Abstract
This review focuses on the monophyletic group of animal RNA viruses united in the order Nidovirales. The order includes the distantly related coronaviruses, toroviruses, and roniviruses, which possess the largest known RNA genomes (from 26 to 32 kb) and will therefore be called ‘large’ nidoviruses in this review. They are compared with their arterivirus cousins, which also belong to the Nidovirales despite having a much smaller genome (13–16 kb). Common and unique features that have been identified for either large or all nidoviruses are outlined. These include the nidovirus genetic plan and genome diversity, the composition of the replicase machinery and virus particles, virus-specific accessory genes, the mechanisms of RNA and protein synthesis, and the origin and evolution of nidoviruses with small and large genomes. Nidoviruses employ single-stranded, polycistronic RNA genomes of positive polarity that direct the synthesis of the subunits of the replicative complex, including the RNA-dependent RNA polymerase and helicase. Replicase gene expression is under the principal control of a ribosomal frameshifting signal and a chymotrypsin-like protease, which is assisted by one or more papain-like proteases. A nested set of subgenomic RNAs is synthesized to express the 3′-proximal ORFs that encode most conserved structural proteins and, in some large nidoviruses, also diverse accessory proteins that may promote virus adaptation to specific hosts. The replicase machinery includes a set of RNA-processing enzymes some of which are unique for either all or large nidoviruses. The acquisition of these enzymes may have improved the low fidelity of RNA replication to allow genome expansion and give rise to the ancestors of small and, subsequently, large nidoviruses.
Keywords: RNA viruses, Nidoviruses, Coronaviruses, Arteriviruses, Roniviruses, Astroviruses, Virus evolution, RNA replication
1. Introduction: nidoviruses as a distinct group of ssRNA+ viruses
RNA viruses have evolved a remarkable diversity of genomes and replicative mechanisms. Although the size variation of RNA virus genomes is quite low (ranging from ∼3 to 32 kb), four distinct classes have been defined, three occupied solely by RNA viruses and one shared with DNA viruses, to order the great diversity of RNA viruses (Baltimore, 1971). DNA viruses, on the other hand, form only two distinct classes, although their genome sizes can differ by as much as ∼3 orders of magnitude. The small size of RNA virus genomes is fundamentally linked to the low fidelity of their RNA synthesis, which is mediated by a cognate replication complex containing an RNA-dependent RNA polymerase (RdRp) that lacks proofreading/repair mechanisms. RNA viruses may generate as much as one mutation per genome per replication round (Drake and Holland, 1999), a feature that is counterbalanced by the large population size of their progeny, which has been characterized as ‘quasispecies’ on the basis of its genome variability (Eigen and Schuster, 1977). As a result of this combination of properties, RNA viruses are “primed” to adapt to new environmental conditions, but they are limited in their ability to expand their genome as they must keep their mutation load below the so-called ‘error threshold’, above which the survival of the quasispecies becomes impossible (for reviews, see Domingo and Holland, 1997, Moya et al., 2004).
Among the four classes of RNA viruses, only the class containing viruses with single-stranded genomes of positive (mRNA) polarity (ssRNA+) exploit the full genome size range outlined above (Fig. 1 ). Members of this class are also the most numerous in terms of the number of families and distinct groups recognized by the International Committee for the Taxonomy of Viruses (ICTV) (Fauquet et al., 2005). With genome sizes of 26–32 kb, three groups of phylogenetically related viruses, namely coronaviruses, toroviruses and roniviruses, are found at the upper end of this genome size scale and are clearly separated from other RNA viruses. We will discuss them in comparison with their much smaller arterivirus cousins (genome size of ‘only’ 13–16 kb), which also belong to the order Nidovirales (Cavanagh, 1997, de Vries et al., 1997, Snijder et al., 2005b, Spaan et al., 2005b). The ICTV recognized roniviruses (Walker et al., 2005) and arteriviruses (Snijder et al., 2005a) as distinct families, while the current taxonomic position of coronaviruses and toroviruses as two genera of the Coronaviridae family is being revised by elevating these virus groups to a higher taxonomic rank, that of either subfamily or family (González et al., 2003, Spaan et al., 2005a). Hereafter, we will refer to coronaviruses, toroviruses and roniviruses as ‘large’ nidoviruses and to arteriviruses as ‘small’ nidoviruses, to stress the clear genome size difference.
Fig. 1.
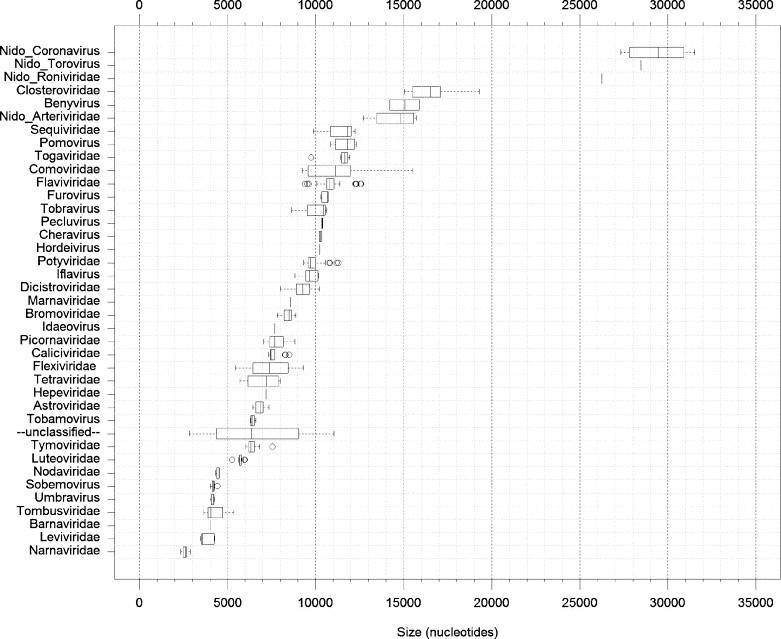
Distribution of genome sizes of ssRNA+ viruses. Box-and-whisker graphs were used to plot the family/group-specific distribution of genome sizes of all ssRNA+ viruses whose genome sequences have been placed in the NCBI Viral Genome Resource (Bao et al., 2004) by December 7, 2005 (Faase and Gorbalenya, unpublished data). Four major groups of nidoviruses are highlighted with the Nido- prefix, and the Coronaviridae family is split into the Nido-Coronavirus and the Nido-Torovirus groups. The box spans from the first to the third quartile and includes the median, indicated by the vertical line. The whiskers extend to the extreme values that are distant from the box at most 1.5 times the interquartile range. Values beyond this distance are indicated by circles (outliers).
Nidoviruses rank among the most complex RNA viruses and their molecular genetics clearly discriminates them from other RNA virus groups. Still, our knowledge about their molecular biology, mostly derived from studies of coronaviruses, is very limited (Lai and Cavanagh, 1997, Lai and Holmes, 2001, Siddell et al., 2005, Ziebuhr, 2004). Nidoviruses attach to their host cell by binding to a receptor on the cell surface, after which fusion of the viral and cellular membrane is presumably mediated by one of the surface glycoproteins. This fusion event (either at the plasma or endosomal membrane) releases the nucleocapsid into the host cell's cytoplasm. After genome uncoating, translation of two replicase open reading frames (ORFs) is initiated by host ribosomes, to yield large polyprotein precursors that undergo autoproteolysis to eventually produce a membrane-bound replicase/transcriptase complex. This complex mediates the synthesis of the genome RNA and a nested set of subgenomic RNAs that direct the synthesis of structural proteins and, optionally, some other proteins (see below). New virus particles are assembled by the association of the new genomes with the cytoplasmic nucleocapsid protein and the subsequent envelopment of the nucleocapsid structure. The viral envelope proteins are inserted into intracellular membranes and targeted to the site of virus assembly (usually membranes between the endoplasmic reticulum and Golgi complex), where they meet up with the nucleocapsid and trigger the budding of virus particles into the lumen of the membrane compartment. The newly formed virions then leave the cell by following the exocytic pathway towards the plasma membrane.
Below, we will discuss (from an evolutionary perspective) the molecular biological properties that unite and separate the large and small nidoviruses. This overview will involve an analysis of the internal and external evolutionary relationships of nidoviruses, which in some cases are very distant. These relationships have originally been delineated using bioinformatics tools and are now more and more supported by the results of functional and structural studies. This fruitful cooperation between theoretical and experimental science was especially instrumental in the dissection of the nidovirus replication apparatus, which is the central theme of this review. Due to the way in which the initial functional assignments were made, the names of many nidovirus replicative proteins were derived from those of viral or cellular homologs (Gorbalenya, 2001, Snijder et al., 2003, Spaan et al., 2005b, Ziebuhr et al., 2000). To conclude, we will broadly discuss our current understanding of the origin and evolution of small and large nidovirus genomes.
2. Nidovirus genome diversity and a common genetic plan
Nidovirus genomes have been studied for ∼20 years, most extensively during the last several years. By today, more than a hundred full-length and over a thousand partial nidovirus genome sequences have been published. Four profoundly separated and unevenly populated genetic clusters have been recognized: corona-, toro-, roni- and arteriviruses. The exact relationship between these clusters is yet to be rigorously resolved (González et al., 2003). The data available from phylogenetic analysis of the most conserved replicase domain of these viruses, the RdRp, with homologs of some other RNA viruses (Gorbalenya et al., 2002) and the analysis of the domain colinearity of their replicase genes (Bredenbeek et al., 1990, Cowley et al., 2000; den Boon et al., 1991a, Draker et al., 2006, Gorbalenya et al., 1989, Gorbalenya, 2001, Snijder et al., 2003) favour clustering coronaviruses and toroviruses (Fig. 2 ). Arteriviruses and roniviruses form two more distant nidovirus lineages, whose relative position as shown in Fig. 2 tree remains poorly supported (Gorbalenya et al., 2002). In fact, comparison of replicase polyproteins (see below) indicates that, contrary to Fig. 2 tree topology, roniviruses may group with corona- and toroviruses to form a supercluster of large nidoviruses, and arteriviruses would then be the first to split from the nidovirus trunk (Fig. 2). Notwithstanding the fact that this revised branching of the arteri- and ronivirus lineages is yet to be independently confirmed, we will in this review consider the large and small nidoviruses as two separate phylogenetic clusters. In addition to the significant genome size variation between the four groups of nidoviruses, also other major differences were documented, involving, for example, virion morphology (see below), host range, and various other biological properties that are outside the scope of this review.
Fig. 2.
