected on a Fluor-S MAX MultiImager (Bio-Rad).
2.5. Enzyme-linked immunosorbent assay (ELISA)
To investigate the binding of the S protein to DPP4, ELISA was carried out. First, 100-ng of human DPP4 (Sino Biological, Beijing, China), human angiotensin-converting enzyme 2 (ACE2, Sino Biological) and bovine serum albumin (BSA) were placed on wells in a 96-well plate. The supernatant was removed from the wells, and 2% skimmed milk in PBS containing 0.1% Tween 20 (PBST) was added to each well, followed by incubation of the plate for 1-h. The blocking solution was removed, and purified SΔTM was put into each well. After incubation at room temperature for 1-h, the wells were washed with PBST 3 times. The monoclonal antibody anti c-Myc (FUJIFILM Wako Pure Chemical) diluted 1,000-fold was added to each well and incubated at room temperature for 1-h, followed by washing each well 3 times with PBST. Anti-mouse IgG antibody-HRP (GE Healthcare Japan) diluted 5,000-fold with PBST was added to each well and incubated at room temperature for 1-h. Each well was washed with TBST followed by the HRP reaction. One hundred microliters of substrate (0.1-mg/ml 3,3′,5,5′-tetramethylbenzidine in 100-mM sodium acetate, pH 6.0, with 0.2% (v/v) of 30% hydrogen peroxide) was reacted in each well, and the plate was incubated for development of the blue coloration at room temperature. The reaction was stopped by the addition of 50 μl of 1 N H2SO4 solution, followed by measurement of the absorbance at 450 nm.
2.6. Preparation of nanovesicles using surfactant treatment or mechanical extrusion
Bm5 cells were infected with a recombinant BmNPV/S, BmNPV/SΔTM containing the S protein gene expression cassette and BmNPV/S/T/M containing the S, E and M protein gene expression cassette at an M.O.I. of 1.0 and cultivated for 4 d. The preparation of nanovesicles by surfactant treatment (SNVs) was performed according to the protocol reported by Zhang et al. (2015). Briefly, 7 × 106 cells were suspended in 700 μl of PBS containing 0.015% (w/v) sodium deoxycholate and cOmplete Mini EDTA-free (Roche Diagnostics, Tokyo Japan) and stirred vigorously. The homogenate was centrifuged at 4000 × g to remove the cell debris and organelles. The supernatant was filtered by a 0.45 μm filter and applied to sucrose density gradient centrifugation (20-60%). The S protein-rich fractions were collected and dialyzed with PBS. Finally, Triton X-100 and sodium deoxycholate were added into the solution to 0.045% (w/v) and 0.05% (w/v), respectively. The preparation of nanovesicles by mechanical extrusion (ENVs) was performed according to the protocol reported by Jang et al. (2013). Briefly, 5 × 106 cells were suspended in PBS and extruded 10 times through a 5 μm polycarbonate track-etched membrane disk (GVS Japan K. K., Tokyo Japan) using a mini-extruder (Avanti Polar Lipids, Alabaster, AL, USA). The filtrate was then subjected to sucrose density gradient centrifugation (20-60%), and the S protein-rich fractions were collected and dialyzed with PBS.
2.7. Transmission electron microscopy
Proteins or nanovesicles were put onto the surface of a film 200 mesh copper grid (Nisshin EM, Tokyo, Japan) and incubated at room temperature for 10 min. The grid was washed 3 times with PBS, and the blocking step was carried out using 1% BSA for 5 min. After the grid was washed with PBS, 100-fold diluted mouse anti c-Myc monoclonal antibody (FUJIFILM Wako pure Chemical) was loaded onto the grid, and the grid was incubated at room temperature for 1 h, then washed with PBS 3 times. The grid was then treated with 100-fold diluted goat anti-mouse IgG+IgM (H+L) polyclonal antibody conjugated with 10 nm gold (BBI, Solutions, Crumlin, UK) for 1 h. Finally, the grid was washed 6 times with PBS, followed by negative staining with phosphotungstic acid (2% v/v). Images were acquired with a transmission electron microscope (TEM, JEM-2100F, JEOL, Tokyo, Japan) operated at 100 kV.
