Abstract
A new disease, termed severe acute respiratory syndrome (SARS), emerged at the end of 2002 and caused profound disturbances in over 30 countries worldwide in 2003. A novel coronavirus was identified as the aetiological agent of SARS and the 30 kb viral genome was deciphered with unprecedented speed in a coordinated manner by the global community. Since then, much progress has been made in the virological and molecular characterization of the proteins encoded by SARS-coronavirus (SARS-CoV) genome, which contains 14 potential open reading frames (ORFs). These investigations can be broadly classified into three groups: (a) studies on the replicase 1a/1b gene products which are important for viral replication, (b) studies on the structural proteins, spike, nucleocapsid, membrane and envelope, which have homologues in all coronaviruses, and are important for viral assembly and (c) expression and functional studies of the “accessory” proteins that are specifically encoded by SARS-CoV. A comparison of the properties of these three groups of SARS-CoV proteins with the knowledge that coronavirologists have generated over more than 30 years of research can help us in the prevention and treatment of SARS in the event of the re-emergence of this new infectious disease.
Keywords: Severe acute respiratory syndrome (SARS), Coronavirus, Viral proteins
1. Introduction
The recent severe acute respiratory syndrome (SARS) epidemic, which affected over 30 countries across five continents, has profoundly disturbed social and economic activities regionally, as well as globally. By the end of the epidemic, which was essentially controlled by isolation, more than 8000 cases have been reported with more than 800 fatalities (World Health Organization, http://www.who.int/csr/sars/country/en/). The concerted efforts of the scientific community led to an extraordinarily rapid identification of a novel coronavirus as the aetiological agent of SARS and the full genome sequencing of the virus (Drosten et al., 2003, Fouchier et al., 2003, Ksiazek et al., 2003, Marra et al., 2003, Peiris et al., 2003, Rota et al., 2003, Ruan et al., 2003). The SARS-coronavirus (SARS-CoV) genome is ∼30 kb in length and contains 14 potential open reading frames (ORFs) (Marra et al., 2003, Thiel et al., 2003).
Coronaviruses are positive-strand RNA viruses and the virion consists of a nucleocapsid core surrounded by an envelope containing three membrane proteins, spike (S), membrane (M) and envelope (E) that are common to all members of the genus (Siddell, 1995, Lai and Holmes, 2001). The RNA is packaged by the nucleocapsid (N) protein into a helical nucleocapsid. The S protein, which forms morphologically characteristic projections on the virion surface, mediates binding to host receptors and membrane fusion. The M protein is a triple-spanning integral membrane protein with a short ectodomain and a large carboxyl-terminus endodomain. More recently, the E protein was shown to play a major role in coronavirus assembly (Bos et al., 1996, Vennema et al., 1996). The genes for these structural proteins and the replicase 1a/1b gene, that is located at the 5′ end of the genome and constitute 2/3 of it, are conserved among the subgroups of coronavirus so is their relative position in the genome (Siddell, 1995, de Vries et al., 1997, Lai and Cavanagh, 1997, Lai and Holmes, 2001). In addition, there are group-specific “accessory” proteins, which are usually dispensable for viral replication but may be important for viral–host interactions. These accessory proteins vary in size and position in the genome.
This review summarizes present knowledge on the SARS-CoV viral proteins: their expression, cellular localization and effects on cellular functions. Expeditious research on SARS-CoV resulted in simultaneous publications from independent laboratories and this leads to a confusing array of nomenclatures used for the various viral proteins. In order to consolidate the information from different publications, this review will adopt the nomenclatures used by Snijder et al., 2003 and Thiel et al., 2003, as they are most consistent with those used for other coronaviruses. Alternate names that have been used in specific publications will be noted in parentheses.
2. Replicase gene (ORFs 1a and 1b)
Analogous to other coronaviruses, the first 2/3 of the SARS-CoV genome encodes the viral replicase genes (ORFs 1a and 1b), which translates into two large polyproteins, pp1a (486 kDa) and pp1ab (790 kDa) (Thiel et al., 2003). Expression of the ORF 1b-encoded region of pp1ab involves ribosomal frameshifting into the −1 frame just upstream of the ORF 1a translation termination codon (Thiel et al., 2003). Proteolytic processings of these polyproteins are mediated by viral cysteine proteinases and produces a minimum of 13 non-structural proteins (also called nsp's), some of which are responsible for replicating the viral genome and/or generating a nested set of subgenomic mRNAs to express all the ORFs downstream of ORF 1b (Ziebuhr et al., 2000). Unlike most coronaviruses, which uses three proteinases for polyprotein processing (Ziebuhr et al., 2000, Gorbalenya, 2001), SARS-CoV is predicted only to have two proteinases, which are a papain-like (accessory) cysteine proteinase (termed as PL2pro), which cleaves at 3 sites, and a 3C-like (main) proteinase (termed 3CLpro or Mpro), which cleaves at 11 sites (Rota et al., 2003, Gao et al., 2003a, Snijder et al., 2003, Thiel et al., 2003). As a result, 16 non-structural proteins (Fig. 1A) are predicted but which of these are essential for the replication of the virus remain to be determined. The proteinase activity of 3CLpro was also experimentally demonstrated as purified 3CLpro was shown to cleave peptides covering all the 11 predicted cleavage sites (Fan et al., 2004). In addition, the three-dimensional structure of 3CLpro was also solved by both crystallography and NMR spectroscopy (Yang et al., 2003, Shi et al., 2004). Both studies reported that 3CLpro exists as a dimer and revealed fine conformational details of its interaction with substrates, thus providing a basis for rational drug design.
Fig. 1.
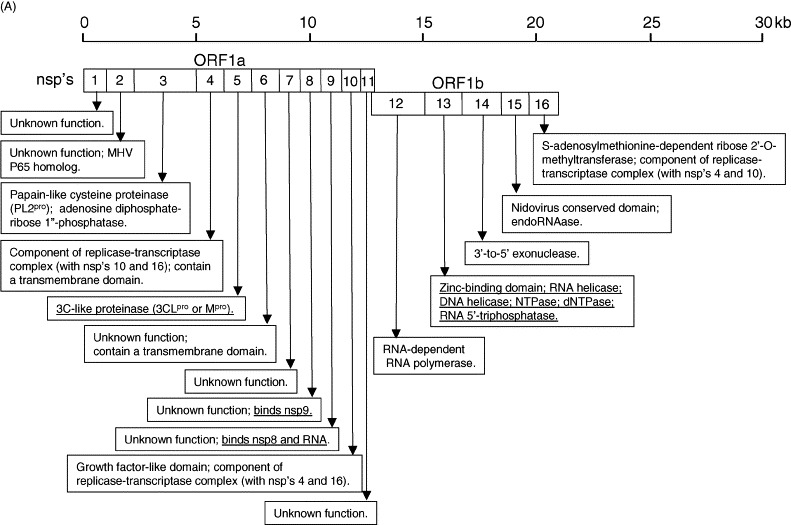
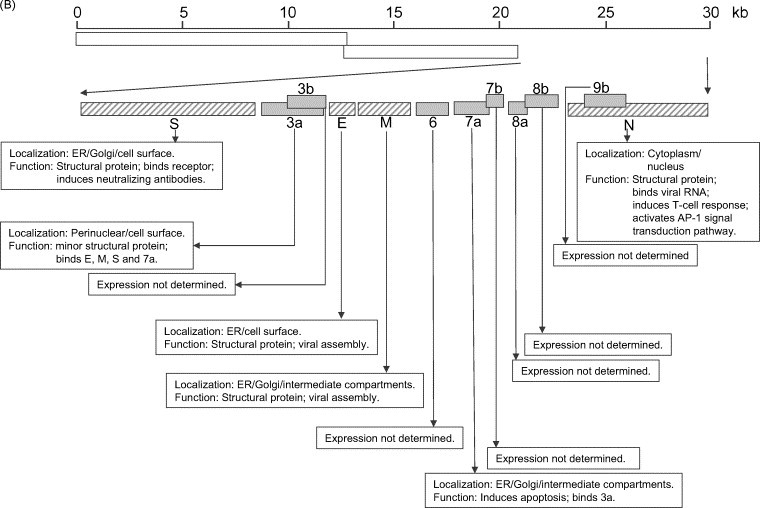
Summary of the SARS-CoV genome organization and viral protein expression. (A) Replicase genes (ORFs 1a and 1b), constituting the first 2/3 of the genome, translate into two large polyproteins, pp1a (486 kDa) and pp1ab (790 kDa). Expression of the ORF 1b-encoded region of pp1ab involves ribosomal frameshifting into the −1 frame just upstream of the ORF 1a translation termination codon. Proteolytic processings of these polyproteins are mediated by viral cysteine proteinases (nsp3, also called PL2pro and nsp5, also called 3CLpro or Mpro) resulting in 16 non-structural proteins (nsp's; open boxes). Putative functional domains present in each nsp's are shown in the text boxes. Functions that have been demonstrated with recombinant proteins are underlined. (B) Open reading frames (ORFs) in the remaining 1/3 of the genome are translated from eight subgenomic mRNAs. Four of these encode the structural proteins (checked boxes), spike (S), membrane (M) and envelope (E) and nucleocapsid (N). Another eight SARS-CoV-unique ORFs (grey solid boxes) encode putative “accessory” proteins with no significance sequence homology to viral proteins of other coronaviruses (3a, 3b, 6, 7a, 7b, 8a, 8b and 9b). The cellular localization and functions of some of these viral proteins have been demonstrated (see text boxes). Also note that S, E, M and ORF 6 are expressed from individual subgenomic mRNAs, while 3a and 3b are predicted to be produced from the same subgenomic mRNA. Similarly, 7a and 7b are also produced from one subgenomic mRNA, and 9b is produced from the same subgenomic mRNA as N. The expression of 3b, 7b and 9b may be via “leaky scanning” by ribosomes or involve a mechanism like internal ribosomal entry. However, it cannot be rule out that they may also be expressed from the synthesis of yet undetected additional subgenomic mRNAs.
