Abstract
Financial resources may limit the number of samples that can be collected and analysed in disease surveillance programmes. When the aim of surveillance is disease detection and identification of case herds, a risk-based approach can increase the sensitivity of the surveillance system. In this paper, the association between two network analysis measures, i.e. ‘in-degree’ and ‘ingoing infection chain’, and signs of infection is investigated. It is shown that based on regression analysis of combined data from a recent cross-sectional study for endemic viral infections and network analysis of animal movements, a positive serological result for bovine coronavirus (BCV) and bovine respiratory syncytial virus (BRSV) is significantly associated with the purchase of animals. For BCV, this association was significant also when accounting for herd size and regional cattle density, but not for BRSV. Examples are given for different approaches to include cattle movement data in risk-based surveillance by selecting herds based on network analysis measures. Results show that compared to completely random sampling these approaches increase the number of detected positives, both for BCV and BRSV in our study population. It is concluded that network measures for the relevant time period based on updated databases of animal movements can provide a simple and straight forward tool for risk-based sampling.
Keywords: In-degree, Ingoing infection chain, Bovine coronavirus, Bovine respiratory syncytial virus
1. Introduction
Surveillance of infectious animal diseases constitutes an important part of the prevention of animal disease and can have several specific purposes, e.g. early detection, declaration of freedom or evaluation of control strategies. However, financial resources may limit the number of samples that can be collected and analysed, and a risk-based approach is then one alternative for increasing the case-finding capacity of the surveillance system. Infectious diseases are seldom homogeneously spread within the population and the benefits of searching “in the most likely place” when monitoring disease, in contrast to overall random sampling, have been previously discussed, e.g. by Cannon (2009) and Stärk et al. (2006).
Many livestock diseases can spread through direct contact between animals, and thus between herds through movements of animals. This is one of the major reasons for registering livestock transports in national databases (Anonymous, 2000). When the aim of surveillance is detection (eradication context, emergence of an exotic disease, etc.), and when the disease is expected to spread through live animal contacts, animal movement data could be used in the selection of herds to be included in surveillance activities. In such cases, herds with many live animal contacts can be assumed to have a higher probability of infection, and sampling of these would therefore increase surveillance sensitivity.
Lately there has been an increasing number of publications analysing livestock movements (Dubé et al., 2009, Martinez-Lopez et al., 2009). For instance, the outbreak of foot-and-mouth disease in the United Kingdom in 2001 was the starting point for a number of studies within this field of research (e.g. see Ortiz-Pelaez et al., 2006). However, although analysis of animal contact patterns has already been suggested for targeting the surveillance of diseases (Christley et al., 2005, Martinez-Lopez et al., 2009, Blickenstorfer et al., 2011, Nöremark et al., 2011), to our knowledge there have been almost no published applications of the use of cattle movement network analysis for implementing a risk-based surveillance so far. There are many different network measures of centrality and, in 1979 in the context of social network analysis, Freeman discussed the importance of using meaningful and intuitively interpretable measures (Freeman, 1979). For surveillance activities that target herds with an increased risk of disease due to ingoing live animal contacts, an intuitive focus would be measures of contacts that have actually occurred, rather than measures describing the relative role of the herd in connecting the entire network (e.g. different measures of betweenness). Inclusion of measures of betweenness may, on the other hand, be more applicable in models simulating spread of disease. There are different network analysis parameters describing incoming contacts that may be applied for risk-based surveillance purposes. For example, the ‘in-degree’ measure (Wasserman et al., 1994) describes the actual number of ingoing animal contacts for a herd. In addition, Nöremark (2010) and Nöremark et al. (2011) described the ‘ingoing infection chain’, which includes secondary contacts in sequences, taking into account the temporal aspect and the order in which these contacts have occurred (Fig. 1 ).
Fig. 1.
