Abstract
Severe acute respiratory syndrome (SARS) is an emerging infectious disease caused by a novel coronavirus (SARS-CoV). The binding of SARS-CoV spike (S) protein to cellular angiotensin-converting enzyme 2 (ACE2) is the first step in SARS-CoV infection. Therefore, we assayed the inhibitory effects of small peptides derived from S protein on the binding of S protein to ACE2 and on the S-protein-pseudotyped retrovirus infectivity. SP-4 (residues 192–203), SP-8 (residues 483–494), and SP-10 (residues 668–679) significantly blocked the interaction between S protein and ACE2 by biotinylated enzyme-linked immunosorbent assay, with IC50 values of 4.30 ± 2.18, 6.99 ± 0.71, and 1.88 ± 0.52 nmol, respectively. Peptide scanning suggested the region spanning residues 660–683 might act as a receptor-binding domain. SP-10 blocked both binding of the S protein and infectivity of S protein-pseudotyped retrovirus to Vero E6 cells. In conclusion, this is the first report of small peptides designed to disrupt the binding of SARS-CoV S protein to ACE2. Our findings suggest that SP-10 may be developed as an anti-SARS-CoV agent for the treatment of SARS-CoV infection.
Keywords: SARS-CoV, Spike protein, Angiotensin-converting enzyme 2, Peptide, Vero E6 cells, Pseudovirus
1. Introduction
Severe acute respiratory syndrome (SARS) is a new emerging human disease, resulting in progressive respiratory failure and death in close to 10% of infected individuals (Ksiazek et al., 2003, Peiris et al., 2003). A novel coronavirus was identified as the etiological agent of SARS and designated as SARS coronavirus (SARS-CoV) (Drosten et al., 2003, Fouchier et al., 2003). The full-length genome sequence of SARS-CoV was elucidated within weeks after the identification of this novel pathogen (Marra et al., 2003, Rota et al., 2003). The angiotensin-converting enzyme 2 (ACE2) was identified as a functional receptor for SARS-CoV (Li et al., 2003). In spite of the remarkable pace of discovery, there are not vaccines or effective therapies currently available.
SARS-CoV spike (S) protein is a large type I transmembrane glycoprotein projected from viral envelope (Bosch et al., 2003). SARS-CoV S protein consists of two functional domains S1 (residues 1–667) and S2 (residues 668–1255) (Wu et al., 2004). S1 contains the receptor-binding site and is responsible for binding to cellular receptors on the target cells (Li et al., 2003). S2 contains two heptad repeat regions (HR1 and HR2), which assemble into a six-helix bundle and result in membrane fusion (Liu et al., 2004, Tripet et al., 2004). Moreover, S protein contains important virus-neutralizing epitopes that elicit neutralizing antibody in the host species (Chen et al., 2005, Hofmann et al., 2004a, Sui et al., 2004). Several reports indicated that blocking the binding of the S protein to cellular receptors can prevent virus entry. For examples, antibody against S protein efficiently neutralizes SARS-CoV and inhibits syncytial formation between S protein and ACE2 (Keng et al., 2005, Sui et al., 2004). Furthermore, soluble S or ACE2 protein blocks S protein mediates infection (Hofmann et al., 2004b, Moore et al., 2004, Wong et al., 2004). These finding suggested that S protein might be an attractive target for drug development.
Recently, small peptides derived from the HR regions of SARS-CoV S protein have been shown to inhibit SARS-CoV infection by the interference of SARS-CoV fusion with target cells (Liu et al., 2004, Ni et al., 2005, Zhu et al., 2004, Yuan et al., 2004). We designed a series of peptides corresponding to SARS-CoV S protein to evaluate the inhibitory potential of small peptides on both the binding of S protein and the infectivity of S-protein-pseudotyped retrovirus to Vero E6 cells in this study. This approach of utilizing compounds that block receptor interaction has proven useful in other viral systems, including human immunodeficiency virus type 1 (HIV-1) and hepatitis C virus (DeClercq, 2001, van Compernolle et al., 2003). Results of this study indicated that SP-10 (residues 668–679) was a potent inhibitor and abolished binding of the S protein to ACE2 and Vero E6 cells.
2. Materials and methods
2.1. Design and synthesis of peptides
The hydrophilicity, surface probability, and chain flexibility of SARS-CoV S protein were calculated by PeptideStructure program of Genetics Computer Group according to Kyte and Doolittle plots, Emini prediction, and Karplus and Schulz prediction, respectively (Emini et al., 1985, Karplus and Schulz, 1985, Kyte and Doolittle, 1982). The peptides of 12 residues from SARS-CoV S protein were synthesized by solid-phase method (CytoMol Corp., Union City, CA, USA). The purity (>97%) and composition of peptides were assessed by high-pressure liquid chromatography and electrospray mass spectrometry. The peptides were dissolved in water to a final concentration of 50 μg/μl and stored at −20 °C.