3. Results
3.1. Expression of SΔTM in silkworm larvae and its purification from the hemolymph
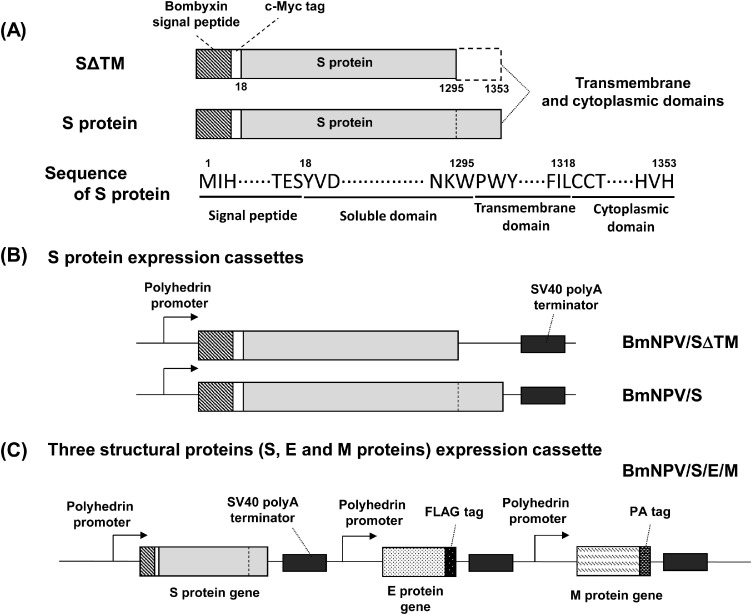
Silkworm larvae have been used for the production of recombinant proteins instead of cultured cells because they can easily express recombinant proteins on a large scale (Kato et al., 2010; Usami et al., 2010). SΔTM with its native transmembrane and cytoplasmic domains removed from its C-terminus (Fig. 1 A) was expressed by BmNPV/SΔTM in silkworm larvae (Fig. 1B). The S protein of MERS-CoV is a class I fusion protein, and therefore, the truncation of its C-terminal domains leads to the secretion of SΔTM into the hemolymph in silkworm larvae. In addition, recombinant BmNPV/S/E/M for the expression of three structural proteins (full-length S, E and M proteins) was also constructed (Fig. 1C).
Fig. 1.
Constructs of structural proteins of MERS-CoV in this study. (A) S proteins constructs expressed in this study. Numbers indicate the amino acid residues in the S protein. (B) S protein expression cassettes in BmNPV/SΔTM and BmNPV/S bacmids. This recombinant BmNPV allows the expression of both S proteins. (C) The expression cassette of three structural proteins (S, E and M proteins) in a single recombinant BmNPV/S/E/M bacmid. This recombinant BmNPV allows the coexpression of S protein with its native transmembrane and cytoplasmic domains with E and M proteins.