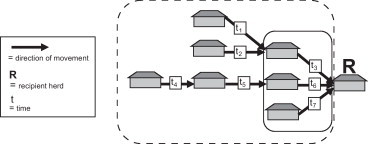
Schematic illustration of the network measures ‘in-degree’ and ‘ingoing infection chain’. The ‘in-degree’ for the recipient herd is 3 (herds included within the solid line), and assuming that t1 and t2 occur before t3, that t4 occurs before t5 and that t5 occurs before t6, the ‘ingoing infection chain’ for the recipient herd is 7 (herds included within the dotted line).
Bovine respiratory syncytial virus (BRSV) and bovine coronavirus (BCV) are examples of pathogens that can spread through live animal contacts and also indirectly, e.g. through visitors and equipment (Elvander et al., 1998, Hägglund et al., 2006, Valarcher and Taylor, 2007, Bidokhti et al., 2009, Ohlson et al., 2010). Identified risk factors for BRSV and BCV infection in Sweden include large herd size (Tråvén et al., 1999, Norström et al., 2000, Ohlson et al., 2010) and being located in southern Sweden (Elvander, 1996, Beaudeau et al., 2010) where the herd density is higher compared to northern parts. Both diseases are distributed worldwide, causing enteric and respiratory disease in beef and dairy cattle (Clark, 1993, Valarcher and Taylor, 2007).
The aim of this study was to elaborate on the potential usefulness of including network analysis measures of animal movements in the design of surveillance programmes aimed at the detection of exotic diseases. In order to investigate potential association between network analysis parameters and the presence of disease, results from a serological survey of BRSV and BCV in Swedish cattle were combined with data of reported animal movements. BRSV and BCV were used as a proxy for exotic diseases, or other serious infections under surveillance, with similar contagiousness and routes of transmission. In other words, the study was not designed to investigate risk factors for these specific diseases. Simulated sampling from the study material was used to visualise and compare risk-based approaches to a random selection strategy.
2. Materials and methods
2.1. Data
Information about movements of individual cattle in Sweden 2006 was retrieved from the database of the Swedish Board of Agriculture (described in more detail by Nöremark (2010) and Nöremark et al. (2011)). Information about herd size, i.e. the number of cattle >1 year of age, and about the geographic location of herds was also included in that database. The regional cattle herd density was calculated for all herds in the study sample by dividing the total number of cattle herds by the total area of their three-digit postal code area.
In addition, results from a cross-sectional serological study investigating spatial patterns of BRSV and BCV in Swedish cattle were used. The design of the cross-sectional study and the analytical methods used are described in detail by Beaudeau et al. (2010). In short, a randomised subset of blood samples collected within the Swedish Bovine Viral Diarrhoea control programme was used. In the original study a total of 2763 samples from young stock >12 months of age in 2137 herds were collected between November 2006 and May 2007. The samples were analyzed for presence of immunoglobulin G antibodies to BRSV and BCV by commercially available indirect enzyme-linked immunosorbent assays (ELISA; SVANOVA Biotech, Uppsala, Sweden). Cut-off was set to a corrected OD of 0.20, which is recommended by the manufacturer for individual samples. At this cut-off, the sensitivity is estimated as 94.6% for BRSV and 84.6% for BCV and specificity to 100% for both tests (SVANOVA manual). In order to get a balanced number of results per herd for the present study, one result was randomly selected from each herd, giving a total of 2137 animals and herds. Of these, 859 (40%) and 899 (42%) had tested positive for BRSV and BCV, respectively.
