Abstract
Caliciviridae are human or non-human pathogenic viruses with a high diversity. Some members of the Caliciviridae, i.e. human pathogenic norovirus or rabbit hemorrhagic disease virus (RHDV), are worldwide emerging pathogens. The norovirus is the major cause of viral gastroenteritis worldwide, accounting for about 85% of the outbreaks in Europe between 1995 and 2000. In the United States, 25 million cases of infection are reported each year. Since its emergence in 1984 as an agent of fatal hemorrhagic diseases in rabbits, RHDV has killed millions of rabbits and has been dispersed to all of the inhabitable continents.
In view of their successful and apparently increasing emergence, the development of antiviral strategies to control infections due to these viral pathogens has now become an important issue in medicine and veterinary medicine. Antiviral strategies have to be based on an understanding of the epidemiology, transmission, clinical symptoms, viral replication and immunity to infection resulting from infection by these viruses. Here, we provide an overview of the mechanisms underlying calicivirus infection, focusing on the molecular aspects of replication in the host cell. Recent experimental data generated through an international collaboration on structural biology, virology and drug design within the European consortium VIZIER is also presented. Based on this analysis, we propose antiviral strategies that may significantly impact on the epidemiological characteristics of these highly successful viral pathogens.
Keywords: Calicivirus, Antiviral therapy, Viral enzymes, VIZIER
1. The Caliciviridae: emerging pathogens with a high diversity
The family Caliciviridae includes viruses that cause a broad spectrum of disease in a wide variety of mammalian, avian, and marine animals. This may range from acute gastroenteritis in humans (norovirus and sapovirus) to lethal encephalitis in immune deficient mice (murine norovirus), through haemorrhagic disease in rabbits and upper airway infection in cats (feline calicivirus). Although caliciviruses infect humans and non-humans, thus far there is no evidence for a zoonotic calicivirus reservoir.
1.1. Morphology and physicochemical properties
The caliciviruses (CV) are small round viruses ranging from 27 to 32 nm in diameter. They are some of the smallest animal pathogenic viruses, and were originally referred to as the SRSV (small round structured viruses). Their surface displays a cup-like structure from which the name “calicivirus” was derived (calyx, cup; Fig. 1 ) (Clarke and Lambden, 2000, Green, 2007).
Fig. 1.
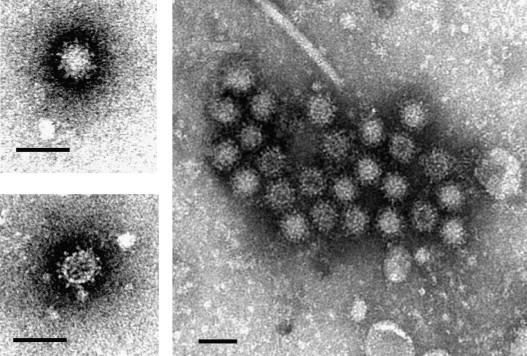
Morphology of the human pathogenic norovirus. Clinical stool samples were negatively stained with 2% phosphotungstate, pH 7.0, and examined by transmission electron microscopy. The typical cup-like surface of the virions is visible. Scale bar = 50 nm.
The cup-like surface of the virions is the capsid protein, consisting of 180 monomers known as virion protein 1 (VP1) which contains two domains: the shell (S), and the protruding (P) domain, being itself subdivided into P1 and P2 subdomains (Chen et al., 2006, Green, 2007). The P2 subdomain is located on the external part of the capsomeres, radiating out of the virion surface creating the cup-like shape. Structural comparison of the capsid proteins reveals genus-specificity within the P1 and P2 subdomains (Chen et al., 2004). The P2 subdomain determines viral antigenicity and attachment of virions to cellular receptors (Chakravarty et al., 2005).
The viral capsid plays an important role in the resistance of virions to environmental changes. It resists high concentrations of chlorine, from 3.65 to 6.25 mg/L (residual concentration of 0.5–1.0 mg/L) (Keswick et al., 1985). This concentration significantly reduces the infectivity of other viral agents of gastroenteritis, i.e. poliovirus and rotavirus. Additionally, the viral capsid confers high resistance to physicochemical agents such as heat (60 °C for 30 min), acid (pH 2.7 for 3 h at room temperature), and organic solvents (20% ether at 4 °C for 18 h) (Green et al., 2001). These physicochemical properties contribute significantly to the highly contagious nature of these viruses.
1.2. Classification of the Caliciviridae
Classification of the Caliciviridae relies largely on phylogenetic analysis of the structural and non-structural viral genes, mainly due to the lack of cell culture systems to cultivate the viruses and thus to enable serological tests to be performed. The Caliciviridae comprises four accepted and two tentative genera, including human and non-human pathogenic viruses (Fauquet et al., 2005, Green, 2007, Green et al., 2000, Mayo, 2002). Non-human pathogenic viruses cluster exclusively in the accepted genera Vesivirus, Lagovirus, and the tentative genera Beco/Nabovirus, and Recovirus, whereas the genera norovirus and sapovirus include both human and non-human pathogens.
Phylogenetic analysis based on the complete VP1 sequence shows that noroviruses cluster tightly in five genogroups. The human pathogenic norovirus belongs to genogroups (G) I, II and IV. Genogroups I and II contain so far 8 and 17 genotypes, respectively (Zheng et al., 2006). The non-human pathogenic strains belong to norovirus GIII and V (Oliver et al., 2007a, Oliver et al., 2007b, Oliver et al., 2003, Park et al., 2007). Genogroup II also contains the bovine and porcine noroviruses (Mauroy et al., 2008, Wang et al., 2007, Wang et al., 2005, Zheng et al., 2006). Genogroup IV, contains the feline (lion) and canine noroviruses (Martella et al., 2007, Martella et al., 2008). Genogroup V contains the murine noroviruses (MNV) (Wobus et al., 2006). These MNV are readily propagated in RAW264.7 murine macrophage cells, as well as murine BV-2 cells and various murine cell lines (Cox et al., 2009, Henderson, 2008, Muller et al., 2007, Mumphrey et al., 2007, Thackray et al., 2007, Wobus et al., 2004). Hence, MNV is an interesting surrogate for pathogenicity, multiplication cycle, and immunity studies that may also reflect properties of noroviruses. Infection in mice is usually asymptomatic. However, in immune deficient mice, i.e. (RAG2/STAT1−/−) MNV type 1 induces lethal encephalitis (Hsu et al., 2007, Karst et al., 2003, Mumphrey et al., 2007). The human pathogenic norovirus (HuNV) is the major cause of human viral gastroenteritis (Wilhelmi et al., 2003) and food-borne non-bacterial gastroenteritis (Lindqvist et al., 2001, Lopman et al., 2002) worldwide. These viruses are highly contagious (Teunis et al., 2008) causing a self-limiting gastroenteritis with a higher prevalence during the winter time in all age groups. They are excreted for weeks after infection in asymptomatic patients (Kirkwood and Streitberg, 2008, Murata et al., 2007, Siebenga et al., 2008).
The genus Sapovirus contains five genogroups. The human pathogenic strains belong to sapovirus GI, II IV and V (Farkas et al., 2004, Gallimore et al., 2006, Hansman et al., 2007, Numata et al., 1997, Schuffenecker et al., 2001), whereas the non-human pathogenic strains belong to GIII (Fauquet et al., 2005, Green et al., 2000, Guo et al., 2001a). Human pathogenic sapoviruses are distributed globally. They cause viral gastroenteritis in all age groups (Gallimore et al., 2006, Humphrey et al., 1984, Johansson et al., 2005, Lopman et al., 2002, Nakata et al., 1985, Sakuma et al., 1981) including outbreaks in hospitals (Johansson et al., 2005, Simor et al., 1990), child day care centers (Matson et al., 1989), and elderly persons homes (Humphrey et al., 1984). The majority of human pathogenic sapovirus strains belong to GI and GII.
Viruses in the genus Lagovirus are causative agents of hemorrhagic disease and hepatitis in the European rabbit (Oryctolagus cuniculus), causing Rabbit haemorrhagic disease. An antigenically related but distinct virus infects the European hare causing brown hare syndrome (Laurent et al., 1997). A non-pathogenic strain designated rabbit calicivirus has been isolated (Capucci et al., 1996) and there is also compelling evidence of circulating non-pathogenic strains that either emerged originally as ancestral lineages of the recognized virulent extant viruses (Moss et al., 2002) or still circulate in wild rabbits as widely divergent non-pathogenic viruses (Forrester et al., 2007). Other non-pathogenic strains arose as variants of virulent virus (Forrester et al., 2003).
The genus Vesivirus includes viruses that infect a broad variety of animals, including felines, reptiles, amphibians, fish and even nematodes (Matson et al., 1996, Smith and Boyt, 1990, Smith et al., 1998). The prototype of this genus is vesicular exanthema of swine virus, VESV, which displays sequence homology with San Miguel Sea Lion Virus (SMSV) (Neill et al., 1998, Smith et al., 1973). Feline calicivirus (FCV), an important veterinary virus, infects domestic and wild cat species causing rhinitis and an ulcerative disease of the upper airways (Radford et al., 2007). Systemic infections with FCV have also been described (Coyne et al., 2006, Foley et al., 2006, Hurley et al., 2004, Pedersen et al., 2000).
The recently recognized tentative genera Beco/Nabovirus and Recovirus include viruses that infect bovines and non-human primates, respectively (Fig. 2 ). Becovirus (bovine enteric calicivirus) and nabovirus (Newbury agent-1 and NB, Nebraska) have been isolated from calf faeces (Oliver et al., 2006, Park et al., 2008, Simmonds et al., 2008, Smiley et al., 2002). Tulane virus was isolated from the faeces of rhesus macaques, being for the time being the only recognized virus in the genus Recovirus (rhesus enteric calicivirus) (Farkas et al., 2008).
Fig. 2.
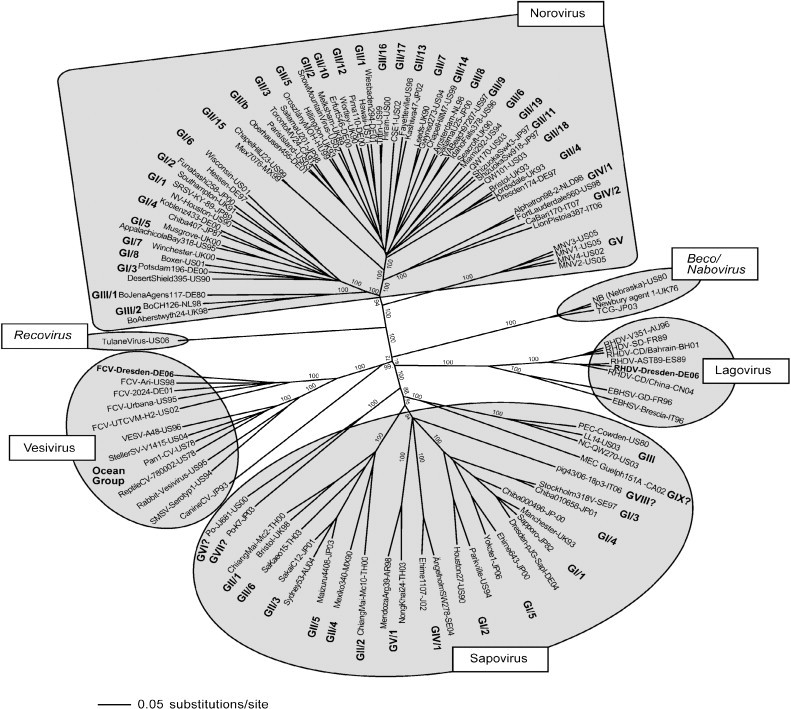
Classification of the Caliciviridae. Phylogenetic analysis of strains belonging to the so far defined genera (Norovirus, Sapovirus, Lagovirus and Vesivirus) and the two tentative genera Beco/Nabovirus and Recovirus, as indicated. The complete capsid sequence of the strains was aligned with Clustal X (Gibson et al., 1999) and a phylogenetic analysis performed with the neighbour-joining method using PAUP (Swofford, 2002). A bootstrap analysis with 1000 replicates was performed. The bootstrap values are indicated. The strains belonging to non-defined genogroups (GVI. VII, VIII and IX of sapovirus) are indicated by a question mark. The strains FCV/Dresden/2006/GE strain (Vesivirus, GenBank accession No. DQ424892), and RHDV/Dresden/2006/GE (Lagovirus, GenBank accession no. EF363035) generated within the consortium VIZIER are highlighted in bold. The tentative Genera Recovirus and Beco/Nabovirus are indicated.
1.3. Genome organization of the Caliciviridae
Caliciviridae genomes consist of a single stranded positive-sense polyadenylated RNA molecule that averages 7500 nucleotides in length and containing either two or three open reading frames, depending on the particular genus (Fig. 3 ). At the 5′-terminus of the genome, the so-called virion protein genome-linked (VPg) is linked to the viral RNA. The ORF1 encodes non-structural (NS) proteins present in all Caliciviridae in the following order: an N-terminal protein with unknown function (NS1-2), the NTPase (NS3), a small protein of about 20–30 kDa (NS4), the VPg (NS5), the viral protease (NS6), and the viral RNA-dependent RNA polymerase (RdRp, NS7). In the genera Sapovirus, Lagovirus and the tentative genera Beco/Nabovirus, the VP1 is encoded within the ORF1, whereas it is encoded in a separate ORF2 in the genera Norovirus, Vesivirus, and Recovirus. The 3-terminus of the genome, encodes a structural virion protein 2 (VP2), which is followed by an untranslated region, and a poly(A)-tail. Since they share some similarities with picornaviruses, the nomenclature of the non-structural proteins of the Caliciviridae was based on that of the picornaviruses, with the viral polymerase being called 3D-like, the 3C-like viral protease, the VPg, the 3A-like and the 2C-like protein. The increased knowledge on calicivirus replication and genome organization in the last years has led to a new nomenclature where the viral structural proteins are designated NS1/NS2 (N-terminal protein), NS3 (NTPase), NS4, NS5 (VPg), NS6 (protease), NS7 (RNA-dependent RNA polymerase, RdRp). The structural proteins are designated VP1 and VP2. In the sapovirus GI, IV and V, an additional ORF encoding a putative small basic protein has been described (Clarke and Lambden, 2000, Hansman et al., 2005b).
Fig. 3.
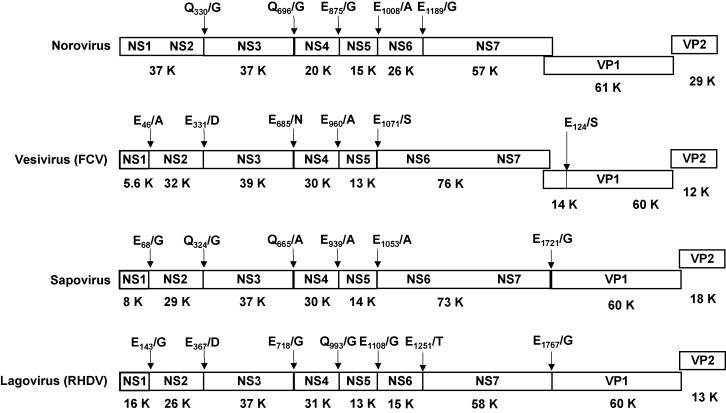
Comparison of the organization of the viral genome of the six calicivirus genera. The complete genome of the following strains is shown: Norovirus GII.4 (Hu/NLV/Dresden174/pUS-NorII/1997/GE strain, GenBank accession no. AY741811), vesivirus (feline calicivirus, FCV/Dresden/2006/GE strain, GenBank accession No. DQ424892), sapovirus GI.1 (Hu/SV/pJG-SapI/DE strain, GenBank accession no. AY694184), and lagovirus (RHDV strain RHDV/Dresden/2006/GE, GenBank accession no. EF363035). The cleavage sites in the viral polyprotein precursor are indicated, as well as the putative molecular weight in kDa (K) of the resulting non-structural proteins.
1.4. Multiplication cycle of the Caliciviridae
The multiplication cycles of the Caliciviridae remains poorly understood, mainly because of the limited possibilities of virus cultivation. Indeed, among the six calicivirus genera only the feline calicivirus and the murine norovirus can be easily propagated in cell systems, both being non-human pathogens. For the human pathogenic viruses, there are no suitable in vitro culture systems. Recently, a human norovirus was isolated and viral RNA was propagated for five sequential passages in small epithelial cells (INT-407) cultivated in suspension (Straub et al., 2007). It remains unclear whether or not this system is suitable for isolation of the various norovirus genotypes.
Another approach to studying the molecular biology of calicivirus replication makes use of replicon systems, such as for the Norwalk-like virus (Asanaka et al., 2005, Chang et al., 2006), and porcine enteric calicivirus (Chang et al., 2002, Chang et al., 2004), but also reverse genetics systems for the murine norovirus (Chaudhry et al., 2007, Ward et al., 2007), the feline calicivirus (Sosnovtsev et al., 2005, Thumfart and Meyers, 2002) and Tulane virus (Farkas et al., 2008). Those systems provide future opportunities for comparative analysis of the multiplication cycles of the different genera.
FCV has been used during the past two decades as a surrogate to investigate the replication strategy of the Caliciviridae. Many studies have brought insights into the replication of the vesivirus genome (Sosnovtsev and Green, 1995, Sosnovtsev and Green, 2000, Sosnovtsev et al., 2005, Sosnovtsev et al., 2003, Sosnovtsev et al., 1998, Sosnovtseva et al., 1999). Lately, additional knowledge has been obtained from studies on the murine norovirus in RAW264.7 murine macrophages in vitro (Sosnovtsev et al., 2006, Wobus et al., 2004, Wobus et al., 2006).