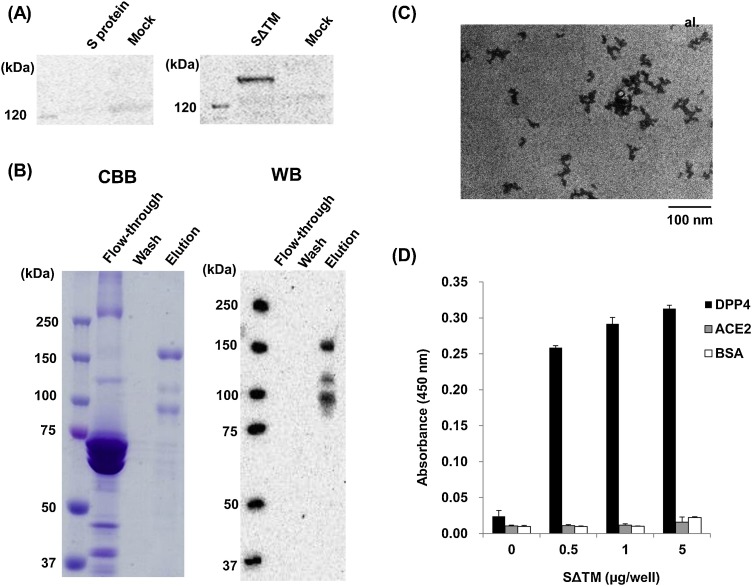
When the S and SΔTM proteins were expressed in silkworm, it was not observed in the hemolymph, but only SΔTM was observed (Fig. 2 A). This result indicates that SΔTM was secreted into the hemolymph because of the removal of its C-terminal domains. SΔTM was purified from the hemolymph using affinity chromatography. Three main bands were observed in the elution fraction by CBB staining and by western blotting (Fig. 2B). These results indicate that these three bands came from expressed SΔTM. The estimated molecular weight of expressed SΔTM is approximately 141.9 kDa, and SΔTM has 25 putative N-glycosylation sites. However, how many N-glycosylation sites in the S protein are N-glycosylated is unknown. These results suggest that one band over 150 kDa may be intact SοTM and that two bands below 150 kDa may be processed SΔTM. From a larva, 8.3 μg of SΔTM was purified. Small particles were observed in the elution fraction by TEM in Fig. 2C, indicating that SΔTM can form such small particles even in the absence of its C-terminal domains. Using Sf-9 cells Coleman et al. (2014) also reported the formation of the particles consisting of the expressed S proteins. The purified SΔTM showed specific binding to human DPP4 (Fig. 2D) by ELISA, not to human angiotensin converting enzyme 2 (ACE2) which is a receptor for S protein of Severe Acute Respiratory Syndrome coronavirus (SARS-CoV). These results indicate that SΔTM expressed in silkworm larvae has biological activity.
Fig. 2.
Expression and purification of SΔTM in silkworm larvae. (A) Expression of S protein and SΔTM in silkworm larvae. Each expressed proteins in hemolymph were detected by western blot using a mouse anti-c-Myc monoclonal antibody. (B) Purification of SΔTM from hemolymph using affinity chromatography. Purified SΔTM was detected by CBB staining and western blotting. (C) TEM image of purified SΔTM. Proteins were stained with phosphotungstic acid as a negative stain. (D) ELISA to analyze the binding of purified SΔTM to its receptor. Human DPP4, human ACE2 and BSA were immobilized onto wells in a 96-well plate. ELISA was performed according to the protocol described in the Materials and Methods.
3.2. Preparation of S protein-displaying MERS-CoV-LPs
To prepare S protein-displaying MERS-CoV-LPs, full-length S, M and E proteins were coexpressed in Bm5 cells using a single recombinant BmNPV/S/E/M (Fig. 1C). Full-length S protein was not secreted into the culture supernatant at 4 d postinfection when the S protein was expressed in Bm5 cells (Fig. 3 A). In the case of the coexpression of the S, M and E proteins, each protein was observed in the cell lysates at 3 and 4 d postinfection (Fig. 3B). However, only the M and E proteins were observed in the culture supernatant on the same days. The cell viability was almost 100% from 1 to 4 d postinfection. The S protein was not detected, although the cell supernatant of the coexpressing Bm5 cells was concentrated by ultracentrifugation (data not shown). In addition, when the three proteins were coexpressed in silkworm larvae, S protein-displaying MERS-CoV-LPs were not generated (data not shown). These results indicate that the M and E proteins may form MERS-CoV-LPs secreted into the cell culture supernatant, but the S protein was not contained in the MERS-CoV-LPs.
Fig. 3.