2.2. Statistical analyses
Network analysis of the cattle movement data was performed, including calculation of ‘in-degree’ and ‘ingoing infection chain’ for all herds in the study sample. Both measures were set to reflect all reported movements of cattle during 2006, excluding transports for slaughter (Nöremark et al., 2011). In addition, possible associations between animals testing positive for BRSV or BCV antibodies and the measures based on network analysis were investigated using logistic regression. In the regression models, the outcome was the dichotomized test result (0 = negative, 1 = positive) as regards BRSV and BCV. In addition to the in-degree and ingoing infection chain measures, herd size and regional cattle herd density were explanatory variables that were also investigated. The main effects models were decided based on univariable regression of each variable followed by a check of the correlation between explanatory variables, and multivariable regression with backward elimination of non-significant variables. The network analysis parameters were investigated separately in different models. For best fit of the models, both ‘in-degree’ and ‘ingoing infection chain’ were included in categorized form. The categories were 0, >0 to <5 and ≥5 for ‘in-degree’, and 0, >0 to <25 and ≥25 for ‘ingoing infection chain’. Fifty-six herds had missing information about live animal contacts and these were included in categories ‘in-degree’ ≥5 and ‘ingoing infection chain’ ≥25. Plausible interactions were tested one by one, and variables and interaction terms were included in the final model if they had a P-value around or below 0.05 (Wald test). Model fit was assessed by applying the Hosmer–Lemeshow goodness-of-fit test and investigation of the influence of covariate patterns (Hosmer and Lemeshow, 2000). Thirty observations were excluded from these analyses due to missing values for one or more variables.
For comparison of different strategies of sample selection, different approaches were used to select 100 results from the total of 2137 in the study sample. First, a total random sample was selected. Second, random samples amongst herds with certain levels of ‘in-degree’ or ‘ingoing infection chain’ were selected. The categories were >0 (n = 1134), and ≥5 for ‘in-degree’ (n = 172), and ≥25 for ‘ingoing infection chain’ (n = 178). Each of these sampling strategies was simulated with 10,000 repetitions, and the median, and the 5th and 95th percentiles, were calculated from the output distribution of the number of positives for BRSV or BCV from these simulations. In addition, the number of positive results in the 100 herds with the highest ‘in-degree’ and the highest ‘ingoing infection chain’ was assessed. The number of test-positive results included in the samples from the risk-based approaches was compared to the random selection approach.
2.3. Software
Data management, random selection, simulations and statistical analyses were performed using STATA/SE 11.1 (Stata Co., College Station, TX, USA). Network analysis was performed using the Python module NetworkX 0.99, and Perl 5.8.7, as described by Nöremark et al. (2011). Input to the calculation of regional densities was managed through the use of Arc GIS 9.2 (ESRI Co., Redlands, CA, USA) and a digitized map ‘Sverige 1000 plus’ version 5/2004 (Statistics Sweden).
3. Results
Based on univariable regression analysis (see Table 1 for detailed results), all potential explanatory variables were significantly associated (P < 0.001) with the outcomes (i.e. testing positive for BRSV or BCV). In the multivariable models (see Table 2 for detailed results), only ‘herd size’ and ‘regional cattle density’ were significantly associated with testing positive for BRSV. In other words, when adjusting for these covariates, neither ‘in-degree’ nor ‘ingoing infection chain’ could be shown to be associated to testing positive for BRSV. For the outcome testing positive to BCV, the significant covariates kept in the final models were ‘herd size’, ‘regional cattle density’ and also ‘in-degree’ or ‘ingoing infection chain’. No interactions between main effects were significant and these were therefore excluded from the final models.
Table 1.
Results from univariable logistic regression analyses of the combined data from a cross-sectional study for bovine coronavirus (BCV) and bovine respiratory syncytial virus (BRSV) with network analysis measures of animal movements in 2137 Swedish dairy herds.
