Abstract
Human immunodeficiency virus (HIV) is often acquired in individuals already infected with hepatitis B virus (HBV) as a result of shared routes of transmission. Since current options for the treatment of HIV and HBV infections are limited, there is an essential need for the development of effective therapies against HIV/HBV co-infections. RNA interference (RNAi) has been used as a powerful tool to silence genes in cells and animals. In this study, we developed a small interfering RNA generation system that expressed two different siRNAs to target the HBs gene of HBV and the gp120 gene of HIV in Bel-7402 and HEK293T cells, respectively. Our results demonstrated that the two siRNA molecules could simultaneously inhibit the expression of HBs and gp120 by 81% and 89%, respectively. In addition, dual siRNA molecules significantly decreased the production of HBs, and simultaneously inhibited the replication of HBV and HIV. This dual siRNA generation system not only proved to be a novel approach for studying functions of multiple genes simultaneously, but also provides a potential approach for the treatment and prevention of HIV and HBV co-infection.
Keywords: HBV, HIV, Dual siRNA, Viral replication
1. Introduction
The prevalence of hepatitis B virus (HBV) in patients infected with human immunodeficiency virus (HIV) is common. Co-infection with HIV and hepatotropic viruses causes complex interactions (Hadler et al., 1991, McNair et al., 1992). HIV-induced impairment of the cell-mediated immunity causes higher replication of hepatotropic viruses (Benhamou et al., 1999). Additionally, HBV leads to enhanced transcription of HIV through an NF-κB element in the long terminal repeat of HIV (Gómez-Gonzalo et al., 2001). More than 95% of adults spontaneously recover from an acute HBV infection (Beasley, 1988), an outcome that is defined by clearance of the hepatitis B surface antigen (HBsAg) from blood. Those persistently infected with HBV are at increased risk of development of cirrhosis and hepatocellular carcinoma (Hsu et al., 2002). HIV-1 causes multidimensional immunosuppression, associated with reduced frequency of spontaneous recovery from HBV infection (Colin et al., 1999). The impact of co-infections on the clinical outcome of HIV-infected patients is unclear and the available treatment options against these viruses are also limited (Shlomai and Shaul, 2003).
RNA interference (RNAi) is an innate cellular process activated by a double-stranded RNA duplex in cells from Caenorhabditis elegans to mammals (Fire et al., 1998). Specific inhibition of cellular mRNA by RNAi can be triggered in mammalian cells by the introduction of synthetic 21–23-nucleotide double-stranded small interfering RNA (siRNA) (Elbashir et al., 2001, Paul et al., 2002) or, alternatively, by the transcription of siRNA from a DNA construct driven by the RNA polymerase cassette (Brummelkamp et al., 2002). RNAi is initiated by degradation of single-stranded RNA of identical sequences. Therefore, RNAi approach can be used to silence gene expression by directly targeting its specific sequence of mRNA. In addition to the widely used strategies for inhibiting gene expression in research work, RNAi approach has been used in therapeutic studies of human diseases including cancer and viral infectious diseases (Cioca et al., 2003, Zhang et al., 2003, Verma et al., 2003). The RNAi approach has been reported as an ideal tool to inhibit infectious virus replication in host cells because siRNA can target and silence important genes of the virus.
It has been shown that siRNA could specifically inhibit human immunodeficiency virus (HIV) replication and virus propagation through targeting major genes in the HIV life cycle, including p24, nef, rev, tat, and vif (Coburn and Cullen, 2002, Jacque et al., 2002, Lee et al., 2002, Capodici et al., 2002). RNAi has also been used in the inhibition of replication of hepatitis B virus (HBV) or hepatitis C virus (HCV), which causes chronic liver disease including cirrhosis and hepatocellular carcinoma (Hamasaki et al., 2003, McCaffrey et al., 2003, Shlomai and Shaul, 2003, Kapadia et al., 2003). It has been demonstrated that siRNA effectively protects human cells against poliovirus infection (Gitlin et al., 2002) and that siRNA could block retroviral infection in chick embryos and inhibit the growth of the Rous sarcoma virus and HIV in cell culture (Hu et al., 2002). siRNA primarily prevented accumulation of the viral RNAs synthesized in the late stage of the infection, but did not degrade the RNA genome of the virus in the early stage of the infection. siRNA molecules generated against the HCV replicon inhibited the HCV mRNA transcripts and protein expression (Kapadia et al., 2003). It has been found that siRNA inhibited severe acute respiratory syndrome associated coronavirus (SARS-CoV) gene expression and replication in cultured cells (He et al., 2003).