Expression of the S protein and coexpression of the S, E and M proteins in Bm5 cells using a single recombinant BmNPV. (A) Expression of the S protein in Bm5 cells. (B) Coexpression of the S, E and M proteins in Bm5 cells. Bm5 cells were infected with each recombinant BmNPV at M.O.I. 1 and cultivated for 4 d. At 4 d after infection, the cell viability was almost 100%.
3.3. Preparation of S protein-displaying nanovesicles by surfactant treatment or mechanical extrusion
S protein-displaying nanovesicles were prepared by surfactant treatment or by mechanical extrusion because S, E and M protein-displaying MERS-CoV-LPs could not be generated in Bm5 cells. The S protein- or S, E and M protein-expressing Bm5 cells were collected 4 d after recombinant BmNPV infection. SNVs and ENVs were partially purified by sucrose density gradient centrifugation (Fig. 4 ). The S protein in SNVs prepared from S protein-expressing Bm5 cells was observed in fractions 3 to 6, while the S protein in ENVs was observed in fractions 5 to 9 (Fig. 4A). The S, E and M proteins in SNVs were observed in fractions 3 to 6, while those in ENVs were observed in fractions 5 to 8 (Fig. 4B).
Fig. 4.
Sucrose density gradient centrifugation of SNVs and ENVs using Bm5 cells expressing the S protein (A) and Bm5 cells expressing the three structural proteins (S, E and M proteins) (B). SNVs and EMVs were prepared by surfactant treatment and mechanical extrusion, respectively, according to the protocol described in Materials and Methods. Each protein was detected by western blot.
The morphology of SNVs and ENVs was investigated by immuno-TEM (Fig. 5 ). In SNVs and ENVs, gold nanoparticles were observed on the surfaces of nanovesicles. The SNVs were broken and shapeless (Fig. 5A and B), while the ENVs were clear and round (Fig. 5C and D). SNVs had a broad peak distribution (Fig. 5E), but ENVs showed a sharp peak at approximately 140 nm (Fig. 5F). This result indicates that ENVs were more uniform than SNVs and that the mechanical extrusion method is favorable for the preparation of nanovesicles displaying the S protein from insect cells. In addition, the coexpression of the M and E proteins with the S protein did not have any influence on the morphology and size of SNVs and ENVs.
Fig. 5.
Immuno-TEM analysis of nanovesicles. (A) SNVs from S protein-expressing Bm5 cells, (B) SNVs from three structural protein-expressing Bm5 cells, (C) ENVs from S protein-expressing Bm5 cells, (D) ENVs from three structural proteins-expressing Bm5 cells. Immuno-TEM was performed by the protocol described in Materials and Methods. DLS analysis of SNVs (E) and ENVs (F). Green and blue lines show nanovesicles prepared from S protein-expressing and three structural protein-expressing Bm5 cells, respectively. Red lines indicate Bm5 cells (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).
4. Discussion
In this study, S protein-displaying MERS-CoV-LPs composed of the S, E and M proteins were not generated in Bm5 cells and silkworm larvae. The S protein was expressed by the fusion of the bombyxin signal peptide from B. mori instead of its native peptide (Fig. 1). Therefore, we tried to express full-length S protein with its native signal peptide for the preparation of S protein-displaying MERS-CoV-LPs but failed. In mammalian cells, the endoplasmic reticulum retrieval signal (ERRS) of the S protein of coronaviruses, which occurs in the cytoplasmic domain at the C-terminus of the protein, is required for the accumulation of S protein in the postmedial Golgi compartment and its return to the ER-Golgi intermediate compartment (ERGIC), followed by the budding of coronaviruses and VLPs (McBride et al., 2007; Ujike et al., 2016). We also coexpressed the S protein fused with the signal peptide and the transmembrane and cytoplasmic domains of ERGIC-53 from B. mori, which is a non-glycosylated type I membrane protein of 53 kDa resided in ERGIC, instead of its native domains with the E and M proteins in Bm5 cells and silkworm larvae. However, S protein-displaying MERS-CoV-LPs were not produced. On the other hands, it was previously reported by Wang et al. that the successful production of S protein-displaying MERS-CoV-LPs composed of the S, E and M proteins in Sf-9 cells (Wang et al., 2017). It is unknown why Bm5 cells and silkworms cannot produce S protein-displaying MERS-CoV-LPs by the coexpression of S, E and M proteins. The localization of S protein in Bm5 cells may clarify why S protein was not displayed on particles composing of M and E proteins.