Caliciviruses display in terms of genome structure and replication the following common characteristics:
-
-
Naked genomic length RNA is infectious per se, being a positive stranded RNA linked to the VPg at the 5′-terminus and bearing a poly(A)-tail at the 3′-terminus.
-
-
The ORF1 encodes a polyprotein that is further processed co-translationally by the viral protease, leading to at least 6 non-structural proteins involved in the replication of the viral genome.
-
-
The viral RdRp and the VPg play an essential role in the transcription and replication of the genomic, antigenomic RNA and subgenomic RNA.
-
-
The VPg acts as a cap-like structure and interacts with the initiation factors eIF4E and eIF3 (Goodfellow et al., 2005, Daughenbaugh et al., 2006).
-
-
In the noroviruses, vesiviruses and possibly the recoviruses, the structural proteins (VP1 and VP2) are encoded by a subgenomic RNA.
-
-
In the sapoviruses, lagoviruses and possibly the beco/naboviruses, the VP1 is released from the ORF1-encoded polyprotein precursor after cleavage by the viral protease.
-
-
The VP2 is a small basic protein that is essential for virion generation (Sosnovtsev et al., 2005).
Schematically, the replication of caliciviruses is thought to contain eight steps (Fig. 4 ). After attachment of the viral capsid to the cellular receptor, the virion is internalized. The RNA genome is then packaged in endocytic vesicles. The VPg linked to the genome interacts with the cellular translation machinery (the eukaryotic initiation factor 4 (eIF4) complex and the 40S ribosomal subunit (Daughenbaugh et al., 2003, Daughenbaugh et al., 2006). Translation of the polyprotein and co-translational processing releases the non-structural proteins and their precursors, most importantly the protease-polymerase precursor (NS6-7propol), the protease (NS6pro), the RNA-dependent RNA polymerase (NS7pol), the putative NTPase (NS3NTPase), and the NS4-NS5VPg proteins. These proteins are putatively involved in the enzymatic complex that replicates the genome, and synthesizes the antigenomic RNA.
Fig. 4.
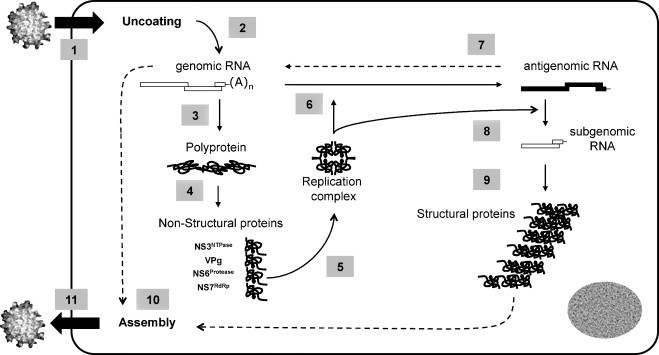
The multiplication cycle of the calicivirus. The replication of the calicivirus can be schematically subdivided into eleven steps. After attachment of the protein core to the cellular receptor, the virion is internalized in the cell (step 1). Uncoating of the viral genome (step 2) is followed by translation of the polyprotein precursor (step 3) and a co-translational processing releasing the non-structural proteins (step 4). Those proteins assemble in a replication complex (step 5) that synthesizes the antigenomic RNA (step 6), being itself used as a template for synthesis of the genomic RNA (step 7). The newly synthesized genomic RNA is translated as a polyprotein precursor (step 3) or being used for packaging in the assembled viral protein core (step 10). The antigenomic RNA is also the template for synthesis of subgenomic RNA (step 8). In noroviruses, vesiviruses and possibly recoviruses, the subgenomic RNA is translated as structural proteins, VP1 and VP2 (step 9). In sapoviruses, lagoviruses and possibly beco/naboviruses, the VP1 is released from the polyprotein precursor after processing by the viral protease. At a still not defined time in the multiplication cycle, assembly of the structural proteins as well as packaging of the genomic RNA occurs (step 10), followed by release of the mature virion from the cell (step 11).
This antigenomic RNA is the template for the subgenomic RNA, a peculiarity of the caliciviruses. In the noroviruses and vesiviruses, transcription of subgenomic RNA yields high amounts of structural proteins, the VP1 and VP2. In FCV, the VP1 is additionally cleaved by the viral protease releasing a leader protein (LC, leader of the capsid protein) and the capsid (Matsuura et al., 2000, Sosnovtsev et al., 1998). In the sapoviruses, lagoviruses and beco/naboviruses, the VP1 is released from the polyprotein precursor after processing by the viral protease. Experimental data on feline calicivirus describes a termination/reinitiation mechanism for VP2 translation (Pöyry et al., 2007). This mechanism involves a so-called “termination upstream ribosomal binding site” (TURBS), that may regulate the ratio of VP1/VP2 production during replication (Luttermann and Meyers, 2007). A similar mechanism of termination-reinitiation has been recently reported in the murine norovirus (Napthine et al., 2009), resembling the termination-initiation mechanisms observed in other viruses, i.e. Influenza virus.
The antigenomic RNA is also the template for the genomic RNA, being subsequently translated in mature non-structural proteins. At a given time point in the multiplication cycle, packaging of the genomic RNA occurs, followed by release of the mature virion from the cell.
1.5. Calicivirus infection: epidemiology, transmission and clinical features
In the following section, we focus on the epidemiology and transmission of the human pathogenic caliciviruses (HuCV, norovirus and sapovirus), since these viruses are of prime interest for antiviral development.
1.5.1. Epidemiology of HuCV
The HuCVs are one of the most common causes of acute gastroenteritis worldwide, infecting both children and adults. In a recent report from CDC on outbreaks occurring in the USA from 1991 to 2002, norovirus infections were recognized as the most common cause of viral gastroenteritis in all ages in the US, accounting for about 33% of all outbreaks (Lynch et al., 2006, Widdowson et al., 2005). The majority of outbreaks were caused by strains belonging to GII.4 (Koopmans, 2005) suggesting that this genogroup displays preferential features in terms of infectivity and/or transmission efficiency. Alternatively it could reflect increased virulence through enhanced activity of the viral enzymes, or shut-off of cellular host factors in the course of infection. Another important aspect relates to the dynamics of emergence in the noroviruses that correlates tightly with the propensity of disease dissemination through the community and over time, with new strains emerging through mutation and/or recombination. This is mainly supported by the emergence and circulation of norovirus and sapovirus recombinant strains (Gallimore et al., 2004, Jiang et al., 1999a, Katayama et al., 2004, Phan et al., 2006, Reuter et al., 2002, Rohayem et al., 2005, Vinje et al., 2000). A further aspect relates to zoonosis, since noroviruses and sapoviruses include strains that are human or non-human pathogenic. A high prevalence of antibodies binding norovirus and sapovirus and neutralizing Tulane virus have been recently detected in captive juvenile macaques (Farkas et al., 2010). Additionally, sera collected from norovirus outbreak patients displayed a neutralizing effect on Tulane virus infectivity in vitro (Farkas et al., 2010). This data, although preliminary, may indicate a possible cross-species transmission of human and/or non-human pathogenic calicivirus, although a cross-reactivity of the sera is not to be excluded. Moreover, cross-species transmission of the bovine norovirus Newbury2/76/UK strain has been recently invalidated by the fact that the carbohydrate ligand required for attachment of the virus is absent from human tissues (Zakhour et al., 2009). All in all, the experimental and epidemiological data generated so far are not strongly in favor of a cross-species transmission.
1.5.2. Transmission and host resistance
Transmission of HuCV occurs via the fecal-oral route, but virus can be transmitted via aerosols, as observed during explosive vomiting (Koopmans, 2005, Koopmans and Duizer, 2004, Koopmans et al., 2002). The environmental resistance of HuCV combined with their high infectivity (with an infectious dose of 10–100 viral particles), facilitates their efficient transmission. Human susceptibility to infection by noroviruses is also dependent on the blood group of the individual (A, B, O). The histo-blood group antigens (HBGA) are one of the recognized binding receptors for noroviruses (Hennessy et al., 2003, Hutson et al., 2002, Lindesmith et al., 2003, Marionneau et al., 2002, Rockx et al., 2005). HBGA are present on the surface of erythrocytes and mucosal epithelial cells or as free oligosaccharides in milk, saliva and gut fluid (Marionneau et al., 2001). A recent study underlies the difference of HBGA present in saliva and milk. In human milk, the HBGA are synthesized in higher quantities and display smaller molecular weights and different backbones, although HBGAs of high molecular weight may also be found (Huang et al., 2009). This variability in HBGA content and nature seem to play an important role in modulating the specific binding of human pathogenic norovirus to the HBGA present in saliva and/or milk, and subsequent transmission.
Binding of the VP1 protein to the HBGA has been shown to occur in the P2 subdomain (Cao et al., 2007, Tan et al., 2009, Tan et al., 2003). The differential binding of the P2 subdomain to the HBGA induces a differential susceptibility of humans to infection with the different norovirus genogroups. Binding of the P2 subdomain is modulated by the phenotype of the fucosyl-transferase (FUT) enzyme. The fucosyl-transferase (FUT) conjugates a fucosyl residue to the N-acetyl-glucosamine of the HBGA. Individuals lacking a functional FUT2 enzyme are called non-secretors, as they do not secrete the ABO HBGA in their saliva and the gut fluid. Individuals harboring a functional FUT2 enzyme are called secretors, displaying the A, B or Lewisb phenotypes. This is related to the expression of the enzymes A and B type 1 that conjugate the precursor H type 1 (HBGA) with a glucosyl residue, or through the FUTa1-4 enzyme with a fucosyl residue. Non-secretors (Lewisa) are resistant to infection with the majority of norovirus stains. The non-secretor phenotype is present in about 20% of the European and North American population (Marionneau et al., 2001).
It should however be kept in mind that neither sapovirus genogroup I nor sapovirus GV virus-like particles display such a binding to the ABO HBGA (Shirato-Horikoshi et al., 2006), indicating a different cellular receptor and possibly mechanism of internalization for noroviruses and sapoviruses.
2. Development of antivirals against caliciviruses
Based on our knowledge of the calicivirus genome organization, molecular biology, transmission and replication, potential antiviral strategies are now at least conceivable. In the following section, we will describe potential calicivirus targets for drug development. Possible antiviral approaches using the experimental data on the structure and function of the viral enzymes of replication that was generated through a successful collaboration within the European consortium VIZIER are also presented.
2.1. Targeting the attachment and internalization of the virions
In the first steps of viral infection, attachment of the virions to the cell surface and interaction with a specific receptor leads to its internalization, followed by uncoating of the viral genome and release into the cytoplasm. To inhibit viral infection of the cell, it may therefore be possible to inhibit binding of the virion and/or internalization and/or release of the viral genome.
Recent work on the feline calicivirus identified the specific receptor for internalization of the virion (Makino et al., 2006). FCV attaches to a-2-6-linked sialic acids, but uses the junctional adhesion molecule 1 (JAM-1), a member of the immunoglobulin superfamily, for internalization in Crandell-Rees feline kidney (CRFK) cells. JAM-1 is expressed in tight junctions in a variety of cells. An anti JAM-1 antibody reduced FCV binding to CRFK cells and thus reduced infectious virus yields (Makino et al., 2006). This advance in the characterization of the specific receptor may represent an interesting antiviral approach by targeting the calicivirus receptor (i.e. JAM-1) using appropriate monoclonal antibodies. However, considering the broad diversity of host cells (enteric cell, respiratory epithelium, dendritic cells, or neuronal cells), important hurdles remain. For instance, the murine norovirus has been recently reported to bind a ganglioside GD1a present on the surface of murine macrophages (Taube et al., 2009). This ganglioside is thought to mediate attachment, but internalization probably requires additional factors, for example, an integrin. A recent study has reported on the involvement of a cholesterol dependent pathway for entry of murine norovirus in RAW264.7 cells, with the clathrin and caveolae pathways being not required for cell entry (Gerondopoulos et al., 2010). Whether or not this phenotype is common to other noroviruses is unclear. It is also unknown whether GD1a is involved alone in binding of the virion or other membrane proteins interact with it. In this case, inhibition of binding to one receptor may lead to inhibition of viral entry in the cell, through a mechanism of steric or allosteric blockade of the interaction between virus and receptor.
There is still little known about the internalization receptor of lagoviruses and sapoviruses. The attachment of HuCV to ABO HBGA, heparan sulfate, and sialyl-Lewis x (Rydell et al., 2009, Tamura et al., 2004) has been demonstrated in vitro using virus-like particles (VLPs). The lack of a cell culture system to reproduce the human pathogenic calicivirus, is impeding further work in this subject area.
An interesting antiviral approach may be to block the interaction of the P2-subdomain of norovirus with the HBGA. In a recent study (Feng and Jiang, 2007), Feng et al. screened a compound library using a saliva-based receptor binding assay for blocking the interaction of secreted HBGA and VLPs of the norovirus strain VA387. VLPs display similar antigenic properties to native virions and have been used as tools to investigate virus–host interactions and immune responses in animal models, and for screening potential anti-norovirus candidates (Green et al., 1993, Huang et al., 2003, Huang et al., 2005a, Jiang et al., 1992). In 14 of the 5000 compounds screened, an EC50 value of less than 15 μM was found, and in 2 compounds, an EC50 < 5 μM. Because HuCV are thought to replicate in intestinal cells, inhibition of attachment of HuCV to the HBGA receptor may represent an interesting strategy to inhibit infection of the cell. If applied early in the course of infection, it may also block propagation of the virions within the tissue. This kind of treatment may be suitable in a preventive set-up in the course of spatial limited outbreaks occurring for example on cruise ships.
A similar approach relies on trapping the norovirus in glycosylated hydrogels (Zhang et al., 2006). The trapping capacity of hydrogel was estimated to be about 24 μg VLPs per mg. Although promising, this approach is more suitable to prevent transmission of norovirus from person-to-person.
Because, in contrast to noroviruses, the human pathogenic sapoviruses do not rely on attachment to HBGA, additional data concerning virus binding and entry will be needed to bring the proof-of-principle for an attachment-inhibition-based antiviral approach in caliciviruses.
2.2. Targeting uncoating and release of the viral genome
Many viruses enter the cell and the cytoplasm through endocytic vesicles (Pelkmans and Helenius, 2003, Smith and Helenius, 2004). In mammalian cells, various endocytosis pathways have been described: macropinocytosis, phagocytosis, endocytosis via caveolae/lipid rafts, and clathrin-mediated endocytosis. Following endocytosis, uncoating of the viral genome may depend on lowering of the pH in the endocytic vesicles. The pH change leads to conformational change of the viral capsid, with exposition of motives located on the subdomains (Bhella et al., 2008). In an experimental set-up, the low pH-environment of the endosome induced structural changes of the FCV capsid leading to release of the viral genomic RNA (Stuart and Brown, 2006). Hence, a possible antiviral approach may be to use compounds or short peptides that inhibit rearrangements of the viral surface related to pH change. Another approach may rely on inducing pH-modifications in the different compartments of the cell, and subsequently the intra-cellular pH-gradient. Two different compounds are known to inhibit the reduction of the endosomal pH: a weak basic compound (chloroquine), and a macrolide antibiotic that selectively inhibits the vacuolar type of vesicular proton ATPase (Bafilomycin A1). Bafilomycin A1 and chloroquine increase in a concentration-dependent manner the pH of the endosomal compartment and may be used to inhibit uncoating of the viral genome by inhibiting acidification of the endosome. Interestingly, chloroquine and Bafilomycin A1 inhibit FCV infectivity in vitro (Stuart and Brown, 2006). Moreover, Chlorpromazine, a compound known to inhibit clathrin-mediated endocytosis, inhibits internalization of FCV in vitro (Stuart and Brown, 2006). It remains unclear whether or not other caliciviruses display similar entry and uncoating mechanisms. Recent data on murine norovirus revealed that infection of murine RAW264.7 was not inhibited by reagents that raise endosomal pH (Gerondopoulos et al., 2010). Nevertheless, and because all three substances are commercially available, they represent a promising approach to control infection by FCV.
2.3. Targeting the viral genomic RNA
After its release into the cytoplasm, the viral mRNA is recruited to the endoplasmic reticulum for translation to produce the viral polyprotein. At this stage, inhibition of the function of the whole genomic RNA and/or parts of it stops viral replication. To this end, two strategies have been developed in Caliciviridae, both making use of nucleic acids targeting the 5′-end and/or the 3′-end of the genome. Comparative analysis of the structural features of members of the Caliciviridae has shown that all strains display stem-loops and hairpin structures at the 5′-terminus and the 3-′-terminus of the viral genome, respectively (Simmonds et al., 2008).
Using an experimental approach based on the murine norovirus reverse genetic system, disruption of the 5′-stem loops or the 3′-hairpins strongly impaired viral replication in RAW264.7 murine macrophages in vitro (Simmonds et al., 2008). Moreover, a polypyrimidine tract located at the 3′-terminus of the genome has been incriminated in regulating viral fitness and virulence in a small animal model of infection (Bailey et al., 2010a). This indicates that those structures play an important role in the infectivity of the MNV.