2.2. Expression and purification of recombinant SARS-CoV S protein
The SARS-CoV S gene was cloned into pET-28b(+) (Novagen, Madison, WI, USA) to create the pET-spike expression plasmid (Ho et al., 2004). Recombinant S protein was expressed in Escherichia coli (E. coli) BL21(DE3)pLysS strain by transforming the pET-spike to produce an N-terminal fusion with six histidine residues. The expression and purification of recombinant SARS-CoV S protein were performed as described previously (Ho et al., 2004). Protein was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and quantified with a Bradford assay (Bio-Rad, Hercules, CA, USA). Recombinant S protein produced by baculovirus expression system was purchased from Protein Science (Meriden, CT, USA).
2.3. Biotinylation of recombinant S protein
Recombinant E. coli-expressed or baculovirus-expressed S protein was mixed with Sulfo-NHS-LS-biotin (Pierce, Rockford, IL, USA) in a ratio of 1:10. After a 2 h-incubation on ice, the unincorporated biotin was removed by centricon-10 filtration (Millipore, Bedford, MA, USA), and the biotinylated S protein was stored at 4 °C until further analysis. Sulfo-NHS-LS-biotin should be prepared freshly by dissolving in water.
2.4. Biotinylated enzyme-linked immunosorbent assay (ELISA)
Microtiter plates (MaxiSorp Nunc-Immum™ plates, Nunc, Denmark) were coated at 4 °C overnight with 50 μl of spike, ACE2 (R&D Systems, Minneapolis, MN, USA), or bovine serum albumin (BSA) (Sigma, St. Louis, MO, USA), which was diluted in 0.05 M carbonate buffer (16 mM Na2CO2, 34 mM NaHCO3, pH 9.6). The wells were rinsed with 200 μl washing buffer (0.5% Tween 20 in phosphate-buffered saline (PBS) (137 mM NaCl, 1.4 mM KH2PO4, 4.3 mM Na2HPO4, 2.7 mM KCl, pH 7.2)), and blocked with 200 μl blocking buffer (5% BSA in washing buffer) by incubating at 37 °C for 30 min. The absorbed protein in each well was challenged with 50 μl diluted biotinylated protein and incubated at 37 °C for 1 h. After three washes with washing buffer, 50 μl diluted peroxidase-conjugated avidin was added to each well and incubated at 37 °C for 1 h. Following three washes, 50 μl chromogenic substrate, 2,2′-azinobis(3-ethylbenzthiazoline-sulfonic acid) (Sigma, St. Louis, MO, USA), was added to each well and incubated at 37 °C for 15 min. The absorbance was read at 405 nm in an ELISA plate reader (Anthos Labtec Instruments, Austria).
For the competition assay, biotinylated S protein (1 nmol) was mixed with 10 nmol peptide and incubated at 37 °C with shaking. After a 2 h-incubation, the mixture was added to wells, which were coated with ACE2, and incubated at 37 °C for 1 h. Following three washes, peroxidase-conjugated avidin and chromatic substrate were sequentially added. The absorbance was read at 405 nm in an ELISA plate reader. The inhibition was calculated by [1 − (OD value of mixture containing peptide and spike/OD value of mixture containing spike only)] × 100.
2.5. Immunofluorescence assay (IFA)
Vero E6 cells (104 cells) were seeded in 24-well plates containing glass coverslips and incubated at 37 °C for 1 day. The coverslips were then rinsed with PBS, fixed with 3.7% PBS-buffed formaldehyde at room temperature for 30 min, and blocked with 1% BSA at 37 °C for 1 h. After four washes with PBS, biotin-labeled S protein was added to each coverslip and incubated at 4 °C overnight. Following four washes with PBS, diluted fluorescence-conjugated streptavidin (Chemicon, Temecula, CA, USA) was added and incubated at 37 °C for 90 min in the dark. The coverslips were then washed four times with PBS, placed onto glass slides, mounted with fluoromount G (Electron Microscopy Sciences, Hatfield, PA, USA), and observed under a confocal microscope (Leica, Germany).
2.6. Infection with S-protein-pseudotyped retrovirus
Recombinant retroviruses expressing a luciferase reporter gene and pseudotyped with S proteins were produced as described previously (Li et al., 2005, Sui et al., 2005). Briefly, 293T cells were cotransfected with a plasmid pcDNA-spike encoding S protein, a plasmid pCMVΔR8.2 encoding HIV-1 Gag-Pol, and a plasmid pHIV-Luc encoding the firefly luciferase reporter gene under control of the HIV-1 long terminal repeat. Forty-eight hours later, viral supernatants were harvested and stored at −80 °C. S-protein-pseudotyped retroviruses were then mixed with various amounts of SP-10 and incubated at 37 °C with shaking. After a 2 h-incubation, the mixture was added to ACE2-expression Vero E6 cells in a 96-well plate. Forty-eight hours postinfection, cells were harvested and the luciferase activity was assayed as previously described (Hsiang et al., 2005). Relative infectivity was presented as comparison with the relative luciferase unit (RLU) relative to untreated cells.
2.7. Statistical analysis
Data were presented as mean ± standard error. Student's t-test was used for comparisons between two experiments. A value of p < 0.05 was considered statistically significant.