To prepare S protein-displaying nanovesicles, we adopted two methods, surfactant treatment (Zhang et al., 2015) and mechanical extrusion using an extruder (Gangadaran et al., 2018; Jang et al., 2013). These methods allow the easy and efficient production of virus-like nanovesicles and exosome-mimic vesicles from mammalian cells for vaccine development and drug delivery systems. In this study, we successfully produced three recombinant proteins, S, E and M, of MERS-CoV-displaying SNVs and ENVs by surfactant treatment or mechanical extrusion, respectively, which were VLP mimetic nanovesicles. For the application of these VLP mimetic nanovesicles composed of multiple structural proteins of viruses, their properties, including stability, morphology and functionality, should be investigated in detail.
Funding
This project was financially supported by JSPS KAKENHI Grant-in-Aid for Scientific Research (A) (Grant No. 16H02544).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Chan J.F., Li K.S., To K.K., Cheng V.C., Chen H., Yuen K.Y. Is the discovery of the novel human betacoronavirus 2c EMC/2012 (HCoV-EMC) the beginning of another SARS-like pandemic? J. Infect. 2012;65:477–489. doi: 10.1016/j.jinf.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlton Hume H.K., Vidigal J., Carrondo M.J.T., Middelberg A.P.J., Roldão A., Lua L.H.L. Synthetic biology for bioengineering virus-like particle vaccines. Biotechnol. Bioeng. 2019;116:919–935. doi: 10.1002/bit.26890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman C.M., Liu Y.V., Mu H., Taylor J.K., Massare M., Flyer D.C., Smith G.E., Frieman M.B. Purified coronavirus spike protein nanoparticles induce coronavirus neutralizing antibodies in mice. Vaccine. 2014;32:3169–3174. doi: 10.1016/j.vaccine.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti D., Zhao J., Pedotti M., Simonelli L., Agnihothram S., Fett C., Fernandez-Rodriguez B., Foglierini M., Agatic G., Vanzetta F., Gopal R., Langrish C.J., Barrett N.A., Sallusto F., Baric R.S., Varani L., Zambon M., Perlman S., Lanzavecchia A. Prophylactic and postexposure efficacy of a potent human monoclonal antibody against MERS coronavirus. Proc. Natl. Acad. Sci. U. S. A. 2015;112:10473–10478. doi: 10.1073/pnas.1510199112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., Yang Y., Zhou Y., Lu L., Li F., Jiang S. MERS-CoV spike protein: a key target for antivirals. Expert Opin. Ther. Targets. 2017;21:131–143. doi: 10.1080/14728222.2017.1271415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuenmayor J., Gòdia F., Cervera L. Production of virus-like particles for vaccines. N. Biotechnol. 2017;39:174–180. doi: 10.1016/j.nbt.2017.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangadaran P., Hong C.M., Oh J.M., Rajendran R.L., Kalimuthu S., Son S.H., Gopal A., Zhu L., Baek S.H., Jeong S.Y., Lee S.W., Lee J., Ahn B.C. In vivo non-invasive imaging of radio-labeled exosome-mimetics derived from red blood cells in mice. Front. Pharmacol. 2018;9:817. doi: 10.3389/fphar.2018.00817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson D.G. Enzymatic assembly of overlapping DNA fragments. Methods Enzymol. 2011;498:349–361. doi: 10.1016/B978-0-12-385120-8.00015-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo P., Huang J., Zhao Y., Martin C.R., Zare R.N., Moses M.A. Nanomaterial preparation by extrusion through nanoporous membranes. Small. 2018;14 doi: 10.1002/smll.201703493. [DOI] [PubMed] [Google Scholar]