Both regions are preferentially targeted by nucleic acid based approaches as both bear highly conserved sequences within genera (i.e. in FCV, Table 1 ). The nucleic acid-based strategies are the antisense approach and the post-transcriptional gene silencing (or RNA-interference) approach. There are important prerequisites for the successful implementation of a nucleic acid-based antiviral approach. First, the target sequence has to be involved in viral replication. Additionally, it must be accessible for oligonucleotide hybridization, and most importantly when dealing with RNA viruses with high genomic variability it must be conserved among different viral species. Hence, the regions that are preferentially targeted reside within the 5′- and 3′-UTRs of the viral genome, where conserved regions may span entire genera.
Table 1.
Sequences of the 5′-UTR and 3′-UTR of feline calicivirus strains displaying 100% identity to the FCV/Dresden/2006/GE strain (GenBank accession no. DQ424892) used for the antiviral nucleic acid based approach. The nucleotide positions as well as the GenBank accession no. of the strains are indicated.
2.3.1. The antisense approach
The antisense approach uses DNA-oligonucleotides targeting conserved regions of the viral genome. It has been applied in various models of viral infections with positive-stranded and negative-stranded RNA viruses (Gabriel et al., 2008, Lupfer et al., 2008, Paessler et al., 2008, Stein, 2008, Stein et al., 2008, Stein and Shi, 2008).
In the Caliciviridae, peptide-linked phosphorodiamidiate morpholino oligomers (PPMOs) targeting a sequence near the 5′-UTR of the murine norovirus genome were effective in reducing translation of viral proteins in vitro (Bok et al., 2008). Our preliminary investigations have made use of the PPMO-approach to inhibit multiplication of the feline calicivirus in vitro. Seven PPMOs (kindly provided by Dr. David Stein, AVI Biopharma) were designed to target the viral genome in particular the 5′-UTR or the 3′-UTR of the feline calicivirus genome (Table 2 ). In a cell proliferation assay, the lowest EC50 observed was of 20.4 μM, with a selectivity index of 9.8 (Table 3 ).
Table 2.
Sequence characteristics of the oligonucleotides used for antiviral nucleic acid based approach.
| Oligonucleotide | Sequence (5′–3′ orientation) | Length (nt) | Target (nt position) |
|---|---|---|---|
| PPMO-1 | GAC ATT GTC TCA AAT TTC TTT TAC | 24 | 1–24 |
| PPMO-2 | GCA CAT GCT CAA ACT TCG AAC AC | 23 | 5294–5313 |
| PPMO-3 | CCC ATG TAG GAG GCA GTA AGA GG | 23 | 7237–7259 |
| PPMO-4 | CCC TCI TGI GIT TAG GCG CTA GAG CGG | 24 | 1497–1520 |
| PPMO-5a | GTA AAA GAA ATT TGA GAC AAT GTC | 24 | 1–24 |
| PPMO-6 | CCG CTC TAG CGC CTA ACC CCA GIG | 24 | 7658–7680 |
| PPMO-7 | TGC TCT GTC TAC AGT AGT GTC A | 22 | 6159–6180 |
| siR19 | ACU CUG AGC UUC GUG CUU ATT | 21 | 29–47 |
| UAA GCA CGA AGC UCA GAG UTT | 21 | ||
| siR20 | CCU GCG CUA ACG UGCUUA ATT | 21 | 5321–5339 |
| UUA AGC ACG UUA GCG CAG GTT | 21 | ||
Table 3.
Antiviral activity and cell toxicity of the oligonucleotides used in the nucleic acid based approach. The EC50 and CC50 values are measured in a colorimetric cell-proliferation assay, as described by others (Oxford et al., 1999).
| Oligonucleotide | EC50 (μM) | CC50 (μM) | SI (CC50/EC50) |
|---|---|---|---|
| PPMO-1 | 20.4 | >200 | 9.8 |
| siR19 | 0.44 | >2.5 | 5.7 |
Phosphorodiamidiate morpholino oligomers (PMO) targeting the 5′-UTR of the FCV genome were used in 3 clinical trials relating to FCV outbreaks. From day 1 of infection, PMOs were administered parenterally to 112 kittens for 7 days in different dosages (0.7–5.0 mg/kg). The PMO displayed a dose-dependent effect. Of the 59 animals treated, 47 survived the infection. In comparison, in 31 animals not treated with PMO, only 3 survived the infection (Smith et al., 2008).
Although preliminary, these experimental data indicate that PPMOs may represent an interesting approach for treatment of calicivirus infections.
2.3.2. Post-transcriptional silencing using RNA-interference
Another approach targeting the viral genome is post-transcriptional silencing using sequence specific RNA-Interference. The use of RNA interference to silence exogenous RNA such as viral genomic RNA has been extensively reported (Haasnoot et al., 2007). Different approaches using siRNA have been employed in viral infection models in vivo and in vitro (Bitko and Barik, 2001, Boden et al., 2003, von Eije et al., 2008, Ge et al., 2003, Ge et al., 2004, Tompkins et al., 2004, Zhang et al., 2005, Bitko et al., 2005).
Major problems with the RNAi-approach reside in the design of the siRNA, and the delivery of the molecules to the targeted cell, and furthermore to the cellular compartment where the targeted RNA is located. Another problem relates to the stability of the siRNA as well as to its bioavailability (Li et al., 2006). A further problem relates to the off-target effects observed when the seed sequences of the siRNA cross-react with host cellular mRNA (Jackson et al., 2003, Lin et al., 2005). Nevertheless, one major advantage of the RNAi-based strategy is the faster and more efficient development of lead compounds compared to classical small molecules. This is particularly of interest for RNA viruses with high genomic variability, such as the Caliciviridae, and more particularly the human pathogenic norovirus. Indeed, in the case of emergence and pandemics of new norovirus variants, the rapid optimization of siRNA molecules may facilitate accelerated design and synthesis of RNAi-compounds and thus, control of the infection. Moreover, through judicious design of the RNAi-molecules, as well as the correct choice of the targeted sequence, broad-spectrum activity of the antiviral can be achieved. In Caliciviridae, a conserved sequence is of interest, for example, specific regions within the 5′-UTR in FCV. However, this region may be problematic because of binding of cellular translation products to the potential antiviral molecules, possibly causing steric blocking of the siRNA.
As observed for small molecules, the administration of siRNA may also induce the emergence of resistant mutants. Therefore, another strategy would be to target cellular factors that are indispensable for viral replication, but dispensable for cellular metabolism.
In a preliminary set-up, we have used small interfering RNA molecules that target the 5′-UTR and the subgenomic region of the FCV genome (FCV/Dresden/2006/GE strain, GenBank accession No. DQ424892). In a prophylaxis model of infection, CRFK cells were transfected with siRNA, followed by infection of the cells. The siRNA completely inhibited viral multiplication (Fig. 5 ). In a cell proliferation assay, the EC50 was of 440 nM that is about 50-fold lower than the EC50 observed with PPMOs targeting the same sequence (Table 3). Although provisional, this data suggests that post-transcriptional gene silencing may be a potential antiviral approach through targeting of the viral genome.
Fig. 5.
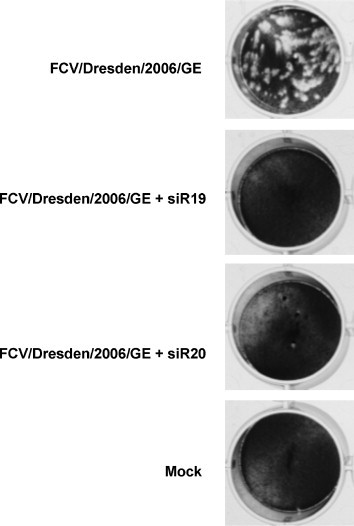
Inhibition of calicivirus particle formation in CRFK-monolayer after infection with the FCV/Dresden/2006/GE strain (GenBank accession No. DQ424892). Two siRNA (siR19 and siR20) targeting conserved sequences in the 5′-UTR and 3′-UTR, respectively, were used. The siRNA (330 nM) was transfected into the CRFK cells 5 h prior to infection with FCV. Plaque assays were performed as described by others (Gray, 1999), yielding a viral titre of 500 PFU/ml. The siRNA completely inhibited viral multiplication in the CRFK cells.
In summary, in comparison to small molecules, the nucleic-acid based approach offers many advantages that may render it attractive as an antiviral strategy. Its strengths rely on its selectivity, and on the possibility of rapid optimization of lead compounds, in particular in the case of emerging viruses with high genomic variability. Nevertheless, several pre-requisites have to be fulfilled before its implementation as a therapeutic tool. Clearly, two major hurdles would be its bioavailability and its specific delivery to the infected cells.
2.4. Targeting the translation of the viral genome
Translation of the viral genome is the first step in the replicative cycle of the positive-sense RNA viruses. Initiation of translation has been reported to be dependent on the interaction of the VPg with cellular translation factors, such as the eIF4F initiation complex.
This complex is a key player in the initiation of translation of eukaryotic mRNA. It assembles the pre-initiation complex to the mRNA via its interaction with the cap-structure and the eIF3 (Gingras et al., 1999, Kapp and Lorsch, 2004). The eIF4F complex consists of an RNA helicase (eIF4A), a factor directly interacting with the cap-structure of the mRNA (eIF4E), and a binding protein (eIF4G) to other cellular translation modulators such as the poly(A)-binding protein (PABP) and eIF3.
In FCV and MNV, the VPg interacts with the initiation factors eIF4E and eIF3, whereas the VPg of NV interacts with the eIF4E (Goodfellow et al., 2005). In both FCV and MNV, translation of the polyprotein precursor depends on the presence of a functional component of the eIF4F complex, the RNA helicase. Indeed, treatment of FCV infected cells with a small molecule inhibitor of eIF4A (hippuristanol) blocks translation of viral proteins. Hippuristanol interacts with the C-terminus of eIF4A with subsequent inhibition of its NTPase hence helicase activity (Bordeleau et al., 2006). Importantly, hippuristanol is expected to have toxic effects on the cells due to the inhibition of host cell – as well as calicivirus – protein synthesis.
In a similar way, pateamine stimulates both helicase and NTPase activities of eIF4A, dysregulating its function within the translation complex (Bordeleau et al., 2006). In this sense, using inhibitors of components of the eIF4F complex, i.e. hippuristanol (Bordeleau et al., 2006), or dysregulators of the eIF4A function, i.e. pateamine (Low et al., 2005) is a promising approach.
Another important regulator of translation is the interaction of the 5′- and 3′-untranslated regions of the viral genome with cellular host factors involved in protein translation. At both ends of the calicivirus genome, untranslated regions (UTR) of variable size between genogroups and genera are present. Although experimental evidence for the direct interaction of these structures with the translation machinery is still lacking, disruption of the structures in a model of infection in vitro led to strong impairment of the replication of murine norovirus. These data support the concept of an important role for the structured termini of the calicivirus genome in the viral multiplication cycle. These properties of the calicivirus genome being highly structured are very common amongst many other positive-strand RNA viruses (Goodfellow et al., 2000, Haasnoot et al., 2002) such as picornaviruses (i.e. the CRE-element (Goodfellow et al., 2003), or the IRES (Pelletier and Sonenberg, 1988, Xiang et al., 1995)) or flaviviruses (Tsukiyama-Kohara et al., 1992, Wang et al., 1993, Gritsun and Gould, 2006a, Gritsun and Gould, 2006b, Gritsun and Gould, 2007, Proutski et al., 1997a, Proutski et al., 1997b, Proutski et al., 1999).
The 5′- and 3′-UTR of the viral genomes are highly structured sequences that fold in a functional conformation. In order to ensure appropriate folding and subsequent interaction with viral structural and non-structural proteins, cellular chaperones such as the polypyrimidine tract-binding protein (PTB) are required (Herschlag, 1995, Treiber and Williamson, 2001). The PTB is expressed in the nucleus and cytoplasm acting as a regulator of splicing pathways (Ghetti et al., 1992, Lin and Patton, 1995). The PTB was postulated to work as a chaperone for the viral genomic RNA, implicated in folding to secondary structures located at the 5′-terminus of the genome. In FCV, the PTB interacts specifically with sequences located at the 5′-UTR of the genome (Karakasiliotis et al., 2006), and is required for multiplication of the FCV in host cells. In murine norovirus, the PTB binds to a polypyrimidine tract located within the 3′-UTR of the viral genome (Bailey et al., 2010a). This interaction was reported not be essential for virus replication, although being an important regulator of viral fitness and virulence (Bailey et al., 2010a).
Host cell factors such as Unr (Boussadia et al., 2003), poly(rC)-binding protein (PCBP) (Parsley et al., 1997), La autoantigen (Meerovitch et al., 1993), hnRNP A1 (Huang and Lai, 2001) and poly(A)-binding protein (PABP) interact with the viral genome of a variety of RNA viruses. The host cellular factors modulate translation and processing of the viral polyproteins, but also the translation of host cell mRNA. In human pathogenic Norwalk virus, PTB (Gutierrez-Escolano et al., 2003), La autoantigen, hnRNP and PCB-2 interact with both the 5′-UTR and the 3′-UTR (for La and PTB) (Gutierrez-Escolano et al., 2003), although no functional roles of those proteins in norovirus replication have been described so far.
Taken together, this knowledge on translation initiation in the Caliciviridae offers interesting possibilities (i.e. peptides, nucleic acids) to target the interaction of the viral genome and/or the VPg with cellular host factors of translation, with subsequent inhibition of viral protein synthesis.
2.5. Targeting replicative viral enzymes
2.5.1. The viral chymotrypsin-like protease
The calicivirus chymotrypsin-like protease plays a central role in viral replication. The molecular mechanisms underlying the catalytic activity and specificity of the norovirus, sapovirus, murine norovirus and feline calicivirus protease have been extensively investigated. Knowledge on the regulation of calicivirus polyprotein processing is an important prerequisite for the development of antiviral drugs to control calicivirus infection.
The viral protease is involved in early steps of replication. Its active form is the viral protein NS6pro, or a protein precursor consisting of the viral protease and polymerase NS6proNS7pol. The cleavage pattern of the calicivirus polyprotein precursor by the viral protease has been investigated both in cell-free expression systems (Belliot et al., 2003, Liu et al., 1999, Seah et al., 2003) and in mammalian cells (Chang et al., 2006, Seah et al., 1999, Seah et al., 2003, Sosnovtsev et al., 2006). Processing of human Norwalk virus in Huh-7 cells (Chang et al., 2006) and of a murine norovirus strain in the macrophage-like cell line RAW264.7 (Sosnovtsev et al., 2006) have shown that both the NS6proNS7pol precursor and the NS6pro are present in infected cells. A similar observation was reported for a related non-human pathogenic calicivirus strain, the rabbit hemorrhagic disease virus (RHDV, genus Lagovirus). However, in another human pathogenic calicivirus, the sapovirus (SV), NS6proNS7pol is the only protein detected in a cell-free system (Oka et al., 2005a, Oka et al., 2005b). In the case of the pathogenic feline calicivirus (FCV), the NS6proNS7pol precursor is detected in mammalian cells (Sosnovtsev et al., 2002, Sosnovtsev et al., 1998, Sosnovtseva et al., 1999). In accordance with our observations (Scheffler et al., 2007), evidence for autocatalytic cleavage of norovirus NS6pro from the polyprotein precursor has been presented (Belliot et al., 2003, Chang et al., 2006, Liu et al., 1999, Seah et al., 1999, Sosnovtsev et al., 2006).
The cleavage specificity and efficiency of norovirus and sapovirus NS6pro and NS6proNS7pol were examined in vitro using peptides bearing the cleavage sites of the polyprotein. In norovirus, Gln-Gly is required between the P1 and P1′ positions framing the scissile bond of the peptide to be cleaved by the protease. In sapovirus, cleavage specificity and efficiency seem to be modulated by the positions P4, P1 and P2′ of the scissile bond.
The cleavage specificity observed is explained by studies on the structure of the norovirus protease (Nakamura et al., 2005, Zeitler et al., 2006) and sapovirus protease (Robel et al., 2008). In the sapovirus NS6pro, the catalytic triad consists of Cys116-His31-Glu52. His131 is located at the bottom of the S1 pocket. Its role is to interact with the P1 side chain of the substrate, as observed in norovirus NS6pro (see below). Another His residue, His102 plays an important role by holding Glu52 in its proper position in the triad. This feature does not exist in norovirus 3C-like protease.
The crystal structure of norovirus NS6pro has revealed that five residues govern the specificity of substrate binding to the protease: His157 and Ala160 in the S1 pocket, and Ile109, Arg112, and Val114 in the S2 pocket (Nakamura et al., 2005). In the S1 pocket, the His157 residue interacts with the Gln (P1) residue at the scissile bond (Zeitler et al., 2006). His157 is important for substrate binding since its mutation severely reduces the activity of NS6pro (Someya et al., 2002). In other RNA viruses (human rhinovirus, hepatitis A virus, poliovirus, and coronavirus), similar observations were made, with the His at the S1 site interacting through its imidazole ring with the carboxamide chains of the P1 residue (Allaire et al., 1994, Anand et al., 2002, Anand et al., 2003, Bergmann et al., 1997, Matthews et al., 1994, Mosimann et al., 1997, Yang et al., 2003). Furthermore, the presence of a small residue (Gly or Ala) at the P1′ positions of the scissile bonds in norovirus polyprotein is consistent with the limited volume allowed for the P1′ residue in the S1 pocket of norovirus protease (Nakamura et al., 2005). In contrast, the S2 hydrophobic pocket of the norovirus protease is large enough to accommodate the side chain of a bulky hydrophobic amino acid such as Phe, Met, or Leu (Nakamura et al., 2005). Similarly, the binding sites for the P3 and P4 residues in norovirus NS6pro are reported to be large enough to accommodate residues of various sizes (i.e., Ser, Gly, His, or Thr in the P3 pocket, and Phe, Met, Ala, or Leu in the P4 pocket) (Nakamura et al., 2005).