3. Results
3.1. Recombinant S protein binds to ACE2 in a dose-dependent manner
In order to investigate the binding ability of S protein to ACE2, we expressed and purified the SARS-CoV S protein from E. coli. The 138-kDa recombinant S protein was expressed in soluble form, and the amount of recombinant S protein recovered was approximately 0.2–0.3 mg/100 ml of bacterial culture (Ho et al., 2004). Biotinylated ELISA and Western blot show the S protein can bind to Vero E6 cell lysate (Ho et al., 2004). Since ACE2 is the functional receptor for SARS-CoV (Li et al., 2003), we designed an ELISA assay with ACE2 instead of Vero E6 cell lysate coated onto the plate. The biotinylated S protein was added to the ELISA plate coated with ACE2 and the interaction was measured using peroxidase-conjugated avidin and peroxidase substrate.
The ACE2 was coated on ELISA plates and challenged with biotin-labeled S protein. The binding ability of S protein to ACE2 was evaluated by the OD value. As shown in Fig. 1 , free biotin or biotin-labeled BSA did not bind to ACE2 in this assay. Additionally, S protein did not bind to the wells coated with the control protein BSA (data not shown). These results demonstrated the specificity of biotinylated ELISA. S protein bound to ACE2 in a dose-dependent manner. The interaction between S protein and ACE2 displayed a sigmoidal curve between 0.001 and 10 μg/ml S protein, indicating that S protein bound to ACE2 cooperatively. Moreover, the binding ability of S protein to ACE2 was saturated when the concentration of S protein exceeded 10 μg/ml, suggesting that one molecule of recombinant S protein probably bound to 1.15 molecules of ACE2. These results revealed the specificity of biotinylated ELISA in analyzing the receptor-binding ability of S protein. It also suggested that recombinant S protein could serve as a probe to analyze the interaction between SARS-CoV and cellular receptors.
Fig. 1.
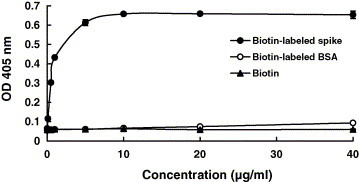
Analysis of SARS-CoV S protein and ACE2 interaction by biotinylated ELISA. The wells were coated with 1 ng of ACE2 and challenged with various amounts of biotin-labeled S protein (●), biotin-labeled BSA (○) or biotin (▴). Following three washes, peroxidase-conjugated avidin and chromatic substrate were sequentially added. The absorbance was read at 405 nm in an ELISA plate reader. Values are mean ± standard error of four independent assays.
3.2. Small peptides block the binding of S protein to ACE2
We designed 14 small peptides derived from S protein to analyze their inhibitory effects on the interaction of S protein and ACE2. Interaction of viral proteins with cellular receptors is mostly determined by hydrophobic interaction, surface charge distribution, and geometry (Dimitrov, 2004). In an attempt to design the small peptides, we analyzed the hydrophilicity, surface probability, and chain flexibility of SARS-CoV S protein according to Kyte and Doolittle plots, Emini prediction, and Karplus and Schulz prediction, respectively (Emini et al., 1985, Karplus and Schulz, 1985, Kyte and Doolittle, 1982). Based on these criteria, we designed 14 synthetic peptides of 12 residues (Table 1 ). The hydrophilic index of 14 small peptides ranged from 1.3344 to −0.3012, with an average of 0.042. We employed the Chou–Fasman method which suggested that SP-3 and SP-14 form an α-helical structure while SP-4, SP-8, and SP-10 form a β-sheet (Chou and Fasman, 1978).
Table 1.
Sequences and properties of synthetic peptides used in this study
| Peptidea | Amino acid sequence | Molecular weight (Da) | Average of hydrophilic index per residueb | Predicted secondary structurec |
|---|---|---|---|---|
| SP-1 (23–34) | DDVQAPNYTQHT | 1388.53 | 1.2319 | – |
| SP-2 (34–45) | TSSMRGVYYPDE | 1404.62 | 0.7761 | – |
| SP-3 (84–95) | KDGIYFAATEKS | 1329.67 | 0.0608 | α-Helix |
| SP-4 (192–203) | GFLYVYKGYQPI | 1447.86 | 0.2798 | β-Sheet |
| SP-5 (292–303) | SFEIDKGIYQTS | 1387.69 | 0.6203 | – |
| SP-6 (426–437) | RNIDATSTGNYN | 1325.43 | 1.2273 | – |
| SP-7 (444–455) | RHGKLRPFERDI | 1524 | 1.3344 | – |
| SP-8 (483–494) | FYTTTGIGYQPY | 1410.68 | 0.4155 | β-Sheet |
| SP-9 (581–592) | VSVITPGTNASS | 1132.26 | −0.3012 | – |
| SP-10 (668–679) | STSQKSIVAYTM | 1315.6 | −0.0427 | β-Sheet |
| SP-11 (740–751) | SFCTQLNRALSG | 1296.62 | −0.1002 | – |
| SP-12 (1027–1038) | CGKGYHLMSFPQ | 1367.86 | 0.1773 | – |
| SP-13 (1066–1078) | HEGKAYFPREGV | 1389.75 | 0.3499 | – |
| SP-14 (1125–1136) | PELDSFKEELDK | 1449.81 | 1.2488 | α-Helix |
Numbers in the brackets are the beginning and end residues for each peptide corresponding to S protein.
The hydrophilic index is calculated according to Kyte and Doolittle plots.