Another protein that is likely to be important for viral replication is the SARS-CoV helicase (also called nsp13 in Snijder et al., 2003 or nsp10 in Gao et al., 2003a and Tanner et al., 2003). Recombinant SARS-CoV helicase has multiple enzymatic activities, including RNA helicase, DNA helicase, NTPase, dNTPase and an RNA 5′-triphosphatase activities (Tanner et al., 2003, Thiel et al., 2003, Ivanov et al., 2004). In addition, the crystal structure of SARS-CoV nsp9, which has no designated function, has been solved and it has been shown to bind RNA as well as another non-structural protein, SARS-CoV nsp8 (Campanacci et al., 2003, Sutton et al., 2004). The SARS-CoV nsp9 may have a similar function as the nsp9 protein of mouse hepatitis virus (MHV), a Group 2 coronavirus, which colocalized and interacted with other proteins of the replication complex (Bost et al., 2000, Brockway et al., 2003). For the remaining non-structural proteins produced from pp1a or pp1ab, putative activities have been predicted based on the presence of functional domains in their sequences or by their structural similarities to other proteins (Gao et al., 2003a, Snijder et al., 2003, von Grotthuss et al., 2003; Fig. 1A).
3. Structural proteins (S, E, M and N)
The S protein of coronavirus is important for binding to cellular receptor and for mediating the fusion of viral and host membranes, both of these processes being critical for virus entry into host cells (Gallagher and Buchmeier, 2001). As such, S is known to be responsible for inducing host immune responses and virus neutralization by antibodies (Holmes, 2003, Navas-Martin and Weiss, 2003). For SARS-CoV, it has been demonstrated that prior infection provided protective immunity in a mouse model and the passive transfer of neutralizing antibodies to naive mice also protected them from infection (Subbarao et al., 2004). Importantly, there was no enhancement of SARS-CoV infection in mice upon re-infection or after the administration of immune serum, unlike the case for one coronavirus, the feline infectious peritonitis virus (Olsen, 1993). A DNA vaccine encoding the S protein alone induced T cell and neutralizing antibody responses and protected mice from SARS-CoV infection (Yang et al., 2004), suggesting the S is indeed the primary target for viral neutralization in SARS-CoV infection. This finding was also confirmed by several studies that use surrogate/carrier viruses to express S in mice or primates (Gao et al., 2003b, Bisht et al., 2004, Buchholz et al., 2004, Bukreyev et al., 2004). From these studies, it is clear that humoral response against S plays an important role in controlling and clearing SARS-CoV infection.
In addition, a host cell receptor, the carboxypeptidase angiotensin-converting enzyme-2 (ACE-2), which is an essential regulator of heart function, has also been identified (Li et al., 2003). At least three independent laboratories subsequently showed that a domain in the N-terminus of S, approximately 300–510 amino acids, is the receptor binding domain (Xiao et al., 2003, Babcock et al., 2004, Wong et al., 2004). Importantly, syncytia formation/membrane fusion and viral replication can be specifically inhibited by an anti-ACE-2 antibody (Li et al., 2003) or a fragment containing the receptor binding domain (Wong et al., 2004) or antibodies recognizing the receptor binding domain (Sui et al., 2004, Chou et al., 2004).
The coronavirus S protein is a class I virus fusion protein and contains two regions with a 4, 3 hydrophobic (heptad) repeat in S2 domain or C-terminus half of the protein (de Groot et al., 1987, Bosch et al., 2003). These domains (termed as HR1 and HR2) are thought to play an important role in defining the oligomeric structure of S and hence mediating the fusion between viral and cellular membranes (Eckert and Kim, 2001). For the SARS-CoV, HR2 is located close to the transmembrane anchor (1148–1193 amino acids) and HR1 is ∼140 amino acids upstream of it (900–1005 amino acids) (Ingallinella et al., 2004). Biochemical studies have shown that peptides corresponding to the HR1 and HR2 of SARS-CoV S protein can associate into an anti-parallel six-helix bundles with structural features typical of class I fusion proteins, suggesting that the membrane fusion and cell entry mechanisms exploited by SARS-CoV are similar to that for other coronaviruses (Bosch et al., 2004, Ingallinella et al., 2004, Liu et al., 2004b, Tripet et al., 2004, Yuan et al., 2004, Zhu et al., 2004). In the full-length S protein, the HR1–HR2 structure brings the fusion peptide, predicted to be near the N-terminus of HR1 (Bosch et al., 2004), in close proximity to the transmembrane domain and this drives the fusion between viral and cellular membranes, and allows the virus to entry the cell. Indeed, peptides from HR1 or HR2 can inhibit viral replication in Vero E6 culture, presumably by interfering with the formation of the six-helix bundle (Bosch et al., 2004, Liu et al., 2004b, Yuan et al., 2004, Zhu et al., 2004).
Studies on the profile of antibodies in SARS patients showed that antibodies against M and E are generally low or not present in SARS patient's sera (Wang et al., 2003, Guo et al., 2004, Leung et al., 2004, Tan et al., 2004b) as these proteins are embedded in the viral envelope. However, it is clear that M and E are important for viral assembly as demonstrated by the formation of virus-like particles in insect cells expressing these proteins (Ho et al., 2004). This result is consistent with previous studies on coronaviruses, which showed that M and E are sufficient for the assembly of viral particles (Bos et al., 1996, Vennema et al., 1996). By using a proteomic approach, a novel phosphorylated site of M was also identified (Zeng et al., 2004a), but the importance of this for the function of M has not been defined.