Because of the specificity of cleavage of the scissile bonds of the polyprotein of the various calicivirus genera, peptidomimetics or substrate-like peptides may constitute an interesting strategy for the inhibition of viral replication. Another possible approach may be to use inhibitors that block the protease active site. For the implementation of such an approach at least two important barriers have to be crossed. These are (i) the large size of the peptidic compounds, and their possible poor bioavailability and (ii) the limited in vivo stability of peptides. Nevertheless, because of the extensive knowledge generated on the structure and function of the chymotrypsin-like protease, targeting the viral protease may be one of the most promising approaches to inhibit calicivirus replication.
2.5.2. The RNA-dependent RNA polymerase
The RdRp is involved in synthesis of the genomic, subgenomic and antigenomic RNA of the calicivirus. Structural analysis of the norovirus, sapovirus and RHDV RdRp has revealed a right-hand structure typical for template-dependent polymerases (Biswal et al., 2005, Choi et al., 2004, Ferrer-Orta et al., 2004, Ng et al., 2002, Ng et al., 2004). Interestingly, structural similarities that exist in the shortened loop section of RHDV, sapovirus and norovirus NS7pol (Ng et al., 2002) suggest similar mechanisms for activation of RNA elongation in Caliciviridae. Nevertheless, sapovirus NS7pol displays functional similarities to norovirus NS7pol, but not to vesivirus (FCV) and lagovirus (RHDV) RdRp. This relates mainly to the ability of both sapovirus and norovirus NS7pol to initiate RNA synthesis de novo on heteropolymeric templates, with a primer-dependent initiation of RNA synthesis on homopolymeric templates, and a terminal transferase activity with a preference for CTP (Rohayem et al., 2006b, Fullerton et al., 2007).
The potential strategies to inhibit the calicivirus RdRp fall in two categories: nucleoside analogue inhibitors and non-nucleoside inhibitors.
Nucleoside analogs all display similar mechanisms of inhibition of the RdRp activity. The nucleosides penetrate the cell and undergo phosphorylation by triphosphate, giving nucleotides. They are then incorporated in the polymerized nucleic acid chain elongated by the RdRp, leading to chain termination. In a preliminary investigation and in order to validate both polymerases of the HuCV as targets for antiviral design, we have tested five different nucleoside analogs for their potential as inhibitors of the activity of the HuCV polymerases. As shown in Fig. 6 , both the 2′-arauridine-5′-triphosphate (TriLink technologies) and the 3′-deoxyuridine-5′-triphosphate (TriLink technologies) displayed potent inhibitor activities against the HuCV polymerases at a concentration of 50 μM. In contrast, other nucleoside analogs did not show any activity against the HuCV polymerase. This provisional data is in favor of using the HuCV RNA-dependent RNA polymerase as a target for the development of antiviral drugs (Fig. 6).
Fig. 6.
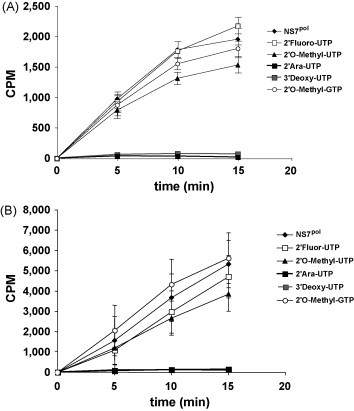
Inhibition of the activity of norovirus and sapovirus RNA-dependent RNA polymerases by nucleoside analogues. (A) Assessment of the activity of the norovirus RNA-dependent RNA polymerase was determined as previously described (Rohayem et al., 2006a, Rohayem et al., 2006b). (B) Assessment of the activity of the sapovirus RNA-dependent RNA polymerase NS7pol was determined as previously described (Fullerton et al., 2007). 2′-Fluor-UTP, 2′-fluorouridine-5′-triphosphate. 2′-O-Methyl-UTP, 2′-O-methyluridine-5′-triphosphate. 2′Ara-UTP, 2′-arauridine-5′-triphosphate; 3′-deoxy-UTP; 3′-deoxyuridine-5′-triphosphate. 2′-O-Methyl-GTP, 2′-O,methylguanidine-5′-triphosphate. The mean and the standard error of the mean of three independent experiments are shown. CPM, counts per minute. Each of the compounds was incubated at a concentration of 50 μM.
Another substance of interest is ribavirin. Ribavirin (1-α-d-ribofuranosyl-1,2,4-triazole-3-carboxamide) efficiently inhibits norovirus replication in vitro (Chang and George, 2007). Ribavirin is a synthetic guanosine analogue, has a broad range of antiviral activity against RNA but also DNA viruses. In the NV replicon system, Ribavirin was reported to display an EC50 of 40 μM (Chang and George, 2007). This value was 100 μM when MNV-infected RAW264.7 cells were treated (Chang and George, 2007). In feline calicivirus, Ribavirin was however not effective in vivo (Povey, 1978a, Povey, 1978b). Although the experimental data in vitro is encouraging, and because Ribavirin has a lot of adverse effects and a very narrow therapeutic index, it is at this time point not suitable for the treatment of a disease such as viral gastroenteritis with almost no lethal outcome.
2.5.3. The VPg
The VPg is putatively linked to the genomic RNA in caliciviruses, although experimental evidence of this linkage is still lacking for non-human pathogenic caliciviruses. The VPg is involved through uridylylation in a protein-primed dependent initiation of RNA synthesis of antigenomic and presumably genomic RNA. Uridylylation is thought to occur at tyrosine 24 in the protein, as shown through mutation analysis in the FCV reverse genetics system (Mitra et al., 2004). Uridylylation has been experimentally proven for noroviruses (Belliot et al., 2008, Rohayem et al., 2006b), and for lagoviruses (Machin et al., 2001).
Uridylylation is thought to involve the viral RdRp, the VPg, as well as the polyadenylated viral genome. Whether or not the antigenomic viral RNA is involved remains unknown, but by analogy with poliovirus it could potentially be.
Prevention of uridylylation of the viral genome could also be an interesting antiviral approach, through inhibition of the interaction of the VPg with the RdRp, and/or with the viral genome, resulting in the shut-off of synthesis of the viral genomic RNA.
2.5.4. The NTPase
Another possible target for antiviral therapy is the calicivirus NTPase. The NTPase (NS3) displays the canonical motives of viral NTPases as described for picornaviruses and flaviviruses, for instance. The occurrence of the three motives A (GXXGXGKS/T), B (DD/E) and C (KXXXFXSXXXXXS/TTN) classifies the calicivirus NS3NTPase in the superfamily 3 of RNA helicases (Pfister and Wimmer, 2001), although experimental evidence in vitro and in cell culture for its putative RNA unwinding activity is still lacking. The recombinant NS3NTPase displays NTPase activity in vitro, but does not bind RNA, nor does it unwind RNA–RNA or RNA–DNA-duplexes (Pfister and Wimmer, 2001). Here, future experimental evidence may help in designing innovative antiviral strategies targeting this important enzyme of calicivirus replication.
By analogy with the flavivirus NTPase/helicase that has been extensively studied and characterized functionally and structurally, inhibitors of the NTPase/helicase may act by (i) competitive inhibition and blockade of the NTP binding site, or (ii) by conformational changes through immobilization of the switch region, (iii) by competitive inhibition of RNA binding, or (iv) by inhibition of unwinding via steric blockade of translocation of the NTPase/helicase along the template RNA.
2.6. Targeting assembly of the replication complex
The viral enzymes of replication interact in a complex of multiple components, constituting the replication complex. This complex may be transient, or relatively stable; and the ability of the viral enzyme to interact with any one partner of interest may be significantly influenced by other complex members. In caliciviruses, the replication complex is predicted to involve mainly three viral enzymes: the RNA-dependent RNA polymerase, the viral protease, and the NTPase/helicase. In addition to the viral genomic/antigenomic and/or subgenomic/anti-subgenomic RNA, structural viral proteins may be included, but also the VPg, or the N-terminal protein. Experimental data using noroviruses has shown that the N-terminal protein interacts in a vesicular complex with the VAP-A-Protein (vesicle-associated membrane protein-associated protein A) leading to inhibition of intracellular protein trafficking (Ettayebi and Hardy, 2003). In FCV, the interaction of NS2 with NS3NTPase, NS4, NS5VPg and NS6-7propol has been demonstrated in vitro (Kaiser et al., 2006). The FCV NS6-7propol was also reported to interact with the NS5VPg, interacting itself with the structural protein VP1 (Kaiser et al., 2006).
So far, the determinants of assembly and structure of the replication complex, as well at its role in regulating the life cycle of the virus remain poorly understood. Moreover, our recent results generated within the VIZIER-Consortium indicate that small molecules inhibiting viral replication in mammalian cells might possibly target the replication complex, specifically the interaction of the viral enzymes within the complex. The effect of small molecules would mainly be to destabilize the complex, subsequently inhibiting the replication of the viral genome, hence the multiplication of the virus within the cells. Therefore, knowledge of the functional and structural determinants of interaction and assembly of the replication complex could provide a cornerstone for developing new antiviral strategies.
Our work within the VIZIER-Consortium has contributed to decipher the activity of the norovirus RdRp, showing that the active form of the viral RdRp of noroviruses is a homodimer displaying a positive cooperativity (Hogbom et al., 2009). Our results also show that homodimerization of norovirus NS7 monomers is concentration-dependent, and that the apparent KD of homodimerization lies within the nM range. Moreover, the monomers display a positive cooperativity in vitro. Similar observations on the NS6-7propol of FCV cultivated in cell systems were reported (Kaiser et al., 2006).
Here, the development of antiviral strategies aiming at destabilizing the interaction between the NS7pol monomers is of interest. Another approach may rely on targeting the interaction of the structural proteins within the replication complex.
2.7. Targeting the post-replicative steps
Following replication of the viral genome, further steps such as the assembly of the capsid protein, budding of the virions in the endoplasmic reticulum, transport and maturation of the virions in the ER and the Golgi complex, and vesicle fusion lead to release of the mature virions. Recent experimental data on the feline calicivirus has shown that upon infection, 3 viral non-structural proteins (p32, p30, and p39) co-localize to the ER, leading to a morphological changes of the ER membranes (Bailey et al., 2010b). It remains however unclear whether these rearrangements are incriminated in the assembly of the replication complex, or not.
For other caliciviruses, these steps are poorly understood. Therefore, antiviral strategies targeting the post-replicative steps of the genome will not be developed, until substantial knowledge of those steps is generated.
3. Vaccines and other approaches
Immunity to human pathogenic caliciviruses is very poorly understood, mainly because of a lack of cell culture systems to investigate neutralization of virus multiplication by serum or intestinal secretory antibodies. However, volunteer challenge studies have established that human pathogenic caliciviruses and particularly noroviruses induce two forms of immunity: a short-term and a long-term immunity (Atmar et al., 2008, Parrino et al., 1977, Wyatt et al., 1974). Short-term immunity is virus-specific; individuals infected with a norovirus strain are resistant to re-infection with the same strain for up to 14 weeks following primary infection. Long-term immunity, in contrast, confers resistance to infection that lasts about 34 months following primary infection. Another interesting aspect relates to the type of cell-mediated immunity following infection with the human pathogenic calicivirus, i.e. norovirus. Volunteer challenge studies have shown that the cell-mediated immune response to norovirus infection displays a Th-1 dominant pattern (Lindesmith et al., 2005). In animal models, important insights into norovirus immunity to infection as well as pathogenesis have been generated recently based on the surrogate MNV (Liu et al., 2009, Bailey et al., 2008, McCartney et al., 2008, Mumphrey et al., 2007).
Virus-like particles that mimic structurally and antigenically the native capsid protein have been extensively used to explore the function and reactivity of noroviruses and sapoviruses (Guo et al., 2001b, Hansman et al., 2005a, Hansman et al., 2006, Jiang et al., 1999b, Oka et al., 2006). VLPs are produced to relatively high yields in baculovirus recombinant expression systems or in the human endothelial kidney cell line HEK-293T (Taube et al., 2005) assembling in the absence of the VP2, although the VP2 seems to enhance VLPs stability (Green et al., 1997, Jiang et al., 1992). Furthermore, feeding of volunteers and laboratory animals with norovirus virus-like particles generated in tomato plants (Huang et al., 2005b) or potato plants (Tacket et al., 2000) led to an immune response (IgG and IgA) with variable intensity, depending on the adjuvant used. These and other previous data on vaccination (Guerrero et al., 2001, Ball et al., 1998) to control infection by human caliciviruses is encouraging and deserves further development and evaluation.
For FCV, a vaccine is available (Radford et al., 2006). The development of an efficient vaccine was possible because FCV can be amplified in CRFK cells, and a small model (felines) is also available. FCV vaccines use either attenuated or inactivated virus. Furthermore, dual-strain FCV vaccines consisting of virulent and attenuated strains have already been successfully implemented in clinical trials (Huang et al., 2010). VLPs have also been effective in generating neutralizing antibodies in rabbits against various FCV strains, indicating that they could represent an interesting alternative to attenuated or inactivated viruses for production of FCV vaccines (Di Martino et al., 2007).
In a recent study (LoBue et al., 2006), multivalent norovirus VLPs adjuvanted with alphavirus or CpG DNA were administrated to mice. The VLPs induced a mucosal and systemic immune response, and a reduction of viral titer in immunized mice following MNV challenge. Sera from immunized mice proved to be protective against MNV when transferred to immunodeficient mice. Similarly, VLPs representing RHDV and bearing a T cell epitope from ovalbumin were shown to be able to elicit a T cell immune response in dendritic cells, and to induce a cellular immune response in mice (Crisci et al., 2009).
These data are encouraging and highlight the potential of the VLP-based approach for the development of calicivirus vaccines.
Another experimental approach is based on inhibition of norovirus replication by interferon. Using a reverse genetics system for NV, Chang and George (2007) have shown that IFN-α and IFN-γ inhibit translation and replication of calicivirus in a dose-dependent manner in vitro. As a correlate, the susceptibility of MNV to IFN was observed in vitro (Changotra et al., 2009, Wobus et al., 2004) and also in vivo (Karst et al., 2003). Although the observed EC50 was in the micromolar range, these data are of interest in the context of the antiviral effect of IFN on caliciviruses.
A more general approach for treatment of human pathogenic calicivirus infection has relied on the substitution of electrolytes and water loss by per os or per enteral rehydration, representing hence a symptomatic treatment of the infection (Anderson, 2010), but not an antiviral approach. The positive effect of nitazoxanide in controlling norovirus infections has also been described (Rossignol and El-Gohary, 2006).
4. Conclusions and future prospects
The possibility to cultivate members of three genera, noroviruses, vesiviruses, and recoviruses within the family Caliciviridae, is an important step towards better understanding of the replication cycle of these viruses and also the development of effective antiviral strategies. Nevertheless, there is clearly a need for culture systems to propagate the human pathogenic variants of noroviruses and sapoviruses in mammalian cells. This has hampered the development of antiviral approaches as well as vaccines, but also the in-depth study of the pathogenicity of the human pathogenic calicivirus, accounting for the slow progress in the field.
One of the potential problems caused by the treatment of calicivirus infections will be, as observed for other RNA viruses with a positive stranded genome, the emergence of drug resistance. The major problem lies in the high genomic variability of the strains in particular for human pathogenic noroviruses, as well as the accelerating emergence of new variants. Therefore, antiviral strategies should aim at targeting conserved regions in the viral genome and possibly not exposed to immunological pressure leading to immune escape. Because of the short duration of the disease, treatment of calicivirus infection may focus on the containment of the infection in the context of outbreaks, as well as long-term shedding of the virions after infection, as observed following infection with noroviruses.
Acknowledgements
This work was supported by the European Consortium “Vizier” (www.vizier-europe.org) as well as the Jürgen Manchot Stiftung (for Mirko Bergmann, Julia Gebhardt and Jacques Rohayem).