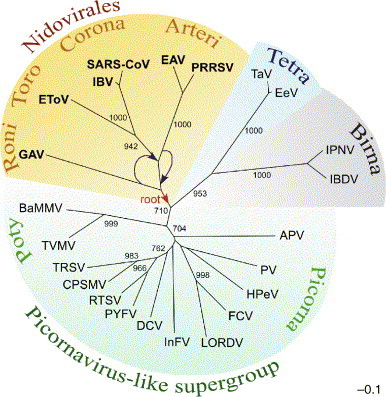
RdRp-based RNA virus tree that includes nidoviruses. The most conserved part of RdRps from representative viruses in the Picornaviridae, Dicistroviridae, Sequiviridae, Comoviridae, Caliciviridae, Potyviridae, Coronaviridae, Roniviridae, Arteriviridae, Birnaviridae, Tetraviridae and unclassified insect viruses was aligned. An unrooted neighbour-joining tree was inferred using the ClustalX1.81 software. For details of the analysis, see Gorbalenya et al. (2002). All bifurcations with support in >700 out of 1000 bootstraps are indicated. Different groups of viruses are highlighted with different colours. The tree was modified from (Gorbalenya et al., 2002, Snijder et al., 2005b, Spaan et al., 2005b). Virus families/groups and abbreviations of viruses included in the analysis are as follows: Coronaviridae: avian infectious bronchitis virus (IBV), severe acute respiratory syndrome virus (SARS-CoV) and Equine torovirus (EToV); Arteriviridae, Equine arteritis virus (EAV) and Porcine reproductive and respiratory syndrome virus strain VR-2332 (PRRSV); Roniviridae: Gill-associated virus (GAV); Picornaviridae, human poliovirus type 3 Leon strain (PV) and parechovirus 1 (HPeV); Iflavirus, infectious flacherie virus (InFV); unclassified insect viruses, Acyrthosiphon pisum virus (APV); Dicistroviridae, Drosophila C virus (DCV); Sequiviridae, Rice tungro spherical virus (RTSV) and Parsnip yellow fleck virus (PYFV); Comoviridae, Cowpea severe mosaic virus (CPSMV) and Tobacco ringspot virus (TRSV); Caliciviridae, Feline calicivirus F9 (FCV) and Lordsdale virus (LORDV); Potyviridae, Tobacco vein mottling virus (TVMV) and Barley mild mosaic virus (BaMMV); Tetraviridae, Thosea asigna virus (TaV) and Euprosterna elaeasa virus (EeV); Birnaviridae, infectious pancreatic necrosis virus (IPNV) and infectious bursal disease virus (IBDV). A plausible direction to the root of the nidovirus domain is indicated (red arrow). As discussed in the text, arteriviruses rather than roniviruses might have been the first to branch off from the nidovirus trunk. This revised topology of the two lineages is also indicated (blue arrows).
The overwhelming majority of the available nidovirus genome sequences have been reported for corona- and arteriviruses, which were the first to be fully sequenced and analyzed (Boursnell et al., 1987, den Boon et al., 1991a). Comparative sequence analysis and other studies of viruses from each of these families revealed three major groups in coronaviruses, called groups 1, 2 and 3, and four comparably distant genetic clusters in arteriviruses (González et al., 2003, Gorbalenya et al., 2004, Snijder et al., 2005a, Spaan et al., 2005a). So far, only very few sequences, including one full-length genome sequence for each virus group, have been reported for toroviruses (Draker et al., 2006) and roniviruses (Cowley and Walker, 2002) and, hence, information on the genetic variability of these nidovirus taxa is extremely limited. Although toroviruses and roniviruses have not been studied in great detail, the features listed below are thought to be common to nidovirus genomes and their expression (Cavanagh, 1997, Lai and Holmes, 2001, Snijder et al., 2005b, Spaan et al., 2005b).
Nidoviruses have linear, single-stranded RNA genomes of positive polarity that contain a 5′ cap structure and a 3′ poly (A) tail. The genomes of nidoviruses include untranslated regions (UTR) at their 5′ and 3′ genome termini. These flank an array of multiple genes whose number may vary in and between the families of the nidovirus order (Fig. 3 ). Roniviruses have only four ORFs, toroviruses have six ORFs, arteriviruses may have between 9 and 12 ORFs and coronaviruses may encode from 9 to 14 ORFs. In all nidoviruses, the two most 5′ and largest ORFs, ORF1a and ORF1b, occupy between two-thirds and three-quarters of the genome. They overlap in a small area containing a −1 ribosomal frameshift (RFS) signal that directs translation of ORF1b by a fraction of ribosomes that have started protein synthesis at the ORF1a initiator AUG. These two ORFs encode the subunits of the replicase machinery, which are produced by autoproteolytic processing of the pp1a and pp1ab polyproteins encoded by ORF1a and ORF1a/b, respectively.
Fig. 3.
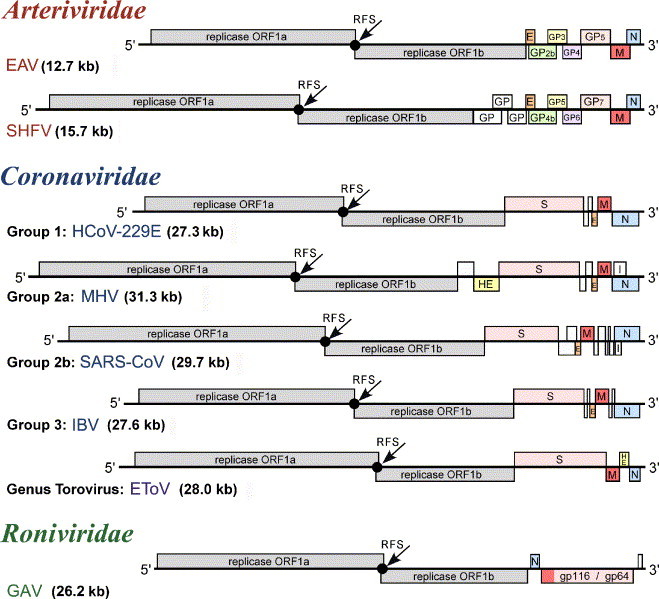
Genome organization of selected nidoviruses. The genomic ORFs of viruses representing major lineages of nidoviruses are indicated and the names of the replicase and main virion genes are given. References to the nomenclature of accessory genes can be found in the text. Genomes of large and small nidoviruses are drawn to different scales. This figure was updated from Fig. 39.2 presented in Siddell et al. (2005).
The ORFs located downstream of ORF1b encode nucleocapsid and envelope protein(s), the numbers of which differ between the major nidovirus branches. Family and group-specific ORFs in this region may encode additional virion and non-structural (“accessory”) proteins (see below). These ORFs located in the 3′-part of the genome are expressed from a nested set of subgenomic RNAs (see below), a property that was reflected in the name of the virus order (nidus in Latin means nest) (Cavanagh, 1997). In large nidoviruses, compared to small nidoviruses, each of the two areas occupied by either the replicase ORFs or the 3′-proximal ORFs is expanded proportionally.
3. Replicase machinery: the conserved nidovirus backbone
What is truly unique for nidoviruses? Polycistronic genome organization may not be this feature as some other RNA viruses possessing either large genomes, e.g., the Closteroviridae (see review of Dolja et al., 2006), or much smaller genomes, e.g., the Luteoviridae (Miller et al., 1997) and Astroviridae (Jiang et al., 1993), employ similar genome organizations. The presence of a common ‘leader’ sequence at the 5′-end of all viral mRNA species (Baric et al., 1983, de Vries et al., 1990, Lai et al., 1984, Spaan et al., 1983) was initially considered to be a possible nidovirus hallmark. However, this property subsequently proved to be not universally conserved amongst nidoviruses (Cowley et al., 2002, van Vliet et al., 2002). Currently, only the organization and composition of the multidomain replicase gene can be used to discriminate nidoviruses from other RNA viruses. The power of this criterion was originally recognized upon large-scale comparisons of ssRNA+ virus genomes enabling the higher-order clustering of different virus families in several super-families/groups (Goldbach, 1986, Strauss and Strauss, 1988), including a Coronavirus-like supergroup (now known as nidoviruses) (Gorbalenya and Koonin, 1993a, Snijder et al., 1993). Thus, for example, replicase ORF1b encodes two domains that have not been identified in other RNA virus families and, therefore, qualify as genetic markers of this virus order. These are:
-
I.
The (putative) multinuclear zinc-binding domain (ZBD) (Cowley et al., 2000, Gorbalenya et al., 1989, Seybert et al., 2005, Snijder et al., 1990a, van Dinten et al., 2000);
-
II.
The uridylate-specific endoribonuclease (NendoU) domain (Bhardwaj et al., 2004, Ivanov et al., 2004b, Posthuma et al., 2006, Snijder et al., 1990a, Snijder et al., 2003).
These two domains are part of a conserved array of domains whose sequential arrangement can be abbreviated as NH2_TM1-TM2-3CLpro-TM3-RFS-RdRp-ZBD-HEL1-NendoU_COOH (Gorbalenya, 2001) (see also below). This array is specific for nidoviruses, although it includes domains that may also be common to other RNA virus families. Some of these domains, which may appear adjacent in the above domain constellation, may actually be separated by other, less conserved domains in specific nidovirus taxa. The conserved nidovirus-specific domain order was suggested to be intimately linked to essential and conserved aspects of the nidovirus replicative cycle (Gorbalenya, 2001). The domains forming the above constellation proved to be essential for nidovirus RNA synthesis and the production of progeny virions in reverse genetics experiments using the equine arterivirus (EAV) and/or various coronaviruses (see below).
The ORF1a-encoded N-terminal part of this array of domains consists of the 3C-like proteinase (3CLpro) and flanking trans-membrane (TM) domains (TM1-TM2-3CLpro-TM3) (Gorbalenya et al., 1989) (Fig. 4 ). The 3CLpro has a chymotrypsin-like fold and a substrate specificity resembling that of picornavirus 3C proteinases (Anand et al., 2002, Barrette-Ng et al., 2002, Draker et al., 2006, Hegyi and Ziebuhr, 2002, Snijder et al., 1996, Ziebuhr et al., 2003, Ziebuhr and Siddell, 1999) (Smits et al., in press). The 3CLpro is responsible for the processing of the downstream, most conserved part of the replicase polyproteins and, because of this property, may also be called “main proteinase” (Mpro) (reviewed by (Ziebuhr et al., 2000)). The enzyme cleaves the C-terminal half of pp1a and the ORF1b-encoded part of pp1ab at 8–11 sites having a glutamine or glutamic acid at the P1 position and a small residue (usually glycine, alanine or serine) at the P1’ position (reviewed in (Ziebuhr et al., 2000, Ziebuhr, 2005)). From an evolutionary perspective, the nidovirus-wide conservation of this specificity is most remarkable. The 3CLpros of the four main nidovirus branches are profoundly diverged and (according to conventional criteria) do not group together, although they have become significantly separated from their viral and cellular homologs. Even the catalytic center, which is uniformly conserved in cellular homologs, has evolved into quite different forms, including a Cys–His catalytic dyad in roni- and coronaviruses, a Ser–His–Asp catalytic triad in arteriviruses, and a Ser–His-based catalytic center in toroviruses (Anand et al., 2002, Anand et al., 2003, Barrette-Ng et al., 2002, Draker et al., 2006, Snijder et al., 1996, Ziebuhr et al., 1997, Ziebuhr et al., 2003). The crystal structures of coronavirus and arterivirus 3CLpros revealed that both groups of enzymes have a three-domain structure, with domains I and II forming a (chymotrypsin-like) two-ß-barrel fold consisting of 12 or 13 ß-strands (Anand et al., 2002, Anand et al., 2003, Barrette-Ng et al., 2002, Yang et al., 2003). The C-terminal domains III of arterivirus and coronavirus 3CLpros differ from each other, both in size and structure, and roni- and torovirus 3CLpro may again have other versions of the C-terminal domain. Outside nidoviruses, only plant potyviruses encode a 3CLpro that has an extra C-terminal domain, a property which correlates with a special sequence resemblance between the catalytic domains of the potyvirus and corona-/ronivirus 3CLpros (Ziebuhr et al., 2003). The three conserved hydrophobic domains encoded by ORF1a (TMs in the above formula), two of which characteristically flank the 3CLpro at either side (Gorbalenya, 2001, Snijder and Meulenberg, 1998, Ziebuhr, 2005), appear to anchor the nidovirus replication complex to intracellular membranes (Prentice et al., 2004).