| Variable | Odds ratio | SE | P-Value | 95% conf interval | |
|---|---|---|---|---|---|
| Outcome | |||||
| Explanatory variable | |||||
| BRSV positive | |||||
| Indegree 0 | Baseline | ||||
| Indegree 1–4 | 1.282 | 0.119 | 0.007 | 1.069 | 1.537 |
| Indegree ≥5 | 2.588 | 0.518 | 0.000 | 1.748 | 3.832 |
| Ing inf chain 0 | Baseline | ||||
| Ing inf chain 1–24 | 1.347 | 0.125 | 0.001 | 1.123 | 1.615 |
| Ing inf chain ≥25 | 1.706 | 0.329 | 0.006 | 1.170 | 2.489 |
| Herd size | 1.015 | 0.002 | 0.000 | 1.012 | 1.018 |
| Regional cattle density | 1.025 | 0.003 | 0.000 | 1.019 | 1.030 |
| BCV positive | |||||
| Indegree 0 | Baseline | ||||
| Indegree 1–4 | 1.613 | 0.149 | 0.000 | 1.345 | 1.933 |
| Indegree ≥5 | 4.898 | 1.068 | 0.000 | 3.194 | 7.509 |
| Ing inf chain 0 | Baseline | ||||
| Ing inf chain 1–24 | 1.645 | 0.152 | 0.000 | 1.372 | 1.973 |
| Ing inf chain ≥25 | 3.824 | 0.780 | 0.000 | 2.565 | 5.704 |
| Herd size | 1.021 | 0.002 | 0.000 | 1.017 | 1.025 |
| Regional cattle density | 1.014 | 0.003 | 0.000 | 1.009 | 1.019 |
Table 2.
Results from multivariable logistic regression analyses of combined data from a cross-sectional study for bovine coronavirus (BCV) and bovine respiratory syncytial virus (BRSV) and network analysis measures of animal movements in 2137 Swedish dairy herds.
| Variable | Odds ratio | SE | P-Value | 95% conf interval | |
|---|---|---|---|---|---|
| Outcome | |||||
| Explanatory variable | |||||
| BRSV positivea | |||||
| Herd size | 1.016 | 0.002 | 0.000 | 1.010 | 1.020 |
| Regional cattle density | 1.026 | 0.003 | 0.000 | 1.020 | 1.030 |
| BCV positiveb | |||||
| Indegree 0 | Baseline | ||||
| Indegree 1–4 | 1.334 | 0.131 | 0.003 | 1.104 | 1.619 |
| Indegree ≥5 | 1.824 | 0.356 | 0.002 | 1.244 | 2.675 |
| Herd size | 1.021 | 0.002 | 0.000 | 1.017 | 1.025 |
| Regional cattle density | 1.015 | 0.003 | 0.000 | 1.010 | 1.021 |
| BCV positiveb | |||||
| Ing inf chain 0 | Baseline | ||||
| Ing inf chain 1–24 | 1.333 | 0.131 | 0.003 | 1.099 | 1.616 |
| Ing inf chain ≥25 | 1.784 | 0.333 | 0.000 | 1.238 | 2.571 |
| Herd size | 1.021 | 0.010 | 0.000 | 1.017 | 1.025 |
| Regional cattle density | 1.015 | 0.003 | 0.000 | 1.010 | 1.021 |
Hosmer Lemeshow goodness-of-fit, prob > χ2 = 0.051.
Hosmer Lemeshow goodness-of-fit, prob > χ2 > 0.480.
In the comparison of selection strategies, all risk-based approaches detected more positive cases compared to total random sampling. However, for BRSV, the only strategies where the median values or number of detected positives were above the 95% percentile of the random sampling distribution were the sampling strategies based on ‘in-degree’ (Fig. 2 ). For BCV on the other hand, all risk-based approaches except random sampling of herds with >0 contacts (i.e. ‘in-degree’ >0 and ‘ingoing infection chain’ >0), had median numbers of detected positives above the 95% percentile of the random sampling distribution (Fig. 3 ). Notice that the more narrow distributions for the selections strategies ‘in-degree’ ≥5 and ‘ingoing infection chain’ ≥25, compared to total random sampling, is a result of the smaller number of herds in these categories (relative to the sample size of 100).
Fig. 2.