References
- Allaire M., Chernaia M.M., Malcolm B.A., James M.N. Picornaviral 3C cysteine proteinases have a fold similar to chymotrypsin-like serine proteinases. Nature. 1994;369:72–76. doi: 10.1038/369072a0. [DOI] [PubMed] [Google Scholar]
- Anand K., Palm G.J., Mesters J.R., Siddell S.G., Ziebuhr J., Hilgenfeld R. Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra alpha-helical domain. Embo J. 2002;21:3213–3224. doi: 10.1093/emboj/cdf327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand K., Ziebuhr J., Wadhwani P., Mesters J.R., Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003;300:1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- Anderson E.J. Prevention and treatment of viral diarrhea in pediatrics. Expert Rev. Anti Infect. Ther. 2010;8:205–217. doi: 10.1586/eri.10.1. [DOI] [PubMed] [Google Scholar]
- Asanaka M., Atmar R.L., Ruvolo V., Crawford S.E., Neill F.H., Estes M.K. Replication and packaging of Norwalk virus RNA in cultured mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 2005 doi: 10.1073/pnas.0408529102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atmar R.L., Opekun A.R., Gilger M.A., Estes M.K., Crawford S.E., Neill F.H., Graham D.Y. Norwalk virus shedding after experimental human infection. Emerg. Infect. Dis. 2008;14:1553–1557. doi: 10.3201/eid1410.080117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey D., Karakasiliotis I., Vashist S., Chung L.M., Reese J., McFadden N., Benson A., Yarovinsky F., Simmonds P., Goodfellow I. Functional analysis of RNA structures present at the 3′ extremity of the murine norovirus genome: the variable polypyrimidine tract plays a role in viral virulence. J. Virol. 2010;84:2859–2870. doi: 10.1128/JVI.02053-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey D., Kaiser W.J., Hollinshead M., Moffat K., Chaudhry Y., Wileman T., Sosnovtsev S.V., Goodfellow I.G. Feline calicivirus p32, p39 and p30 proteins localize to the endoplasmic reticulum to initiate replication complex formation. J. Gen. Virol. 2010;91:739–749. doi: 10.1099/vir.0.016279-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey D., Thackray L.B., Goodfellow I.G. A single amino acid substitution in the murine norovirus capsid protein is sufficient for attenuation in vivo. J. Virol. 2008;82:7725–7728. doi: 10.1128/JVI.00237-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball J.M., Hardy M.E., Atmar R.L., Conner M.E., Estes M.K. Oral immunization with recombinant Norwalk virus-like particles induces a systemic and mucosal immune response in mice. J. Virol. 1998;72:1345–1353. doi: 10.1128/jvi.72.2.1345-1353.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belliot G., Sosnovtsev S.V., Chang K.O., McPhie P., Green K.Y. Nucleotidylylation of the VPg protein of a human norovirus by its proteinase-polymerase precursor protein. Virology. 2008;374:33–49. doi: 10.1016/j.virol.2007.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belliot G., Sosnovtsev S.V., Mitra T., Hammer C., Garfield M., Green K.Y. In vitro proteolytic processing of the MD145 norovirus ORF1 nonstructural polyprotein yields stable precursors and products similar to those detected in calicivirus-infected cells. J. Virol. 2003;77:10957–10974. doi: 10.1128/JVI.77.20.10957-10974.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann E.M., Mosimann S.C., Chernaia M.M., Malcolm B.A., James M.N. The refined crystal structure of the 3C gene product from hepatitis A virus: specific proteinase activity and RNA recognition. J. Virol. 1997;71:2436–2448. doi: 10.1128/jvi.71.3.2436-2448.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhella D., Gatherer D., Chaudhry Y., Pink R., Goodfellow I.G. Structural insights into calicivirus attachment and uncoating. J. Virol. 2008;82:8051–8058. doi: 10.1128/JVI.00550-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswal B.K., Cherney M.M., Wang M., Chan L., Yannopoulos C.G., Bilimoria D., Nicolas O., Bedard J., James M.N.G. Crystal structures of the RNA dependent RNA polymerase genotype 2a of hepatitis C virus reveal two conformations and suggest mechanisms of inhibition by non-nucleoside inhibitors. J. Biol. Chem. 2005;280:18202–18210. doi: 10.1074/jbc.M413410200. [DOI] [PubMed] [Google Scholar]
- Bitko V., Barik S. Phenotypic silencing of cytoplasmic genes using sequence-specific double-stranded short interfering RNA and its application in the reverse genetics of wild type negative-strand RNA viruses. BMC Microbiol. 2001;1:34. doi: 10.1186/1471-2180-1-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitko V., Musiyenko A., Shulyayeva O., Barik S. Inhibition of respiratory viruses by nasally administered siRNA. Nat. Med. 2005;11:50–55. doi: 10.1038/nm1164. [DOI] [PubMed] [Google Scholar]
- Boden D., Pusch O., Lee F., Tucker L., Ramratnam B. Human immunodeficiency virus type 1 escape from RNA interference. J. Virol. 2003;77:11531–11535. doi: 10.1128/JVI.77.21.11531-11535.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bok K., Cavanaugh V.J., Matson D.O., Gonzalez-Molleda L., Chang K.O., Zintz C., Smith A.W., Iversen P., Green K.Y., Campbell A.E. Inhibition of norovirus replication by morpholino oligomers targeting the 5′-end of the genome. Virology. 2008;380:328–337. doi: 10.1016/j.virol.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordeleau M.E., Cencic R., Lindqvist L., Oberer M., Northcote P., Wagner G., Pelletier J. RNA-mediated sequestration of the RNA helicase eIF4A by Pateamine A inhibits translation initiation. Chem. Biol. 2006;13:1287–1295. doi: 10.1016/j.chembiol.2006.10.005. [DOI] [PubMed] [Google Scholar]
- Boussadia O., Niepmann M., Creancier L., Prats A.C., Dautry F., Jacquemin-Sablon H. Unr is required in vivo for efficient initiation of translation from the internal ribosome entry sites of both rhinovirus and poliovirus. J. Virol. 2003;77:3353–3359. doi: 10.1128/JVI.77.6.3353-3359.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S., Lou Z., Tan M., Chen Y., Liu Y., Zhang Z., Zhang X.C., Jiang X., Li X., Rao Z. Structural basis for the recognition of blood group trisaccharides by norovirus. J. Virol. 2007;81:5949–5957. doi: 10.1128/JVI.00219-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capucci L., Fusi P., Lavazza A., Pacciarini M.L., Rossi C. Detection and preliminary characterization of a new rabbit calicivirus related to rabbit hemorrhagic disease virus but nonpathogenic. J. Virol. 1996;70:8614–8623. doi: 10.1128/jvi.70.12.8614-8623.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarty S., Hutson A.M., Estes M.K., Prasad B.V. Evolutionary trace residues in noroviruses: importance in receptor binding, antigenicity, virion assembly, and strain diversity. J. Virol. 2005;79:554–568. doi: 10.1128/JVI.79.1.554-568.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang K.O., George D.W. Interferons and ribavirin effectively inhibit Norwalk virus replication in replicon-bearing cells. J. Virol. 2007;81:12111–12118. doi: 10.1128/JVI.00560-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang K.O., Kim Y., Green K.Y., Saif L.J. Cell-culture propagation of porcine enteric calicivirus mediated by intestinal contents is dependent on the cyclic AMP signaling pathway. Virology. 2002;304:302–310. doi: 10.1006/viro.2002.1665. [DOI] [PubMed] [Google Scholar]
- Chang K.O., Sosnovtsev S.V., Belliot G., Kim Y., Saif L.J., Green K.Y. Bile acids are essential for porcine enteric calicivirus replication in association with down-regulation of signal transducer and activator of transcription 1. Proc. Natl. Acad. Sci. U.S.A. 2004;101:8733–8738. doi: 10.1073/pnas.0401126101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang K.O., Sosnovtsev S.V., Belliot G., King A.D., Green K.Y. Stable expression of a Norwalk virus RNA replicon in a human hepatoma cell line. Virology. 2006 doi: 10.1016/j.virol.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Changotra H., Jia Y., Moore T.N., Liu G., Kahan S.M., Sosnovtsev S.V., Karst S.M. Type I and type II interferons inhibit the translation of murine norovirus proteins. J. Virol. 2009;83:5683–5692. doi: 10.1128/JVI.00231-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry Y., Skinner M.A., Goodfellow I.G. Recovery of genetically defined murine norovirus in tissue culture by using a fowlpox virus expressing T7 RNA polymerase. J. Gen. Virol. 2007;88:2091–2100. doi: 10.1099/vir.0.82940-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R., Neill J.D., Estes M.K., Prasad B.V. X-ray structure of a native calicivirus: structural insights into antigenic diversity and host specificity. Proc. Natl. Acad. Sci. U.S.A. 2006;103:8048–8053. doi: 10.1073/pnas.0600421103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R., Neill J.D., Noel J.S., Hutson A.M., Glass R.I., Estes M.K., Prasad B.V. Inter- and intragenus structural variations in caliciviruses and their functional implications. J. Virol. 2004;78:6469–6479. doi: 10.1128/JVI.78.12.6469-6479.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K.H., Groarke J.M., Young D.C., Kuhn R.J., Smith J.L., Pevear D.C., Rossmann M.G. The structure of the RNA-dependent RNA polymerase from bovine viral diarrhea virus establishes the role of GTP in de novo initiation. Proc. Natl. Acad. Sci. U.S.A. 2004;101:4425–4430. doi: 10.1073/pnas.0400660101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke I.N., Lambden P.R. Organization and expression of calicivirus genes. J. Infect. Dis. 2000;181(Suppl. 2):S309–S316. doi: 10.1086/315575. [DOI] [PubMed] [Google Scholar]
- Coyne K.P., Jones B.R., Kipar A., Chantrey J., Porter C.J., Barber P.J., Dawson S., Gaskell R.M., Radford A.D. Lethal outbreak of disease associated with feline calicivirus infection in cats. Vet. Rec. 2006;158:544–550. doi: 10.1136/vr.158.16.544. [DOI] [PubMed] [Google Scholar]
- Cox C., Cao S., Lu Y. Enhanced detection and study of murine norovirus-1 using a more efficient microglial cell line. Virol. J. 2009;6:196. doi: 10.1186/1743-422X-6-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisci E., Almanza H., Mena I., Cordoba L., Gomez-Casado E., Caston J.R., Fraile L., Barcena J., Montoya M. Chimeric calicivirus-like particles elicit protective anti-viral cytotoxic responses without adjuvant. Virology. 2009;387:303–312. doi: 10.1016/j.virol.2009.02.045. [DOI] [PubMed] [Google Scholar]
- Daughenbaugh K.F., Fraser C.S., Hershey J.W., Hardy M.E. The genome-linked protein VPg of the Norwalk virus binds eIF3, suggesting its role in translation initiation complex recruitment. Embo J. 2003;22:2852–2859. doi: 10.1093/emboj/cdg251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daughenbaugh K.F., Wobus C.E., Hardy M.E. VPg of murine norovirus binds translation initiation factors in infected cells. Virol. J. 2006;3:33. doi: 10.1186/1743-422X-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Martino B., Marsilio F., Roy P. Assembly of feline calicivirus-like particle and its immunogenicity. Vet. Microbiol. 2007;120:173–178. doi: 10.1016/j.vetmic.2006.10.021. [DOI] [PubMed] [Google Scholar]
- Ettayebi K., Hardy M.E. Norwalk virus nonstructural protein p48 forms a complex with the SNARE regulator VAP-A and prevents cell surface expression of vesicular stomatitis virus G protein. J. Virol. 2003;77:11790–11797. doi: 10.1128/JVI.77.21.11790-11797.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas T., Dufour J., Jiang X., Sestak K. Detection of norovirus-, sapovirus- and rhesus enteric calicivirus-specific antibodies in captive juvenile macaques. J. Gen. Virol. 2010;91:734–738. doi: 10.1099/vir.0.015263-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas T., Sestak K., Wei C., Jiang X. Characterization of a rhesus monkey calicivirus representing a new genus of Caliciviridae. J. Virol. 2008;82:5408–5416. doi: 10.1128/JVI.00070-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas T., Zhong W.M., Jing Y., Huang P.W., Espinosa S.M., Martinez N., Morrow A.L., Ruiz-Palacios G.M., Pickering L.K., Jiang X. Genetic diversity among sapoviruses. Arch. Virol. 2004;149:1309–1323. doi: 10.1007/s00705-004-0296-9. [DOI] [PubMed] [Google Scholar]
- Fauquet C.M., Mayo M.A., Manioloff J., Desselberger U., Ball L.A., editors. Virus Taxonomy, VIIIth Report of the International Committee on Taxonomy of Viruses. Elsevier/Academic Press; London: 2005. [Google Scholar]
- Feng X., Jiang X. Library screen for inhibitors targeting norovirus binding to histo-blood group antigen receptors. Antimicrob. Agents Chemother. 2007;51:324–331. doi: 10.1128/AAC.00627-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer-Orta C., Arias A., Perez-Luque R., Escarmis C., Domingo E., Verdaguer N. Structure of foot-and-mouth disease virus RNA-dependent RNA polymerase and its complex with a template-primer RNA. J. Biol. Chem. 2004;279:47212–47221. doi: 10.1074/jbc.M405465200. [DOI] [PubMed] [Google Scholar]
- Foley J., Hurley K., Pesavento P.A., Poland A., Pedersen N.C. Virulent systemic feline calicivirus infection: local cytokine modulation and contribution of viral mutants. J. Feline Med. Surg. 2006;8:55–61. doi: 10.1016/j.jfms.2005.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrester N.L., Boag B., Moss S.R., Turner S.L., Trout R.C., White P.J., Hudson P.J., Gould E.A. Long-term survival of New Zealand rabbit haemorrhagic disease virus RNA in wild rabbits, revealed by RT-PCR and phylogenetic analysis. J. Gen. Virol. 2003;84:3079–3086. doi: 10.1099/vir.0.19213-0. [DOI] [PubMed] [Google Scholar]
- Forrester N.L., Trout R.C., Gould E.A. Benign circulation of rabbit haemorrhagic disease virus on Lambay Island, Eire. Virology. 2007;358:18–22. doi: 10.1016/j.virol.2006.09.011. [DOI] [PubMed] [Google Scholar]
- Fullerton S.W., Blaschke M., Coutard B., Gebhardt J., Gorbalenya A., Canard B., Tucker P.A., Rohayem J. Structural and functional characterization of sapovirus RNA-dependent RNA polymerase. J. Virol. 2007;81:1858–1871. doi: 10.1128/JVI.01462-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel G., Nordmann A., Stein D.A., Iversen P.L., Klenk H.D. Morpholino oligomers targeting the PB1 and NP genes enhance the survival of mice infected with highly pathogenic influenza A H7N7 virus. J. Gen. Virol. 2008;89:939–948. doi: 10.1099/vir.0.83449-0. [DOI] [PubMed] [Google Scholar]
- Gallimore C.I., Green J., Lewis D., Richards A.F., Lopman B.A., Hale A.D., Eglin R., Gray J.J., Brown D.W. Diversity of noroviruses cocirculating in the north of England from 1998 to 2001. J. Clin. Microbiol. 2004;42:1396–1401. doi: 10.1128/JCM.42.4.1396-1401.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallimore C.I., Iturriza-Gomara M., Lewis D., Cubitt D., Cotterill H., Gray J.J. Characterization of sapoviruses collected in the United Kingdom from 1989 to 2004. J. Med. Virol. 2006;78:673–682. doi: 10.1002/jmv.20592. [DOI] [PubMed] [Google Scholar]
- Ge Q., Filip L., Bai A., Nguyen T., Eisen H.N., Chen J. Inhibition of influenza virus production in virus-infected mice by RNA interference. Proc. Natl. Acad. Sci. U.S.A. 2004;101:8676–8681. doi: 10.1073/pnas.0402486101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Q., McManus M.T., Nguyen T., Shen C.H., Sharp P.A., Eisen H.N., Chen J. RNA interference of influenza virus production by directly targeting mRNA for degradation and indirectly inhibiting all viral RNA transcription. Proc. Natl. Acad. Sci. U.S.A. 2003;100:2718–2723. doi: 10.1073/pnas.0437841100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerondopoulos A., Jackson T., Monaghan P., Doyle N., Roberts L.O. Murine norovirus-1 cell entry is mediated through a non-clathrin, non-caveolae, dynamin and cholesterol dependent pathway. J. Gen. Virol. 2010 doi: 10.1099/vir.0.016717-0. (Epub Feb 10) [DOI] [PubMed] [Google Scholar]
- Ghetti A., Pinol-Roma S., Michael W.M., Morandi C., Dreyfuss G. hnRNP I, the polypyrimidine tract-binding protein: distinct nuclear localization and association with hnRNAs. Nucleic Acids Res. 1992;20:3671–3678. doi: 10.1093/nar/20.14.3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson T., Higgins D., Thompson J. IGBMC; Strasbourg: 1999. ClustalX, Heidelberg: EMBL, Cork: UCC. [Google Scholar]