The secondary structure of peptide is calculated according to Chou–Fasman prediction.
To evaluate the inhibitory effects of the 14 small peptides on the interaction between S protein and ACE2, we mixed 10 nmol of each peptide with 1 nmol of biotin-labeled S protein, incubated the mixture at 37 °C for 2 h, and added to ACE2-coated wells. As shown in Fig. 2 , pre-incubation of biotinylated S protein with BSA slightly inhibited the binding of S protein to ACE2, with the inhibitory percentage of 7%. Pre-incubation of biotinylated S protein with the peptide (endoG, EGWRRRREDARAAL) derived from human endonuclease G inhibited the binding of S protein to ACE2, with the inhibitory percentage of 30%. However, SP-4, SP-8, SP-10, SP-11, and SP-12 significantly blocked the binding of S protein to ACE2, exhibiting 50–90% inhibition at 10 nmol. Therefore, these results suggested that SP-4, SP-8, SP-10, SP-11, and SP-12 were potential inhibitors of the interaction between S protein and ACE2.
Fig. 2.
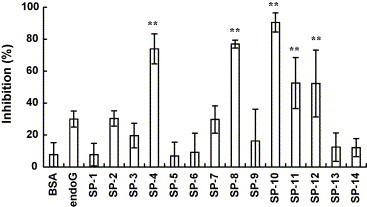
Inhibitory effects of peptides on the SARS-CoV S protein and ACE2 interaction by competitive biotinylated ELISA. Biotin-labeled S protein (1 nmol) was mixed with 10 nmol of synthetic peptides or BSA and incubated at 37 °C with shaking. After a 2-h incubation, the mixtures were added to wells, which were coated with 1 ng of ACE2 and incubated at 37 °C for 1 h. Following three washes, peroxidase-conjugated avidin and chromatic substrate were sequentially added. The absorbance was read at 405 nm in an ELISA plate reader. The results are expressed as inhibition described in Section 2. Values are mean ± standard error of six independent assays. **p < 0.01, compared with BSA.
3.3. A novel putative receptor-binding region of S protein is identified by a peptide-scanning method
SP-4, SP-8, and SP-10 blocked the ACE2 and S protein interaction in a dose-dependent manner (Fig. 3 ). The IC50 values of SP-4, SP-8, and SP-10 were 4.3 ± 2.18, 6.99 ± 0.71, and 1.88 ± 0.52 nmol, respectively. SP-11 and SP-12 were excluded because they bound not only S protein but also BSA (data not shown). To further identify the biologically active peptides, the peptide-scanning method that involved synthesizing overlapping peptides of 12 residues covering additional amino acids on both the amino and carboxy ends of SP-10 was used. The inhibitory potential of small overlapping peptides on the SARS-CoV S protein and ACE2 interaction was analyzed by competitive biotinylated ELISA. SP-10-2 (residues 660–671), SP-10-3 (residues 664–675), SP-10-4 (residues 672–683) had inhibitory activities, with the IC50 values of 6.21 ± 2.13, 5.47 ± 0.41, and 2.07 ± 1.01 nmol, respectively (Table 2 ). However, SP-10-1 (residues 648–659) and SP-10-5 (residues 676–687) did not block the ACE2 and S protein interaction. These findings suggested that the region spanning residues 660–683 of SARS-CoV S protein may interact with ACE2.
Fig. 3.
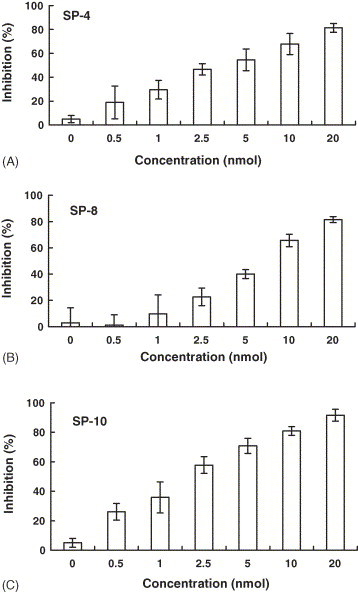
Inhibitory effects of SP-4, SP-8, and SP-10 on the SARS-CoV S protein and ACE2 interaction by competitive biotinylated ELISA. Biotin-labeled S protein (1 nmol) was mixed with various amounts of SP-4 (A), SP-8 (B) or SP-10 (C), and incubated at 37 °C with shaking. After a 2-h incubation, the mixtures were added to wells, which were coated with 1 ng of ACE2, and incubated at 37 °C for 1 h. Following three washes, peroxidase-conjugated avidin and chromatic substrate were sequentially added. The absorbance was read at 405 nm in an ELISA plate reader. The results are expressed as inhibition described in Section 2. Values are mean ± standard error of six independent assays.
Table 2.