The N protein has been shown to be very abundant in SARS-CoV infected Vero E6 cells (Krokhin et al., 2003, Rota et al., 2003) and several independent studies have shown that >90% of sera obtained from convalescent SARS patients have antibodies against N (Shi et al., 2003, Wang et al., 2003, Guo et al., 2004, Leung et al., 2004, Tan et al., 2004b). In addition, as N is not glycosylated, easily expressed in bacteria and highly immunogenic, it is an ideal candidate for development of enzyme-linked immunosorbent assays for the detection of SARS-CoV infection, either for detection of anti-N antibodies (Shi et al., 2003, Guan et al., 2004a) or for direct antigen detection (Che et al., 2004, Lau et al., 2004). In addition, it was reported that the SARS-CoV N can induce specific T-cell responses (Gao et al., 2003b, Kim et al., 2004), as have been observed with other coronaviruses (Siddell, 1995), but how important is this for protective immunity remains to be determined.
Other molecular aspects of N have also been reported, including self-dimerization (He et al., 2004, Surjit et al., 2004a), RNA-binding capabilities (Huang et al., 2004), cleavage by caspase 3 (Ying et al., 2004) and its ability to activate signal transduction pathways (He et al., 2003). In addition, the N protein of SARS-CoV was shown to induce apoptosis and actin reorganization in mammalian cells under stressed conditions (Surjit et al., 2004b). Interestingly, Mizutani et al. (2004) showed that the p38MARK pathway is activated in SARS-CoV infected Vero E6 cells, but it is not clear if this is directly/entirely due to the expression of N. The N proteins of other coronaviruses, including avian infectious bronchitis virus, porcine transmissible gastroenteritis virus and mouse hepatitis virus, are localized to both cytoplasm and nucleolus, and the presence of N in the nucleolus may be important for the synthesis of viral RNA (Hiscox et al., 2001, Wurm et al., 2001). For SARS-CoV N protein, it has been reported to be found in the cytoplasm and nucleus of SARS-CoV infected cells (Chang et al., 2004, Zeng et al., 2004a).
4. Group-specific genes
Eight subgenomic mRNAs are produced in SARS-CoV infected Vero E6 cells and these are used to express the ORFs besides the replicase 1a/1b (Snijder et al., 2003, Thiel et al., 2003). These include the S (ORF 2), E (ORF 4), M (ORF 5) and N (ORF 9) and another eight ORFs that encode putative proteins with no significance sequence homology to viral proteins of other coronaviruses (ORF 3a, 3b, 6, 7a, 7b, 8a, 8b and 9b) (Fig. 1B). Of these SARS-CoV-unique ORFs, two of them (3a and 7a) have been shown to be expressed during SARS-CoV infection (Fielding et al., 2004, Tan et al., 2004c, Yu et al., 2004, Zeng et al., 2004b) and antibodies against another four of them (3b, 7b, 8a and 9b which were termed as ORF 4, 9, 10 and 13, respectively, in Guo et al., 2004) have been detected in the sera of convalescent patients, suggesting that these proteins were expressed during infection in vivo.
3a (also termed as ORF 3 in Marra et al., 2003 and Guo et al., 2004, as X1 in Rota et al., 2003 and as U274 in Tan et al., 2004b, Tan et al., 2004c) is the largest of these unique ORFs and consists of 274 amino acids and contains three putative transmembrane domains. Three groups independently reported the expression of 3a in SARS-CoV infected cells (Tan et al., 2004c, Yu et al., 2004, Zeng et al., 2004b) and it is also detected in a SARS-CoV infected patient's lung specimen (Yu et al., 2004). Antibodies against 3a were also found in convalescent patients (Guo et al., 2004, Tan et al., 2004b, Yu et al., 2004, Zeng et al., 2004b). 3a is localized in the perinuclear region and is also transported to the cell surface, where it can undergo internalization (Tan et al., 2004c, Yu et al., 2004). It is intriguing to find that SARS-CoV has evolved to express a viral protein with endocytotic properties, as endocytosis has been shown to play important roles in the replication of a number of viruses as well as their adaptation to the host cells (Marsh and Pelchen-Matthews, 2000). The transportation of 3a to the cell surface depends on a region in the cytoplasmic domain that contains two different sorting motifs, a YxxΦ (where x is any amino acids and Φ is an amino acid with a bulky hydrophobic sidechain) upstream of a ExD (diacidic) motif (Tan et al., 2004c). The diacidic motif is required for efficient ER export (Nishimura and Balch, 1997) while the YxxΦ motif has been implicated in directing protein localization to various intracellular compartments (Bonifacino and Traub, 2003). The juxtaposition of these two motifs appears to be important for the transport of proteins to the plasma membrane (Bannykh et al., 1998).
The topology of 3a was determined experimentally: its N-terminus is facing the extracellular matrix and its C-terminus is facing the cytoplasm (Tan et al., 2004c). Interestingly, when Liu et al. (2004a) used phage-display technology to characterize B cell epitopes recognized by antibodies from SARS patients, they found one consensus motif VKIXN, which corresponded uniquely to 18–22 amino acids of the N-terminus ectodomain of 3a. Taken together, these data strongly suggest that 3a could play an important immunological role as it is clearly presented to the host immune system during infection. 3a can also interact specifically with M and E, which are two key players in the viral assembly of coronaviruses, as well as with the S protein (Tan et al., 2004c, Zeng et al., 2004b); hence it may also be important for viral assembly and/or release of virus from infected cells. It is tempting to postulate that 3a is a novel structural protein as only the coronavirus structural proteins, S, hemagglutinin-esterase (HE) and E, have been shown to be transported to the plasma membrane/cell surface (Kienzle et al., 1990, Parker et al., 1990, Smith et al., 1990, Vennema et al., 1990). Indeed, Zeng et al. (2004b) could detect disulfide-linked complexes of S and 3a in the medium of SARS-CoV infected cells, indicating that 3a was secreted together with S, possibly through the formation of virus particles. However, it is necessary to confirm this finding with highly purified SARS-CoV virions as viral proteins could also be released into the medium through cells lysis.
Mutations in 3a were observed in a Singapore isolate after three passages in cell culture, resulting in several different forms of 3a in infected cells (Tan et al., 2004c). This may not necessarily be a cell culture adaptation, but may indicate the presence of quasi-species, as comparative analysis of the different isolates of SARS-CoV also showed high frequency of mutations in the 3a gene (Chen et al., 2003, Tan et al., 2004b, Yeh et al., 2004, Zeng et al., 2004b). In fact, sequence comparison of isolates from different clusters of infection showed that both S and 3a showed positive selections during virus evolution, implying that these proteins play important roles in the virus life cycle and/or disease development (3a was termed as X1 in Guan et al., 2004b, Yeh et al., 2004, Zeng et al., 2004b).
The other ORF that has been shown to be expressed in SARS-CoV infected cells is 7a, which contains a cleavable signal peptide at the N-terminus and a transmembrane domain near the C-terminus (7a was also known as ORF 8 in Marra et al., 2003, as X4 in Rota et al., 2003 and as U122 in Fielding et al., 2004, Tan et al., 2004c). An endoplasmic reticulum (ER) retrieval motif (KRKTE), which is important for transport of proteins back to the ER (Teasdale and Jackson, 1996), is located at the C-terminus cytoplasmic domain of 7a and mediates the recycling of 7a between the ER and Golgi apparatus such that 7a is present in the intermediate compartments, where coronaviruses are known to assemble and bud (Fielding et al., 2004). Interestingly 7a can also interact with 3a, which can interact with M, E and S, suggesting that these viral proteins may form complexes during infection (Tan et al., 2004c).
In addition, the over-expression of 7a induces apoptosis via a caspase-dependent pathway, and in cell-lines derived from different organs, including lung, kidney and liver (Tan et al., 2004a). Although there are other factors that contribute to the induction of apoptosis during SARS-CoV infection, the ability of 7a to induce apoptosis in different cell-types is consistent with the clinical observation of apoptosis in different organs infected by SARS-CoV and suggests that the expression of 7a during infection may be one of the underlying mechanisms for the pathogenesis of SARS-CoV infection.