- Gingras A.C., Raught B., Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- Goodfellow I., Chaudhry Y., Gioldasi I., Gerondopoulos A., Natoni A., Labrie L., Laliberte J.F., Roberts L. Calicivirus translation initiation requires an interaction between VPg and eIF 4 E. EMBO Rep. 2005;6:968–972. doi: 10.1038/sj.embor.7400510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodfellow I., Chaudhry Y., Richardson A., Meredith J., Almond J.W., Barclay W., Evans D.J. Identification of a cis-acting replication element within the poliovirus coding region. J. Virol. 2000;74:4590–4600. doi: 10.1128/jvi.74.10.4590-4600.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodfellow I.G., Kerrigan D., Evans D.J. Structure and function analysis of the poliovirus cis-acting replication element (CRE) RNA. 2003;9:124–137. doi: 10.1261/rna.2950603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray J. Assays for virus infection. In: Cahn J., editor. Virus Culture: A Practical Approach. Oxford University Press; Oxford: 1999. [Google Scholar]
- Green K.Y. Caliciviridae: the noroviruses. In: Knipe D.M., Howley P.M., Griffin D.E., Lamb R.A., Martin M.A., Roizman B., Straus S.E., editors. Fields Virology. Lippincott Williams and Wilkins; 2007. [Google Scholar]
- Green K.Y., Ando T., Balayan M.S., Berke T., Clarke I.N., Estes M.K., Matson D.O., Nakata S., Neill J.D., Studdert M.J., Thiel H.J. Taxonomy of the caliciviruses. J. Infect. Dis. 2000;181(Suppl. 2):S322–S330. doi: 10.1086/315591. [DOI] [PubMed] [Google Scholar]
- Green K.Y., Chanock R.M., Kapikian A.Z. Human caliciviruses. In: Knipe D.M., Howley P.M., Griffin D.E., Lamb R.A., Martin M.A., Roizman B., Straus S.E., editors. Fields Virology. Lippincott Williams and Wilkins; 2001. [Google Scholar]
- Green K.Y., Kapikian A.Z., Valdesuso J., Sosnovtsev S., Treanor J.J., Lew J.F. Expression and self-assembly of recombinant capsid protein from the antigenically distinct Hawaii human calicivirus. J. Clin. Microbiol. 1997;35:1909–1914. doi: 10.1128/jcm.35.7.1909-1914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green K.Y., Lew J.F., Jiang X., Kapikian A.Z., Estes M.K. Comparison of the reactivities of baculovirus-expressed recombinant Norwalk virus capsid antigen with those of the native Norwalk virus antigen in serologic assays and some epidemiologic observations. J. Clin. Microbiol. 1993;31:2185–2191. doi: 10.1128/jcm.31.8.2185-2191.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gritsun T.S., Gould E.A. The 3′ untranslated region of tick-borne flaviviruses originated by the duplication of long repeat sequences within the open reading frame. Virology. 2006;354:217–223. doi: 10.1016/j.virol.2006.03.052. [DOI] [PubMed] [Google Scholar]
- Gritsun T.S., Gould E.A. The 3′ untranslated regions of Kamiti River virus and cell fusing agent virus originated by self-duplication. J. Gen. Virol. 2006;87:2615–2619. doi: 10.1099/vir.0.81950-0. [DOI] [PubMed] [Google Scholar]
- Gritsun T.S., Gould E.A. Direct repeats in the flavivirus 3′ untranslated region; a strategy for survival in the environment? Virology. 2007;358:258–265. doi: 10.1016/j.virol.2006.09.033. [DOI] [PubMed] [Google Scholar]
- Guerrero R.A., Ball J.M., Krater S.S., Pacheco S.E., Clements J.D., Estes M.K. Recombinant Norwalk virus-like particles administered intranasally to mice induce systemic and mucosal (fecal and vaginal) immune responses. J. Virol. 2001;75:9713–9722. doi: 10.1128/JVI.75.20.9713-9722.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M., Evermann J.F., Saif L.J. Detection and molecular characterization of cultivable caliciviruses from clinically normal mink and enteric caliciviruses associated with diarrhea in mink. Arch. Virol. 2001;146:479–493. doi: 10.1007/s007050170157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M., Qian Y., Chang K.O., Saif L.J. Expression and self-assembly in baculovirus of porcine enteric calicivirus capsids into virus-like particles and their use in an enzyme-linked immunosorbent assay for antibody detection in swine. J. Clin. Microbiol. 2001;39:1487–1493. doi: 10.1128/JCM.39.4.1487-1493.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Escolano A.L., Vazquez-Ochoa M., Escobar-Herrera J., Hernandez-Acosta J. La, PTB, and PAB proteins bind to the 3(′) untranslated region of Norwalk virus genomic RNA. Biochem. Biophys. Res. Commun. 2003;311:759–766. doi: 10.1016/j.bbrc.2003.10.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haasnoot J., Westerhout E.M., Berkhout B. RNA interference against viruses: strike and counterstrike. Nat. Biotechnol. 2007;25:1435–1443. doi: 10.1038/nbt1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haasnoot P.C., Olsthoorn R.C., Bol J.F. The Brome mosaic virus subgenomic promoter hairpin is structurally similar to the iron-responsive element and functionally equivalent to the minus-strand core promoter stem-loop C. RNA. 2002;8:110–122. doi: 10.1017/s1355838202012074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansman G.S., Katayama K., Oka T., Natori K., Takeda N. Mutational study of sapovirus expression in insect cells. Virol. J. 2005;2:13. doi: 10.1186/1743-422X-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansman G.S., Oka T., Katayama K., Takeda N. Enhancement of sapovirus recombinant capsid protein expression in insect cells. FEBS Lett. 2006;580:4047–4050. doi: 10.1016/j.febslet.2006.06.040. [DOI] [PubMed] [Google Scholar]
- Hansman G.S., Oka T., Katayama K., Takeda N. Human sapoviruses: genetic diversity, recombination, and classification. Rev. Med. Virol. 2007;17:133–141. doi: 10.1002/rmv.533. [DOI] [PubMed] [Google Scholar]
- Hansman G.S., Takeda N., Oka T., Oseto M., Hedlund K.O., Katayama K. Intergenogroup recombination in sapoviruses. Emerg. Infect. Dis. 2005;11:1916–1920. doi: 10.3201/eid1112.050722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson K.S. Murine norovirus, a recently discovered and highly prevalent viral agent of mice. Lab. Anim. (NY) 2008;37:314–320. doi: 10.1038/laban0708-314. [DOI] [PubMed] [Google Scholar]
- Hennessy E.P., Green A.D., Connor M.P., Darby R., MacDonald P. Norwalk virus infection and disease is associated with ABO histo-blood group type. J. Infect. Dis. 2003;188:176–177. doi: 10.1086/375829. [DOI] [PubMed] [Google Scholar]
- Herschlag D. RNA chaperones and the RNA folding problem. J. Biol. Chem. 1995;270:20871–20874. doi: 10.1074/jbc.270.36.20871. [DOI] [PubMed] [Google Scholar]
- Hogbom M., Jager K., Robel I., Unge T., Rohayem J. The active form of the norovirus RNA-dependent RNA polymerase is a homodimer with cooperative activity. J. Gen. Virol. 2009;90:281–291. doi: 10.1099/vir.0.005629-0. [DOI] [PubMed] [Google Scholar]
- Hsu C.C., Riley L.K., Livingston R.S. Molecular characterization of three novel murine noroviruses. Virus Genes. 2007;34:147–155. doi: 10.1007/s11262-006-0060-1. [DOI] [PubMed] [Google Scholar]
- Huang C., Hess J., Gill M., Hustead D. A dual-strain feline calicivirus vaccine stimulates broader cross-neutralization antibodies than a single-strain vaccine and lessens clinical signs in vaccinated cats when challenged with a homologous feline calicivirus strain associated with virulent systemic disease. J. Feline Med. Surg. 2010;12:129–137. doi: 10.1016/j.jfms.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P., Morrow A.L., Jiang X. The carbohydrate moiety and high molecular weight carrier of histo-blood group antigens are both required for norovirus-receptor recognition. Glycoconj. J. 2009;26:1085–1096. doi: 10.1007/s10719-009-9229-x. [DOI] [PubMed] [Google Scholar]
- Huang P., Farkas T., Marionneau S., Zhong W., Ruvoen-Clouet N., Morrow A.L., Altaye M., Pickering L.K., Newburg D.S., LePendu J., Jiang X. Noroviruses bind to human ABO, Lewis, and secretor histo-blood group antigens: identification of 4 distinct strain-specific patterns. J. Infect. Dis. 2003;188:19–31. doi: 10.1086/375742. [DOI] [PubMed] [Google Scholar]
- Huang P., Farkas T., Zhong W., Tan M., Thornton S., Morrow A.L., Jiang X. Norovirus and histo-blood group antigens: demonstration of a wide spectrum of strain specificities and classification of two major binding groups among multiple binding patterns. J. Virol. 2005;79:6714–6722. doi: 10.1128/JVI.79.11.6714-6722.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P., Lai M.M. Heterogeneous nuclear ribonucleoprotein a1 binds to the 3′-untranslated region and mediates potential 5′-3′-end cross talks of mouse hepatitis virus RNA. J. Virol. 2001;75:5009–5017. doi: 10.1128/JVI.75.11.5009-5017.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z., Elkin G., Maloney B.J., Beuhner N., Arntzen C.J., Thanavala Y., Mason H.S. Virus-like particle expression and assembly in plants: hepatitis B and Norwalk viruses. Vaccine. 2005;23:1851–1858. doi: 10.1016/j.vaccine.2004.11.017. [DOI] [PubMed] [Google Scholar]
- Humphrey T.J., Cruickshank J.G., Cubitt W.D. An outbreak of calicivirus associated gastroenteritis in an elderly persons home. A possible zoonosis? J. Hyg. (Lond.) 1984;93:293–299. doi: 10.1017/s0022172400064822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley K.E., Pesavento P.A., Pedersen N.C., Poland A.M., Wilson E., Foley J.E. An outbreak of virulent systemic feline calicivirus disease. J. Am. Vet. Med. Assoc. 2004;224:241–249. doi: 10.2460/javma.2004.224.241. [DOI] [PubMed] [Google Scholar]
- Hutson A.M., Atmar R.L., Graham D.Y., Estes M.K. Norwalk virus infection and disease is associated with ABO histo-blood group type. J. Infect. Dis. 2002;185:1335–1337. doi: 10.1086/339883. [DOI] [PubMed] [Google Scholar]
- Jackson A.L., Bartz S.R., Schelter J., Kobayashi S.V., Burchard J., Mao M., Li B., Cavet G., Linsley P.S. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- Jiang X., Espul C., Zhong W.M., Cuello H., Matson D.O. Characterization of a novel human calicivirus that may be a naturally occurring recombinant. Arch. Virol. 1999;144:2377–2387. doi: 10.1007/s007050050651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X., Wang M., Graham D.Y., Estes M.K. Expression, self-assembly, and antigenicity of the Norwalk virus capsid protein. J. Virol. 1992;66:6527–6532. doi: 10.1128/jvi.66.11.6527-6532.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X., Zhong W., Kaplan M., Pickering L.K., Matson D.O. Expression and characterization of Sapporo-like human calicivirus capsid proteins in baculovirus. J. Virol. Methods. 1999;78:81–91. doi: 10.1016/s0166-0934(98)00169-4. [DOI] [PubMed] [Google Scholar]
- Johansson P.J., Bergentoft K., Larsson P.A., Magnusson G., Widell A., Thorhagen M., Hedlund K.O. A nosocomial sapovirus-associated outbreak of gastroenteritis in adults. Scand. J. Infect. Dis. 2005;37:200–204. doi: 10.1080/00365540410020974. [DOI] [PubMed] [Google Scholar]
- Kaiser W.J., Chaudhry Y., Sosnovtsev S.V., Goodfellow I.G. Analysis of protein–protein interactions in the feline calicivirus replication complex. J. Gen. Virol. 2006;87:363–368. doi: 10.1099/vir.0.81456-0. [DOI] [PubMed] [Google Scholar]
- Kapp L.D., Lorsch J.R. The molecular mechanics of eukaryotic translation. Annu. Rev. Biochem. 2004;73:657–704. doi: 10.1146/annurev.biochem.73.030403.080419. [DOI] [PubMed] [Google Scholar]
- Karakasiliotis I., Chaudhry Y., Roberts L.O., Goodfellow I.G. Feline calicivirus replication: requirement for polypyrimidine tract-binding protein is temperature-dependent. J. Gen. Virol. 2006;87:3339–3347. doi: 10.1099/vir.0.82153-0. [DOI] [PubMed] [Google Scholar]
- Karst S.M., Wobus C.E., Lay M., Davidson J., Virgin H.W.t. STAT1-dependent innate immunity to a Norwalk-like virus. Science. 2003;299:1575–1578. doi: 10.1126/science.1077905. [DOI] [PubMed] [Google Scholar]
- Katayama H., Okuma K., Furumai H., Ohgaki S. Series of surveys for enteric viruses and indicator organisms in Tokyo Bay after an event of combined sewer overflow. Water Sci. Technol. 2004;50:259–262. [PubMed] [Google Scholar]
- Keswick B.H., Satterwhite T.K., Johnson P.C., DuPont H.L., Secor S.L., Bitsura J.A., Gary G.W., Hoff J.C. Inactivation of Norwalk virus in drinking water by chlorine. Appl. Environ. Microbiol. 1985;50:261–264. doi: 10.1128/aem.50.2.261-264.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkwood C.D., Streitberg R. Calicivirus shedding in children after recovery from diarrhoeal disease. J. Clin. Virol. 2008;43:346–348. doi: 10.1016/j.jcv.2008.08.001. [DOI] [PubMed] [Google Scholar]
- Koopmans M. Outbreaks of viral gastroenteritis: what's new in 2004? Curr. Opin. Infect. Dis. 2005;18:295–299. doi: 10.1097/01.qco.0000171922.55075.ad. [DOI] [PubMed] [Google Scholar]
- Koopmans M., Duizer E. Foodborne viruses: an emerging problem. Int. J. Food Microbiol. 2004;90:23–41. doi: 10.1016/S0168-1605(03)00169-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopmans M., von Bonsdorff C.H., Vinje J., de Medici D., Monroe S. Foodborne viruses. FEMS Microbiol. Rev. 2002;26:187–205. doi: 10.1111/j.1574-6976.2002.tb00610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent S., Vautherot J.F., Le Gall G., Madelaine M.F., Rasschaert D. Structural, antigenic and immunogenic relationships between European brown hare syndrome virus and rabbit haemorrhagic disease virus. J. Gen. Virol. 1997;78(Pt 11):2803–2811. doi: 10.1099/0022-1317-78-11-2803. [DOI] [PubMed] [Google Scholar]
- Li C.X., Parker A., Menocal E., Xiang S., Borodyansky L., Fruehauf J.H. Delivery of RNA interference. Cell Cycle. 2006;5:2103–2109. doi: 10.4161/cc.5.18.3192. [DOI] [PubMed] [Google Scholar]
- Lin C.H., Patton J.G. Regulation of alternative 3′ splice site selection by constitutive splicing factors. RNA. 1995;1:234–245. [PMC free article] [PubMed] [Google Scholar]
- Lin X., Ruan X., Anderson M.G., McDowell J.A., Kroeger P.E., Fesik S.W., Shen Y. siRNA-mediated off-target gene silencing triggered by a 7 nt complementation. Nucleic Acids Res. 2005;33:4527–4535. doi: 10.1093/nar/gki762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindesmith L., Moe C., Lependu J., Frelinger J.A., Treanor J., Baric R.S. Cellular and humoral immunity following Snow Mountain virus challenge. J. Virol. 2005;79:2900–2909. doi: 10.1128/JVI.79.5.2900-2909.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindesmith L., Moe C., Marionneau S., Ruvoen N., Jiang X., Lindblad L., Stewart P., LePendu J., Baric R. Human susceptibility and resistance to Norwalk virus infection. Nat. Med. 2003;9:548–553. doi: 10.1038/nm860. [DOI] [PubMed] [Google Scholar]
- Lindqvist R., Andersson Y., Lindback J., Wegscheider M., Eriksson Y., Tidestrom L., Lagerqvist-Widh A., Hedlund K.O., Lofdahl S., Svensson L., Norinder A. A one-year study of foodborne illnesses in the municipality of Uppsala, Sweden. Emerg. Infect. Dis. 2001;7:588–592. doi: 10.3201/eid0707.010742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G., Kahan S.M., Jia Y., Karst S.M. Primary high-dose murine norovirus 1 infection fails to protect from secondary challenge with homologous virus. J. Virol. 2009;83:6963–6968. doi: 10.1128/JVI.00284-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B.L., Viljoen G.J., Clarke I.N., Lambden P.R. Identification of further proteolytic cleavage sites in the Southampton calicivirus polyprotein by expression of the viral protease in E. coli. J. Gen. Virol. 1999;80(Pt 2):291–296. doi: 10.1099/0022-1317-80-2-291. [DOI] [PubMed] [Google Scholar]
- LoBue A.D., Lindesmith L., Yount B., Harrington P.R., Thompson J.M., Johnston R.E., Moe C.L., Baric R.S. Multivalent norovirus vaccines induce strong mucosal and systemic blocking antibodies against multiple strains. Vaccine. 2006;24:5220–5234. doi: 10.1016/j.vaccine.2006.03.080. [DOI] [PubMed] [Google Scholar]
- Lopman B.A., Brown D.W., Koopmans M. Human caliciviruses in Europe. J. Clin. Virol. 2002;24:137–160. doi: 10.1016/s1386-6532(01)00243-8. [DOI] [PubMed] [Google Scholar]
- Low W.K., Dang Y., Schneider-Poetsch T., Shi Z., Choi N.S., Merrick W.C., Romo D., Liu J.O. Inhibition of eukaryotic translation initiation by the marine natural product pateamine A. Mol. Cell. 2005;20:709–722. doi: 10.1016/j.molcel.2005.10.008. [DOI] [PubMed] [Google Scholar]