- Health Protection Agency (HPA), UK Novel Coronavirus Investigation team Evidence of person-to-person transmission within a family cluster of novel coronavirus infections, United Kingdom, February 2013. Euro Surveill. 2013;18:20427. doi: 10.2807/ese.18.11.20427-en. [DOI] [PubMed] [Google Scholar]
- Jiang L., Wang N., Zuo T., Shi X., Poon K.M., Wu Y., Gao F., Li D., Wang R., Guo J., Fu L., Yuen K.Y., Zheng B.J., Wang X., Zhang L. Potent neutralization of MERS-CoV by human neutralizing monoclonal antibodies to the viral spike glycoprotein. Sci. Transl. Med. 2014;6 doi: 10.1126/scitranslmed.3008140. [DOI] [PubMed] [Google Scholar]
- Jang S.C., Kim O.Y., Yoon C.M., Choi D.S., Roh T.Y., Park J., Nilsson J., Lötvall J., Kim Y.K., Gho Y.S. Bioinspired exosome-mimetic nanovesicles for targeted delivery of chemotherapeutics to malignant tumors. ACS Nano. 2013;7:7698–7710. doi: 10.1021/nn402232g. [DOI] [PubMed] [Google Scholar]
- Kato T., Manoha S.L., Tanaka S., Park E.Y. High-titer preparation of Bombyx mori nucleopolyhedrovirus (BmNPV) displaying recombinant protein in silkworm larvae by size exclusion chromatography and its characterization. BMC Biotechnol. 2009;9:55. doi: 10.1186/1472-6750-9-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T., Kajikawa M., Maenaka K., Park E.Y. Silkworm expression system as a platform technology in life science. Appl. Microbiol. Biotechnol. 2010;85:459–470. doi: 10.1007/s00253-009-2267-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C., Li Y., Wang L., Zhao G., Tao X., Tseng C.T., Zhou Y., Du L., Jiang S. Intranasal vaccination with recombinant receptor-binding domain of MERS-CoV spike protein induces much stronger local mucosal immune responses than subcutaneous immunization: implication for designing novel mucosal MERS vaccines. Vaccine. 2014;32:2100–2108. doi: 10.1016/j.vaccine.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C., Wang L., Tao X., Zhang N., Yang Y., Tseng C.K., Li F., Zhou Y., Jiang S., Du L. Searching for an ideal vaccine candidate among different MERS coronavirus receptor-binding fragments-the importance of immunofocusing in subunit vaccine design. Vaccine. 2014;32:6170–6176. doi: 10.1016/j.vaccine.2014.08.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride C.E., Li J., Machamer C.E. The cytoplasmic tail of the severe acute respiratory syndrome coronavirus spike protein contains a novel endoplasmic reticulum retrieval signal that binds COPI and promotes interaction with membrane protein. J. Virol. 2007;81:2418–2428. doi: 10.1128/JVI.02146-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memish Z.A., Mishra N., Olival K.J., Fagbo S.F., Kapoor V., Epstein J.H., Alhakeem R., Durosinloun A., Al Asmari M., Islam A., Kapoor A., Briese T., Daszak P., Al Rabeeah A.A., Lipkin W.I. Middle East respiratory syndrome coronavirus in bats, Saudi Arabia. Emerg. Infect. Dis. 2013;19:1819–1823. doi: 10.3201/eid1911.131172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi P., Zhang P., Liu G. Bio-inspired virus-like nanovesicle for effective vaccination. Hum. Vaccin. Immunother. 2016;12:2090–2091. doi: 10.1080/21645515.2016.1157244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minkner R., Park E.Y. Purification of virus-like particles (VLPs) expressed in the silkworm Bombyx mori. Biotechnol. Lett. 2018;40:659–666. doi: 10.1007/s10529-018-2516-5. [DOI] [PubMed] [Google Scholar]