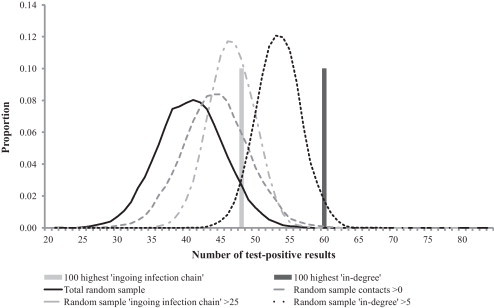
Number of test-positive results, i.e. actual numbers (bars) or output distributions, in different samples of 100 test results as regards BRSV. The samples were selected through different selection strategies from a total of 2137 test results.
Fig. 3.
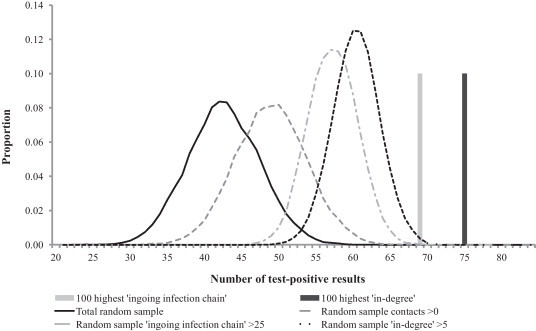
Number of test-positive results, i.e. actual numbers (bars) or output distributions, in different samples of 100 test results as regards BCV. The samples were selected through different selection strategies from a total of 2137 test results.
4. Discussion
Results from this study show that network analysis parameters representing animal purchase can be associated with the presence of infectious diseases (such as BCV or BRSV) in cattle. The ‘in-degree’ measure, which takes only direct contacts into account, was slightly more strongly associated with testing positive to BCV, compared to the ‘ingoing infection chain’ measure, where secondary contacts through sequence of movements are also incorporated. However, when comparing the exact values of influence of the two network analysis measures, it should be kept in mind that results can be expected to be highly dependent on the cut-offs used in the categorization of these parameters.
For BRSV, none of the network analysis parameters were found to be significantly associated when herd size and regional cattle density were accounted for. Nevertheless, these measures could still be useful for risk-based sampling because buying animals can be a risk regardless of whether a herd also has other characteristics associated with disease introduction. This was illustrated in the comparison of selection strategies, where sampling strategies based on ‘in-degree’ detected more BRSV positives compared to random sampling. In fact, controlling for potential confounders such as herd size may not be appropriate in risk-based sampling, as pointed out by Willeberg and co-authors in this issue of Preventive Veterinary Medicine (Willeberg et al., 2012). In many European countries, animal movements are continuously recorded, and obtaining network measures for the relevant time period based on these can provide a simple and straight forward tool for risk-based sampling, and also when information about other herd characteristics is missing. In a recent study, live animal contacts were recognized as a major risk for the spread of emerging infectious animal diseases and an increased need for surveillance was also identified (Wentholt et al., 2012). Especially in the first stages of an outbreak, or for diseases where animals do not show clear clinical symptoms, we see benefits from targeting herds with high measures of live animal contacts.
Although BCV and BRSV were primarily used as general examples of contagious diseases, some of the findings in this study can be worthwhile mentioning in specific relation to BCV and BRSV. Both diseases can spread through other types of contacts, e.g. visitors such as veterinarians; and the relative importance of animal trade and farm visits has not been investigated in relation to these infections in Swedish beef cattle. Previous studies conducted in Norway and Sweden have evaluated the association between herd-level characteristics and BCV and BRSV infections in dairy herds. The identified risk factors were similar for both viruses: large herd size was found to be a risk factor compared with small herd size (Tråvén et al., 1999, Norström et al., 2000), as was artificial insemination (AI) by farm personnel compared with AI by external technicians, conventional compared with organic management (Bidokhti et al., 2009), and not providing boots for visitors (Ohlson et al., 2010). However, in the present study, animal trade seems to be more important for BCV than BRSV. This is an interesting finding that may be explained by the slightly different epidemiology of these two viral diseases. Signs of diarrhoea (the main symptom of BCV) might not be recorded by the farmer as easily as coughing (the main symptom of BRSV), so the risk of selling animals with ongoing BRSV infection might therefore be lower. The fact that BCV is shed via faeces (Clark, 1993) could also contribute, as it is difficult to clean out and more voluminous than nasal discharge, which is the primary means of transmission for BRSV (Van der Poel et al., 1994).