- Lupfer C., Stein D.A., Mourich D.V., Tepper S.E., Iversen P.L., Pastey M. Inhibition of influenza A H3N8 virus infections in mice by morpholino oligomers. Arch. Virol. 2008;153:929–937. doi: 10.1007/s00705-008-0067-0. [DOI] [PubMed] [Google Scholar]
- Luttermann C., Meyers G. A bipartite sequence motif induces translation reinitiation in feline calicivirus RNA. J. Biol. Chem. 2007;282:7056–7065. doi: 10.1074/jbc.M608948200. [DOI] [PubMed] [Google Scholar]
- Lynch M., Painter J., Woodruff R., Braden C. Surveillance for foodborne-disease outbreaks—United States, 1998–2002. MMWR Surveill. Summ. 2006;55:1–42. [PubMed] [Google Scholar]
- Machin A., Martin Alonso J.M., Parra F. Identification of the amino acid residue involved in rabbit hemorrhagic disease virus VPg uridylylation. J. Biol. Chem. 2001;276:27787–27792. doi: 10.1074/jbc.M100707200. [DOI] [PubMed] [Google Scholar]
- Makino A., Shimojima M., Miyazawa T., Kato K., Tohya Y., Akashi H. Junctional adhesion molecule 1 is a functional receptor for feline calicivirus. J. Virol. 2006;80:4482–4490. doi: 10.1128/JVI.80.9.4482-4490.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marionneau S., Cailleau-Thomas A., Rocher J., Le Moullac-Vaidye B., Ruvoen N., Clement M., Le Pendu J. ABH and Lewis histo-blood group antigens, a model for the meaning of oligosaccharide diversity in the face of a changing world. Biochimie. 2001;83:565–573. doi: 10.1016/s0300-9084(01)01321-9. [DOI] [PubMed] [Google Scholar]
- Marionneau S., Ruvoen N., Le Moullac-Vaidye B., Clement M., Cailleau-Thomas A., Ruiz-Palacois G., Huang P., Jiang X., Le Pendu J. Norwalk virus binds to histo-blood group antigens present on gastroduodenal epithelial cells of secretor individuals. Gastroenterology. 2002;122:1967–1977. doi: 10.1053/gast.2002.33661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martella V., Campolo M., Lorusso E., Cavicchio P., Camero M., Bellacicco A.L., Decaro N., Elia G., Greco G., Corrente M., Desario C., Arista S., Banyai K., Koopmans M., Buonavoglia C. Norovirus in captive lion cub (Panthera leo) Emerg. Infect. Dis. 2007;13:1071–1073. doi: 10.3201/eid1307.070268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martella V., Lorusso E., Decaro N., Elia G., Radogna A., D’Abramo M., Desario C., Cavalli A., Corrente M., Camero M., Germinario C.A., Banyai K., Di Martino B., Marsilio F., Carmichael L.E., Buonavoglia C. Detection and molecular characterization of a canine norovirus. Emerg. Infect. Dis. 2008;14:1306–1308. doi: 10.3201/eid1408.080062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matson D.O., Berke T., Dinulos M.B., Poet E., Zhong W.M., Dai X.M., Jiang X., Golding B., Smith A.W. Partial characterization of the genome of nine animal caliciviruses. Arch. Virol. 1996;141:2443–2456. doi: 10.1007/BF01718642. [DOI] [PubMed] [Google Scholar]
- Matson D.O., Estes M.K., Glass R.I., Bartlett A.V., Penaranda M., Calomeni E., Tanaka T., Nakata S., Chiba S. Human calicivirus-associated diarrhea in children attending day care centers. J. Infect. Dis. 1989;159:71–78. doi: 10.1093/infdis/159.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura Y., Tohya Y., Onuma M., Roerink F., Mochizuki M., Sugimura T. Expression and processing of the canine calicivirus capsid precursor. J. Gen. Virol. 2000;81:195–199. doi: 10.1099/0022-1317-81-1-195. [DOI] [PubMed] [Google Scholar]
- Matthews D.A., Smith W.W., Ferre R.A., Condon B., Budahazi G., Sisson W., Villafranca J.E., Janson C.A., McElroy H.E., Gribskov C.L. Structure of human rhinovirus 3C protease reveals a trypsin-like polypeptide fold, RNA-binding site, and means for cleaving precursor polyprotein. Cell. 1994;77:761–771. doi: 10.1016/0092-8674(94)90059-0. [DOI] [PubMed] [Google Scholar]
- Mauroy A., Scipioni A., Mathijs E., Miry C., Ziant D., Thys C., Thiry E. Noroviruses and sapoviruses in pigs in Belgium. Arch. Virol. 2008;153:1927–1931. doi: 10.1007/s00705-008-0189-4. [DOI] [PubMed] [Google Scholar]
- Mayo M.A. A summary of taxonomic changes recently approved by ICTV. Arch. Virol. 2002;147:1655–1663. doi: 10.1007/s007050200039. [DOI] [PubMed] [Google Scholar]
- McCartney S.A., Thackray L.B., Gitlin L., Gilfillan S., Virgin Iv H.W., Colonna M. MDA-5 recognition of a murine norovirus. PLoS Pathog. 2008;4:e1000108. doi: 10.1371/journal.ppat.1000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meerovitch K., Svitkin Y.V., Lee H.S., Lejbkowicz F., Kenan D.J., Chan E.K., Agol V.I., Keene J.D., Sonenberg N. La autoantigen enhances and corrects aberrant translation of poliovirus RNA in reticulocyte lysate. J. Virol. 1993;67:3798–3807. doi: 10.1128/jvi.67.7.3798-3807.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra T., Sosnovtsev S.V., Green K.Y. Mutagenesis of tyrosine 24 in the VPg protein is lethal for feline calicivirus. J. Virol. 2004;78:4931–4935. doi: 10.1128/JVI.78.9.4931-4935.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosimann S.C., Cherney M.M., Sia S., Plotch S., James M.N. Refined X-ray crystallographic structure of the poliovirus 3C gene product. J. Mol. Biol. 1997;273:1032–1047. doi: 10.1006/jmbi.1997.1306. [DOI] [PubMed] [Google Scholar]
- Moss S.R., Turner S.L., Trout R.C., White P.J., Hudson P.J., Desai A., Armesto M., Forrester N.L., Gould E.A. Molecular epidemiology of Rabbit haemorrhagic disease virus. J. Gen. Virol. 2002;83:2461–2467. doi: 10.1099/0022-1317-83-10-2461. [DOI] [PubMed] [Google Scholar]
- Muller B., Klemm U., Mas Marques A., Schreier E. Genetic diversity and recombination of murine noroviruses in immunocompromised mice. Arch. Virol. 2007;152:1709–1719. doi: 10.1007/s00705-007-0989-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumphrey S.M., Changotra H., Moore T.N., Heimann-Nichols E.R., Wobus C.E., Reilly M.J., Moghadamfalahi M., Shukla D., Karst S.M. Murine norovirus 1 infection is associated with histopathological changes in immunocompetent hosts, but clinical disease is prevented by STAT1-dependent interferon responses. J. Virol. 2007;81:3251–3263. doi: 10.1128/JVI.02096-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata T., Katsushima N., Mizuta K., Muraki Y., Hongo S., Matsuzaki Y. Prolonged norovirus shedding in infants <or=6 months of age with gastroenteritis. Pediatr. Infect. Dis. J. 2007;26:46–49. doi: 10.1097/01.inf.0000247102.04997.e0. [DOI] [PubMed] [Google Scholar]
- Nakamura K., Someya Y., Kumasaka T., Ueno G., Yamamoto M., Sato T., Takeda N., Miyamura T., Tanaka N. A norovirus protease structure provides insights into active and substrate binding site integrity. J. Virol. 2005;79:13685–13693. doi: 10.1128/JVI.79.21.13685-13693.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata S., Chiba S., Terashima H., Nakao T. Prevalence of antibody to human calicivirus in Japan and Southeast Asia determined by radioimmunoassay. J. Clin. Microbiol. 1985;22:519–521. doi: 10.1128/jcm.22.4.519-521.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napthine, S., Lever, R.A., Powell, M.L., Jackson, R.J., Brown, T.D., Brierley, I., 2009. Expression of the VP2 protein of murine norovirus by a translation termination-reinitiation strategy. PLoS One 4 (Epub Dec 22). [DOI] [PMC free article] [PubMed]
- Neill J.D., Meyer R.F., Seal B.S. The capsid protein of vesicular exanthema of swine virus serotype A48: relationship to the capsid protein of other animal caliciviruses. Virus Res. 1998;54:39–50. doi: 10.1016/s0168-1702(98)00013-6. [DOI] [PubMed] [Google Scholar]
- Ng K.K., Cherney M.M., Vazquez A.L., Machin A., Alonso J.M., Parra F., James M.N. Crystal structures of active and inactive conformations of a caliciviral RNA-dependent RNA polymerase. J. Biol. Chem. 2002;277:1381–1387. doi: 10.1074/jbc.M109261200. [DOI] [PubMed] [Google Scholar]
- Ng K.K., Pendas-Franco N., Rojo J., Boga J.A., Machin A., Alonso J.M., Parra F. Crystal structure of norwalk virus polymerase reveals the carboxyl terminus in the active site cleft. J. Biol. Chem. 2004;279:16638–16645. doi: 10.1074/jbc.M400584200. [DOI] [PubMed] [Google Scholar]
- Numata K., Hardy M.E., Nakata S., Chiba S., Estes M.K. Molecular characterization of morphologically typical human calicivirus Sapporo. Arch. Virol. 1997;142:1537–1552. doi: 10.1007/s007050050178. [DOI] [PubMed] [Google Scholar]
- Oka T., Hansman G.S., Katayama K., Ogawa S., Nagata N., Miyamura T., Takeda N. Expression of sapovirus virus-like particles in mammalian cells. Arch. Virol. 2006;151:399–404. doi: 10.1007/s00705-005-0613-y. [DOI] [PubMed] [Google Scholar]
- Oka T., Katayama K., Ogawa S., Hansman G.S., Kageyama T., Miyamura T., Takeda N. Cleavage activity of the sapovirus 3C-like protease in Escherichia coli. Arch. Virol. 2005;150:2539–2548. doi: 10.1007/s00705-005-0591-0. [DOI] [PubMed] [Google Scholar]
- Oka T., Katayama K., Ogawa S., Hansman G.S., Kageyama T., Ushijima H., Miyamura T., Takeda N. Proteolytic processing of sapovirus ORF1 polyprotein. J. Virol. 2005;79:7283–7290. doi: 10.1128/JVI.79.12.7283-7290.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver S.L., Asobayire E., Charpilienne A., Cohen J., Bridger J.C. Complete genomic characterization and antigenic relatedness of genogroup III, genotype 2 bovine noroviruses. Arch. Virol. 2007;152:257–272. doi: 10.1007/s00705-006-0856-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver S.L., Asobayire E., Dastjerdi A.M., Bridger J.C. Genomic characterization of the unclassified bovine enteric virus Newbury agent-1 (Newbury1) endorses a new genus in the family Caliciviridae. Virology. 2006;350:240–250. doi: 10.1016/j.virol.2006.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver S.L., Dastjerdi A.M., Wong S., El-Attar L., Gallimore C., Brown D.W., Green J., Bridger J.C. Molecular characterization of bovine enteric caliciviruses: a distinct third genogroup of noroviruses (Norwalk-like viruses) unlikely to be of risk to humans. J. Virol. 2003;77:2789–2798. doi: 10.1128/JVI.77.4.2789-2798.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver S.L., Wood E., Asobayire E., Wathes D.C., Brickell J.S., Elschner M., Otto P., Lambden P.R., Clarke I.N., Bridger J.C. Serotype 1 and 2 bovine noroviruses are endemic in cattle in the United kingdom and Germany. J. Clin. Microbiol. 2007;45:3050–3052. doi: 10.1128/JCM.02015-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxford J., Kelly L., Davies S., Lambkin R. Antiviral testing. In: Cahn J., editor. Virus Culture: A Practical Approach. Oxford University Press; Oxford: 1999. [Google Scholar]
- Paessler S., Rijnbrand R., Stein D.A., Ni H., Yun N.E., Dziuba N., Borisevich V., Seregin A., Ma Y., Blouch R., Iversen P.L., Zacks M.A. Inhibition of alphavirus infection in cell culture and in mice with antisense morpholino oligomers. Virology. 2008;376:357–370. doi: 10.1016/j.virol.2008.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.I., Jeong C., Kim H.H., Park S.H., Park S.J., Hyun B.H., Yang D.K., Kim S.K., Kang M.I., Cho K.O. Molecular epidemiology of bovine noroviruses in South Korea. Vet. Microbiol. 2007;124:125–133. doi: 10.1016/j.vetmic.2007.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.I., Jeong C., Park S.J., Kim H.H., Jeong Y.J., Hyun B.H., Chun Y.H., Kang M.I., Cho K.O. Molecular detection and characterization of unclassified bovine enteric caliciviruses in South Korea. Vet. Microbiol. 2008;130:371–379. doi: 10.1016/j.vetmic.2008.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrino T.A., Schreiber D.S., Trier J.S., Kapikian A.Z., Blacklow N.R. Clinical immunity in acute gastroenteritis caused by Norwalk agent. N. Engl. J. Med. 1977;297:86–89. doi: 10.1056/NEJM197707142970204. [DOI] [PubMed] [Google Scholar]
- Parsley T.B., Towner J.S., Blyn L.B., Ehrenfeld E., Semler B.L. Poly (rC) binding protein 2 forms a ternary complex with the 5′-terminal sequences of poliovirus RNA and the viral 3CD proteinase. RNA. 1997;3:1124–1134. [PMC free article] [PubMed] [Google Scholar]
- Pedersen N.C., Elliott J.B., Glasgow A., Poland A., Keel K. An isolated epizootic of hemorrhagic-like fever in cats caused by a novel and highly virulent strain of feline calicivirus. Vet. Microbiol. 2000;73:281–300. doi: 10.1016/S0378-1135(00)00183-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelkmans L., Helenius A. Insider information: what viruses tell us about endocytosis. Curr. Opin. Cell Biol. 2003;15:414–422. doi: 10.1016/s0955-0674(03)00081-4. [DOI] [PubMed] [Google Scholar]
- Pelletier J., Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- Pfister T., Wimmer E. Polypeptide p41 of a Norwalk-like virus is a nucleic acid-independent nucleoside triphosphatase. J. Virol. 2001;75:1611–1619. doi: 10.1128/JVI.75.4.1611-1619.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan T.G., Okitsu S., Muller W.E., Kohno H., Ushijima H. Novel recombinant sapovirus, Japan. Emerg. Infect. Dis. 2006;12:865–867. doi: 10.3201/eid1205.051608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Povey R.C. Effect of orally administered ribavirin on experimental feline calicivirus infection in cats. Am. J. Vet. Res. 1978;39:1337–1341. [PubMed] [Google Scholar]
- Povey R.C. In vitro antiviral efficacy of ribavirin against feline calicivirus, feline viral rhinotracheitis virus, and canine parainfluenza virus. Am. J. Vet. Res. 1978;39:175–178. [PubMed] [Google Scholar]
- Pöyry T.A., Kaminski A., Connell E.J., Fraser C.S., Jackson R.J. The mechanism of an exceptional case of reinitiation after translation of a long ORF reveals why such events do not generally occur in mammalian mRNA translation. Genes Dev. 2007;21:3149–3162. doi: 10.1101/gad.439507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proutski V., Gaunt M.W., Gould E.A., Holmes E.C. Secondary structure of the 3′-untranslated region of yellow fever virus: implications for virulence, attenuation and vaccine development. J. Gen. Virol. 1997;78(Pt 7):1543–1549. doi: 10.1099/0022-1317-78-7-1543. [DOI] [PubMed] [Google Scholar]
- Proutski V., Gould E.A., Holmes E.C. Secondary structure of the 3′ untranslated region of flaviviruses: similarities and differences. Nucleic Acids Res. 1997;25:1194–1202. doi: 10.1093/nar/25.6.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proutski V., Gritsun T.S., Gould E.A., Holmes E.C. Biological consequences of deletions within the 3′-untranslated region of flaviviruses may be due to rearrangements of RNA secondary structure. Virus Res. 1999;64:107–123. doi: 10.1016/s0168-1702(99)00079-9. [DOI] [PubMed] [Google Scholar]
- Radford A.D., Coyne K.P., Dawson S., Porter C.J., Gaskell R.M. Feline calicivirus. Vet. Res. 2007;38:319–335. doi: 10.1051/vetres:2006056. [DOI] [PubMed] [Google Scholar]
- Radford A.D., Dawson S., Coyne K.P., Porter C.J., Gaskell R.M. The challenge for the next generation of feline calicivirus vaccines. Vet. Microbiol. 2006;117:14–18. doi: 10.1016/j.vetmic.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Reuter G., Farkas T., Berke T., Jiang X., Matson D.O., Szucs G. Molecular epidemiology of human calicivirus gastroenteritis outbreaks in Hungary, 1998 to 2000. J. Med. Virol. 2002;68:390–398. doi: 10.1002/jmv.10216. [DOI] [PubMed] [Google Scholar]
- Robel I., Gebhardt J., Mesters J.R., Gorbalenya A., Coutard B., Canard B., Hilgenfeld R., Rohayem J. Functional characterization of the cleavage specificity of the sapovirus chymotrypsin-like protease. J. Virol. 2008;82:8085–8093. doi: 10.1128/JVI.00693-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockx B.H., Vennema H., Hoebe C.J., Duizer E., Koopmans M.P. Association of histo-blood group antigens and susceptibility to norovirus infections. J. Infect. Dis. 2005;191:749–754. doi: 10.1086/427779. [DOI] [PubMed] [Google Scholar]
- Rohayem J., Jager K., Robel I., Scheffler U., Temme A., Rudolph W. Characterization of norovirus 3Dpol RNA-dependent RNA polymerase activity and initiation of RNA synthesis. J. Gen. Virol. 2006;87:2621–2630. doi: 10.1099/vir.0.81802-0. [DOI] [PubMed] [Google Scholar]