The IC50 values of overlapping peptides covering additional amino acids on both the amino and carboxy ends of SP-10
| Peptidea | Amino acid sequence | IC50 (nmol)b |
|---|---|---|
| SP-10-1 (648–659) | CDIPIGAGICAS | >20 |
| SP-10-2 (660–671) | YHTVSLLRSTSQ | 6.21 ± 2.13 |
| SP-10-3 (664–675) | SLLRSTSQKSIV | 5.47 ± 0.41 |
| SP-10 (668–679) | STSQKSIVAYTM | 1.88 ± 0.52 |
| SP-10-4 (672–683) | KSIVAYTMSLGA | 2.07 ± 1.01 |
| SP-10-5 (676–687) | AYTMSLGADSSI | >20 |
Numbers in the brackets are the beginning and end residues for each peptide corresponding to S protein.
The IC50 value of each peptide was determined as the quantity of peptide required to inhibit the interaction between S protein and ACE2 at 50%. Values are mean ± standard error of four independent assays.
3.4. SP-10 blocks both the binding of S protein and the infectivity of S-protein-pseudotyped retrovirus to Vero E6 cells
The inhibitory potential of SP-10 on the SARS-CoV S protein and Vero E6 cell interaction was further analyzed by IFA and S-protein-pseudotyped retrovirus infectivity. Vero E6 cells were treated with BSA, biotin-labeled S protein or SP-10/biotin-labeled S protein mixture, and stained with fluorescence-conjugated streptavidin. BSA-treated cells showed negative result, whereas the biotinylated S protein-treated Vero E6 cells showed the strong fluorescence (Fig. 4A). Treatment of Vero E6 cells with SP-10/biotin-labeled S protein mixture diminished the cell-associated fluorescence in a dose-dependent manner. Because the S protein expressed in E. coli is not glycosylated, we further analyzed the inhibitory potential of SP-10 using glycosylated S protein purified from baculovirus. Similar results were obtained by using the recombinant S protein produced by baculovirus expression system (Fig. 4B). These results indicated that SP-10 was capable of blocking the binding of S protein to Vero cells.
Fig. 4.
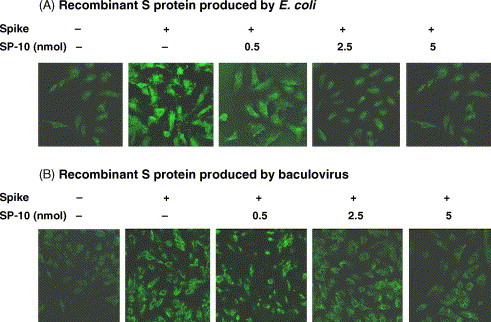
Inhibitory effect of SP-10 on the SARS-CoV S protein and Vero E6 cell interaction by IFA. Vero E6 cells were cultured on glass coverslips and incubated with 20 μg/ml BSA, 20 μg/ml biotin-labeled S protein, 0.5 nmol SP-10/20 μg/ml biotin-labeled S protein mixture, 2.5 nmol SP-10/20 μg/ml biotin-labeled S protein mixture, or 5 nmol SP-10/20 μg/ml biotin-labeled S protein mixture. Recombinant S protein was purified from E. coli (A) or baculovirus (B). After a 16 h-incubation at 4 °C, cells were stained with fluorescence-conjugated streptavidin and evaluated under a confocal microscope. Magnification, 400×.
In addition to IFA, Vero E6 cells transfected with the plasmid encoding human ACE2 were infected with vector-pseudotyped retrovirus, S-protein-pseudotyped retrovirus, or SP-10/S-protein-pseudotyped retrovirus mixture. The luciferase activity was assayed 48 h postinfection. SP-10 inhibited the S-protein-pseudotyped retrovirus infectivity in a dose-dependent manner (Fig. 5 ). These results indicated that SP-10 was a novel anti-SARS-CoV peptide that was able to block the SARS-CoV S protein binding to Vero E6 cells.
Fig. 5.
Inhibitory effect of SP-10 on the SARS-CoV S-protein-pseudotyped retrovirus infectivity. S-protein-pseudotyped retroviruses were mixed with various amounts of SP-10 and incubated at 37 °C with shaking. After a 2-h incubation, the mixtures were inoculated with Vero E6 cells transfected with the plasmid encoding human ACE2. The luciferase activity of cell lysate was assayed 2 days postinfection. Relative infectivity is presented as comparison with the RLU relative to untreated cells. Values are mean ± standard error of four independent assays.
4. Discussion
In this study, we analyzed peptide mimics of the S protein to block its interaction with Vero E6 cells by biotinylated ELISA, IFA, and S-protein-pseudotyped retrovirus infectivity. SP-4 (residues 192–203), SP-8 (residues 483–494), and SP-10 (residues 660–683) exhibited the inhibitory potentials. Additional mapping showed that SP-10-2 (residues 660–671), SP-10-3 (residues 664–675), and SP-10-4 (residues 672–683) had inhibitory activities. The receptor-binding regions of SARS-CoV S protein have been defined in several reports. For examples, Xiao et al. (2003) identified that the receptor-binding site of S protein is localized between amino acid residues 203 and 537. Wong et al. (2004) indicated that the 193-amino acid fragment of S protein (residues 318–510) binds ACE2 efficiently. Babcock et al. (2004) suggested that amino acids 270–510 of S protein are required for interaction with receptor. By analyzing a series of peptides derived from S protein, we identified that peptides spanning residues 192–203, 483–494, and 660–683 of S protein efficiently blocked the binding of S protein to ACE2. Therefore, our findings refined and extended the possible domains involved in receptor interaction.