The investigated covariates in this study were on herd level and the outcomes were based on just one single animal. Although both BRSV and BCV are highly infectious with high within-herd seroprevalence (Verhoeff et al., 1984, Alenius et al., 1991, Hägglund et al., 2006, Bidokhti et al., 2009, Ohlson et al., 2009), herd sensitivity can thus be expected to be less than 100% and to vary to some extent. This means that on herd level some of our observations probably were false negatives. Because all serological results used here came from analysis of samples from young stock, presence of antibodies can be assumed to reflect a relatively recent infection. A truly positive animal could nevertheless be a false positive on herd level, i.e. if the animal was born elsewhere and had gone through infection before introduction to its current herd. The unique identities of the tested animals were not available to us and individual level factors, such as place of birth and time spent in the current herd, could therefore not be investigated. On-farm biosecurity measures and frequency of categories of visitors are examples of higher level covariates that could also be of interest.
Although the number of detected positives was highest when the sampling was based on the 100 herds with the highest ‘in-degree’ in this study, this does not disqualify selection based on ‘ingoing infection chain’. The usefulness of these measures in the design of future surveillance activities will depend on the epidemiology of the disease studied. For example, ‘ingoing infection chain’ is expected to be more useful when included in risk-based surveillance of diseases such as bovine paratuberculosis, where infected animals often show few or no clinical symptoms. Many of the Swedish cattle herds have few direct contacts and ‘in-degree’ and thus ‘ingoing infection chain’ do not always correspond (Nöremark et al., 2011), e.g. a holding with low ‘in-degree’ can have a high ‘ingoing infection chain’. With a focus on ‘in-degree’ only, such herds may be excluded from sampling.
One possible alternative would be to combine the two measures, and another improvement could be adding different weights to the contacts in the ‘ingoing infection chain’ depending on the number of animals for each contact and on how many steps away the source herds are from the recipient herd. Also, the time periods for which the network measures are calculated need to be adjusted depending on the disease studied and on the age category of the tested animals. For some diseases, the more recent contacts will be the most interesting whereas for others, with long incubation periods, e.g. such as scrapie or paratuberculosis, trade events several years back may be still be of great importance. Moreover, the measures out-degree and outgoing infection chain (Dubé et al., 2008) can be used to identify outgoing contacts when the target of the surveillance is herds with a high risk of spreading disease.
5. Conclusion
For diseases where live animal trade constitute a main risk for disease introduction, and where reliable animal movement data is available, including network analysis parameters in the selection of herds can increase the surveillance sensitivity compared to total random sampling.
Conflict of interest
There are no conflicts of interest.
Acknowledgment
Jenny Frössling and Maria Nöremark were financially supported by the Swedish Civil Contingencies Agency.
References
- Alenius S., Niskanen R., Juntti N., Larsson B. Bovine coronavirus as the causative agent of winter dysentery: serological evidence. Acta Vet. Scand. 1991;32(2):163–170. doi: 10.1186/BF03546976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anonymous, 2000. Regulation (EC) No 1760/2000 of the European Parliament and of the Council of 17 July 2000 establishing a system for the identification and registration of bovine animals. Off. J. L (11.8.2000), pp. 1–10.