Fig. 4.
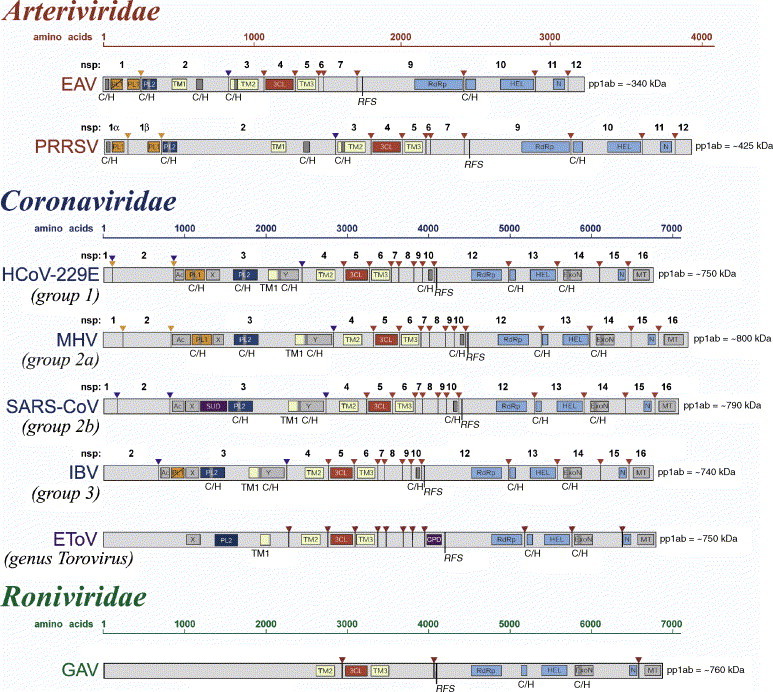
Domain organization of the replicase pp1ab polyprotein for selected nidoviruses. Shown are currently mapped domains in the pp1ab replicase polyproteins of viruses representing major lineages of nidoviruses. Arrows represent sites in pp1ab that are cleaved by papain-like proteinases (orange and blue) or chymotrypsin-like (3CLpro) proteinase (red). For viruses with the full cleavage site map available (arteriviruses and coronaviruses), the proteolytic cleavage products are numbered. A tentative cleavage site map for EToV is from an unpublished work by A.E. Gorbalenya. Within the cleavage products, the location and names of domains that have been identified as structurally and functionally related are highlighted. These include diverse domains with conserved Cys and His residues (C/H), putative transmembrane domains (TM), domains with conserved features (AC, X and Y), and domains that have been associated with proteolysis (PL1, PL2, and 3CL), RNA-dependent RNA synthesis (RdRp), helicase (HEL), exonuclease (ExoN), uridylate-specific endoribonuclease (N; NendoU in the main text), methyl transferase (MT) and cyclic phosphodiesterase (CPD) activities. Note that due to space limitations, the domain names in this figure may be abbreviated derivatives from those used in the main text. Polyproteins of large and small nidoviruses are drawn to different scales. This figure was updated from Fig. 39.4 presented in Siddell et al. (2005).
The four ORF1a-encoded domains (TM1-TM2-3CLpro-TM3) are linked through an RFS to the ORF1b-encoded array of conserved nidovirus domains (Fig. 4). The RFS includes a 'slippery’ heptanucleotide sequence, at which the ribosome makes the actual −1 frameshift. This 'slippery’ heptanucleotide is conserved among corona-, toro- and arteriviruses, but not in roniviruses which may have evolved a different sequence to perform the same function (Brierley et al., 1987, Brierley, 1995, Cowley et al., 2000). An elaborate RNA pseudoknot, which seems to be structurally conserved among nidoviruses, is located immediately downstream of the slippery sequence (Baranov et al., 2005, Brierley et al., 1991, Cowley et al., 2000, den Boon et al., 1991a, Draker et al., 2006, Herold and Siddell, 1993, Plant et al., 2005, Snijder et al., 1990a).
In the ORF1b-encoded polyprotein, the C-terminal part of pp1ab, four conserved domains have been identified in all nidoviruses (from N- to C-terminus): RdRp, ZBD, 5′-to-3′ helicase belonging to superfamily 1 (HEL1), and NendoU (Fig. 4). They comprise the most conserved sequences delineated among nidoviruses. The RdRp is the largest conserved domain, occupying the C-terminal region of a larger protein whose size varies significantly among nidoviruses (Bredenbeek et al., 1990, Cowley et al., 2000, den Boon et al., 1991a; Gorbalenya et al., 1989). The nidovirus RdRps cluster with orthologs encoded by viruses comprising the Picornavirus-like supergroup (Gorbalenya et al., 1989, Gorbalenya et al., 2002, Koonin, 1991) (Fig. 2). The RdRps of nidoviruses carry a notable replacement of a Gly residue (with one exception, by a Ser residue (Gorbalenya et al., 1989, Stephensen et al., 1999)) that is otherwise conserved among ssRNA+ viruses and precedes two key catalytic Asp residues in the sequence that is known as ‘the GDD motif’, an RdRp hallmark (Kamer and Argos, 1984). The RdRp domain is essential for RNA replication (van Marle et al., 1999b) and the in vitro RdRp activity of this domain was recently verified for SARS coronavirus (SARS-CoV) (Cheng et al., 2005).
Immediately downstream of the RdRp, the pp1ab polyproteins of all nidoviruses contain a protein that includes ZBD and HEL1 (Bredenbeek et al., 1990, Cowley et al., 2000, den Boon et al., 1991a, Gorbalenya et al., 1988, Gorbalenya et al., 1989) (Fig. 4). This relative position of RdRp and HEL domains is exceptional, as the helicase domain resides upstream of the RdRp in the replicase polyproteins of all other families of ssRNA+ viruses that have these domains in their repertoire (Gorbalenya et al., 1989). Distant homologs of the nidovirus HEL1 domain were identified in viruses of the alphavirus supergroup, and they are widespread among cellular organisms in all three kingdoms of life (Gorbalenya and Koonin, 1993b). Homologs of the ZBD, which is characterized by 12–13 conserved Cys/His residues, were not described. The ZBD appears to be required for the multiple enzymatic activities of the HEL1 domain, including NTPase, RNA 5′-triphosphatase and nucleic acid duplex unwinding (Bautista et al., 2002, Heusipp et al., 1997, Ivanov et al., 2004a, Ivanov and Ziebuhr, 2004, Seybert et al., 2000b, Seybert et al., 2000a, Seybert et al., 2005, Tanner et al., 2003). The arteri- and coronavirus ZBD-HEL1 proteins are capable of separating (probably in a processive manner) extended regions of up to several hundred base pairs of double-stranded RNA (and DNA) in the 5′-to-3′ direction (Ivanov et al., 2004a, Ivanov and Ziebuhr, 2004, Seybert et al., 2000b, Seybert et al., 2000a). Both domains were implicated in arterivirus genomic and subgenomic RNA synthesis (Seybert et al., 2005; van Dinten et al., 1997, van Dinten et al., 2000).
The most C-terminal domain of the conserved constellation of nidovirus replicase domains is NendoU (Bhardwaj et al., 2004, Ivanov et al., 2004b, Snijder et al., 2003). It is produced as part of a larger protein that includes another domain, which, depending on the nidovirus family, is either located upstream or downstream of NendoU in pp1ab (Fig. 4). Outside nidoviruses, distant homologs of NendoU were identified in some prokaryotes and eukaryotes that form a small protein family prototyped by the Xenopus laevis XendoU (Gioia et al., 2005, Laneve et al., 2003, Snijder et al., 2003). NendoU is a Mn2+-dependent enzyme that produces 2′-3′ cyclic phosphate ends (Ivanov et al., 2004b) and it may function as homohexamer (Guarino et al., 2005). Both the SARS-CoV and human coronavirus 229E (HCoV-229E) NendoU domains efficiently cleave double-stranded RNA at specific uridylate-containing sequences, although they also are able to process (albeit less specifically) single-stranded RNA at each uridylate present in the sequence. This domain is essential for RNA synthesis and/or the production of virus progeny in corona- and arteriviruses (Ivanov et al., 2004b, Posthuma et al., 2006; Hertzig et al., unpublished data).
Apart from the domains discussed above, sequence conservation among the most distant nidovirus branches could be barely identified in ORF1a and even some parts of ORF1b. This may be due to technical reasons related to weak conservation in a heavily biased sequence set or due to different origins of the respective domains in the main nidovirus branches. Most recently, structural studies have started to contribute to the mapping of putative functional domains in nidovirus replicase polyproteins (Bartlam et al., 2005, Egloff et al., 2004, Sutton et al., 2004, Zhai et al., 2005) and the nidovirus-specific domain constellation may be further expanded in the future.
The first candidate to be added to the constellation is another proteinase. Besides the conserved Mpro (3CLpro), arteri-, corona- and toroviruses encode so-called accessory proteinases (one to four, depending on the virus) that autocatalytically process the large N-proximal regions of the replicase polyproteins in which they reside ((Baker et al., 1989, Snijder et al., 1992) (reviewed by Ziebuhr et al., 2000); see also Draker et al., 2006). They belong to the same superfamily of papain-like (PLpro) cysteine proteinases despite often having little sequence resemblance and very different sizes (den Boon et al., 1995, Gorbalenya et al., 1991) (Fig. 4). Three nidoviruses, EAV, simian hemorrhagic fever virus (SHFV) and avian infectious bronchitis virus (IBV), encode PLpros that are not proteolytically active indicating that PLpros may have other, non-proteolytic function(s) in the nidovirus replicative cycle (den Boon et al., 1995, Ziebuhr et al., 2001). Nidovirus PLpro domains may be linked with zinc finger (Zf) domains, whose type and relative position vary among nidoviruses (Herold et al., 1999, Snijder et al., 1995, Tijms et al., 2001). A similar association with a Zf was not reported for the numerous homologs encoded by ssRNA+ viruses, although nidovirus accessory proteinases do share the substrate specificity towards small aliphatic residues at the P1 and P1’ positions that is prevailing among RNA virus-encoded homologs (Dong and Baker, 1994, Snijder et al., 1992) (for reviews, see Gorbalenya and Snijder, 1996, Ziebuhr et al., 2000). The Zf-PLpro-containing nsp1 protein of the arterivirus EAV was shown to be dispensable for replication but absolutely required for subgenomic RNA synthesis (Tijms et al., 2001). Interestingly, this protein provided a functional replacement for a remotely related PLpro in closterovirus RNA replication (Peng et al., 2002). The PLpros of coronaviruses contain a Zn-ribbon domain connecting two catalytic domains (Herold et al., 1999). A similar organization was also found in a herpesvirus-associated enzyme with which some coronavirus PLpros share deubiqitinating activity (Barretto et al., 2005, Lindner et al., 2005, Sulea et al., 2005). In roniviruses, the corresponding N-terminal pp1a/pp1ab region of about 2000 residues has not yet been functionally characterized, and no putative PLpro domain has been identified (Fig. 4).
4. Replicase machinery: group-specific domains
In addition to the domains that form the nidovirus-specific constellation, a number of other conserved (replicative) domains have been identified in some but not all nidoviruses. These domains reside in the pp1a and pp1ab polyproteins or, in one case, in a separate ORF.