We have previously established a dual small interfering RNA (siRNA) expression system, which could simultaneously generate two different siRNA molecules specifically targeting two genes of HBV (Wu et al., 2005). In this study, we extended our study by using this system to produce simultaneously two siRNA duplexes that targeted the S gene of HBV and the gp120 gene of HIV-1, respectively. To study the effects of dual RNAi on HBV gene expression and replication in a cell culture model, we used a derivative of the human HepG2 hepatoma cell line, HepG2.2.1.5, which has been stably transformed with several copies of the HBV genome and is used as an in vitro model for HBV replication. To study the effects of dual RNAi on HIV-1 gene expression and replication in mammalian cells, we used two HIV-1 expression vector pNL4-3 and pNL4-3.luc.R-E-. pNL4-3 is an HIV-based infectious vector and upon transfection this clone directed the production of infectious virus particles in a wide variety of cells. pNL4-3.luc.R-E- is a non-infectious HIV-1 recombinant clone, in which firefly luciferase gene was inserted into the pNL4-3 nef and two frameshifts (5′ Env and Vpr aa 26) rendered this clone Env− and Vpr− and allowed only a single cycle of replication to transfect HEK 293T. The effects of dual siRNA molecules on gene expression and replication of HBV and HIV-1 were investigated and discussed in this study.
2. Materials and methods
2.1. Cell culture and transfection
Human embryo kidney cell line HEK293T and two human hepatoma cell lines, Bel-7402 and HepG2.2.1.5, were maintained in Dulbecco's modified Eagle's medium (GIBCO/BRL) supplemented with 100 units/ml penicillin, 100 μg/ml streptomycin, and 10% heat-inactivated fetal bovine serum at 37 °C under 5% CO2. African green monkey kidney cell line (COS-7) were maintained in RPMI 1640 medium (GIBCO/BRL) supplemented with 100 units/ml penicillin, 100 μg/ml streptomycin, and 10% heat-inactivated fetal bovine serum at 37 °C under 5% CO2. Cells were seeded onto 24-well plates at a density of 1.0 × 105 or 4.0 × 105 cells per 24-well plate or 6-well plate and grown to the confluence reaching approximately 60% at the time of transfection. Cells were transfected with 0.1 μg or 0.4 μg of plasmid pCMV–HBs together with 0.45 μg or 1.2 μg pSliencer-2.1-U6-siRNA, using Sofast™ transfection reagent (Xiamen Sunma Biotechnology Co., Ltd., China) according to the protocol provided by the manufacturer. The cells were harvested at 48 h after transfection.
2.2. Plasmid construction
HBSsiRNA2 (pSilencer-2.1-U6-HBS2) was a plasmid that expressed siRNA molecules targeting HBs gene and inhibited HBV replication (Wu et al., 2005). The plasmid pNL4-3.luc.R-E- was non-infectious HIV-1 recombinant clone, which firefly luciferase gene was inserted into the pNL4-3 nef and two frameshifts (5′ Env and Vpr aa 26) rendered this clone Env− and Vpr− and allowed only a single cycle of replication and the plasmid pNL4-3 is an HIV-based infectious vector and upon transfection this clone directed the production of infectious virus particles in a wide variety of cells (NIH, MD). HBs gene was cloned into the HindIII and SacI sites of vector pCMV–tag2A (Stratagene) to yield plasmid pCMV–HBs. A pair of primers P1 (sense), P2 (antisense) were used to amplify the HIV gp120 gene, the PCR products were then cloned into SalI and BglII sites of pLucF to generate plasmid pLucF–gp120 (Fig. 1A), in which the HIVgp120 was fused in frame with the luciferase gene and the expression of the fusion gene was driven by the CMV promoter (Fig. 1A). All primers used in this study are listed in Table 1 .
Fig. 1.
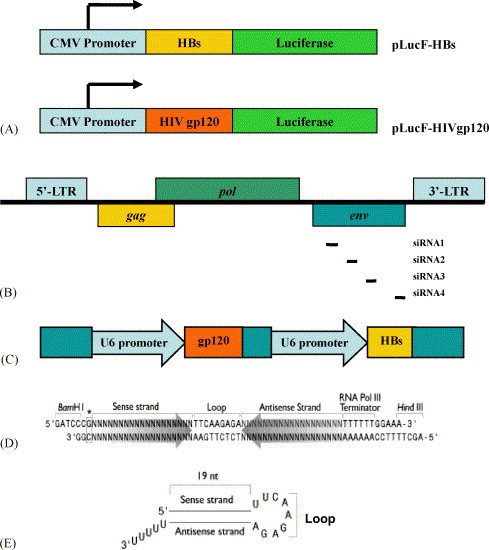
Schematic diagrams of luciferase fusion genes, siRNA targeting sites and dual siRNA expression cassettes: (A) diagram of the reporter fusion vector, which contains targeted sequences of HBs and the luciferase reporter gene driven by the CMV promoter; (B) diagram of the reporter fusion vector, which contains targeted sequences of HIVgp120 gene and the luciferase reporter gene driving by the CMV promoter; (C) diagram of dual siRNA expression cassettes; (D) sequences and structures of the design map for pSilencer-2.1-U6; (E) predicted folding hairpin structure of a siRNA molecule that is expressed from the cassette.
Table 1.