Group-specific genes in coronaviruses, also called “accessory” proteins, are usually dispensable for viral replication in cell culture systems but may be important for viral–host interactions and thus contribute to viral stability and/or pathogenesis in vivo. For example, although the 7b gene of feline coronavirus is easily lost upon virus adaptation to cell culture, it is strictly maintained in naturally occurring strains and its loss was correlated with reduced virulence (Herrewegh et al., 1995). Recent studies also showed that some of these group-specific genes are not essential for viral replication in cell culture, but their deletion, by reverse genetics, is attenuating in the natural host (de Haan et al., 2002, Ortego et al., 2003). It has not yet been established which of these SARS-CoV-unique ORFs are essential for viral replication and/or for viral–host interactions.
5. Future directions
Thus far, characterization of the three groups of viral proteins of the SARS-CoV revealed that for the first two groups of proteins, i.e. the replicase genes produced from cleavage of the polyprotein pp1a/pp1ab and structural proteins (S, M, E and N), there are significance similarities in their properties when compared to their homologues in other coronaviruses. Nevertheless, detailed analysis have revealed important differences but whether these contributed to severe clinical manifestations of SARS-CoV infection in contrast to the mild diseases caused by most human or animal coronaviruses, remains to be determined.
As for the third group of viral proteins, i.e. the group-specific genes encoded by the sequences between S and N, it is difficult to compare to other coronaviruses as these “accessory” proteins are generally not well characterized. However, it is interesting to note that the part of viral genome that encodes these “accessory” proteins appears to be prone to large insertions or deletions. For example, in some strains of MHV, ORF 4 is interrupted and becomes ORFs 4a and 4b, while in others, ORF 2a is completely deleted (Schwarz et al., 1990, Weiss et al., 1993). Shen et al. (2003) also reported an insertion of six nucleotides in 3b gene of the infectious bronchitis virus after continuous passages in Vero E6 cells and this resulted in a truncated 3b protein of 34 amino acids with only the first 17 amino acids being homologous to the original full-length 3b protein of 64 amino acids. For SARS-CoV, Guan et al. (2003) analyzed SARS-CoV isolates obtained from animals in a live-market in Guangdong and found that all the animal isolates contain a 29-nucleotides sequence which is absent in most human isolates. As a result of this, the ORF 8a (termed as ORF 10 in Guan et al., 2003) and 8b (termed as ORF 11 in Guan et al., 2003) in the human isolates becomes one ORF encoding a protein of 122 amino acids, whose N-terminus is identical to ORF 8a and C-terminus is identical to ORF 8b. Another extensive study of 63 SARS-CoV isolates obtained from early, middle and late phases of the SARS epidemic in China also showed that there are major deletions in this region of the viral genome (The Chinese SARS Molecular Epidemiology Consortium, 2004). Interestingly, the clustering of patients with different patterns of deletion was correlated with the different phases of the epidemic. Although these mutations do not appear to have any adverse effect on the survival of the virus, it is conceivable that the different variants of ORF 8a/8b have different stabilities and/or functions, and hence would contribute differently to viral pathogenesis in vivo. Another study also reported an in frame deletion of 45 nucleotides in ORF 7b in the Frankfurt isolate after three passages in tissue culture (Thiel et al., 2003). Since SARS-CoV appeared to have only recently crossed species from animal to human and that it has been observed that the profiles of these mutations correlated with clusters of infection, it is likely that the properties of these unique ORFs are changing as the virus undergoes adaptive evolution and these mutations could cause SARS-CoV infection to become more benign but more persistent.
As a full-length infectious clone of SARS-CoV has been assembled (Yount et al., 2003), the use of reverse genetics would certainly reveal more about the contributions of individual viral proteins. Although SARS-CoV caused little or no disease in mice, it is able to replicate in the respiratory tract of the mice to a high level (Subbarao et al., 2004, Wentworth et al., 2004). Together with the technologies to create transgenic or knockout mice, the mouse model will contribute to the developments of vaccines and anti-viral therapeutics against SARS-CoV infection and may also help us understand why certain cohorts of patients are more vulnerable to the disease while others only develop mild symptoms. In addition, experimentally SARS-CoV infected cats, ferrets and primates can develop at least some of the clinical symptoms observed in SARS-CoV infected patients (Martina et al., 2003, Kuiken et al., 2004). Thus, these animal models will be essential for studying virus-host interactions and for delineating the precise contributions of the viral proteins to SARS-CoV infection, replication and pathogenesis.
Acknowledgements
We thank members of the Collaborative Anti-Viral Research group, Institute of Molecular and Cell Biology, for critically reading the manuscript. We apologize to any investigators whose work we have inadvertently omitted.
References
- Babcock G.J., Esshaki D.J., Thomas W.D., Jr., Ambrosino D.M. Amino acids 270 to 510 of the severe acute respiratory syndrome coronavirus spike protein are required for interaction with receptor. J. Virol. 2004;78:4552–4560. doi: 10.1128/JVI.78.9.4552-4560.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannykh S.I., Nishimura N., Balch W.E. Getting into the Golgi. Trends Cell Biol. 1998;8:21–25. doi: 10.1016/s0962-8924(97)01184-7. [DOI] [PubMed] [Google Scholar]
- Bisht H., Roberts A., Vogel L., Bukreyev A., Collins P.L., Murphy B.R., Subbarao K., Moss B. Severe acute respiratory syndrome coronavirus spike protein expressed by attenuated vaccinia virus protectively immunizes mice. Proc. Natl. Acad. Sci. U.S.A. 2004;101:6641–6646. doi: 10.1073/pnas.0401939101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino J.S., Traub L.M. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem. 2003;72:395–447. doi: 10.1146/annurev.biochem.72.121801.161800. [DOI] [PubMed] [Google Scholar]