Two such recently identified domains, the 3′-to-5′ exoribonuclease (ExoN) and the ribose-2′-O-methyltransferase (O-MT), are of a special interest as they are only conserved in the ORF1b-encoded portion of pp1ab of large nidoviruses (Snijder et al., 2003) (Fig. 4). Like the members of the nidovirus-specific array of replicase domains, the ExoN and O-MT were shown to be indispensable for viral RNA synthesis and/or the production of virus progeny in HCoV-229E (Hertzig et al., unpublished data; Minskaia et al., in press) and SARS-CoV (Almazan et al., unpublished data).
The ExoN domain occupies the N-terminal part of a protein located between the ZBD–HEL1 protein and the NendoU-containing cleavage product and has no equivalent in the arterivirus pp1ab (Cowley et al., 2000, de Vries et al., 1997) (Fig. 4) or in the genomes of any other RNA viruses (Snijder et al., 2003). This domain is very distantly related to numerous cellular proteins (Snijder et al., 2003) that form the extremely diverse DEDD superfamily of exonucleases, which are involved in many aspects of nucleic acid metabolism including proofreading, repair, and recombination (Moser et al., 1997, Zuo and Deutscher, 2001). Unlike their cellular homologs, the nidovirus ExoNs contain either one (corona- and toroviruses) or two copies (roniviruses) of a Zf domain (Snijder et al., 2003). Using bacterially expressed forms of SARS-CoV ExoN, the predicted exoribonuclease activity was recently established and characterized in vitro (Minskaia et al., in press).
The O-MT domain (Feder et al., 2003, Snijder et al., 2003, von Grotthuss et al., 2003) is proteolytically released as a mature protein from the most C-terminal part of pp1ab. In toroviruses, this protein may additionally include the NendoU domain that is located immediately upstream of the O-MT domain (Fig. 4). In the arterivirus pp1ab, the equivalent position (downstream of NendoU) is occupied by an arterivirus-specific protein (nsp12) of uncharacterized function (Fig. 3). O-MT homologs were identified in the genera Flavivirus and Alphavirus of the ssRNA+ families Flaviviridae and Togaviridae, respectively, and in members of the ssRNA- Mononegavirales. All RNA virus-encoded O-MTs belong to a large protein family containing numerous DNA-encoded homologs, which is prototyped by the RrmJ protein (Feder et al., 2003, Ferron et al., 2002, Koonin, 1993, Snijder et al., 2003, von Grotthuss et al., 2003). The O-MT activity has not yet been verified for nidoviruses.
Two other RNA-processing domains, ADP-ribose-1″-phosphatase (ADRP) and cyclic phosphodiesterase (CPD) (Martzen et al., 1999), are conserved in smaller subsets of nidoviruses (Fig. 4). The ORF1a-encoded ADRP (originally called X domain (Gorbalenya et al., 1991)) was identified upstream of PL2pro in the largest multidomain coronavirus protein (nsp3) (Snijder et al., 2003) and in a similar position in the torovirus pp1a/pp1ab (Draker et al., 2006). The ADRP is also part of the replicase polyprotein of all mammalian viruses of the alphavirus-like supergroup (Koonin et al., 1992). It has a peculiar, but not ubiquitous, distribution in cellular organisms of all kingdoms of life and a yet-to-be-identified function (Pehrson and Fuji, 1998, Allen et al., 2003, Martzen et al., 1999). The predicted ADRP activity was verified in vitro for bacterially expressed versions of this domain from SARS-CoV, HCoV-229E, and porcine transmissible gastroenteritis virus (TGEV) (Putics et al., 2005, Putics et al., 2006, Putics et al., in press) and the crystal structure of the SARS-CoV ADRP has been solved (Saikatendu et al., 2005). Substitutions of putative coronavirus ADRP active-site residues resulted in viruses that did not display apparent defects in RNA synthesis and grew to titers comparable to those of the wild-type virus (Putics et al., 2005). The CPD domain is encoded in ORF1a immediately upstream of the ORF1a/ORF1b junction in toroviruses (Fig. 4), and, surprisingly, by ORF2 of a phylogenetically compact subset of subgroup 2a coronaviruses (Snijder et al., 1991) (Fig. 3), which thus express this gene from a separate subgenomic mRNA. Some dsRNA rotaviruses are the only other RNA viruses that encode a (distantly related) CPD domain, whose homologs are abundant in cellular organisms (Mazumder et al., 2002, Snijder et al., 2003). Deletion of the CPD-encoding ORF2 did not have a noticeable effect on replication of mouse hepatitis virus (MHV) in tissue culture (Schwarz et al., 1990). In contrast, a mutation in MHV ORF2 was tolerated in vitro but caused attenuation in the natural host (Sperry et al., 2005).
The coronavirus nsp3 contains, in addition to the ADRP and PLpro, many other domains, some of which are group- or virus-specific and have not been functionally characterized (Ziebuhr et al., 2001; Gorbalenya, unpublished observations). Among those is the SARS-CoV unique domain (SUD) (Snijder et al., 2003) (Fig. 4). Also, two upstream proteins, nsp1 and nsp2, appear to exist in a variety of forms, each specific for some coronaviruses (Snijder et al., 2003). MHV reverse genetics data demonstrated that coronaviruses tolerate specific substitutions and even deletions in this region of the replicase gene. For example, cleavage of the nsp1|nsp2 site was shown to be dispensable for viral replication and even deletion of the C-terminal part of nsp1 and the entire nsp2, respectively, had only minor effects on viral replication (Brockway and Denison, 2005, Denison et al., 2004, Graham et al., 2005). Taken together, the data suggest that coronavirus replicases have evolved to include a number of non-essential functions that may provide a selective advantage only in the host (see also Section 6).
In addition to the CPD of toroviruses, a variable number of other domains are proteolytically released from the nidovirus pp1a/pp1ab region delimited by TM3 and the RFS site (Fig. 4). Some of these domains seem to be conserved in corona- and toroviruses (Gorbalenya, unpublished observations), but no conservation that extends to arteriviruses and/or roniviruses has been reported. In coronaviruses, this region encompasses the nsps 7–10. Genetic analysis implicated nsp10 in RNA synthesis (Sawicki et al., 2005) and recent crystallographic studies showed that nsp9 is an RNA-binding protein with a new version of the OB-fold (Egloff et al., 2004) that structurally resembles the domain II of coronavirus 3CLpros (Sutton et al., 2004). Furthermore, nsp7 and nsp8 form a hexadecameric supercomplex that is capable of encircling RNA and may operate as a cofactor in viral RNA synthesis (Zhai et al., 2005).
The interaction between coronaviruses nsp7 and nsp8 is likely to be one of many other protein–protein interactions from the various subunits that are proteolytically released from pp1a/pp1ab. They may facilitate the formation of the replication complex and, probably, are critical to the functioning of this complex. For instance, the RdRp (nsp12) was proposed to interact with the main protease (nsp5) and nsp8 and nsp9 in the coronavirus MHV (Brockway et al., 2003) and a strong interaction between nsp2 and nsp3 was documented for the arterivirus EAV (Wassenaar et al., 1997). The full picture of the network of interactions has only started to emerge and we will briefly return to this topic in the following chapters.
5. Family specific virion morphology and major virion components
Nidoviruses have evolved enveloped virions with different morphology to mediate entry into the cell and many extracellular functions essential for virus reproduction and spread (reviewed by Lai and Holmes, 2001, Snijder and Meulenberg, 1998). Both the number and composition of structural proteins vary between viruses of the four major branches and may even vary among viruses of the same branch. Weak sequence similarities between functionally equivalent proteins have been reported only for relatively closely related toroviruses and coronaviruses (den Boon et al., 1991b, Snijder et al., 1990b). The structural proteins of arteriviruses and roniviruses may have originated from unrelated ancestors or they have diverged beyond the point where ancestral relationships with other nidovirus orthologs could be recognized from sequence comparisons.
Given these differences, it is no wonder that virions of the four major nidovirus branches have (strikingly) different architectures, although they all have an external lipid bilayer with associated proteins (envelope) that encloses the internal nucleocapsid structure (Enjuanes et al., 2000, Spaan et al., 2005b) (Fig. 5 ). Coronaviruses, toroviruses and the significantly smaller arteriviruses have spherical virions. In addition, elongated rod-shaped virions are observed inside the torovirus-infected cells, and ronivirus particles were shown to be rod-shaped. Virions of the Coronaviridae and the Roniviridae families carry large projections that protrude from the envelope (peplomers). These oligomeric structures provide them with the characteristic “crown” observed by electron microscopy, which inspired the name of the coronavirus family. Ronivirus envelopes are also studded with prominent peplomers (Walker et al., 2005), but only relatively small and indistinct projections have been observed on the arterivirus particle surface (Snijder et al., 2005a).
Fig. 5.
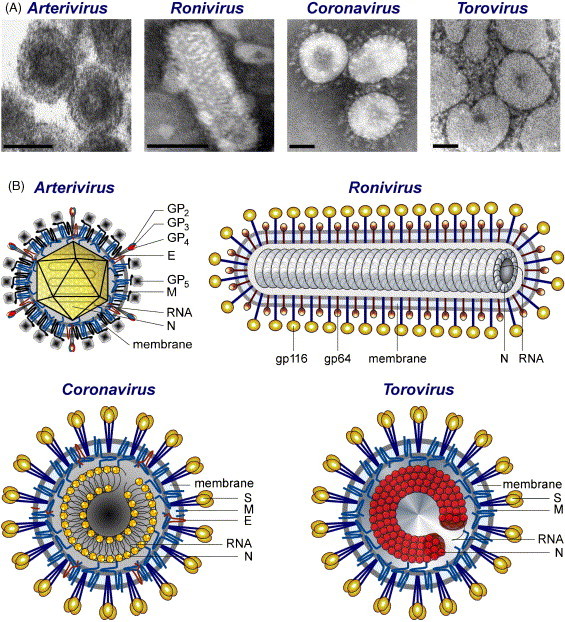
Architecture of particles of members of the order Nidovirales: electron micrographs (A) and schematic representations (B). N, nucleocapsid protein; S, spike protein; M, membrane protein; E, envelope protein; HE, hemagglutinin-esterase. Coronavirus M protein interacts with the N protein. In arterivirus, GP5 and M are major envelope proteins, while GP2, GP3, GP4, and E are minor envelope proteins. Toro- and roniviruses lack the E protein present in corona- and arteriviruses. An equivalent of the M protein has not yet been identified in roniviruses (although see also Fig. 3). This figure was reproduced from Fig. 20.1 presented in Snijder et al. (2005b). Images were reproduced (with permission) from (Snijder and Muelenberg, 1998) (arterivirus), (Spann et al., 1995) (ronivirus), Dr. Fred Murphy, Centers for Disease Control and Prevention (CDC), Atlanta, USA (coronavirus) and (Weiss et al., 1983) (torovirus).
The major envelope proteins are spike (S) and membrane (M) in coronaviruses and toroviruses, GP5 and M in arteriviruses, and gp116 and gp64 in roniviruses. Only the S and M protein species of corona- and toroviruses share very limited sequence similarities, indicating possible common origins. S proteins differ in size, but all have a highly exposed globular domain and a stem portion containing heptad repeats organized in a coiled-coil structure (Bosch et al., 2004, de Groot et al., 1987, Ingallinella et al., 2004, Liu et al., 2004, Snijder et al., 1990b, Supekar et al., 2004, Xu et al., 2004). At least in coronaviruses, the S protein forms trimers (Delmas and Laude, 1990). The four glycoproteins of arteriviruses are organized into two structures: one formed by a disulfide-linked heterodimer of GP5 with the M protein and another by a heterotrimer of minor structural proteins GP2, GP3, and GP4 (reviewed by Siddell et al., 2005). The M proteins of coronaviruses, toroviruses and arteriviruses have a triple-spanning membrane topology with the amino-terminus being located on the outside of the virus and the carboxy-terminus residing on the inside. In virions of the coronavirus TGEV, the M protein may have a quadruple-spanning membrane topology leading to the exposure of both ends of the protein on the surface of the virion (Escors et al., 2001a). The two envelope proteins of the Roniviridae seem to be generated by post-translational processing of a precursor glycoprotein whose N-terminal region has the predicted triple-spanning membrane topology typical of the M proteins of all other nidoviruses (Cowley and Walker, 2002) (Fig. 5).