Primers used in this study
| Name | Sequences (5′–3′) |
|---|---|
| P1 | TACGTGTCGACTGGGTCACAGTCTATTATGG |
| P2 | ATAACAGATCTGTCCACTGATGGGAGGGGCA |
| P3 | GCTGATGACGTCAGTGGAAAGACGCG |
| P4 | TCAGCGAATTCACGCCAAGCTTTTCC |
| P5 | GCGGGGTTTTTCTTGTTGAC |
| P6 | CTACGAACCACTGAACAAAT |
| P7 | GGGTCACAGTCTATTATGGG |
| P8 | ATTATCATTCTCCCGCTACT |
| P9 | ATCCTGCTGCTATGCCTCATCTT |
| P10 | ACAGTGGGGAAAGCCCTACGAA |
| Probe | TGGCTAGTTTACTAGTGCCATTTTG |
2.3. Generation of siRNA expression vectors
Four regions of the HIV gp120 gene were selected as the targeted sequences of siRNA in this study (Fig. 1B). To construct single siRNA expression vector, two 64 nt primers, each containing a 19 nt target sequence in the sense and antisense forms from different regions of the HIV gp120 gene as indicated below, were synthesized (Invitrogen).
-
•
5′-CATAATGTTTGGGCCACAC-3′ (HIVsiRNA1) (5966–5984);
-
•
5′-GGCTGGTTTTGCGATTCTA-3′ (HIVsiRNA2) (6430–6447);
-
•
5′-AAGAATCCGTATCCAGAGA-3′ (HIVsiRNA3) (6687–6705);
-
•
5′-CATGTGGCAGAAAGTGGA-3′ (HIVsiRNA4) (7045–7063).
Sense and antisense primers were then cloned into pSilence-2.1-U6 plasmid (Amibion) at BamHI and HindIII sites after annealing according to the manufacturer's instructions.
To generate the dual siRNA expression plasmid, two primers P3 (sense) and P4 (antisense) were designed to amplify a DNA fragment containing U6 promoter and HIVsiRNA3 expression cassette from recombinant plasmid pSilencer-2.1-U6-HIV3. The PCR product was then cloned into AatII and EcoRI sites of plasmid pSilencer-2.1-U6-HBS2 to generate recombinant plasmid HBV–HIVsiRNA, which carries two independent siRNA expression cassettes (Fig. 1C).
2.4. Measurement of luciferase activity
Bel-7402 cells or 293T cells were co-transfected with reporter plasmids and siRNA expression plasmids. Cells were washed with PBS and lysed with luciferase cell culture lysis reagent (Promega). About 10 μl of the cell lysates and 100 μl of luciferase assay substrate (Promega) were mixed and fluorescence intensity was detected by the luminometer (Turner T20/20). Assays were performed in triplicate, and expressed as means ± S.D. relative to vector control as 100%.
2.5. Hepatitis B surface antigen (HBsAg) assay
Bel-7402 cells and HepG2.2.1.5 cells were transfected with siRNA expression plasmids, the level of HBsAg protein in culture media from transfected cells were then determined by enzyme-linked immunosorbent assay using a HBV diagnostic kit (Shanghai Kehua Biotech Co., Ltd.). Assays were performed in triplicate independent experiments.
2.6. In vitro HIV-1 replication assay
COS-7 cells were co-transfected with pNL4-3 and siRNA expression plasmids, and the level of p24 protein in culture media from transfected cells was then determined by enzyme-linked immunosorbent assay kit from Innogenetics (Ghent, Belgium).
2.7. RNA isolation and RT-PCR assay
HepG2.2.1.5 cells were transfected with siRNA expression plasmids or 293T cells were co-transfected with pNL4-3.luc.R-E- and siRNA expression plasmids, total RNA were then extracted from transfected cells by Trizol Reagent (Invitrogen) according to the method described in the manufacturer's manual. Reverse transcription was performed with total RNA as the template. The cDNAs were amplified for 25 cycles of 94 °C for 1 min, 60 °C for 1 min, 72 °C for 1 min with HBs or HIV gp120 gene-specific primers, P5 (sense), P6 (antisense) or P7 (sense), P8 (antisense).
2.8. Assay for core-associated DNA of HBV by real-time PCR
To detect the efficiency of siRNA in inhibiting HBV replication in treated and control cells, intracellular core-associated HBV DNA was extracted by a previously described method (Pugh et al., 1988). Briefly, 1 × 105 treated or control cells were lysed and centrifuged at 25 °C. Magnesium chloride was added to the supernatant. DNA not protected by HBV core was degraded with deoxyribonuclease, DNase I (Invitrogen). The lysates were then treated with proteinase K and extracted with phenol/chloroform. Core-associated HBV DNA was recovered by ethanol precipitation, and quantified by the HBV DNA in real-time PCR kit as described by the manufacturer (PG BIOTECH, Shenzhen, China) using PCR primers P9 and P10 and probe. PCR reaction was analyzed by PE Gene Amp 7700 (Perkin-Elmer).