- Bos E.C., Luytjes W., van der Meulen H.V., Koerten H.K., Spaan W.J. The production of recombinant infectious DI-particles of a murine coronavirus in the absence of helper virus. Virology. 1996;218:52–60. doi: 10.1006/viro.1996.0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch B.J., van der Zee R., de Haan C.A., Rottier P.J. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 2003;77:8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch B.J., Martina B.E., Van Der Zee R., Lepault J., Haijema B.J., Versluis C., Heck A.J., De Groot R., Osterhaus A.D., Rottier P.J. Severe acute respiratory syndrome coronavirus (SARS-CoV) infection inhibition using spike protein heptad repeat-derived peptides. Proc. Natl. Acad. Sci. U.S.A. 2004;101:8455–8460. doi: 10.1073/pnas.0400576101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bost A.G., Carnahan R.H., Lu X.T., Denison M.R. Four proteins processed from the replicase gene polyprotein of mouse hepatitis virus colocalize in the cell periphery and adjacent to sites of virion assembly. J. Virol. 2000;74:3379–3387. doi: 10.1128/jvi.74.7.3379-3387.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockway S.M., Clay C.T., Lu X.T., Denison M.R. Characterization of the expression, intracellular localization, and replication complex association of the putative mouse hepatitis virus RNA-dependent RNA polymerase. J. Virol. 2003;77:10515–10527. doi: 10.1128/JVI.77.19.10515-10527.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchholz U.J., Bukreyev A., Yang L., Lamirande E.W., Murphy B.R., Subbarao K., Collins P.L. Contributions of the structural proteins of severe acute respiratory syndrome coronavirus to protective immunity. Proc. Natl. Acad. Sci. U.S.A. 2004;101:9804–9809. doi: 10.1073/pnas.0403492101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukreyev A., Lamirande E.W., Buchholz U.J., Vogel L.N., Elkins W.R., St Claire M., Murphy B.R., Subbarao K., Collins P.L. Mucosal immunisation of African green monkeys (Cercopithecus aethiops) with an attenuated parainfluenza virus expressing the SARS coronavirus spike protein for the prevention of SARS. Lancet. 2004;363:2122–2127. doi: 10.1016/S0140-6736(04)16501-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campanacci V., Egloff M.P., Longhi S., Ferron F., Rancurel C., Salomoni A., Durousseau C., Tocque F., Bremond N., Dobbe J.C., Snijder E.J., Canard B., Cambillau C. Structural genomics of the SARS coronavirus: cloning, expression, crystallization and preliminary crystallographic study of the Nsp9 protein. Acta. Crystallogr. D. Biol. Crystallogr. 2003;59:1628–1631. doi: 10.1107/S0907444903016779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang M.S., Lu Y.T., Ho S.T., Wu C.C., Wei T.Y., Chen C.J., Hsu Y.T., Chu P.C., Chen C.H., Chu J.M., Jan Y.L., Hung C.C., Fan C.C., Yang Y.C. Antibody detection of SARS-CoV spike and nucleocapsid protein. Biochem. Biophys. Res. Commun. 2004;314:931–936. doi: 10.1016/j.bbrc.2003.12.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che X.Y., Qiu L.W., Pan Y.X., Wen K., Hao W., Zhang L.Y., Wang Y.D., Liao Z.Y., Hua X., Cheng V.C., Yuen K.Y. Sensitive and specific monoclonal antibody-based capture enzyme immunoassay for detection of nucleocapsid antigen in sera from patients with severe acute respiratory syndrome. J. Clin. Microbiol. 2004;42:2629–2635. doi: 10.1128/JCM.42.6.2629-2635.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.L., Ou H.Y., Zhang R., Zhang C.T. ZCURVE_CoV: a new system to recognize protein coding genes in coronavirus genomes, and its applications in analyzing SARS-CoV genomes. Biochem. Biophys. Res. Commun. 2003;307:382–388. doi: 10.1016/S0006-291X(03)01192-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou C.-F., Shen S., Tan Y.-J., Fielding B.C., Tan T.H.P., Fu J., Xu Q., Lim S.G., Hong W. A novel cell-based binding assay system reconstituting interaction between SARS-CoV S protein and its cellular receptor. J. Virol. Methods. 2004;123:41–48. doi: 10.1016/j.jviromet.2004.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot R.J., Luytjes W., Horzinek M.C., van der Zeijst B.A., Spaan W.J., Lenstra J.A. Evidence for a coiled-coil structure in the spike proteins of coronaviruses. J. Mol. Biol. 1987;196:963–966. doi: 10.1016/0022-2836(87)90422-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan C.A.M., Masters P.S., Shen X., Weiss S., Rottier P.J.M. The group-specific murine coronavirus genes are not essential, but their deletion, by reverse genetics, is attenuating in the natural host. Virology. 2002;296:177–189. doi: 10.1006/viro.2002.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries A.A.F., Horzinek M.C., Rottier P.J.M., de Groot R.J. The genome organization of the Nidovirales: similarities and differences between Arteri-, Toro-, and Coronaviruses. Semin. Virol. 1997;8:33–47. doi: 10.1006/smvy.1997.0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., Berger A., Burguiere A.M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.C., Muller S., Rickerts V., Sturmer M., Vieth S., Klenk H.D., Osterhaus A.D., Schmitz H., Doerr H.W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- Eckert D.M., Kim P.S. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 2001;70:777–810. doi: 10.1146/annurev.biochem.70.1.777. [DOI] [PubMed] [Google Scholar]
- Fan K., Wei P., Feng Q., Chen S., Huang C., Ma L., Lai B., Pei J., Liu Y., Chen J., Lai L. Biosynthesis, purification, and substrate specificity of severe acute respiratory syndrome coronavirus 3C-like proteinase. J. Biol. Chem. 2004;279:1637–1642. doi: 10.1074/jbc.M310875200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fielding B.C., Tan Y.-J., Shen S., Tan T.H.P., Ooi E.-E., Lim S.G., Hong W., Goh P.-Y. Characterization of a unique group-specific protein (U122) of the Severe Acute Respiratory Syndrome (SARS) coronavirus. J. Virol. 2004;78:7311–7318. doi: 10.1128/JVI.78.14.7311-7318.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouchier R.A., Kuiken T., Schutten M., van Amerongen G., van Doornum G.J., van den Hoogen B.G., Peiris M., Lim W., Stohr K., Osterhaus A.D. Aetiology: Koch's postulates fulfilled for SARS virus. Nature. 2003;423:240. doi: 10.1038/423240a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher T.M., Buchmeier M.J. Coronavirus spike proteins in viral entry and pathogenesis. Virology. 2001;279:371–374. doi: 10.1006/viro.2000.0757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F., Ou H.Y., Chen L.L., Zheng W.X., Zhang C.T. Prediction of proteinase cleavage sites in polyproteins of coronaviruses and its applications in analyzing SARS-CoV genomes. FEBS Lett. 2003;553:451–456. doi: 10.1016/S0014-5793(03)01091-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W., Tamin A., Soloff A., D’Aiuto L., Nwanegbo E., Robbins P.D., Bellini W.J., Barratt-Boyes S., Gambotto A. Effects of a SARS-associated coronavirus vaccine in monkeys. Lancet. 2003;362:1895–1896. doi: 10.1016/S0140-6736(03)14962-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E. Big nidovirus genome. When count and order of domains matter. Adv. Exp. Med. Biol. 2001;494:1–17. [PubMed] [Google Scholar]
- Guan Y., Zheng B.J., He Y.Q., Liu X.L., Zhuang Z.X., Cheung C.L., Luo S.W., Li P.H., Zhang L.J., Guan Y.J., Butt K.M., Wong K.L., Chan K.W., Lim W., Shortridge K.F., Yuen K.Y., Peiris J.S., Poon L.L. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- Guan M., Chen H.Y., Foo S.Y., Tan Y.-J., Goh P.Y., Wee S.H. Recombinant protein-based enzyme-linked immunosorbent assay and immunochromatographic tests for detection of immunoglobulin G antibodies to severe acute respiratory syndrome (SARS) coronavirus in SARS patients. Clin. Diagn. Lab. Immunol. 2004;11:287–291. doi: 10.1128/CDLI.11.2.287-291.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y., Peiris J.S., Zheng B., Poon L.L., Chan K.H., Zeng F.Y., Chan C.W., Chan M.N., Chen J.D., Chow K.Y., Hon C.C., Hui K.H., Li J., Li V.Y., Wang Y., Leung S.W., Yuen K.Y., Leung F.C. Molecular epidemiology of the novel coronavirus that causes severe acute respiratory syndrome. Lancet. 2004;363:99–104. doi: 10.1016/S0140-6736(03)15259-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J.P., Petric M., Campbell W., McGeer P.L. SARS coronavirus peptides recognized by antibodies in the sera of convalescent cases. Virology. 2004;324:251–256. doi: 10.1016/j.virol.2004.