Coronaviruses, including SARS-CoV, have an internal core shell that seems to be formed by the helical nucleocapsid and the M protein (Escors et al., 2001b). The torovirus nucleocapsid resembles a toroid, which inspired the name of this nidovirus (reviewed by Snijder and Horzinek, 1993), while the nucleocapsid of roniviruses has an extended, helical organization. Unlike the three other groups, arteriviruses probably have an icosahedral core shell (reviewed by Snijder and Meulenberg, 1998). In all nidoviruses, the nucleocapsid includes only a single protein species that is uniformly named nucleocapsid protein (N), even though the N proteins of viruses from the four major branches are probably not evolutionarily related. Unlike its arterivirus counterpart (Molenkamp et al., 2000), the N protein of coronaviruses was reported to be essential for efficient sg RNA synthesis and genome replication (Almazan et al., 2004, Schelle et al., 2005). The crystal structures of the capsid-forming core domain of the arterivirus porcine reproductive and respiratory syndrome virus (PRRSV) N protein (Doan and Dokland, 2003) and the N-terminal RNA-binding domain of the SARS-CoV N protein (Huang et al., 2004) have been solved.
Nidovirus particles can include other proteins that may be dispensable in some nidoviruses or are non-essential at all. One protein, called E, is ubiquitous in viruses of the corona- and arterivirus branches (Fig. 3). This protein is essential for arteriviruses, but not for all coronaviruses. Relative to the major structural proteins, the E protein is present in a reduced number of copies (around 20) in coronavirions. Deletion of the E gene may lead either to a block of virus maturation, preventing virus release and spread (group 1 coronavirus TGEV) (Ortego et al., 2002), or to a ten to one hundred thousand-fold reduction in virus titers (group 2 coronaviruses MHV and SARS-CoV) (Fischer et al., 1998, Kuo and Masters, 2003; DeDiego et al., unpublished observations). The coronavirus E protein belongs to a family of proteins known as viroporins, which modify membrane permeability (Gonzalez and Carrasco, 2003) by forming ion channels in the virion envelope. The E protein may also modify the plasma membrane of infected cells, which could facilitate the release of virus progeny (Wilson et al., 2004). Additional proteins may be present in virions of some coronaviruses and/or toroviruses (Enjuanes et al., 2000, Spaan et al., 2005b, Tan et al., 2005) and they will be discussed in the following chapter.
6. Virus-specific accessory genes: fast adaptability to a new niche?
In addition to the family specific structural protein genes, the 3′-proximal region of the genome may include ORFs encoding proteins that we will call “accessory” (Fig. 3). They are specific for either a single species or a few viruses that form a compact phylogenetic cluster. Based on both studies with naturally occurring (deletion) mutants and targeted mutagenesis studies, many of these genes are now known to be dispensable for virus growth in cell culture, and, as a result, may be called “non-essential” (Spaan et al., 2005a). The proteins encoded by these genes may be used to build virions or, alternatively, may function in infected cells or the infected host as a whole. Some functionally dispensable ORF1a-encoded replicase domains may also qualify as “accessory” (see Section 4). The CPD may be the most compelling example of accessory proteins encoded in different genome regions but belonging to the same class. This domain is part of the replicase polyproteins in toroviruses, but is encoded by ORF2 in specific group 2 coronaviruses (but not in other coronaviruses) (see Section 4). Below we will restrict our description to the accessory genes located in the 3′-proximal region of the genome.
Only the two nidovirus branches with the largest genomes (coronaviruses and toroviruses) seem to encode accessory genes (Fig. 3). They have been acquired relatively late during nidovirus evolution, as suggested by their distribution within these branches. The absence of accessory genes in “small” arteriviruses cannot be currently rationalized. One might argue that it is related to the tight gene organization (with extensive overlaps between individual ORFs) in the arterivirus 3′-proximal genome regions (reviewed by Snijder and Meulenberg, 1998). Although this organization complicates the insertion of new genes without disrupting the functionality of the existing gene set, it seems to be compatible with either the insertion of an accessory gene downstream of the most 3′-proximal ORF or evolution of an accessory gene that fully overlaps with an existing gene in an alternative reading frame. Both these possibilities have been used to evolve accessory genes in coronaviruses (see below) but not in arteriviruses.
In coronaviruses, accessory genes are typically found in variable numbers (2–8), while in the torovirus genome only one accessory gene, encoding the hemagglutinin-esterase (HE), has been identified (Spaan et al., 2005a) (Fig. 3). Coronavirus accessory genes may occupy any intergenic position in the conserved array of the four genes encoding the major structural proteins (5′-S-E-M-N-3′), or they may flank this gene array or overlap with a gene in this array (Enjuanes et al., 2000, Spaan et al., 2005a). Historically, some of the accessory genes were considered to be group-specific markers in coronaviruses (Lai and Cavanagh, 1997). With the recent expansion of our knowledge about the coronavirus diversity (Fouchier et al., 2004, Lau et al., 2005, Li et al., 2005, Marra et al., 2003, Poon et al., 2005, Rota et al., 2003, van der Hoek et al., 2004, Woo et al., 2005), they have been found not in every new virus of a respective group.
Group 1 CoVs may have two to three accessory genes located between the S and E genes and up to two others downstream of the N gene. Viruses of group 2 form the most diverse coronavirus cluster and they may have between three and eight accessory genes. In this cluster, MHV, human coronavirus OC43 (HCoV-OC43), and bovine coronavirus (BCoV) form a phylogenetically compact subset that is characterized by two accessory genes inserted between ORF1b and the S gene, encoding the CPD and HE functions, respectively (original group 2-specific markers), and other two genes located between, and partially overlapping with, the S and E genes, and yet another gene I that is internal to the N gene. From this set, homologs of only three proteins are encoded by the recently identified human coronavirus HKU1 (HCoV-HKU1) (Woo et al., 2005), which is the closest known relative of the above virus cluster. In contrast, the most distant group 2 member, SARS-CoV (Lau et al., 2005, Li et al., 2005, Marra et al., 2003, Rota et al., 2003), has seven or eight unique accessory genes, two between the S and E genes, four to five between the M and N genes, plus ORF 9b, which entirely overlaps with the N gene in an alternative reading frame. In the group 3 avian coronaviruses, prototyped by IBV, bipartite accessory genes were identified between the S and E genes (gene 3), and between the M and N genes (gene 5), respectively (Boursnell et al., 1987, Cavanagh, 2005). Except for CPD and HE, no homologs of nidovirus accessory genes have been identified in other RNA viruses. The CPD homologs were discussed in Section 4, while the coronavirus/torovirus HE proteins are homologs of the HE protein of the ssRNA- influenza C virus (Luytjes et al., 1988, Snijder et al., 1991). The recently solved tertiary structure of an 81-residue luminal domain of the SARS-CoV 7a protein revealed a compact Ig-like fold (Nelson et al., 2005).
Many, possibly all, accessory proteins are non-essential for nidovirus replication in cell culture, but they may play an important role for replication in vivo and/or virulence. For the group 1 coronavirus TGEV, deletion of gene 7 had little effect on virus replication in cell culture but, in vivo, reduced the virus titers in target organs (lung and gut) by more than 100-fold, thus considerably decreasing virus virulence (Ortego et al., 2003). Also, the deletion of either of the two accessory gene clusters (3abc or 7ab) in feline infectious peritonitis virus (FIPV) produced mutants with an attenuated phenotype in the cat. These mutants did not cause any clinical symptoms upon infection with a dose considered to be fatal in the case of a wt virus infection (Haijema et al., 2004). For the group 2 MHV, four accessory genes proved to be non-essential in vitro and in vivo, but their deletion produced mutants with an attenuated phenotype upon infection of the natural host (de Haan et al., 2002). The I gene was shown to be not essential for viral replication in mice (Fischer et al., 1997). Characterization of field isolates and reverse genetics experiments with SARS-CoV mutants lacking one or more accessory genes (3a, 3b, 6, 7a, 7b, 8a, 8b, and 9b) showed that these genes are not essential for virus replication in cell culture (e.g., Vero cells) and in the lung of infected animal model hosts (e.g., mouse) (Song et al., 2005, Yount et al., 2005). However, deletion of some of these accessory genes may have an attenuating effect in humans. This may be true for gene 3a, which encodes a SARS-CoV-specific structural protein (Ito et al., 2005). Other deletions, as those observed in SARS-CoV ORF 8a/b, leading to the loss of 29, 82, or 415 nts, may have been the result of the adaptation of SARS-CoV to civet cats and humans, after the virus was transferred from the natural reservoir, possibly bats. The deletion of 29 nt present in most of the human isolates has also been observed in civet cats, while the full size ortholog of ORF8a/b was identified in the SARS-like coronavirus isolated from bats (He et al., 2004, Lau et al., 2005, Li et al., 2005). Mutants of the group 3 IBV, in which gene 3ab was deleted, replicate equally well as the wild-type virus in vitro. Using isogenic IBV generated by reverse genetics, in which the expression of genes 3 and 5 was prevented, it was shown that these genes are not required for replication in tissue culture or chicken tracheal organ cultures (Casais et al., 2005, Hodgson et al., 2006). Inactivation of gene 5 also has no effect on the yield of IBV progeny in embryonic eggs, although this experiment, due to the use of a non-pathogenic virus strain, did not rule out a possible role for this gene in vivo (Casais et al., 2005).
When present on the virus surface, another accessory structural protein found in coronaviruses, HE, may increase viral infectivity by binding sialic acid moieties on the cell surface (Kazi et al., 2005). In fact, it has been shown that the coronavirus HE presumably promotes virus spread and entry in vivo by facilitating the reversible attachment of the virus to O-acetylated sialic acids (Lissenberg et al., 2005). While HE may provide a strong selective advantage during natural infection, many laboratory strains of mouse hepatitis virus (MHV) no longer produce this protein. Apparently, their HE genes were inactivated during cell culture adaptation. Additional experiments suggested that wild-type HE reduces the in vitro propagation efficiency, indicating that the expression of “luxury” proteins may come at a fitness penalty. Probably, under natural conditions the costs of maintaining HE are outweighed by the benefits.
The variability of the HE gene within toroviruses is also remarkable. The HE gene is present in porcine, bovine, equine, and putative human toroviruses (Cornelissen et al., 1997, Duckmanton et al., 1999, Kroneman et al., 1998). Most newly characterized bovine torovirus variants seem to have emerged from a genetic exchange, during which the 3′ end of the HE gene, the N gene, and the 3′ nontranslated region of a bovine virus (Breda type strain) have been swapped for those of a porcine torovirus. Moreover, some porcine and bovine torovirus variants carried chimeric HE genes, which apparently resulted from recombination events, presumably through a double template switch, involving hitherto unknown toroviruses. From these observations it has been inferred that toroviruses may be even more promiscuous than other mammalian nidoviruses, the coronaviruses and arteriviruses (Smits et al., 2003). The extensive evolution of the torovirus HE gene may have been a host-adaptation strategy or a way to evade the host immune response.