3. Results
3.1. The expression of HBs–luciferase or HIVgp120–luciferase fusion genes was inhibited by the treatment of single siRNA or dual siRNA expression systems
In our previous study, a siRNA expressing plasmid HBSsiRNA2 was selected to inhibit specifically the expression of HBs gene (Wu et al., 2005). To efficiently screen siRNA molecules that target HIVgp120, selected targeting DNA sequences were fused in frame with the luciferase gene to generated plasmid pLucF–gp120 (Fig. 1A, Table 2 ), in which luciferase activity represented the level of HIVgp120 mRNA expression. Cells were co-transfected with pLucF–gp120 and four single siRNA expression vectors, respectively; luciferase activities were then determined from transfected cells. HIVsiRNA3 strongly inhibited luciferase activities by 88% comparing to that of vector control, while HIVsiRNA1, HIVsiRNA2, and HIVsiRNA4 showed no inhibitory effect (Fig. 2A, Table 2), indicating this siRNA3 could efficiently degrade the mRNA of HIVgp120–luciferase fusion gene.
Table 2.
Suppression efficiency of siRNAs in this stuy (%)
Fig. 2.
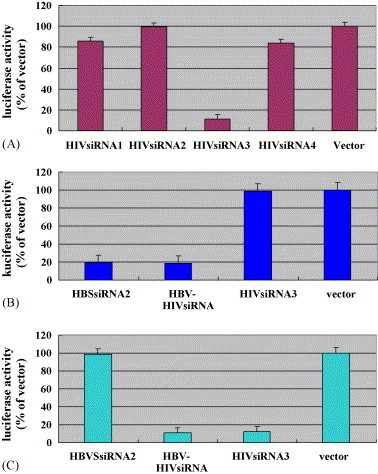
Quantitative analysis of luciferase activity in cells transfected with siRNA expressing plasmids: (A) HEK293T cells were co-transfected with pLucF–gp120 and pSliencer-2.1-U6-siRNA plasmids (HIVsiRNA1, HIVsiRNA2, HIVsiRNA3, HIVsiRNA4). pSliencer-2.1-U6 plasmid was used as a control; (B) Bel-7402 cells were co-transfected with pLucF–HBs plasmid and pSliencer-2.1-U6-siRNA (HBSsiRNA2, HBV–HIVsiRNA, HIVsiRNA3) plasmids. pSliencer-2.1-U6 vector was used as a control; (C) HEK293T cells were co-transfected with pLucF–gp120 and pSliencer-2.1-U6-siRNA (HBSsiRNA2, HBV–HIVsiRNA, HIVsiRNA3) plasmids. pSliencer-2.1-U6 was used as vector control. About 48 h after transfection, cells were lysed and luciferase activities were determined by luminometer.
To determine the effects of dual siRNAs on the inhibition of HBs–luciferase and HIVgp120–luciferase fusion gene expression, cells were co-transfected with pLucF–HBs (Fig. 1A) or pLucF–gp120 and the dual siRNA expression plasmid HBV–HIVsiRNA (Fig. 1C). Luciferase activity assays indicated that the reduction rate of luciferase activity caused by HBV–HIVsiRNA treatment was 81% to HBs of HBV (Fig. 2B) and 89% to gp120 of HIV (Fig. 2C), respectively, indicating there was the same reduction in luciferase activity by dual siRNA duplexes (HBV–HIVsiRNA) comparing to that of single siRNA expression vectors (HBSsiRNA2 or HIVsiRNA3).
3.2. Single or dual siRNA expression system had a similar inhibitory effect on the production of HBsAg protein
HepG2.2.1.5 cells were transfected with pSilencer2.1-U6-siRNA or HBV–HIVsiRNA, which expressed either single or dual siRNAs, respectively. HBsAg concentrations in the culture media of transfected and control cells were measured at 2 days after transfection by ELISA using HBsAg diagnostic kit. HepG2.2.1.5 cells transfected with HBSsiRNA2 or HBV–HIVsiRNA reduced the level of HBsAg production by 82% and 84%, respectively (Fig. 3 ; Table 2). This result suggested that HBSsiRNA2 and HBV–HIVsiRNA had a similar effect on the production of the viral protein HBsAg.
Fig. 3.
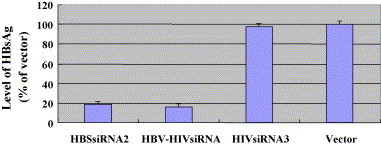
Analysis of the effects of siRNAs on HBsAg protein expression. HepG2.2.1.5 cells were transfected with pSliencer-2.1-U6-siRNA (HBSsiRNA2, HBV–HIVsiRNA, HIVsiRNA3). pSliencer-2.1-U6 was used as vector control. HBsAg levels were determined by ELISA using a HBV diagnostic kit.
3.3. Single or dual siRNA generation system inhibited HIV-1 replication
To determine whether siRNA molecules generated from single and dual siRNA expression systems had any affect on HIV-1 replication, COS-7 cells were co-transfected with HBV–HIVsiRNA or HIVsiRNA3 and plasmid pNL4-3, an HIV-based infectious vector. Upon transfection this clone directed the production of infectious virus particles in a wide variety of cells. The level of p24 protein in culture media from transfected cells was then determined by an enzyme-linked immunosorbent assay kit from Innogenetics (Ghent, Belgium) at 2 days after transfection. Results indicated that p24 (Fig. 4 ) were significantly decreased by 82% and 79%, respectively, after treatment with HBV–HIVsiRNA and HIVsiRNA3. These results demonstrated that HIVsiRNA generated from single or dual siRNA expression vectors had a similar effect on the viral genome replication.