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He R., Leeson A., Andonov A., Li Y., Bastien N., Cao J., Osiowy C., Dobie F., Cutts T., Ballantine M., Li X. Activation of AP-1 signal transduction pathway by SARS coronavirus nucleocapsid protein. Biochem. Biophys. Res. Commun. 2003;311:870–876. doi: 10.1016/j.bbrc.2003.10.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He R., Dobie F., Ballantine M., Leeson A., Li Y., Bastien N., Cutts T., Andonov A., Cao J., Booth T.F., Plummer F.A., Tyler S., Baker L., Li X. Analysis of multimerization of the SARS coronavirus nucleocapsid protein. Biochem. Biophys. Res. Commun. 2004;316:476–483. doi: 10.1016/j.bbrc.2004.02.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrewegh A.A., Vennema H., Horzinek M.C., Rottier P.J., de Groot R.J. The molecular genetics of feline coronaviruses: comparative sequence analysis of the ORF 7a/7b transcription unit of different biotypes. Virology. 1995;212:622–631. doi: 10.1006/viro.1995.1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiscox J.A., Wurm T., Wilson L., Britton P., Cavanagh D., Brooks G. The coronavirus infectious bronchitis virus nucleoprotein localizes to the nucleolus. J. Virol. 2001;75:506–512. doi: 10.1128/JVI.75.1.506-512.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y., Lin P.H., Liu C.Y., Lee S.P., Chao Y.C. Assembly of human severe acute respiratory syndrome coronavirus-like particles. Biochem. Biophys. Res. Commun. 2004;318:833–838. doi: 10.1016/j.bbrc.2004.04.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes K.V. SARS coronavirus: a new challenge for prevention and therapy. J. Clin. Invest. 2003;111:1605–1609. doi: 10.1172/JCI18819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q., Yu L., Petros A.M., Gunasekera A., Liu Z., Xu N., Hajduk P., Mack J., Fesik S.W., Olejniczak E.T. Structure of the N-terminal RNA-binding domain of the SARS CoV nucleocapsid protein. Biochemistry. 2004;43:6059–6063. doi: 10.1021/bi036155b. [DOI] [PubMed] [Google Scholar]
- Ingallinella P., Bianchi E., Finotto M., Cantoni G., Eckert D.M., Supekar V.M., Bruckmann C., Carfi A., Pessi A. Structural characterization of the fusion-active complex of severe acute respiratory syndrome (SARS) coronavirus. Proc. Natl. Acad. Sci. U.S.A. 2004;101:8709–8714. doi: 10.1073/pnas.0402753101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov K.A., Thiel V., Dobbe J.C., van der Meer Y., Snijder E.J., Ziebuhr J. Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J. Virol. 2004;78:5619–5632. doi: 10.1128/JVI.78.11.5619-5632.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kienzle T.E., Abraham S., Hogue B.G., Brian D.A. Structure and orientation of expressed bovine coronavirus hemagglutinin-esterase protein. J. Virol. 1990;64:1834–1838. doi: 10.1128/jvi.64.4.1834-1838.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.W., Lee J.H., Hung C.F., Peng S., Roden R., Wang M.C., Viscidi R., Tsai Y.C., He L., Chen P.J., Boyd D.A., Wu T.C. Generation and characterization of DNA vaccines targeting the nucleocapsid protein of severe acute respiratory syndrome coronavirus. J. Virol. 2004;78:4638–4645. doi: 10.1128/JVI.78.9.4638-4645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krokhin O., Li Y., Andonov A., Feldmann H., Flick R., Jones S., Stroeher U., Bastien N., Dasuri K.V., Cheng K., Simonsen J.N., Perreault H., Wilkins J., Ens W., Plummer F., Standing K.G. Mass spectrometric characterization of proteins from the SARS virus: a preliminary report. Mol. Cell. Proteomics. 2003;2:346–356. doi: 10.1074/mcp.M300048-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A.E., Humphrey C.D., Shieh W.J., Guarner J., Paddock C.D., Rota P., Fields B., DeRisi J., Yang J.Y., Cox N., Hughes J.M., LeDuc J.W., Bellini W.J., Anderson L.J., SARS Working Group A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- Kuiken T., van den Hoogen B.G., van Riel D.A., Laman J.D., van Amerongen G., Sprong L., Fouchier R.A., Osterhaus A.D. Experimental human metapneumovirus infection of cynomolgus macaques (Macaca fascicularis) results in virus replication in ciliated epithelial cells and pneumocytes with associated lesions throughout the respiratory tract. Am. J. Pathol. 2004;164:1893–1900. doi: 10.1016/S0002-9440(10)63750-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M.M.C., Cavanagh D. The molecular biology of coronaviruses. Adv. Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M.M.C., Holmes K.V. Coronaviruses. In: Knipe D.M., Howley P.M., editors. Fields Virology. fourth ed. Lippincott; Philadelphia, USA: 2001. pp. 1163–1185. [Google Scholar]
- Lau S.K., Woo P.C., Wong B.H., Tsoi H.W., Woo G.K., Poon R.W., Chan K.H., Wei W.I., Peiris J.S., Yuen K.Y. Detection of severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein in sars patients by enzyme-linked immunosorbent assay. J. Clin. Microbiol. 2004;42:2884–2889. doi: 10.1128/JCM.42.7.2884-2889.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung D.T., Tam F.C., Ma C.H., Chan P.K., Cheung J.L., Niu H., Tam J.S., Lim P.L. Antibody response of patients with severe acute respiratory syndrome (SARS) targets the viral nucleocapsid. J. Infect. Dis. 2004;190:379–386. doi: 10.1086/422040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu I.J., Hsueh P.R., Lin C.T., Chiu C.Y., Kao C.L., Liao M.Y., Wu H.C. Disease-specific B cell epitopes for serum antibodies from patients with severe acute respiratory syndrome (SARS) and serologic detection of SARS antibodies by epitope-based peptide antigens. J. Infect. Dis. 2004;190:797–809. doi: 10.1086/422753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Xiao G., Chen Y., He Y., Niu J., Escalante C.R., Xiong H., Farmar J., Debnath A.K., Tien P., Jiang S. Interaction between heptad repeat 1 and 2 regions in spike protein of SARS-associated coronavirus: implications for virus fusogenic mechanism and identification of fusion inhibitors. Lancet. 2004;363:938–947. doi: 10.1016/S0140-6736(04)15788-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S., Khattra J., Asano J.K., Barber S.A., Chan S.Y., Cloutier A., Coughlin S.M., Freeman D., Girn N., Griffith O.L., Leach S.R., Mayo M., McDonald H., Montgomery S.B., Pandoh P.K., Petrescu A.S., Robertson A.G., Schein J.E., Siddiqui A., Smailus D.E., Stott J.M., Yang G.S., Plummer F., Andonov A., Artsob H., Bastien N., Bernard K., Booth T.F., Bowness D., Czub M., Drebot M., Fernando L., Flick R., Garbutt M., Gray M., Grolla A., Jones S., Feldmann H., Meyers A., Kabani A., Li Y., Normand S., Stroher U., Tipples G.A., Tyler S., Vogrig R., Ward D., Watson B., Brunham R.C., Krajden M., Petric M., Skowronski D.M., Upton C., Roper R.L. The Genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- Marsh M., Pelchen-Matthews A. Endocytosis in viral replication. Traffic. 2000;1:525–532. doi: 10.1034/j.1600-0854.2000.010701.x. [DOI] [PubMed] [Google Scholar]
- Martina B.E., Haagmans B.L., Kuiken T., Fouchier R.A., Rimmelzwaan G.F., Van Amerongen G., Peiris J.S., Lim W., Osterhaus A.D. Virology: SARS virus infection of cats and ferrets. Nature. 2003;425:915. doi: 10.1038/425915a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani T., Fukushi S., Saijo M., Kurane I., Morikawa S. Phosphorylation of p38 MAPK and its downstream targets in SARS coronavirus-infected cells. Biochem. Biophys. Res. Commun. 2004;319:1228–1234. doi: 10.1016/j.bbrc.2004.05.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navas-Martin S., Weiss S.R. SARS: lessons learned from other coronaviruses. Viral Immunol. 2003;16:461–474. doi: 10.1089/088282403771926292. [DOI] [PubMed] [Google Scholar]