- Beaudeau F., Björkman C., Alenius S., Frössling J. Spatial patterns of bovine corona virus and bovine respiratory syncytial virus in the Swedish beef cattle population. Acta Vet. Scand. 2010;52(33):1–7. doi: 10.1186/1751-0147-52-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidokhti M.R., Tråvén M., Fall N., Emanuelson U., Alenius S. Reduced likelihood of bovine coronavirus and bovine respiratory syncytial virus infection on organic compared to conventional dairy farms. Vet. J. 2009;182(3):436–440. doi: 10.1016/j.tvjl.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blickenstorfer S., Schwermer H., Engels M., Reist M., Doherr M.G., Hadorn D.C. Using scenario tree modelling for targeted herd sampling to substantiate freedom from disease. BMC Vet. Res. 2011;7(49):1–9. doi: 10.1186/1746-6148-7-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christley R.M., Pinchbeck G.L., Bowers R.G., Clancy D., French N.P., Bennett R., Turner J. Infection in social networks: using network analysis to identify high-risk individuals. Am. J. Epidemiol. 2005;162:1024–1031. doi: 10.1093/aje/kwi308. [DOI] [PubMed] [Google Scholar]
- Cannon R.M. Inspecting and monitoring on a restricted budget – where best to look? Prev. Vet. Med. 2009;92(1–2):163–174. doi: 10.1016/j.prevetmed.2009.06.009. [DOI] [PubMed] [Google Scholar]
- Clark M.A. Bovine coronavirus. Br. Vet. J. 1993;149(1):51–70. doi: 10.1016/S0007-1935(05)80210-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubé C., Ribble C., Kelton D., McNab B. Comparing network analysis measures to determine potential epidemic size of highly contagious exotic diseases in fragmented monthly networks of dairy cattle movements in Ontario, Canada. Transbound. Emerg. Dis. 2008;55(9–10):382–392. doi: 10.1111/j.1865-1682.2008.01053.x. [DOI] [PubMed] [Google Scholar]
- Dubé C., Ribble C., Kelton D., McNab B. A review of network analysis terminology and its application to foot-and-mouth disease modelling and policy development. Transbound. Emerg. Dis. 2009;56:73–85. doi: 10.1111/j.1865-1682.2008.01064.x. [DOI] [PubMed] [Google Scholar]
- Elvander M. Severe respiratory disease in dairy cows caused by infection with bovine respiratory syncytial virus. Vet. Rec. 1996;138(5):101–105. doi: 10.1136/vr.138.5.101. [DOI] [PubMed] [Google Scholar]
- Elvander M., Baule C., Persson M., Egyed L., Ballagi-Pordany A., Belak S., Alenius S. An experimental study of a concurrent primary infection with bovine respiratory syncytial virus (BRSV) and bovine viral diarrhoea virus (BVDV) in calves. Acta. Vet. Scand. 1998;39:251–264. doi: 10.1186/BF03547797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman L.C. Centrality in social networks conceptual clarification. Soc. Netw. 1978–1979;1(3):215–239. [Google Scholar]
- Hosmer D.W., Lemeshow S. 2nd ed. John Wiley & Sons, Inc.; New York: 2000. Applied Logistic Regression. [Google Scholar]
- Hägglund S., Svensson C., Emanuelson U., Valarcher J.F., Alenius S. Dynamics of virus infections involved in the bovine respiratory disease complex in Swedish dairy herds. Vet. J. 2006;172(2):320–328. doi: 10.1016/j.tvjl.2005.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Lopez B., Perez A.M., Sanchez-Vizcaino J.M. Social network analysis. Review of general concepts and use in preventive veterinary medicine. Transbound. Emerg. Dis. 2009;56:109–120. doi: 10.1111/j.1865-1682.2009.01073.x. [DOI] [PubMed] [Google Scholar]
- Norström M., Skjerve E., Jarp J. Risk factors for epidemic respiratory disease in Norwegian cattle herds. Prev. Vet. Med. 2000;44(1–2):87–96. doi: 10.1016/s0167-5877(99)00113-0. [DOI] [PubMed] [Google Scholar]
- Nöremark, M., 2010. Infection through the farm gate: studies on movements of livestock and on-farm biosecurity, doctoral thesis. Uppsala, Swedish University of Agricultural Sciences. Available at: http://pub.epsilon.slu.se/2227/1/Noremark_M_100210.pdf.