- Mortola E., Roy P. Efficient assembly and release of SARS coronavirus-like particles by a heterologous expression system. FEBS Lett. 2004;576:174–178. doi: 10.1016/j.febslet.2004.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi T., Shimojima T., Fukagawa T., Maenaka K., Park E.Y. Efficient large-scale protein production of larvae and pupae of silkworm by Bombyx mori nuclear polyhedrosis virus bacmid system. Biochem. Biophys. Res. Commun. 2005;326:564–569. doi: 10.1016/j.bbrc.2004.11.060. [DOI] [PubMed] [Google Scholar]
- Rohovie M.J., Nagasawa M., Swartz J.R. Virus-like particles: next-generation nanoparticles for targeted therapeutic delivery. Bioeng. Transl. Med. 2017;2:43–57. doi: 10.1002/btm2.10049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu Y.L., Teoh K.T., Lo J., Chan C.M., Kien F., Escriou N., Tsao S.W., Nicholls J.M., Altmeyer R., Peiris J.S., Bruzzone R., Nal B. The M, E, and N structural proteins of the severe acute respiratory syndrome coronavirus are required for efficient assembly, trafficking, and release of virus-like particles. J. Virol. 2008;82:11318–11330. doi: 10.1128/JVI.01052-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ujike M., Huang C., Shirato K., Makino S., Taguchi F. The contribution of the cytoplasmic retrieval signal of severe acute respiratory syndrome coronavirus to intracellular accumulation of S proteins and incorporation of S protein into virus-like particles. J. Gen. Virol. 2016;97:1853–1864. doi: 10.1099/jgv.0.000494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usami A., Suzuki T., Nagaya H., Kaki H., Ishiyama S. Silkworm as a host of baculovirus expression. Curr. Pharm. Biotechnol. 2010;11:246–250. doi: 10.2174/138920110791112013. [DOI] [PubMed] [Google Scholar]
- Wang Q., Qi J., Yuan Y., Xuan Y., Han P., Wan Y., Ji W., Li Y., Wu Y., Wang J., Iwamoto A., Woo P.C., Yuen K.Y., Yan J., Lu G., Gao G.F. Bat origins of MERS-CoV supported by bat coronavirus HKU4 usage of human receptor CD26. Cell Host Microbe. 2014;16:328–337. doi: 10.1016/j.chom.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Zheng X., Gai W., Zhao Y., Wang H., Wang H., Feng N., Chi H., Qiu B., Li N., Wang T., Gao Y., Yang S., Xia X. MERS-CoV virus-like particles produced in insect cells induce specific humoral and cellular immunity in rhesus macaques. Oncotarget. 2017;8:12686–12694. doi: 10.18632/oncotarget.8475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissmann F., Petzold G., VanderLinden R., Huis In’t Veld P.J., Brown N.G., Lampert F., Westermann S., Stark H., Schulman B.A., Peters J.M. biGBac enables rapid gene assembly for the expression of large multisubunit protein complexes. Proc. Natl. Acad. Sci. U. S. A. 2016;113:E2564–E2569. doi: 10.1073/pnas.1604935113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO, 2019, https://www.who.int/emergencies/mers-cov/en/.
- Zhang P., Chen Y., Zeng Y., Shen C., Li R., Guo Z., Li S., Zheng Q., Chu C., Wang Z., Zheng Z., Tian R., Ge S., Zhang X., Xia N.S., Liu G., Chen X. Virus-mimetic nanovesicles as a versatile antigen-delivery system. Proc. Natl. Acad. Sci. U. S. A. 2015;112:E6129–E6138. doi: 10.1073/pnas.1505799112. [DOI] [PMC free article] [PubMed] [Google Scholar]