Fig. 4.
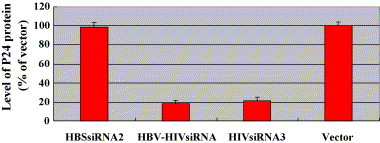
Determination the effects of siRNA on HIV-1 replication. COS-7 cells were co-transfected with plasmid pNL4-3 and pSliencer-2.1-U6-siRNA (HBSsiRNA2, HBV–HIVsiRNA, HIVsiRNA3) plasmids. pSliencer-2.1-U6 plasmid was used as a control. About 48 h after transfection, the level of p24 protein in culture media from transfected cells were determined by enzyme-linked immunosorbent assay kit.
3.4. The expression of HBs and HIVgp120 gene was inhibited by the dual siRNA expression system
We used semi-quantitative RT-PCR analyses to determine the levels of HBs or HIVgp120 mRNA in two different cell lines, 293T (Fig. 5A–C) or HepG2.2.1.5 (Fig. 5D) at 2 days after transfection. Results indicated that the levels of HIVgp120 mRNA were decreased by the treatment of HBV–HIVsiRNA (Fig. 5B, lane 2; Fig. 5C, lane 2) and HIVsiRNA3 (Fig. 5A, lane3; Fig. 5B, lane 1; Fig. 5C, lane 3), but not by HBSsiRNA2 (Fig. 5C, lane 1) and pSliencer-2.1-U6 (Fig. 5B, lane 4; Fig. 5C, lane 5). The levels of HBs mRNA were significantly inhibited by the treatment of HBSsiRNA2 (Fig. 5D, lane 1) or HBV–HIVsiRNA (Fig. 5D, lane 2) in HepG2.2.1.5 cells, but not by HIVsiRNA3 or pSliencer-2.1-U6 (Fig. 5D, lanes 3 and 4).
Fig. 5.
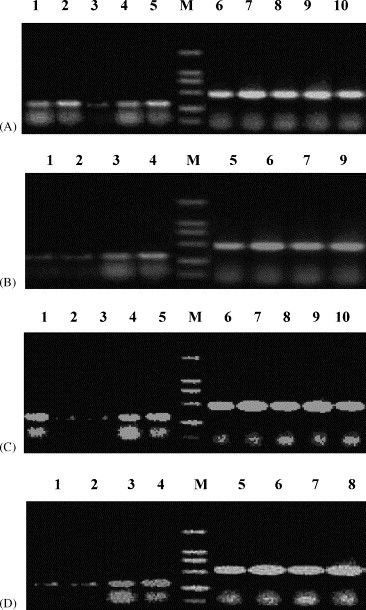
Levels of RNA expression determined by semi-quantitated RT-PCR analysis. Total RNA was used as template to synthesize cDNA. HIVgp120-specific primers were then applied for detection in (A) and (C), and HBs specific primers in (D). (A) HEK293T cells were co-transfected with pCMV–HIVgp120 and HIVsiRNA1 (lane 1), HIVsiRNA2 (lane 2), HIVsiRNA3 (lane 3), HIVsiRNA4 (lane 4), and pSliencer-2.1-U6 (lane 5, as a vector control). Lanes 6–10 were β-actin controls. (B) HEK293T cells were co-transfected with pCMV–HIVgp120 and HIVsiRNA3 (lane 1), HBV–HIVsiRNA (lane 2) or pSliencer-2.1-U6 (lane3, as a vector control). As control, HEK293T cells were also transfected with pCMV–HIVgp120 only (lane 4). Lanes 5–8 were β-actin controls. (C) HEK293T cells were co-transfected with pNL4-3.luc.R-E- and HBSsiRNA2 (lane 1), HBV–HIVsiRNA (lane 2), HIVsiRNA3 (lane 3), or pSliencer-2.1-U6 (lane 4, as a vector control). As control, HEK293T cells were also transfected with pNL4-3.luc.R-E- only (lane 5). Lanes 6–10 were β-actin controls. (D) HepG2.2.1.5 cells were transfected with HBSsiRNA2 (lane 1), HBV–HIVsiRNA (lane 2), HIVsiRNA3 (lane 3) or pSliencer-2.1-U6 (lane 4, as a vector control). Lanes 5–8 were β-actin controls. Lane M indicates DNA markers.
3.5. HBV replication was inhibited by the treatment of single or dual siRNA generation system
HepG2.2.1.5 cells were transfected with plasmids expressing HBSsiRNA2, HBV–HIVsiRNA, and HIVsiRNA3, respectively, and pSliencer-2.1-U6 was used as vector control. HBV core-associated DNA, the replicative intermediate in the transfected cells were measured by a real-time PCR kit. Results showed that the level of core-associated DNA was significantly decreased in the presence of HBSsiRNA2 and HBV–HIVsiRNA, but not in the presence of HIVsiRNA3 or vector control, indicating both single and dual siRNA generation system can inhibit HBV replication in HepG2.2.1.5 cells (Fig. 6 ; Table 2).