It is apparent from the above overview, that coronaviruses are capable of expanding their repertoire of essential genes with many new and non-essential ones specific for a few closely related viruses. Although the analysis of accessory protein functions is technically challenging, and still in its infancy, it seems likely that this genome plasticity might provide the respective nidoviruses with selective advantages and fast adaptability in their natural hosts. As these accessory genes represent only minor extensions to the set of essential genes, this adaptability may be a direct benefit from the coronavirus ability to evolve and replicate a giant-size genome.
7. RNA synthesis in Nidovirales: what makes it special?
The synthesis of the replicase polyproteins is directed by the genome RNA, while the structural and accessory proteins are produced from sg mRNAs whose number may vary between 2 and 9 in different nidoviruses. For years, the detailed analysis of the RNA species involved in the synthesis of sg mRNAs (transcription) and genomic RNA (replication) has been one of the busiest areas of nidovirus research and the data produced often divided the field (for reviews see Brian and Spaan, 1997, Lai and Cavanagh, 1997, Lai and Holmes, 2001, Sawicki and Sawicki, 1995, Snijder and Meulenberg, 1998, van der Most and Spaan, 1995).
In the previous sections, we used the results from a comparative analysis of the replicative apparatus of nidoviruses to discuss the conserved features that distinguish nidoviruses from other viruses and the features that separate large and small nidoviruses. Since the replicase machinery has evolved to mediate RNA synthesis, the above-mentioned specifics must be linked to the unique aspects of nidovirus replication/transcription. Initially, it appeared that copying (and joining) of non-contiguous genome sequences during the synthesis of sg mRNAs (Baric et al., 1983, de Vries et al., 1990, Lai et al., 1984, Spaan et al., 1983) could be one of the major unifying aspects common and specific only to nidoviruses. More recent studies on the relatively well characterized corona- (Zuniga et al., 2004) and arteriviruses (Pasternak et al., 2001, van Marle et al., 1999a) and the poorly characterized toro- (van Vliet et al., 2002) and roniviruses (Cowley et al., 2002) revealed another common theme in nidovirus RNA synthesis, which, however, does not distinguish either all or the large nidoviruses from (all) other RNA viruses.
Nidovirus sg mRNAs are generated as an extensive 3′-coterminal nested set from which ORFs in the 3′-proximal region of the polycistronic genome are expressed. With a few notable exceptions, only the 5′-proximal ORF is expressed from each sg mRNA species and, consequently, the number of sg mRNA species synthesized is roughly proportional to the number of ORFs located downstream of ORF1b. In the genomes of all nidoviruses studied, a copy of a short characteristic AU-rich sequence, at present mostly referred to as transcription-regulating sequence (TRS), is located upstream of ORF1a and immediately upstream of each of the downstream ORFs that can also occupy the 5′-proximal position in one of the sg mRNAs. As their name implies, the TRSs are thought to regulate the transcription of the genome template. Most probably, they act as signals for either attenuation or termination of minus strand RNA synthesis in all nidoviruses, thus producing subgenome-size minus strands that are later utilized as templates in sg mRNA synthesis (Sawicki et al., 2001). Similar mechanisms for attenuation of minus strand RNA synthesis were described for a number of polycistronic plant RNA virus genomes of different families and sizes (Gowda et al., 2001, Miller and Koev, 2000).
In arteriviruses and coronaviruses, the TRSs associated with the ORFs located in the 3′-proximal domain of the genome (known as body TRSs) are believed to direct attenuation of minus strand RNA synthesis which, in a process guided by a base-pairing interaction between complementary sequences, is resumed at the TRS upstream of ORF1a (known as leader TRS) to complete the synthesis of the subgenome-size minus strands. This process has been dubbed ‘discontinuous extension of minus strand RNA’ (for a recent review see Sawicki and Sawicki, 2005). The negative-sense RNAs are subsequently used as templates for the synthesis of complementary sg mRNAs, which have two adjacent parts (known as leader and body) that correspond to two noncontiguous parts of the genome RNA. The sg- and genome-length RNAs share a 5′ leader sequence of 55–92 nt (coronaviruses) or 170–210 nt (arteriviruses) (Lai and Cavanagh, 1997, Snijder and Meulenberg, 1998, Zuniga et al., 2004). A sg mRNA of similar composition, carrying a leader sequence that matches the 5′-terminal 18 nt of the genome, is synthesized to express ORF2 of equine torovirus (Snijder et al., 1990c, van Vliet et al., 2002), whereas the three remaining, smaller torovirus sg mRNAs (Snijder et al., 1990c, van Vliet et al., 2002) and both sg mRNAs of roniviruses (Cowley et al., 2002) lack a common 5′-leader sequence, and are probably produced in a process that does not involve discontinuous RNA synthesis.
The use of continuous versus discontinuous subgenomic RNA synthesis among nidoviruses separates arteri- and coronaviruses from roniviruses, with toroviruses being an intermediate. This grouping is different from the one based on genome size, indicating that the evolution of transcription was a complex process that was not directly linked to genome expansion. Irrespective of the use of discontinuous subgenomic RNA synthesis, nothing in the available data on RNA synthesis explains why the nidovirus replicase machinery must include the large number of (unique) enzymatic and other subunits that has been described in Section 3. Indeed, the relatively complex mechanism for discontinuous RNA synthesis could be considered (in full agreement with the available data!) to be a variant of the replicative similarity-assisted template switching (Pasternak et al., 2001) that operates during RNA recombination in viruses with relatively small genomes and “simple” replicase complexes (Lai, 1992). A much more detailed understanding of the molecular processes involved in nidovirus RNA synthesis will probably be required to find a plausible explanation for this paradox.
8. Mechanisms of evolution: mutations and recombination
Two common evolutionary mechanisms – mutation and recombination – have been implicated in the generation of nidovirus genome diversity. In contemporary coronaviruses, like SARS-CoV and HCoV-OC43, the mutation rate was estimated (Salemi et al., 2004, Vijgen et al., 2005) to be in the range that is typical for other RNA viruses, with much smaller genomes, whose replicase fidelity is known to be low (Castro et al., 2005, Domingo and Holland, 1997, Drake and Holland, 1999). Mutations are accepted at an uneven rate across the nidovirus genome, with the structural protein genes, especially those encoding envelope glycoproteins, and the 5′-proximal half of ORF1a evolving relatively fast and ORF1b evolving relatively slowly (Chouljenko et al., 2001, Song et al., 2005). The observed differences must be linked to the roles played by the respective products of these genome regions in the nidovirus life cycle (see above). In the most conserved parts of the genome, nonsynonymous replacements start to accumulate significantly only at relatively large evolutionary distances separating the major nidovirus groups. In these cases, replacements have sometimes been accepted at places of extraordinary conservation, e.g., in the active site of the 3CLpro, which are not known to have been mutated in their DNA-encoded homologs. Although little is known about the absolute time scale of nidovirus evolution, it was argued that a high mutation rate may have been at work for a very long time to generate the contemporary diversity of the most conserved enzymes in RNA viruses (Koonin and Gorbalenya, 1989).
There is plenty of evidence, especially in corona- (Makino et al., 1986) and toroviruses (Herrewegh et al., 1998), for frequent recombination between contemporary nidoviruses (or their immediate ancestors), either in the field or in experimental settings (for reviews see Brian and Spaan, 1997, Lai, 1996). This recombination commonly involves closely related viruses, e.g., belonging to the same group of coronaviruses, and is believed to occur during replication and to be mediated by replicase-driven template switching in a homologous genome region. As a result, chimeric progeny genomes are produced with a mosaic relationship to their parental templates. This type of recombination, which is common in ssRNA+ viruses (Lai, 1992), could be effective in removing deleterious (point) mutations (Chao, 1988, Worobey and Holmes, 1999) that might otherwise accumulate in the big nidovirus genome and could drive it into extinction. To counteract the adverse consequences of a high mutation rate for their big genomes, nidoviruses may employ yet other, unique repair mechanism(s), possibly involving the diverse RNA-processing enzymes described in Section 3 (see also below).
An aberrant variant of homologous recombination (Lai, 1992) may have also been involved in the expansion and shrinking of the nidovirus genome. Arteri- and coronaviruses have multiple (possibly up to four) copies of PLpro (den Boon et al., 1995, Gorbalenya et al., 1991, Ziebuhr et al., 2001), which are likely to have arisen from duplication of a locus. PLpro duplications may have occurred independently in arteri- and coronaviruses, since the PLpros of these two nidovirus branches do not seem to interleave with each other (Ziebuhr et al., 2000, Ziebuhr et al., 2001). The pattern of distribution of viruses with one and two PLpros in the coronavirus tree suggests that their evolution may have been accompanied by either independent duplication or loss of PLpro domains (Ziebuhr et al., 2001). Duplications were also documented in a region immediately upstream of PL1pro in HCoV-HKU (Woo et al., 2005) and in the 3′-proximal region of the arterivirus SHFV (Godeny et al., 1998). Also, the coronavirus nsp9 was proposed to have originated from a duplication of a 3CLpro domain (Sutton et al., 2004).
Heterologous recombination, directed by template switching in nonhomologous regions, might have promoted the relocation of the CPD and HE genes in an ancestor of either toroviruses or group 2a coronaviruses. Under the alternative scenario, the CPD and HE genes must have been acquired independently by these ancestors (Snijder et al., 1991). It is heterologous RNA recombination that may have generated the diverse gross changes in the nidovirus genome that were discussed in previous chapters. Almost nothing is known about the parameters of heterologous recombination in nidoviruses, including the origins of parental (donor) sequences of which (a part of) the replicase domains must have been acquired. This complete lack of information on the origin of specific sequences extends from the accessory genes in the 3′-part of the coronavirus genome to the non-essential, lineage-specific replicative domains and also includes the essential ExoN and O-MT domains that are specific for large nidoviruses.
9. Origins of the Nidovirales and nidovirus families
The complex genetic plan and the replicase gene of nidoviruses must have evolved from simpler ones. The reconstruction of such ancestral events is in no way a straightforward exercise for a group of viruses in which major internal (i.e., nidovirus-specific) and external relationships (with other viruses and cellular systems) are extremely remote and the direction of evolution may not be readily recovered (Zanotto et al., 1996). Even though tentative, such an effort may have a definitive merit as it puts to a critical test the maturity of our knowledge base and involves across-virus comparisons to produce a large scale picture that otherwise would have been obscured. We offer below a speculative scenario which is based on the assumption that, generally, RNA viruses expand rather than shrink their genomes, which may not be true at any given time point.
It is safe to assume that the last common ancestor (LCA) of the Nidovirales may have had a genome size close to that of the contemporary arteriviruses. In this scenario, the transition from this arterivirus-like nidovirus LCA to the LCA of the large nidoviruses may have been accompanied by the acquisition of the otherwise unique ExoN domain (Snijder et al., 2003) and a (gradual?) genome size enlargement of about 14 kb (Fig. 6 ). The correlation between these two events fits perfectly in the relationship between the quality of genome copying and genome size observed in various biological systems. In this context, the ExoN domain may have been acquired to improve the low fidelity of RdRp-mediated RNA replication through its 3′ → 5′ exonuclease activity (Minskaia et al., in press), which might operate in proof-reading mechanisms similar to those employed by DNA-based life forms (reviewed by Kunkel and Bebenek, 2000, Kunkel and Erie, 2005). This parallel does not imply that the ExoN-containing replicase complex of large nidoviruses may work like a DNA polymerase complex. To perform its vital function for copying large genomes, ExoN may work in concert with other nidovirus RNA processing enzymes, like their distant homologs do in two cellular intron-processing pathways (Fig. 7 ). Based on this parallel, it was suggested that the genetically segregated and down-regulated HEL1, ExoN, NendoU, and O-MT domains (Fig. 3) may provide RNA specificity, and that the relatively abundant CPD and ADRP, when available (Fig. 3, Fig. 4), may modulate the pace of a reaction in a common pathway (Snijder et al., 2003), which, as we propose, could be part of an oligonucleotide-directed repair mechanism.