- Nishimura N., Balch W.E. A di-acidic signal required for selective export from the endoplasmic reticulum. Science. 1997;277:556–558. doi: 10.1126/science.277.5325.556. [DOI] [PubMed] [Google Scholar]
- Olsen C.W. A review of feline infectious peritonitis virus: molecular biology, immunopathogenesis, clinical aspects, and vaccination. Vet. Microbiol. 1993;36:1–37. doi: 10.1016/0378-1135(93)90126-R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortego J., Sola I., Almazan F., Ceriani J.E., Riquelme C., Balasch M., Plana J., Enjuanes L. Transmissible gastroenteritis coronavirus gene 7 is not essential but influences in vivo virus replication and virulence. Virology. 2003;308:13–22. doi: 10.1016/S0042-6822(02)00096-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker M.D., Yoo D., Cox G.J., Babiuk L.A. Primary structure of the S peplomer gene of bovine coronavirus and surface expression in insect cells. J. Gen. Virol. 1990;71:263–270. doi: 10.1099/0022-1317-71-2-263. [DOI] [PubMed] [Google Scholar]
- Peiris J.S., Lai S.T., Poon L.L., Guan Y., Yam L.Y., Lim W., Nicholls J., Yee W.K., Yan W.W., Cheung M.T., Cheng V.C., Chan K.H., Tsang D.N., Yung R.W., Ng T.K., Yuen K.Y., SARS Study Group Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.H., Tong S., Tamin A., Lowe L., Frace M., DeRisi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., Gunther S., Osterhaus A.D., Drosten C., Pallansch M.A., Anderson L.J., Bellini W.J. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- Ruan Y.J., Wei C.L., Ee A.L., Vega V.B., Thoreau H., Su S.T., Chia J.M., Ng P., Chiu K.P., Lim L., Zhang T., Peng C.K., Lin E.O., Lee N.M., Yee S.L., Ng L.F., Chee R.E., Stanton L.W., Long P.M., Liu E.T. Comparative full-length genome sequence analysis of 14 SARS coronavirus isolates and common mutations associated with putative origins of infection. Lancet. 2003;361:1779–1785. doi: 10.1016/S0140-6736(03)13414-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz B., Routledge E., Siddell S.G. Murine coronavirus non-structural protein ns2 is not essential for virus replication in transformed cells. J. Virol. 1990;64:4784–4791. doi: 10.1128/jvi.64.10.4784-4791.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S., Wen Z.L., Liu D.X. Emergence of a coronavirus infectious bronchitis virus mutant with a truncated 3b gene: functional characterization of the 3b protein in pathogenesis and replication. Virology. 2003;311:16–27. doi: 10.1016/S0042-6822(03)00117-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Yi Y., Li P., Kuang T., Li L., Dong M., Ma Q., Cao C. Diagnosis of severe acute respiratory syndrome (SARS) by detection of SARS coronavirus nucleocapsid antibodies in an antigen-capturing enzyme-linked immunosorbent assay. J. Clin. Microbiol. 2003;41:5781–5782. doi: 10.1128/JCM.41.12.5781-5782.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Wei Z., Song J. Dissection study on the severe acute respiratory syndrome 3C-like protease reveals the critical role of the extra domain in dimerization of the enzyme: defining the extra domain as a new target for design of highly specific protease inhibitors. J. Biol. Chem. 2004;279:24765–24773. doi: 10.1074/jbc.M311744200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddell S.G. Plenum Press; New York, USA: 1995. The Coronaviridae. [Google Scholar]
- Smith A.R., Boursnell M.E., Binns M.M., Brown T.D., Inglis S.C. Identification of a new membrane-associated polypeptide specified by the coronavirus infectious bronchitis virus. J. Gen. Virol. 1990;71:3–11. doi: 10.1099/0022-1317-71-1-3. [DOI] [PubMed] [Google Scholar]
- Snijder E.J., Bredenbeek P.J., Dobbe J.C., Thiel V., Ziebuhr J., Poon L.L., Guan Y., Rozanov M., Spaan W.J., Gorbalenya A.E. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 2003;331:991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbarao K., McAuliffe J., Vogel L., Fahle G., Fischer S., Tatti K., Packard M., Shieh W.J., Zaki S., Murphy B. Prior infection and passive transfer of neutralizing antibody prevent replication of severe acute respiratory syndrome coronavirus in the respiratory tract of mice. J. Virol. 2004;78:3572–3577. doi: 10.1128/JVI.78.7.3572-3577.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui J., Li W., Murakami A., Tamin A., Matthews L.J., Wong S.K., Moore M.J., Tallarico A.S., Olurinde M., Choe H., Anderson L.J., Bellini W.J., Farzan M., Marasco W.A. Potent neutralization of severe acute respiratory syndrome (SARS) coronavirus by a human mAb to S1 protein that blocks receptor association. Proc. Natl. Acad. Sci. U.S.A. 2004;101:2536–2541. doi: 10.1073/pnas.0307140101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surjit M., Liu B., Kumar P., Chow V.T., Lal S.K. The nucleocapsid protein of the SARS coronavirus is capable of self-association through a C-terminal 209 amino acid interaction domain. Biochem. Biophys. Res. Commun. 2004;317:1030–1036. doi: 10.1016/j.bbrc.2004.03.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surjit M., Liu B., Jameel S., Chow V.T., Lal S.K. The SARS coronavirus nucleocapsid (N) protein induces actin reorganization and apoptosis in COS-1 cells in the absence of growth factors. Biochem. J. 2004 doi: 10.1042/BJ20040984. (published online on August 5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton G., Fry E., Carter L., Sainsbury S., Walter T., Nettleship J., Berrow N., Owens R., Gilbert R., Davidson A., Siddell S., Poon L.L., Diprose J., Alderton D., Walsh M., Grimes J.M., Stuart D.I. The nsp9 replicase protein of SARS-coronavirus, structure and functional insights. Structure (Camb) 2004;12:341–353. doi: 10.1016/j.str.2004.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y.-J., Fielding B.C., Goh P.-Y., Shen S., Tan T.H.P., Lim S.G., Hong W. Over-expression of 7a, a protein specifically encoded by the Severe Acute Respiratory Syndrome (SARS)-coronavirus, induces apoptosis via a caspase-dependent pathway. J. Virol. 2004;78:14043–14047. doi: 10.1128/JVI.78.24.14043-14047.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y.-J., Goh P.-Y., Fielding B.C., Shen S., Chou C.-F., Fu J.-L., Leong H.N., Leo Y.S., Ooi E.E., Ling A.E., Lim S.G., Hong W. Profile of antibody responses against SARS-coronavirus recombinant proteins and their potential use as diagnostic markers. Clin. Diag. Lab. Immunol. 2004;11:362–371. doi: 10.1128/CDLI.11.2.362-371.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y.-J., Teng E., Shen S., Tan T.H.P., Goh P.-Y., Fielding B.C., Ooi E.-E., Tan H.-C., Lim S.G., Hong W. A novel SARS coronavirus protein, U274, is transported to the cell surface and undergoes endocytosis. J. Virol. 2004;78:6723–6734. doi: 10.1128/JVI.78.13.6723-6734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner J.A., Watt R.M., Chai Y.B., Lu L.Y., Lin M.C., Peiris J.S., Poon L.L., Kung H.F., Huang J.D. The severe acute respiratory syndrome (SARS) coronavirus NTPase/helicase belongs to a distinct class of 5′ to 3′ viral helicases. J. Biol. Chem. 2003;278:39578–39582. doi: 10.1074/jbc.C300328200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teasdale R.D., Jackson M.R. Signal-mediated sorting of membrane proteins between the endoplasmic reticulum and the golgi apparatus. Annu. Rev. Cell Dev. Biol. 1996;12:27–54. doi: 10.1146/annurev.cellbio.12.1.27. [DOI] [PubMed] [Google Scholar]