Fig. 6.
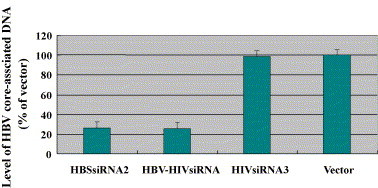
Determination of the effects of siRNAs on the levels of HBV DNA replication by real-time PCR. HepG2.2.1.5 cells were transfected with siRNA (HBSsiRNA2, HBV–HIVsiRNA, and HIVsiRNA3). pSliencer-2.1-U6 was used as a vector control. The levels of HBV core-associated DNA in the transfected cells were measured by a real-time PCR kit.
3.6. Dual siRNA generation system simultaneously inhibited the replication of HIV-1 and the expression of HBV
To determine the effects of dual siRNAs on the simultaneous inhibition of HIV-1 replication and of HBV gene expression, COS-7 cells were co-transfected with plasmid pNL4-3, pCMV–HBs and siRNAs (HBSsiRNA2, HBV–HIVsiRNA, and HIVsiRNA3) or vector as control. The level of p24 protein in transfected cells were then determined by enzyme-linked immunosorbent assay kit and HBsAg concentrations in the transfected cells were measured by ELISA using HBsAg diagnostic kit 2 days after transfection. Results showed that the level of p24 protein and HBsAg was significantly decreased by 85% and 81%, respectively, in the presence of HBV–HIVsiRNA, but not in the presence of vector control (Fig. 7 , Table 2).
Fig. 7.
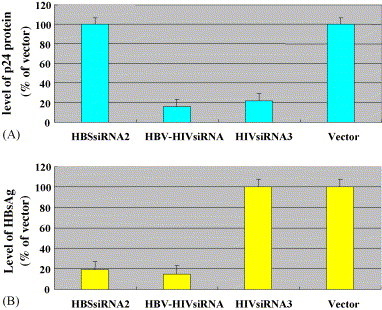
Dual siRNA generation system simultaneously inhibited the replication of HIV-1 and the expression of HBV. COS-7 cells were co-transfected with plasmid pNL4-3, pCMV–HBs and siRNA (HBSsiRNA2, HBV–HIVsiRNA, and HIVsiRNA3) or empty vector as control. The level of p24 protein in culture media from transfected cells was then determined by enzyme-linked immunosorbent assay kit (A) and HBsAg concentrations in the transfected cells were measured by ELISA using HBsAg diagnostic kit at 2 days after transfection (B).
4. Discussions
A number of studies indicated that mammalian RNAi machinery could be programmed to induce effective antiviral responses. Introduction of siRNAs specific for HIV-1 into mammalian cells could lead to the degradation of viral RNA, inhibition of HIV-1 gene expression, and knock-down of viral replication during different stages of the viral life cycle. HIV-1 has a genome with 9 kb in size that contains nine genes encoding for 15 proteins (Peterlin and Luciw, 1988). The first potential target for siRNAs is the viral genomic RNA upon viral entry and uncoating. At least one report demonstrates that siRNA-mediated destruction of incoming HIV-1 can take place (Jacque et al., 2002) and some sequences of the HIV genome have been proven to be effective siRNA targeting sites.
The HBV genome is a partially double-stranded DNA molecule with 3.2 kb in size and is transcribed to generate the four viral RNAs (Seeger and Mason, 2000), the 3.5, 2.4, 2.1, and 0.7 kb transcripts encode for the core protein, HBeAg, polymerase–reverse transcriptase, HBsAg, and X protein, respectively. All viral transcripts utilize a common polyadenylation signal located within the core protein-coding region. The 3.5 kb mRNA not only serves for translation of the core protein/HBeAg and polymerase–reverse transcriptase, but also represents the template for reverse transcription. The level of 3.5 kb pre-genome RNA has been demonstrated to be reduced by specific siRNAs and resulted in reduction of the levels of secreted HBeAg and replicative intermediates converted from the 3.5 kb pre-genome RNA (Hamasaki et al., 2003; McCaffrey, 2003; Shlomai and Shaul, 2003).
The aim of this study is to design a siRNA expression system that expresses dual siRNAs to inhibit both HBV and HIV replication. In order to construct a useful tool in the selection of effective siRNA molecules, we created a quick screening system by fusing the targeted DNA and the luciferase reporter gene together to produce recombinant pLucF plasmid that could express HBs–luciferase or HIVgp120–luciferase fusion mRNAs. Therefore, we can initially select the suitable siRNA duplexes rapidly by simply analyzing the activities of luciferase. By using this approach, we have demonstrated that one of the siRNA molecules (HIVsiRNA3) had significant impact on the expression of HIV–luciferase fusion gene. This provides a quick approach to select effective siRNA in the study of gene expression and function analysis.