- Rohayem J., Munch J., Rethwilm A. Evidence of recombination in the norovirus capsid gene. J. Virol. 2005;79:4977–4990. doi: 10.1128/JVI.79.8.4977-4990.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohayem J., Robel I., Jager K., Scheffler U., Rudolph W. Protein-primed and de novo initiation of RNA synthesis by norovirus 3Dpol. J. Virol. 2006;80:7060–7069. doi: 10.1128/JVI.02195-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossignol J.F., El-Gohary Y.M. Nitazoxanide in the treatment of viral gastroenteritis: a randomized double-blind placebo-controlled clinical trial. Aliment Pharmacol. Ther. 2006;24:1423–1430. doi: 10.1111/j.1365-2036.2006.03128.x. [DOI] [PubMed] [Google Scholar]
- Rydell G.E., Nilsson J., Rodriguez-Diaz J., Ruvoen-Clouet N., Svensson L., Le Pendu J., Larson G. Human noroviruses recognize sialyl Lewis × neoglycoprotein. Glycobiology. 2009;19:309–320. doi: 10.1093/glycob/cwn139. [DOI] [PubMed] [Google Scholar]
- Sakuma Y., Chiba S., Kogasaka R., Terashima H., Nakamura S., Horino K., Nakao T. Prevalence of antibody to human calicivirus in general population of northern Japan. J. Med. Virol. 1981;7:221–225. doi: 10.1002/jmv.1890070306. [DOI] [PubMed] [Google Scholar]
- Scheffler U., Rudolph W., Gebhardt J., Rohayem J. Differential cleavage of the norovirus polyprotein precursor by two active forms of the viral protease. J. Gen. Virol. 2007;88:2013–2018. doi: 10.1099/vir.0.82797-0. [DOI] [PubMed] [Google Scholar]
- Schuffenecker I., Ando T., Thouvenot D., Lina B., Aymard M. Genetic classification of “Sapporo-like viruses”. Arch. Virol. 2001;146:2115–2132. doi: 10.1007/s007050170024. [DOI] [PubMed] [Google Scholar]
- Seah E.L., Marshall J.A., Wright P.J. Open reading frame 1 of the Norwalk-like virus Camberwell: completion of sequence and expression in mammalian cells. J. Virol. 1999;73:10531–10535. doi: 10.1128/jvi.73.12.10531-10535.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seah E.L., Marshall J.A., Wright P.J. Trans activity of the norovirus Camberwell proteinase and cleavage of the N-terminal protein encoded by ORF1. J. Virol. 2003;77:7150–7155. doi: 10.1128/JVI.77.12.7150-7155.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirato-Horikoshi H., Ogawa S., Wakita T., Takeda N., Hansman G.S. Binding activity of norovirus and sapovirus to histo-blood group antigens. Arch. Virol. 2006 doi: 10.1007/s00705-006-0883-z. [DOI] [PubMed] [Google Scholar]
- Siebenga J.J., Beersma M.F., Vennema H., van Biezen P., Hartwig N.J., Koopmans M. High prevalence of prolonged norovirus shedding and illness among hospitalized patients: a model for in vivo molecular evolution. J. Infect. Dis. 2008;198:994–1001. doi: 10.1086/591627. [DOI] [PubMed] [Google Scholar]
- Simmonds P., Karakasiliotis I., Bailey D., Chaudhry Y., Evans D.J., Goodfellow I.G. Bioinformatic and functional analysis of RNA secondary structure elements among different genera of human and animal caliciviruses. Nucleic Acids Res. 2008;36:2530–2546. doi: 10.1093/nar/gkn096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simor A.E., McArthur M., McGeer A., Shurtleff S., Jabar M. Calicivirus gastroenteritis in a geriatric long-term care facility—Ontario. Can. Dis. Wkly. Rep. 1990;16:239–240. 243. [PubMed] [Google Scholar]
- Smiley J.R., Chang K.O., Hayes J., Vinje J., Saif L.J. Characterization of an enteropathogenic bovine calicivirus representing a potentially new calicivirus genus. J. Virol. 2002;76:10089–10098. doi: 10.1128/JVI.76.20.10089-10098.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A.E., Helenius A. How viruses enter animal cells. Science. 2004;304:237–242. doi: 10.1126/science.1094823. [DOI] [PubMed] [Google Scholar]
- Smith A.W., Akers T.G., Madin S.H., Vedros N.A. San Miguel sea lion virus isolation, preliminary characterization and relationship to vesicular exanthema of swine virus. Nature. 1973;244:108–110. doi: 10.1038/244108a0. [DOI] [PubMed] [Google Scholar]
- Smith A.W., Boyt P.M. Caliciviruses of ocean origin: a review. J. Zoo Wildl. Med. 1990;21:3–23. [Google Scholar]
- Smith A.W., Iversen P.L., O’Hanley P.D., Skilling D.E., Christensen J.R., Weaver S.S., Longley K., Stone M.A., Poet S.E., Matson D.O. Virus-specific antiviral treatment for controlling severe and fatal outbreaks of feline calicivirus infection. Am. J. Vet. Res. 2008;69:23–32. doi: 10.2460/ajvr.69.1.23. [DOI] [PubMed] [Google Scholar]
- Smith A.W., Skilling D.E., Cherry N., Mead J.H., Matson D.O. Calicivirus emergence from ocean reservoirs: zoonotic and interspecies movements. Emerg. Infect. Dis. 1998;4:13–20. doi: 10.3201/eid0401.980103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Someya Y., Takeda N., Miyamura T. Identification of active-site amino acid residues in the Chiba virus 3C-like protease. J. Virol. 2002;76:5949–5958. doi: 10.1128/JVI.76.12.5949-5958.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosnovtsev S., Green K.Y. RNA transcripts derived from a cloned full-length copy of the feline calicivirus genome do not require VpG for infectivity. Virology. 1995;210:383–390. doi: 10.1006/viro.1995.1354. [DOI] [PubMed] [Google Scholar]
- Sosnovtsev S.V., Belliot G., Chang K.O., Onwudiwe O., Green K.Y. Feline calicivirus VP2 is essential for the production of infectious virions. J. Virol. 2005;79:4012–4024. doi: 10.1128/JVI.79.7.4012-4024.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosnovtsev S.V., Belliot G., Chang K.O., Prikhodko V.G., Thackray L.B., Wobus C.E., Karst S.M., Virgin H.W., Green K.Y. Cleavage map and proteolytic processing of the murine norovirus nonstructural polyprotein in infected cells. J. Virol. 2006;80:7816–7831. doi: 10.1128/JVI.00532-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosnovtsev S.V., Garfield M., Green K.Y. Processing map and essential cleavage sites of the nonstructural polyprotein encoded by ORF1 of the feline calicivirus genome. J. Virol. 2002;76:7060–7072. doi: 10.1128/JVI.76.14.7060-7072.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosnovtsev S.V., Green K.Y. Identification and genomic mapping of the ORF3 and VPg proteins in feline calicivirus virions. Virology. 2000;277:193–203. doi: 10.1006/viro.2000.0579. [DOI] [PubMed] [Google Scholar]
- Sosnovtsev S.V., Prikhod’ko E.A., Belliot G., Cohen J.I., Green K.Y. Feline calicivirus replication induces apoptosis in cultured cells. Virus Res. 2003;94:1–10. doi: 10.1016/s0168-1702(03)00115-1. [DOI] [PubMed] [Google Scholar]
- Sosnovtsev S.V., Sosnovtseva S.A., Green K.Y. Cleavage of the feline calicivirus capsid precursor is mediated by a virus-encoded proteinase. J. Virol. 1998;72:3051–3059. doi: 10.1128/jvi.72.4.3051-3059.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosnovtseva S.A., Sosnovtsev S.V., Green K.Y. Mapping of the feline calicivirus proteinase responsible for autocatalytic processing of the nonstructural polyprotein and identification of a stable proteinase-polymerase precursor protein. J. Virol. 1999;73:6626–6633. doi: 10.1128/jvi.73.8.6626-6633.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein D.A. Inhibition of RNA virus infections with peptide-conjugated morpholino oligomers. Curr. Pharm. Des. 2008;14:2619–2634. doi: 10.2174/138161208786071290. [DOI] [PubMed] [Google Scholar]
- Stein D.A., Huang C.Y., Silengo S., Amantana A., Crumley S., Blouch R.E., Iversen P.L., Kinney R.M. Treatment of AG129 mice with antisense morpholino oligomers increases survival time following challenge with dengue 2 virus. J. Antimicrob. Chemother. 2008;62:555–565. doi: 10.1093/jac/dkn221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein D.A., Shi P.Y. Nucleic acid-based inhibition of flavivirus infections. Front. Biosci. 2008;13:1385–1395. doi: 10.2741/2769. [DOI] [PubMed] [Google Scholar]
- Straub T.M., Honer zu Bentrup K., Orosz-Coghlan P., Dohnalkova A., Mayer B.K., Bartholomew R.A., Valdez C.O., Bruckner-Lea C.J., Gerba C.P., Abbaszadegan M., Nickerson C.A. In vitro cell culture infectivity assay for human noroviruses. Emerg. Infect. Dis. 2007;13:396–403. doi: 10.3201/eid1303.060549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart A.D., Brown T.D. Entry of feline calicivirus is dependent on clathrin-mediated endocytosis and acidification in endosomes. J. Virol. 2006;80:7500–7509. doi: 10.1128/JVI.02452-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford D.L. Sinauer Associates; Sunderland: 2002. PAUP, Phylogenetic Analysis Using Parsimony (and other methods) [Google Scholar]
- Tacket C.O., Mason H.S., Losonsky G., Estes M.K., Levine M.M., Arntzen C.J. Human immune responses to a novel norwalk virus vaccine delivered in transgenic potatoes. J. Infect. Dis. 2000;182:302–305. doi: 10.1086/315653. [DOI] [PubMed] [Google Scholar]
- Tamura M., Natori K., Kobayashi M., Miyamura T., Takeda N. Genogroup II noroviruses efficiently bind to heparan sulfate proteoglycan associated with the cellular membrane. J. Virol. 2004;78:3817–3826. doi: 10.1128/JVI.78.8.3817-3826.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, M., Xia, M., Chen, Y., Bu, W., Hegde, R.S., Meller, J., Li, X., Jiang, X., 2009. Conservation of carbohydrate binding interfaces: evidence of human HBGA selection in norovirus evolution. PLoS One 4, e5058 (Epub 2009 Apr 1). [DOI] [PMC free article] [PubMed]
- Tan M., Huang P., Meller J., Zhong W., Farkas T., Jiang X. Mutations within the P2 domain of norovirus capsid affect binding to human histo-blood group antigens: evidence for a binding pocket. J. Virol. 2003;77:12562–12571. doi: 10.1128/JVI.77.23.12562-12571.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taube S., Kurth A., Schreier E. Generation of recombinant norovirus-like particles (VLP) in the human endothelial kidney cell line 293T. Arch. Virol. 2005;150:1425–1431. doi: 10.1007/s00705-005-0517-x. [DOI] [PubMed] [Google Scholar]
- Taube S., Perry J.W., Yetming K., Patel S.P., Auble H., Shu L., Nawar H.F., Lee C.H., Connell T.D., Shayman J.A., Wobus C.E. Ganglioside-linked terminal sialic acid moieties on murine macrophages function as attachment receptors for murine noroviruses. J. Virol. 2009;83:4092–4101. doi: 10.1128/JVI.02245-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teunis P.F., Moe C.L., Liu P., Miller S.E., Lindesmith L., Baric R.S., Le Pendu J., Calderon R.L. Norwalk virus: how infectious is it? J. Med. Virol. 2008;80:1468–1476. doi: 10.1002/jmv.21237. [DOI] [PubMed] [Google Scholar]
- Thackray L.B., Wobus C.E., Chachu K.A., Liu B., Alegre E.R., Henderson K.S., Kelley S.T., Virgin H.W.t. Murine noroviruses comprising a single genogroup exhibit biological diversity despite limited sequence divergence. J. Virol. 2007;81:10460–10473. doi: 10.1128/JVI.00783-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thumfart J.O., Meyers G. Feline calicivirus: recovery of wild-type and recombinant viruses after transfection of cRNA or cDNA constructs. J. Virol. 2002;76:6398–6407. doi: 10.1128/JVI.76.12.6398-6407.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompkins S.M., Lo C.Y., Tumpey T.M., Epstein S.L. Protection against lethal influenza virus challenge by RNA interference in vivo. Proc. Natl. Acad. Sci. U.S.A. 2004;101:8682–8686. doi: 10.1073/pnas.0402630101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treiber D.K., Williamson J.R. Beyond kinetic traps in RNA folding. Curr. Opin. Struct. Biol. 2001;11:309–314. doi: 10.1016/s0959-440x(00)00206-2. [DOI] [PubMed] [Google Scholar]
- Tsukiyama-Kohara K., Iizuka N., Kohara M., Nomoto A. Internal ribosome entry site within hepatitis C virus RNA. J. Virol. 1992;66:1476–1483. doi: 10.1128/jvi.66.3.1476-1483.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinje J., Green J., Lewis D.C., Gallimore C.I., Brown D.W., Koopmans M.P. Genetic polymorphism across regions of the three open reading frames of “Norwalk-like viruses”. Arch. Virol. 2000;145:223–241. doi: 10.1007/s007050050020. [DOI] [PubMed] [Google Scholar]
- von Eije K.J., ter Brake O., Berkhout B. Human immunodeficiency virus type 1 escape is restricted when conserved genome sequences are targeted by RNA interference. J. Virol. 2008;82:2895–2903. doi: 10.1128/JVI.02035-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Sarnow P., Siddiqui A. Translation of human hepatitis C virus RNA in cultured cells is mediated by an internal ribosome-binding mechanism. J. Virol. 1993;67:3338–3344. doi: 10.1128/jvi.67.6.3338-3344.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q.H., Costantini V., Saif L.J. Porcine enteric caliciviruses: genetic and antigenic relatedness to human caliciviruses, diagnosis and epidemiology. Vaccine. 2007;25:5453–5466. doi: 10.1016/j.vaccine.2006.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q.H., Han M.G., Cheetham S., Souza M., Funk J.A., Saif L.J. Porcine noroviruses related to human noroviruses. Emerg. Infect. Dis. 2005;11:1874–1881. doi: 10.3201/eid1112.050485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward V.K., McCormick C.J., Clarke I.N., Salim O., Wobus C.E., Thackray L.B., Virgin H.W.t., Lambden P.R. Recovery of infectious murine norovirus using pol II-driven expression of full-length cDNA. Proc. Natl. Acad. Sci. U.S.A. 2007;104:11050–11055. doi: 10.1073/pnas.0700336104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widdowson M.A., Monroe S.S., Glass R.I. Are noroviruses emerging? Emerg. Infect. Dis. 2005;11:735–737. doi: 10.3201/eid1105.041090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelmi I., Roman E., Sanchez-Fauquier A. Viruses causing gastroenteritis. Clin. Microbiol. Infect. 2003;9:247–262. doi: 10.1046/j.1469-0691.2003.00560.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wobus C.E., Karst S.M., Thackray L.B., Chang K.O., Sosnovtsev S.V., Belliot G., Krug A., Mackenzie J.M., Green K.Y., Virgin H.W. Replication of Norovirus in cell culture reveals a tropism for dendritic cells and macrophages. PLoS Biol. 2004;2:e432. doi: 10.1371/journal.pbio.0020432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wobus C.E., Thackray L.B., Virgin H.W.t. Murine norovirus: a model system to study norovirus biology and pathogenesis. J. Virol. 2006;80:5104–5112. doi: 10.1128/JVI.02346-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt R.G., Dolin R., Blacklow N.R., DuPont H.L., Buscho R.F., Thornhill T.S., Kapikian A.Z., Chanock R.M. Comparison of three agents of acute infectious nonbacterial gastroenteritis by cross-challenge in volunteers. J. Infect. Dis. 1974;129:709–714. doi: 10.1093/infdis/129.6.709. [DOI] [PubMed] [Google Scholar]
- Xiang W., Harris K.S., Alexander L., Wimmer E. Interaction between the 5′-terminal cloverleaf and 3AB/3CDpro of poliovirus is essential for RNA replication. J. Virol. 1995;69:3658–3667. doi: 10.1128/jvi.69.6.3658-3667.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Yang M., Ding Y., Liu Y., Lou Z., Zhou Z., Sun L., Mo L., Ye S., Pang H., Gao G.F., Anand K., Bartlam M., Hilgenfeld R., Rao Z. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc. Natl. Acad. Sci. U.S.A. 2003;100:13190–13195. doi: 10.1073/pnas.1835675100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakhour, M., Ruvoën-Clouet, N., Charpilienne, A., Langpap, B., Poncet, D., Peters, T., Bovin, N., Le Pendu, J., 2009. The alphaGal epitope of the histo-blood group antigen family is a ligand for bovine norovirus Newbury2 expected to prevent cross-species transmission. PLoS Pathog 5 (Epub Jul 3). [DOI] [PMC free article] [PubMed]
- Zeitler C.E., Estes M.K., Venkataram Prasad B.V. X-ray crystallographic structure of the Norwalk virus protease at 1.5-A resolution. J. Virol. 2006;80:5050–5058. doi: 10.1128/JVI.80.10.5050-5058.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Yang H., Kong X., Mohapatra S., San Juan-Vergara H., Hellermann G., Behera S., Singam R., Lockey R.F., Mohapatra S.S. Inhibition of respiratory syncytial virus infection with intranasal siRNA nanoparticles targeting the viral NS1 gene. Nat. Med. 2005;11:56–62. doi: 10.1038/nm1174. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Yao Q., Xia C., Jiang X., Wang P.G. Trapping norovirus by glycosylated hydrogels: a potential oral antiviral drug. ChemMedChem. 2006;1:1361–1366. doi: 10.1002/cmdc.200600135. [DOI] [PubMed] [Google Scholar]
- Zheng D.P., Ando T., Fankhauser R.L., Beard R.S., Glass R.I., Monroe S.S. Norovirus classification and proposed strain nomenclature. Virology. 2006;346:312–323. doi: 10.1016/j.virol.2005.11.015. [DOI] [PubMed] [Google Scholar]