Virus entry process is an attractive drug target for blocking infection. Virus envelope protein initially binds to cell surface receptor, leading to its global conformational changes. Subsequently, the fusion peptide is exposed to insert into the target cell membrane, resulting in fusion between the cell membrane and viral envelope, which allow the virus genome to enter the target cells (Liu et al., 2004). Based on this process, fusion-inhibitory peptides have been successfully developed, or under advance clinical trial for various viruses, such as T-20 for HIV-1, C-peptides (P-400 and Pcr-400) for human T-cell leukemia virus, and D4E1 for feline immunodeficiency virus (Kilby et al., 1998, Ma et al., 2002, Pinon et al., 2003). Recently, this strategy of utilizing peptides that block membrane fusion has been applied in SARS-CoV. For examples, HR2-peptide of S protein has been shown to interfere with S protein conformational changes, resulting in inhibition of SARS-CoV fusion with target cells (Liu et al., 2004, Ni et al., 2005, Zhu et al., 2004, Yuan et al., 2004). Peptides derived from HR1 region of S protein have also been identified as potential inhibitors for virus entry (Ni et al., 2005, Yuan et al., 2004). In this study, we designed small peptides derived from S protein to evaluate their inhibitions of S protein binding to ACE2 and Vero E6 cells. This approach of utilizing compounds that block receptor interaction has proven useful in other viral systems, including HIV-1 and hepatitis C virus, where compounds designed to inhibit viral entry through blockade of the viral receptors are finding success in vitro and in clinical trials (DeClercq, 2001, van Compernolle et al., 2003). Because RNA viruses are capable of being randomly and selectively altered due to high replication numbers and the infidelity of the viral RNA polymerase, it is possible that virus would adapt resistance to inhibitors. However, Chakraborti et al. (2005) showed that most of the natural or artificial mutations on S protein did not affect the binding of S protein to ACE2. These findings suggested that the viruses could easily mutate and escape antibodies that do not exhibit the same energy profile of binding to S as ACE2. Therefore, our finding suggested that SP-10 was a potent inhibitor for the inhibition of S protein binding to Vero E6 cells, the first step in viral infection.
In conclusion, this is the first report of small peptides designed to disrupt the interaction of SARS-CoV S protein and ACE2. By analyzing a series of peptides derived from S protein, we identified that peptides spanning residues 192–203, 483–494, and 660–683 of S protein efficiently blocked the binding of S protein to ACE2. A novel putative receptor-binding region of S protein was defined by a peptide-scanning method. Furthermore, SP-10 has been identified as a potent inhibitor for the inhibition of S protein binding to Vero E6 cells. Therefore, SP-10 can be used as a candidate in designing more potent anti-SARS-CoV peptides for drug development in the future.
Acknowledgement
We thank Prof. M. Farzan for providing plasmids for the construction of pseudovirus. This work was supported by grants from National Science Council and China Medical University, Taiwan, ROC.
References
- Babcock G.J., Esshaki D.J., Thomas W.D., Ambrosino D.M. Amino acids 270 to 510 of the severe acute respiratory syndrome coronavirus spike protein are required for interaction with receptor. J. Virol. 2004;78:4552–4560. doi: 10.1128/JVI.78.9.4552-4560.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch B.J., van der Zee R., de Haan C.A., Rottier P.J. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 2003;77:8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborti S., Prabakaran P., Xiao X., Dimitrov D.S. The SARS coronavirus glycoprotein receptor binding domain: fine mapping and functional characterization. Virol. J. 2005;2:73–82. doi: 10.1186/1743-422X-2-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Zhang L., Qin C., Ba L., Yi C.E., Zhang F., Wei Q., He T., Yu W., Yu J., Gao H., Tu X., Gettie A., Farzan M., Yuen K.Y., Ho D.D. Recombinant modified vaccinia virus Ankara expressing the spike glycoprotein of severe acute respiratory syndrome coronavirus induces protective neutralizing antibodies primarily targeting the receptor binding region. J. Virol. 2005;79:2678–2688. doi: 10.1128/JVI.79.5.2678-2688.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou P.Y., Fasman G.D. Prediction of the secondary structure of proteins from their amino acid sequence. Adv. Enzymol. Relat. Areas Mol. Biol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- DeClercq E. New developments in anti-HIV chemotherapy. Curr. Med. Chem. 2001;8:1543–1572. doi: 10.2174/0929867013371842. [DOI] [PubMed] [Google Scholar]