Fig. 6.
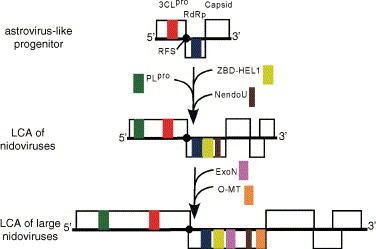
Origin and evolution of the nidovirus genome plan. A tentative evolutionary scenario leading to the origin of the LCAs of nidoviruses and large nidoviruses from a progenitor with an astrovirus-like genome organization is illustrated. The three shown genomes are fictitious, although they are drawn to a relative common size scale and include major replicative domains found in genomes of respective contemporary viruses, as discussed in the text.
Fig. 7.
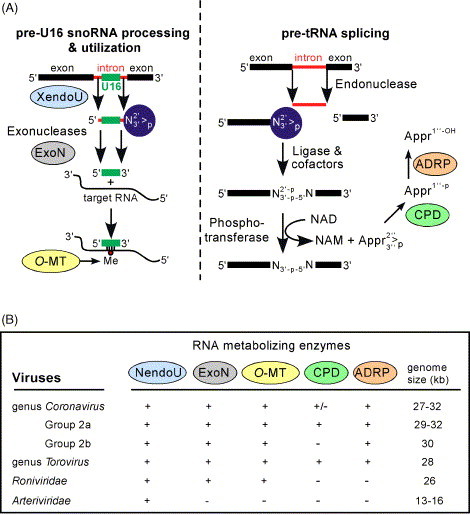
RNA-processing enzymes of nidoviruses: possible cooperation and virus distribution. (A) Cellular pathways for processing of pre-U16 small nucleolar RNA (snoRNA) and pre-tRNA splicing in which homologs of five nidovirus RNA-processing enzymes are involved. Note that both pathways produce intermediates with 2′-3′-cyclic phosphate termini (blue circles), indicating the structural basis for a possible cooperation of the nidovirus homologs in a single pathway (Snijder et al., 2003). (B) Table summarizing the conservation of five (putative) RNA-processing enzymes among representatives of large and small nidoviruses. This figure was updated from Fig. 5 presented in Snijder et al. (2003).
The ExoN function likely was acquired by an ancestral virus that already had a multi-subunit replicase complex whose fidelity was good enough to copy a larger than average-size genome. Based on the genetic diversity among contemporary viruses, we can assume that the domain composition of the replicase polyproteins of the arterivirus-like nidovirus LCA was more complex than that of other known RNA virus families. At the stage at which (presumably) the nidovirus LCA emerged, the two genetic marker domains, ZBD and NendoU, may have been acquired (Fig. 6). They may have been used to build a prototype repair mechanism sufficient to copy a ∼14 kb genome that later, following the acquisition of ExoN, evolved into a more advanced system that was subsequently used in the Coronaviridae/Roniviridae branch. It is likely that extensive evolution producing a number of major virus lineages, which may either have become extinct or eluded identification thus far, preceded the origin of the Nidovirus LCA.
Among contemporary viruses, the Astroviridae (Jiang et al., 1993) may most strongly resemble the reconstructed nidovirus LCA in two major respects (Fig. 6). First, they have a nidovirus-like genetic plan with overlapping ORFs 1a/1b encoding replicase polyproteins and a downstream located ORF for the capsid protein that is expressed from an sg mRNA. Second, the backbone of the astrovirus replicase gene, which is composed of three domains, 3CLpro-RFS-RdRp, matches the central part of the nidovirus-specific domain arrangement. These three domains are key elements in the control of genome expression and replication and therefore the ancestral relationship of Astroviridae and Nidovirales would also be in agreement with functional considerations.
It is also interesting to note that astroviruses are at the upper border of the genome size of RNA virus families that lack a HEL gene (Fig. 1). A remarkably strong positive correlation was noted between an ssRNA+ virus genome size of more than ∼6 kb and the presence of a HEL domain in the replicative polyproteins of such viruses (Gorbalenya and Koonin, 1989). This correlation implies that the acquisition of the HEL domain may have been a turning point in RNA virus evolution that allowed enlargement of RNA virus genomes to sizes of more than 6–7 kb. Mechanistically, the acquisition of a HEL domain may have liberated RNA viruses from constraints related to the unwinding of large double-stranded RNA regions. Efficient control over this reaction may be essential for regulating replication, expression and transport of RNA virus genomes (Kadare and Haenni, 1997). The acquisition of the HEL domain apparently coincided with a major diversification of ssRNA+ viruses to produce more than 30 families and groups that include viruses with different genetic layouts and genomes with sizes ranging from ∼6 to 32 kb.
In conclusion, we may then speculate that an ancestral virus with the genome and replicase gene organization of astroviruses, which already feature the largest genomes among their classmates (Fig. 1), may have acquired a HEL1 domain from a virus of the alphavirus-like supergroup or another source. The HEL1 evolved its 5′ → 3′ polarity and was, in the association with ZBD, inserted downstream of the RdRp domain, all novelties among RNA viruses that may have been essential for the genome expansion. The acquisition of other domains might have followed. To ensure control over the expression of gradually growing replicase polyproteins, the PLpro was acquired, eventually resulting in the LCA of nidoviruses (Fig. 6).
10. Concluding remarks
In stunning contrast to the three-log size variation among the genomes of the contemporary DNA viruses, the entire diversity of the much more numerous RNA viruses is “squeezed” into a bandwidth of just one order of magnitude. Despite this huge scale difference, RNA viruses must have undergone extensive evolution to expand their genomes, as is illustrated by the Nidovirales order (this review) and the Closteroviridae family (see the review by Dolja et al., 2006). The three groups of nidoviruses with large genomes stand alone at the upper edge of the RNA virus genome size scale. The large size gap that separates these viruses from others, which densely populate the bottom half of the genome size scale (Fig. 1), suggests that expansion of viral genomes above ∼15 kb is a formidable challenge that has been rarely met in RNA virus evolution. As speculated in this review, the large nidoviruses may have cleared this barrier with the largest margin because of prior evolution resulting in unprecedented genome expansions in several of their ancestors (Fig. 1, Fig. 6). The comparison of the large corona-, toro- and roniviruses with their much smaller Arteriviridae cousins provides an intellectual framework for rationalizing the genome expansion in the Nidovirales. In this review, the common and unique features that have been recognized in either the large or even all nidoviruses were briefly discussed. Obviously, the progression of our understanding of nidovirus evolution depends on advancements in many research areas, including studies on nidovirus-specific RNA and protein synthesis, interactions of these viruses with their hosts, virus sampling in the field, and diverse evolutionary analyses. These advancements may help to dissect the molecular mechanisms that nidoviruses have evolved to synthesize and express their giant genomes and maintain the necessary balance between fast adaptation to changing environmental conditions on the one hand and genome stability on the other. To better understand this aspect, information on how nidoviruses make use of the unprecedented complexity and special features of their replicase machinery will be of prime interest. Furthermore, a comprehensive understanding of the parameters of nidovirus adaptability may result in the identification of the driving forces that have shaped nidovirus evolution. In this context, it will be essential to continue the sampling of the natural diversity of nidoviruses, also to see whether the genome size gap between the small and large nidoviruses will be filled with viruses with intermediate-size genomes, and whether nidoviruses infecting other phyla, e.g., plants and insects, can be identified. Ultimately, we may hope to learn whether the large nidoviruses (particularly, coronaviruses) have reached the theoretical genome size limit that may have been set for RNA viruses by nature. Finally, and more practically, our progress in understanding the fundamental aspects of RNA virus genome expansion may help to monitor and control infections caused by (known and potentially emerging) nidoviruses.
Acknowledgements
The kind help of Johan Faase in preparing Fig. 1 is gratefully acknowledged. Nidovirus research conducted in the authors’ groups was (partly) supported by the Netherlands Bioinformatics Centre (SP 3.2.2 grant) (A.E.G.), the EU grant FP6 IP Vizier LSHG-CT-2004-511960 (A.E.G. and E.J.S.), the Council for Chemical Sciences of the Netherlands Organisation for Scientific Research (NWO-CW 700.52.306 grant) (to E.J.S. and A.E.G.), the EU grant FP6 SARS-DTV SP22-CT-2004-511064 (E.J.S., A.E.G. and J.Z.), the Deutsche Forschungsgemeinschaft (SFB479-TPA8, Zi 618/3, Zi 618/4) (to J.Z.), the Sino-German Center for Science Promotion (GZ 230) (to J.Z.), EU grants DISSECT No. SP22-CT-2004-511060 and CICYT BIO2004-00636 (L.E.).