- Nöremark M., Håkansson N., Lewerin S.S., Lindberg A., Jonsson A. Network analysis of cattle and pig movements in Sweden: measures relevant for disease control and risk based surveillance. Prev. Vet. Med. 2011;99(2–4):78–90. doi: 10.1016/j.prevetmed.2010.12.009. [DOI] [PubMed] [Google Scholar]
- Ohlson A., Heuer C., Lockhart C., Tråvén M., Emanuelson U., Alenius S. Risk factors for seropositivity to bovine coronavirus and bovine respiratory syncytial virus in dairy herds. Vet. Rec. 2010;167(6):201–206. doi: 10.1136/vr.c4119. [DOI] [PubMed] [Google Scholar]
- Ohlson A., Tråvén M., Emanuelson U., Alenius S. The relationship between pooled and individual milk samples for detecting antibodies to bovine coronavirus and bovine respiratory syncytial virus. Proceedings of 12th Symposium of the International Society for Veterinary Epidemiology and Economics; Durban, South Africa, August 10–14; 2009. [Google Scholar]
- Ortiz-Pelaez A., Pfeiffer D.U., Soares-Magalhães R.J., Guitian F.J. Use of social network analysis to characterize the pattern of animal movements in the initial phases of the 2001 foot and mouth disease (FMD) epidemic in the UK. Prev. Vet. Med. 2006;76(1–2):40–55. doi: 10.1016/j.prevetmed.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Stärk K.D., Regula G., Hernandez J., Knopf L., Fuchs K., Morris R.S., Davies P. Concepts for risk-based surveillance in the field of veterinary medicine and veterinary public health: review of current approaches. BMC Health Serv. Res. 2006;6:20–27. doi: 10.1186/1472-6963-6-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tråvén M., Björnerot L., Larsson B. Nationwide survey of antibodies to bovine coronavirus in bulk milk from Swedish dairy herds. Vet. Rec. 1999;144(19):527–529. doi: 10.1136/vr.144.19.527. [DOI] [PubMed] [Google Scholar]
- Valarcher J.F., Taylor G. Bovine respiratory syncytial virus infection. Vet. Res. 2007;38(2):153–180. doi: 10.1051/vetres:2006053. [DOI] [PubMed] [Google Scholar]
- Van der Poel W.H., Brand A., Kramps J.A., Van Oirschot J.T. Respiratory syncytial virus infections in human beings and in cattle. J. Infect. 1994;29(2):215–228. doi: 10.1016/s0163-4453(94)90866-4. [DOI] [PubMed] [Google Scholar]
- Verhoeff J., Van der Ban M., van Nieuwstadt A.P. Bovine respiratory syncytial virus infections in young dairy cattle: clinical and haematological findings. Vet. Rec. 1984;114:9–12. doi: 10.1136/vr.114.1.9. [DOI] [PubMed] [Google Scholar]
- Wasserman S., Faust K., Iacobucci D., Granovetter M. Cambridge University Press; 1994. Social Network Analysis: Methods and Applications (Structural Analysis in the Social Sciences) [Google Scholar]
- Willeberg P., Nielsen L.R., Salman M. Risk-based surveillance: estimating the effect of unwarranted confounder adjustment. Prev. Vet. Med. 2012 doi: 10.1016/j.prevetmed.2011.12.012. [DOI] [PubMed] [Google Scholar]
- Wentholt M.T., Cardoen S., Imberechts H., Van Huffel X., Ooms B.W., Frewer L.J. Defining European preparedness and research needs regarding emerging infectious animal diseases: results from a Delphi expert consultation. Prev. Vet. Med. 2012;103:81–92. doi: 10.1016/j.prevetmed.2011.09.021. [DOI] [PubMed] [Google Scholar]