- Dimitrov D.S. Virus entry: molecular mechanisms and biomedical application. Nat. Rev. Microbiol. 2004;2:109–122. doi: 10.1038/nrmicro817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., Berger A., Burguiere A.M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.C., Muller S., Rickerts V., Sturmer M., Vieth S., Klenk H.D., Osterhaus A.D., Schmitz H., Doerr H.W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- Emini E.A., Hughes J.V., Perlow D.S., Boger J. Induction of hepatitis A virus-neutralizing antibody by a virus-specific synthetic peptide. J. Virol. 1985;55:836–839. doi: 10.1128/jvi.55.3.836-839.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouchier R.A., Kuiken T., Schutten M., van Amerongen G., van Doornum G.J.J., van den Hoogen B.G., Peiris M., Lim W., Stohr K., Osterhaus A.D.M.E. Koch's postulates fulfilled for SARS virus. Nature. 2003;423:240. doi: 10.1038/423240a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho T.Y., Wu S.L., Cheng S.E., Wei Y.C., Huang S.P., Hsiang C.Y. Antigenicity and receptor-binding ability of recombinant SARS coronavirus spike protein. Biochem. Biophys. Res. Commun. 2004;313:938–947. doi: 10.1016/j.bbrc.2003.11.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann H., Hattermann K., Marzi A., Gramberg T., Geiner M., Krumbiegel M., Kuate S., Uberla K., Niedrig M., Pohlmann S.H. S protein of severe acute respiratory syndrome-associated coronavirus mediates entry into hepatoma cell lines and is targeted by neutralizing antibodies in infected patients. J. Virol. 2004;78:6134–6142. doi: 10.1128/JVI.78.12.6134-6142.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann H., Geier M., Marzi A., Krumbiegel M., Peipp M., Fey G.H., Gramberg T., Pohlmann S. Susceptibility to SARS coronavirus S protein-driven infection correlates with expression of angiotensin converting enzyme 2 and infection can be blocked by soluble receptor. Biochem. Biophys. Res. Commun. 2004;319:1216–1221. doi: 10.1016/j.bbrc.2004.05.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiang C.Y., Wu S.L., Ho T.Y. Morin inhibited 12-O-tetradecanoylphorbol-13-acetate-induced hepatocellular transformation via activator protein 1 signaling pathway and cell cycle progression. Biochem. Pharmcol. 2005;69:1603–1611. doi: 10.1016/j.bcp.2005.03.008. [DOI] [PubMed] [Google Scholar]
- Karplus P.A., Schulz G.E. Prediction of chain flexibility in proteins. A tool for the selection of peptide antigens. Naturwisenschaften. 1985;72:212–213. [Google Scholar]
- Keng C.T., Zhang A., Shen S., Lip K.M., Fielding B.C., Tan T.H., Chou C.F., Loh C.B., Wang S., Fu J., Yang X., Lim S.G., Hong W., Tan Y.J. Amino acids 1055 to 1192 in the S2 region of severe acute respiratory syndrome coronavirus S protein induce neutralizing antibodies: implications for the development of vaccines and antiviral agents. J. Virol. 2005;79:3289–3296. doi: 10.1128/JVI.79.6.3289-3296.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilby J.M., Hopkins S., Venetta T.M., DiMassimo B., Cloud G.A., Lee J.Y., Alldredge L., Hunter E., Lambert D., Bolognesi D., Matthews T., Johnson M.R., Nowak M.A., Shaw G.M., Saag M.S. Potent suppression of HIV-1 replication in humans by T-20, a peptide inhibitor of gp41-mediated virus entry. Nat. Med. 1998;4:1302–1307. doi: 10.1038/3293. [DOI] [PubMed] [Google Scholar]
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A., Humphrey C.D., Shieh W., Guarner J., Paddock C.D., Rota P., Fields B., DeRisi J., Yang J., Cox N., Hughes J.M., LeDuc J.W., Bellini W.J., Anderson L.J. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 1953-1966;348 doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Zhang C., Sui J., Kuhn J.H., Moore M.J., Luo S., Wong S.K., Huang I.C., Xu K., Vasilieva N., Murakami A., He Y., Marasco W.A., Guan Y., Choe H., Farzan M. Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. EMBO J. 2005;24:1634–1643. doi: 10.1038/sj.emboj.7600640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Xiao G., Chen Y., He Y., Niu J., Escalante C.R., Xiong H., Farmar J., Debnath A.K., Tien P., Jiang S. Interaction between heptad repeat 1 and 2 regions in spike protein of SARS-associated coronavirus: implications for virus fusogenic mechanism and identification of fusion inhibitors. Lancet. 2004;363:938–947. doi: 10.1016/S0140-6736(04)15788-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Kennedy-Stoskopf S., Jaynes J.M., Thurmond L.M., Tompkins W.A. Inhibitory activity of synthetic peptide antibiotics on feline immunodeficiency virus infectivity in vitro. J. Virol. 2002;76:9952–9961. doi: 10.1128/JVI.76.19.9952-9961.