- The Chinese SARS Molecular Epidemiology Consortium Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science. 2004;303:1666–1669. doi: 10.1126/science.1092002. [DOI] [PubMed] [Google Scholar]
- Thiel V., Ivanov K.A., Putics A., Hertzig T., Schelle B., Bayer S., Weissbrich B., Snijder E.J., Rabenau H., Doerr H.W., Gorbalenya A.E., Ziebuhr J. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 2003;84:2305–2315. doi: 10.1099/vir.0.19424-0. [DOI] [PubMed] [Google Scholar]
- Tripet B., Howard M.W., Jobling M., Holmes R.K., Holmes K.V., Hodges R.S. Structural characterization of the SARS-coronavirus spike S fusion protein core. J. Biol. Chem. 2004;279:20836–20849. doi: 10.1074/jbc.M400759200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vennema H., Heijnen L., Zijderveld A., Horzinek M.C., Spaan W.J. Intracellular transport of recombinant coronavirus spike proteins: implications for virus assembly. J. Virol. 1990;64:339–346. doi: 10.1128/jvi.64.1.339-346.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vennema H., Godeke G.J., Rossen J.W., Voorhout W.F., Horzinek M.C., Opstelten D.J., Rottier P.J. Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J. 1996;15:2020–2028. doi: 10.1002/j.1460-2075.1996.tb00553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Grotthuss M., Wyrwicz L.S., Rychlewski L. mRNA cap-1 methyltransferase in the SARS genome. Cell. 2003;113:701–702. doi: 10.1016/S0092-8674(03)00424-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Wen J., Li J., Yin J., Zhu Q., Wang H., Yang Y., Qin E., You B., Li W., Li X., Huang S., Yang R., Zhang X., Yang L., Zhang T., Yin Y., Cui X., Tang X., Wang L., He B., Ma L., Lei T., Zeng C., Fang J., Yu J., Wang J., Yang H., West M.B., Bhatnagar A., Lu Y., Xu N., Liu S. Assessment of immunoreactive synthetic peptides from the structural proteins of severe acute respiratory syndrome coronavirus. Clin. Chem. 2003;49:1989–1996. doi: 10.1373/clinchem.2003.023184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss S.R., Zoltick P.W., Leibowitz J.L. The ns4 gene of mouse hepatitis virus (MHV), strain A 59 contains two ORFs and thus differs from ns 4 of the JHM and S strains. Arch. Virol. 1993;129:301–309. doi: 10.1007/BF01316905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wentworth D.E., Gillim-Ross L., Espine N., Bernard K.A. Mice susceptible to SARS coronavirus. Emerg. Infect. Dis. 2004;10:1293–1296. doi: 10.3201/eid1007.031119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S.K., Li W., Moore M.J., Choe H., Farzan M.A. 193-amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. J. Biol. Chem. 2004;279:3197–3201. doi: 10.1074/jbc.C300520200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurm T., Chen H., Hodgson T., Britton P., Brooks G., Hiscox J.A. Localization to the nucleolus is a common feature of coronavirus nucleoproteins, and the protein may disrupt host cell division. J. Virol. 2001;75:9345–9356. doi: 10.1128/JVI.75.19.9345-9356.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X., Chakraborti S., Dimitrov A.S., Gramatikoff K., Dimitrov D.S. The SARS-CoV S glycoprotein: expression and functional characterization. Biochem. Biophys. Res. Commun. 2003;312:1159–1164. doi: 10.1016/j.bbrc.2003.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Yang M., Ding Y., Liu Y., Lou Z., Zhou Z., Sun L., Mo L., Ye S., Pang H., Gao G.F., Anand K., Bartlam M., Hilgenfeld R., Rao Z. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc. Natl. Acad. Sci. U.S.A. 2003;100:13190–13195. doi: 10.1073/pnas.1835675100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z.Y., Kong W.P., Huang Y., Roberts A., Murphy B.R., Subbarao K., Nabel G.J. A DNA vaccine induces SARS coronavirus neutralization and protective immunity in mice. Nature. 2004;428:561–564. doi: 10.1038/nature02463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh S.H., Wang H.Y., Tsai C.Y., Kao C.L., Yang J.Y., Liu H.W., Su I.J., Tsai S.F., Chen D.S., Chen P.J., National Taiwan University SARS Research Team Characterization of severe acute respiratory syndrome coronavirus genomes in Taiwan: molecular epidemiology and genome evolution. Proc. Natl. Acad. Sci. U.S.A. 2004;101:2542–2547. doi: 10.1073/pnas.0307904100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying W., Hao Y., Zhang Y., Peng W., Qin E., Cai Y., Wei K., Wang J., Chang G., Sun W., Dai S., Li X., Zhu Y., Li J., Wu S., Guo L., Dai J., Wang J., Wan P., Chen T., Du C., Li D., Wan J., Kuai X., Li W., Shi R., Wei H., Cao C., Yu M., Liu H., Dong F., Wang D., Zhang X., Qian X., Zhu Q., He F. Proteomic analysis on structural proteins of Severe Acute Respiratory Syndrome coronavirus. Proteomics. 2004;4:492–504. doi: 10.1002/pmic.200300676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yount B., Curtis K.M., Fritz E.A., Hensley L.E., Jahrling P.B., Prentice E., Denison M.R., Geisbert T.W., Baric R.S. Reverse genetics with a full-length infectious cDNA of severe acute respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. U.S.A. 2003;100:12995–13000. doi: 10.1073/pnas.1735582100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C.-J., Chen Y.-C., Hsiao C.-H., Kuo T.-C., Chang S.C., Lu C.-Y., Wei W.-C., Lee C.-H., Huang L.-M., Chang M.-F., Ho H.-N., Lee F.-J.S. Identification of a novel protein 3a from severe acute respiratory syndrome coronavirus. FEBS Lett. 2004;565:111–116. doi: 10.1016/j.febslet.2004.03.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan K., Yi L., Chen J., Qu X., Qing T., Rao X., Jiang P., Hu J., Xiong Z., Nie Y., Shi X., Wang W., Ling C., Yin X., Fan K., Lai L., Ding M., Deng H. Suppression of SARS-CoV entry by peptides corresponding to heptad regions on spike glycoprotein. Biochem. Biophys. Res. Commun. 2004;319:746–752. doi: 10.1016/j.bbrc.2004.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng R., Ruan H.Q., Jiang X.S., Zhou H., Shi L., Zhang L., Sheng Q.H., Tu Q., Xia Q.C., Wu J.R. Proteomic analysis of SARS associated coronavirus using two-dimensional liquid chromatography mass spectrometry and one-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by mass spectroemtric analysis. J. Proteome Res. 2004;3:549–555. doi: 10.1021/pr034111j. [DOI] [PubMed] [Google Scholar]
- Zeng R., Yang R.F., Shi M.D., Jiang M.R., Xie Y.H., Ruan H.Q., Jiang X.S., Shi L., Zhou H., Zhang L., Wu X.D., Lin Y., Ji Y.Y., Xiong L., Jin Y., Dai E.H., Wang X.Y., Si B.Y., Wang J., Wang H.X., Wang C.E., Gan Y.H., Li Y.C., Cao J.T., Zuo J.P., Shan S.F., Xie E., Chen S.H., Jiang Z.Q., Zhang X., Wang Y., Pei G., Sun B., Wu J.R. Characterization of the 3a protein of SARS-associated coronavirus in infected vero E6 cells and SARS patients. J. Mol. Biol. 2004;341:271–279. doi: 10.1016/j.jmb.2004.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Xiao G., Xu Y., Yuan F., Zheng C., Liu Y., Yan H., Cole D.K., Bell J.I., Rao Z., Tien P., Gao G.F. Following the rule: formation of the 6-helix bundle of the fusion core from severe acute respiratory syndrome coronavirus spike protein and identification of potent peptide inhibitors. Biochem. Biophys. Res. Commun. 2004;319:283–288. doi: 10.1016/j.bbrc.2004.04.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr J., Snijder E.J., Gorbalenya A.E. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 2000;81:853–879. doi: 10.1099/0022-1317-81-4-853. [DOI] [PubMed] [Google Scholar]