After screening the effective siRNA by luciferase activity assay, we further studied the effects of selected dual siRNA molecules on HIV and HBV gene expression and viral replication in cell culture models by using HEK293T and a derivative of the human HepG2 hepatoma cell line, HepG2.2.1.5, which carries several copies of the HBV genome on its chromosome. The effects of dual siRNAs on HIV and HBV gene expression and viral replication were studied thoroughly by the analyzing the levels of viral protein production through enzyme-linked immunosorbent assays and the levels of viral RNA expression by semi-quantitated RT-PCR analysis. All results indicated that dual siRNAs had significant inhibitory effects on viral mRNA expression, viral protein production, or the replication of HIV and HBV.
Although HIV and HBV co-infection is a major health problem worldwide, there is no completely effective antiviral treatment. Our dual siRNA approach would provide a possible therapeutic strategy against co-infection of these viruses, although further studies are necessary to determine the antiviral mechanism of dual siRNAs in different stage of viral replication. Our approach could also be used to generate more than two siRNAs duplexes that could silent more genes in order to study the interactions of genes and their functions. Such strategies of constructing multiple-siRNA vectors could confront multiple virus infections. Obviously, this “cocktail” siRNA approach could be applied, especially for those viruses that show a high mutation rate, such as HIV.
Acknowledgments
This research was supported by research grants from the Major State Basic Research Development Program of China (“973” project, no. 2005CB522901), the National Natural Science Foundation of China (no. 30470087 and 30570070), and Ph.D. Program Foundation of Ministry of Education of China (no. 20040486037) to J. Wu, and from the National Natural Science Foundation of China to F. Liu (no. 30570069), and from the Hubei Provincial Science Foundation for Distinguish Youth Scholar to Y. Zhu.
Contributor Information
Ying Zhu, Email: yingzhu@whu.edu.cn.
Jianguo Wu, Email: wu9988@vip.sina.com.
References
- Beasley R.P. Hepatitis B virus. The major etiology of hepatocellular carcinoma. Cancer. 1988;61:1942–1956. doi: 10.1002/1097-0142(19880515)61:10<1942::aid-cncr2820611003>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- Benhamou Y., Bochet M., Di Martino V., Charlotte F., Azria F., Coutellier A., Vidaud M., Bricaire F., Opolon P., Katlama C., Poynard T. Liver fibrosis progression in human immunodeficiency virus and hepatitis C virus coinfected patients. The Multivirc Group. Hepatology. 1999;30:1054–1058. doi: 10.1002/hep.510300409. [DOI] [PubMed] [Google Scholar]
- Brummelkamp T.R., Bernards R., Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- Capodici J., Kariko K., Weissman D. Inhibition of HIV-1 infection by small interferring RNA-mediated RNA interference. J. Immunol. 2002;169:5196–5201. doi: 10.4049/jimmunol.169.9.5196. [DOI] [PubMed] [Google Scholar]
- Cioca D.P., Aoki Y., Kiyosawa K. RNA interference is a functional pathway with therapeutic potential in human myeloid leukemia cell lines. Cancer Gene Ther. 2003;10:125–133. doi: 10.1038/sj.cgt.7700544. [DOI] [PubMed] [Google Scholar]
- Coburn G.A., Cullen B.R. Potent and specific inhibition of human immunodeficiency virus type 1 replication by RNA interference. J. Virol. 2002;76:9225–9231. doi: 10.1128/JVI.76.18.9225-9231.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colin J.F., Cazals-Hatem D., Loriot M.A., Martinot-Peignoux M., Pham B.N., Auperin A., Degott C., Benhamou J.P., Erlinger S., Valla D., Marcellin P. Influence of human immunodeficiency virus infection on chronic hepatitis B in homosexual men. Hepatology. 1999;29:1306–1310. doi: 10.1002/hep.510290447. [DOI] [PubMed] [Google Scholar]
- Elbashir S.M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Fire A., Xu S., Montgomery M.K., Kostas S.A., Driver S.E., Mello C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Gitlin L., Karelsky S., Andino R. Short interfering RNA confers intracellular antiviral immunity in human cells. Nature. 2002;418:430–434. doi: 10.1038/nature00873. [DOI] [PubMed] [Google Scholar]
- Gómez-Gonzalo M., Carretero M., Rullas J., Lara-Pezzi E., Aramburu J., Berkhout B., Alcami J., Lopez-Cabrera M. The hepatitis B virus X protein induces HIV-1 replication and transcription in synergy with T-cell activation signals: functional roles of NK-kappaB/NF-AT and SP1-binding sites in the HIV-1 long terminal repeat promoter. J. Biol. Chem. 2001;276:35435–43443. doi: 10.1074/jbc.M103020200. [DOI] [PubMed] [Google Scholar]