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S., Khattra J., Asano J.K., Barber S.A., Chan S.Y., Cloutier A., Coughlin S.M., Freeman D., Girn N., Griffith O.L., Leach S.R., Mayo M., McDonald H., Montgomery S.B., Pandoh P.K., Petrescu A.S., Robertson A.G., Schein J.E., Siddiqui A., Smailus D.E., Stott J.M., Yang G.S., Plummer F., Andonov A., Artsob H., Bastien N., Bernard K., Booth T.F., Bowness D., Czub M., Drebot M., Fernando L., Flick R., Garbutt M., Gray M., Grolla A., Jones S., Feldmann H., Meyers A., Kabani A., Li Y., Normand S., Stroher U., Tipples G.A., Tyler S., Vogrig R., Ward D., Watson B., Brunham R.C., Krajden M., Petric M., Skowronski D.M., Upton C., Roper R.L. The Genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- Moore M.J., Dorfman T., Li W., Wong S.K., Li Y., Kuhn J.H., Coderre J., Vasilieva N., Han Z., Greenough T.C., Farzan M., Choe H. Retroviruses pseudotyped with the severe acute respiratory syndrome coronavirus spike protein efficiently infect cells expressing angiotensin-converting enzyme 2. J. Virol. 2004;78:10628–10635. doi: 10.1128/JVI.78.19.10628-10635.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni L., Zhu J., Zhang J., Yan M., Gao G.F., Tien P. Design of recombinant protein-based SARS-CoV entry inhibitors targeting the heptad-repeat regions of the spike protein S2 domain. Biochem. Biophys. Res. Commun. 2005;330:39–45. doi: 10.1016/j.bbrc.2005.02.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris J.S.M., Lai S.T., Poon L.L.M., Guan Y., Yam L.Y.C., Lim W., Nicholls J., Yee W.K.S., Yan W.W., Cheung M.T., Cheng V.C.C., Chan K.H., Tsang D.N.S., Yung R.W.H., Ng T.K., Yuen K.Y., members of the SARS study group Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinon J.D., Kelly S.M., Price N.C., Flanagan J.U., Brighty D.W. An antiviral peptide targets a coiled-coil domain of the human T-cell leukemia virus envelope glycoprotein. J. Virol. 2003;77:3281–3290. doi: 10.1128/JVI.77.5.3281-3290.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.H., Tong S., Tamin A., Lowe L., Frace M., DeRisi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., Gunther S., Osterhaus A.D., Drosten C., Pallansch M.A., Anderson L.J., Bellini W.J. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- Sui J., Li W., Murakami A., Tamin A., Matthews L.J., Wong S.K., Moore M.J., Tallarico A.S., Olurinde M., Choe H., Anderson L.J., Bellini W.J., Farzan M., Marasco W.A. Potent neutralization of severe acute respiratory syndrome (SARS) coronavirus by a human mAb to S1 protein that blocks receptor association. Proc. Natl. Acad. Sci. U.S.A. 2004;101:2536–2541. doi: 10.1073/pnas.0307140101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui J., Li W., Roberts A., Mattews L.J., Murakami A., Vogel L., Wong S.K., Subbarao K., Farzan M., Marasco W.A. Evaluation of human monoclonal antibody 80R for immunoprophylaxis of severe acure respiratory syndrome by an animal study, epitope mapping, and analysis of spike variants. J. Virol. 2005;79:5900–5906. doi: 10.1128/JVI.79.10.5900-5906.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripet B., Howard M.W., Jobling M., Holmes R.K., Holmes K.V., Hodges R.S.B. Structural characterization of the SARS-coronavirus spike S fusion protein core. J. Biol. Chem. 2004;279:20836–20849. doi: 10.1074/jbc.M400759200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Compernolle S.E., Wiznycia A.V., Rush J.R., Dhanasekaran M., Baures P.W., Todd S.C. Small molecule inhibition of hepatitis C virus E2 binding to CD81. Virology. 2003;314:371–380. doi: 10.1016/s0042-6822(03)00406-9. [DOI] [PubMed] [Google Scholar]
- Wong S.K., Li W., Moore M.J., Choe H., Farzan M. A 193-amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. J. Biol. Chem. 2004;279:3197–3201. doi: 10.1074/jbc.C300520200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X.D., Shang B., Yang R.F., Yu H., Ma Z.H., Shen X., Ji Y.Y., Lin Y., Wu Y.D., Lin G.M., Tian L., Gan X.Q., Yang S., Jiang W.H., Dai E.H., Wang X.Y., Jiang H.L., Xie Y.H., Zhu X.L., Pei G., Li L., Wu J.R., Sun B. The spike protein of severe acute respiratory syndrome (SARS) is cleaved in virus infected Vero-E6 cells. Cell Res. 2004;14:400–406. doi: 10.1038/sj.cr.7290240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X., Chakraborti S., Dimitrov A.S., Gramatikoff K., Dimitrov D.S. The SARS-CoV S glycoprotein: expression and functional characterization. Biochem. Biophys. Res. Commun. 2003;312:1159–1164. doi: 10.1016/j.bbrc.2003.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan K., Yi L., Chen J., Qu X., Qing T., Rao X., Jiang P., Hu J., Xiong Z., Nie Y., Shi X., Wang W., Ling C., Yin X., Fan K., Lai L., Ding M., Deng H. Suppression of SARS-CoV entry by peptides corresponding to heptad regions on spike glycoprotein. Biochem. Biophys. Res. Commun. 2004;319:746–752. doi: 10.1016/j.bbrc.2004.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Xiao G., Xu Y., Yuan F., Zheng C., Liu Y., Yan H., Cole D.K., Bell J.I., Rao Z., Tien P., Gao G.F. Following the rule: formation of the 6-helix bundle of the fusion core from severe acute respiratory syndrome coronavirus spike protein and identification of potent peptide inhibitors. Biochem. Biophys. Res. Commun. 2004;319:283–288. doi: 10.1016/j.bbrc.2004.04.141. [DOI] [PMC free article] [PubMed] [Google Scholar]