- Hadler S.C., Judson F.N., O’Malley P.M., Altman N.L., Penley K., Buchbinder S., Schable C.A., Coleman P.J., Ostrow D.N., Francis D.P. Outcome of hepatitis B virus infection in homosexual men and its relation to prior human immunodeficiency virus infection. J. Infect. Dis. 1991;163:454–459. doi: 10.1093/infdis/163.3.454. [DOI] [PubMed] [Google Scholar]
- Hamasaki K., Nakao K., Matsumoto K., Ichikawa T., Ishikawa H., Eguchi K. Short interfering RNA-directed inhibition of hepatitis B virus replication. FEBS Lett. 2003;543:51–54. doi: 10.1016/s0014-5793(03)00400-9. [DOI] [PubMed] [Google Scholar]
- He M.L., Zheng B., Peng Y., Peiris J.S., Poon L.L., Yuen K.Y., Lin M.C., Kung H.F., Guan Y. Inhibition of SARS-associated coronavirus infection and replication by RNA interference. JAMA. 2003;290:2665–2666. doi: 10.1001/jama.290.20.2665. [DOI] [PubMed] [Google Scholar]
- Hsu Y.S., Chien R.N., Yeh C.T., Sheen I.S., Chiou H.Y., Chu C.M., Liaw Y.F. Long-term outcome after spontaneous HBeAg seroconversion in patients with chronic hepatitis B. Hepatology. 2002;35:1522–1527. doi: 10.1053/jhep.2002.33638. [DOI] [PubMed] [Google Scholar]
- Hu W.Y., Myers C.P., Kilzer J.M., Pfaff S.L., Bushman F.D. Inhibition of retroviral pathogenesis by RNA interference. Curr. Biol. 2002;12:1301–1311. doi: 10.1016/s0960-9822(02)00975-2. [DOI] [PubMed] [Google Scholar]
- Jacque J.M., Triques K., Stevenson M. Modulation of HIV-1 replication by RNA interference. Nature. 2002;418:435–538. doi: 10.1038/nature00896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapadia S.B., Brideau-Andersen A., Chisari F.V. Interference of hepatitis C virus RNA replication by short interfering RNAs. Proc. Natl. Acad. Sci. U.S.A. 2003;100:2014–2018. doi: 10.1073/pnas.252783999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N.S., Dohjima T., Bauer G., Li H., Li M.J., Ehsani A., Salvaterra P., Rossi J. Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nat. Biotechnol. 2002;20:500–505. doi: 10.1038/nbt0502-500. [DOI] [PubMed] [Google Scholar]
- McCaffrey A.P., Nakai H., Pandey K., Huang Z., Salazar F.H., Xu H., Wieland S.F., Marion P.L., Kay M.A. Inhibition of hepatitis B virus in mice by RNA interference. Nat. Biotechnol. 2003;21:639–644. doi: 10.1038/nbt824. [DOI] [PubMed] [Google Scholar]
- McNair A.N., Main J., Thomas H.C. Interactions of the human immunodeficiency virus and the hepatotropic viruses. Semin. Liver Dis. 1992;12:188–196. doi: 10.1055/s-2007-1007390. [DOI] [PubMed] [Google Scholar]
- Paul C.P., Good P.D., Winer I., Engelke D.R. Effective expression of small interfering RNA in human cells. Nat. Biotechnol. 2002;20:505–508. doi: 10.1038/nbt0502-505. [DOI] [PubMed] [Google Scholar]
- Peterlin B.M., Luciw P.A. Molecular biology of HIV. AIDS. 1988;2(Suppl. 1):S29–S40. doi: 10.1097/00002030-198800001-00005. [DOI] [PubMed] [Google Scholar]
- Pugh J.C., Yaginuma K., Koike K., Summers J. Duck hepatitis B virus (DHBV) particles produced by transient expreesion of DHBV DNA in a human hepatoma cell line are infectious in vitro. J. Virol. 1988;62:3516–3531. doi: 10.1128/jvi.62.9.3513-3516.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger C., Mason W.S. Hepatitis B virus biology. Microbiol. Mol. Biol. Rev. 2000;64:51–68. doi: 10.1128/mmbr.64.1.51-68.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlomai A., Shaul Y. Inhibition of hepatitis B virus expression and replication by RNA interference. Hepatology. 2003;37:764–770. doi: 10.1053/jhep.2003.50146. [DOI] [PubMed] [Google Scholar]
- Verma U.N., Surabhi R.M., Schmaltieg A., Becerra C., Gaynor R.B. Small interfering RNAs directed against beta-catenin inhibit the in vitro and in vivo growth of colon cancer cells. Clin. Cancer Res. 2003;9(4):1291–1300. [PubMed] [Google Scholar]
- Wu K.L., Zhang X., Zhang J., Yang Y., Mu Y.X., Liu M., Lu L., Li Y., Zhu Y., Wu J. Inhibition of hepatitis B virus gene expression by single and dual small interfering RNA treatment. Virus Res. 2005;112:100–107. doi: 10.1016/j.virusres.2005.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Yang N., Mohamed-Hadley A., Rubin S.C., Coukos G. Vector based RNAi; a novel tool for isoform-specific knock-down of VEGF and anti-angiogenesis gene therapy of cancer. Biochem. Biophys. Res. Commun. 2003;303:1169–1178. doi: 10.1016/s0006-291x(03)00495-9. [DOI] [PubMed] [Google Scholar]