References
- Allen M.D., Buckle A.M., Cordell S.C., Lowe J., Bycroft M. The crystal structure of AF1521 a protein from Archaeoglobus fulgidus with homology to the non-histone domain of MacroH2A. J. Mol. Biol. 2003;330:503–511. doi: 10.1016/s0022-2836(03)00473-x. [DOI] [PubMed] [Google Scholar]
- Almazan F., Galan C., Enjuanes L. The nucleoprotein is required for efficient coronavirus genome replication. J. Virol. 2004;78:12683–12688. doi: 10.1128/JVI.78.22.12683-12688.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand K., Palm G.J., Mesters J.R., Siddell S.G., Ziebuhr J., Hilgenfeld R. Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra alpha-helical domain. EMBO J. 2002;21:3213–3224. doi: 10.1093/emboj/cdf327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand K., Ziebuhr J., Wadhwani P., Mesters J.R., Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003;300:1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- Baker S.C., Shieh C.K., Soe L.H., Chang M.F., Vannier D.M., Lai M.M. Identification of a domain required for autoproteolytic cleavage of murine coronavirus gene A polyprotein. J. Virol. 1989;63:3693–3699. doi: 10.1128/jvi.63.9.3693-3699.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltimore D. Expression of animal virus genomes. Bacteriol. Rev. 1971;35:235–241. doi: 10.1128/br.35.3.235-241.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Y.M., Federhen S., Leipe D., Pham V., Resenchuk S., Rozanov M., Tatusov R., Tatusova T. National Center for Biotechnology Information Viral Genomes Project. J. Virol. 2004;78:7291–7298. doi: 10.1128/JVI.78.14.7291-7298.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranov P.V., Henderson C.M., Anderson C.B., Gesteland R.F., Atkins J.F., Howard M.T. Programmed ribosomal frameshifting in decoding the SARS-CoV genome. Virology. 2005;332:498–510. doi: 10.1016/j.virol.2004.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baric R.S., Stohlman S.A., Lai M.M.C. Characterization of replicative intermediate RNA of mouse hepatitis virus—presence of leader RNA sequences on nascent chains. J. Virol. 1983;48:633–640. doi: 10.1128/jvi.48.3.633-640.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrette-Ng I.H., Ng K.K.S., Mark B.L., van Aken D., Cherney M.M., Garen C., Kolodenko Y., Gorbalenya A.E., Snijder E.J., James M.N.G. Structure of arterivirus nsp4—the smallest chymotrypsin-like proteinase with an alpha/beta C-terminal extension and alternate conformations of the oxyanion hole. J. Biol. Chem. 2002;277:39960–39966. doi: 10.1074/jbc.M206978200. [DOI] [PubMed] [Google Scholar]
- Barretto N., Jukneliene D., Ratia K., Chen Z., Mesecar A.D., Baker S.C. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J. Virol. 2005;79:15189–15198. doi: 10.1128/JVI.79.24.15189-15198.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlam M., Yang H., Rao Z. Structural insights into SARS coronavirus proteins. Curr. Opin. Struct. Biol. 2005;15:664–672. doi: 10.1016/j.sbi.2005.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bautista E.M., Faaberg K.S., Mickelson D., McGruder E.D. Functional properties of the predicted helicase of porcine reproductive and respiratory syndrome virus. Virology. 2002;298:258–270. doi: 10.1006/viro.2002.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj K., Guarino L., Kao C.C. The severe acute respiratory syndrome coronavirus Nsp15 protein is an endoribonuclease that prefers manganese as a cofactor. J. Virol. 2004;78:12218–12224. doi: 10.1128/JVI.78.22.12218-12224.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch B.J., Martina B.E.E., van der Zee R., Lepault J., Haijema B.J., Versluis C., Heck A.J.R., De Groot R., Osterhaus A.D.M.E., Rottier P.J.M. Severe acute respiratory syndrome coronavirus (SARS-CoV) infection inhibition using spike protein heptad repeat-derived peptides. Proc. Natl. Acad. Sci. U.S.A. 2004;101:8455–8460. doi: 10.1073/pnas.0400576101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boursnell M.E.G., Brown T.D.K., Foulds I.J., Green P.F., Tomley F.M., Binns M.M. Completion of the sequence of the genome of the coronavirus avian infectious bronchitis virus. J. Gen. Virol. 1987;68:57–77. doi: 10.1099/0022-1317-68-1-57. [DOI] [PubMed] [Google Scholar]
- Bredenbeek P.J., Snijder E.J., Noten F.H., den Boon J.A., Schaaper W.M., Horzinek M.C., Spaan W.J. The polymerase gene of corona- and toroviruses: evidence for an evolutionary relationship. Adv. Exp. Med. Biol. 1990;276:307–316. doi: 10.1007/978-1-4684-5823-7_42. [DOI] [PubMed] [Google Scholar]
- Brian D.A., Spaan W.J.M. Recombination and coronavirus defective interfering RNAs. Semin. Virol. 1997;8:101–111. doi: 10.1006/smvy.1997.0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I. Ribosomal frameshifting viral RNAs. J. Gen. Virol. 1995;76:1885–1892. doi: 10.1099/0022-1317-76-8-1885. [DOI] [PubMed] [Google Scholar]
- Brierley I., Boursnell M.E., Binns M.M., Bilimoria B., Blok V.C., Brown T.D., Inglis S.C. An efficient ribosomal frame-shifting signal in the polymerase-encoding region of the coronavirus IBV. EMBO J. 1987;6:3779–3785. doi: 10.1002/j.1460-2075.1987.tb02713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I., Rolley N.J., Jenner A.J., Inglis S.C. Mutational analysis of the RNA pseudoknot component of a coronavirus ribosomal frameshifting signal. J. Mol. Biol. 1991;220:889–902. doi: 10.1016/0022-2836(91)90361-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockway S.M., Clay C.T., Lu X.T., Denison M.R. Characterization of the expression, intracellular localization, and replication complex association of the putative mouse hepatitis virus RNA-dependent RNA polymerase. J. Virol. 2003;77:10515–10527. doi: 10.1128/JVI.77.19.10515-10527.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockway S.M., Denison M.R. Mutagenesis of the murine hepatitis virus nsp1-coding region identifies residues important for protein processing, viral RNA synthesis, and viral replication. Virology. 2005;340:209–223. doi: 10.1016/j.virol.2005.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casais R., Davies M., Cavanagh D., Britton P. Gene 5 of the avian coronavirus infectious bronchitis virus is not essential for replication. J. Virol. 2005;79:8065–8078. doi: 10.1128/JVI.79.13.8065-8078.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro C., Arnold J.J., Cameron C.E. Incorporation fidelity of the viral RNA-dependent RNA polymerase: a kinetic, thermodynamic and structural perspective. Virus Res. 2005;107:141–149. doi: 10.1016/j.virusres.2004.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh D. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch. Virol. 1997;142:629–633. [PubMed] [Google Scholar]
- Cavanagh D. Coronaviruses in poultry and other birds. Avian Pathol. 2005;34:439–448. doi: 10.1080/03079450500367682. [DOI] [PubMed] [Google Scholar]
- Chao L. Evolution of sex in RNA viruses. J. Theor. Biol. 1988;133:99–112. doi: 10.1016/s0022-5193(88)80027-4. [DOI] [PubMed] [Google Scholar]
- Cheng A., Zhang W., Xie Y., Jiang W., Arnold E., Sarafianos S.G., Ding J. Expression, purification, and characterization of SARS coronavirus RNA polymerase. Virology. 2005;335:165–176. doi: 10.1016/j.virol.2005.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouljenko V.N., Lin X.Q., Storz J., Kousoulas K.G., Gorbalenya A.E. Comparison of genomic and predicted amino acid sequences of respiratory and enteric bovine coronaviruses isolated from the same animal with fatal shipping pneumonia. J. Gen. Virol. 2001;82:2927–2933. doi: 10.1099/0022-1317-82-12-2927. [DOI] [PubMed] [Google Scholar]
- Cornelissen L.A.H.M., Wierda C.M.H., van der Meer F.J., Herrewegh A.A.P.M., Horzinek M.C., Egberink H.F., de Groot R.J. Hemagglutinin-esterase: a novel structural protein of torovirus. J. Virol. 1997;71:5277–5286. doi: 10.1128/jvi.71.7.5277-5286.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowley J.A., Dimmock C.M., Spann K.M., Walker P.J. Gill-associated virus of Penaeus monodon prawns: an invertebrate virus with ORF1a and ORF1b genes related to arteri- and coronaviruses. J. Gen. Virol. 2000;81:1473–1484. doi: 10.1099/0022-1317-81-6-1473. [DOI] [PubMed] [Google Scholar]
- Cowley J.A., Dimmock C.M., Walker P.J. Gill-associated nidovirus of Penaeus monodon prawns transcribes 3′-coterminal subgenomic mRNAs that do not possess 5′-leader sequences. J. Gen. Virol. 2002;83:927–935. doi: 10.1099/0022-1317-83-4-927. [DOI] [PubMed] [Google Scholar]
- Cowley J.A., Walker P.J. The complete genome sequence of gill-associated virus of Penaeus monodon prawns indicates a gene organization unique among nidoviruses. Arch. Virol. 2002;147:1977–1987. doi: 10.1007/s00705-002-0847-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot R.J., Luytjes W., Horzinek M.C., Vanderzeijst B.A.M., Spaan W.J.M., Lenstra J.A. Evidence for a coiled-coil structure in the spike proteins of coronaviruses. J. Mol. Biol. 1987;196:963–966. doi: 10.1016/0022-2836(87)90422-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan C.A.M., Masters P.S., Shen X.L., Weiss S., Rottier P.J.M. The group-specific murine coronavirus genes are not essential, but their deletion, by reverse genetics, is attenuating in the natural host. Virology. 2002;296:177–189. doi: 10.1006/viro.2002.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries A.A.F., Chirnside E.D., Bredenbeek P.J., Gravestein L.A., Horzinek M.C., Spaan W.J.M. All subgenomic messenger-RNAs of equine arteritis virus contain a common leader sequence. Nucleic Acids Res. 1990;18:3241–3247. doi: 10.1093/nar/18.11.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries A.A.F., Horzinek M.C., Rottier P.J.M., de Groot R.J. The genome organization of the Nidovirales: similarities and differences between arteri-, toro-, and coronaviruses. Semin. Virol. 1997;8:33–47. doi: 10.1006/smvy.1997.0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmas B., Laude H. Assembly of coronavirus spike protein into trimers and its role in epitope expression. J. Virol. 1990;64:5367–5375. doi: 10.1128/jvi.64.11.5367-5375.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Boon J.A., Faaberg K.S., Meulenberg J.J., Wassenaar A.L., Plagemann P.G., Gorbalenya A.E., Snijder E.J. Processing and evolution of the N-terminal region of the arterivirus replicase ORF1a protein: identification of two papainlike cysteine proteases. J. Virol. 1995;69:4500–4505. doi: 10.1128/jvi.69.7.4500-4505.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Boon J.A., Snijder E.J., Chirnside E.D., de Vries A.A., Horzinek M.C., Spaan W.J. Equine arteritis virus is not a togavirus but belongs to the coronaviruslike superfamily. J. Virol. 1991;65:2910–2920. doi: 10.1128/jvi.65.6.2910-2920.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Boon J.A., Snijder E.J., Locker J.K., Horzinek M.C., Rottier P.J. Another triple-spanning envelope protein among intracellularly budding RNA viruses: the torovirus E protein. Virology. 1991;182:655–663. doi: 10.1016/0042-6822(91)90606-C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison M.R., Yount B., Brockway S.M., Graham R.L., Sims A.C., Lu X., Baric R.S. Cleavage between replicase proteins p28 and p65 of mouse hepatitis virus is not required for virus replication. J. Virol. 2004;78:5957–5965. doi: 10.1128/JVI.78.11.5957-5965.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolja V.V., Kreuze J.F., Valkonen J.P.T. Comparative and functional genomics of the family Closteroviridae. Virus Res. 2006;117:38–51. doi: 10.1016/j.virusres.2006.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo E., Holland J.J. RNA virus mutations and fitness for survival. Ann. Rev. Microbiol. 1997;51:151–178. doi: 10.1146/annurev.micro.51.1.151. [DOI] [PubMed] [Google Scholar]
- Dong S., Baker S.C. Determinants of the p28 cleavage site recognized by the first papain-like cysteine proteinase of murine coronavirus. Virology. 1994;204:541–549. doi: 10.1006/viro.1994.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake J.W., Holland J.J. Mutation rates among RNA viruses. Proc. Natl. Acad. Sci. U.S.A. 1999;96:13910–13913. doi: 10.1073/pnas.96.24.13910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draker R., Roper R.L., Petric M., Tellier R. The complete sequence of the bovine torovirus genome. Virus Res. 2006;115:56–68. doi: 10.1016/j.virusres.2005.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duckmanton L., Tellier R., Richardson C., Petric M. The novel hemagglutinin-esterase genes of human torovirus and Breda virus. Virus Res. 1999;64:137–149. doi: 10.1016/S0168-1702(99)00088-X. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Egloff M.P., Ferron F., Campanacci V., Longhi S., Rancurel C., Dutartre H., Snijder E.J., Gorbalenya A.E., Cambillau C., Canard B. The severe acute respiratory syndrome-coronavirus replicative protein nsp9 is a single-stranded RNA-binding subunit unique in the RNA virus world. Proc. Natl. Acad. Sci. U.S.A. 2004;101:3792–3796. doi: 10.1073/pnas.0307877101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eigen M., Schuster P. Hypercycle—principle of natural self-organization. A. Emergence of hypercycle. Naturwissenschaften. 1977;64:541–565. doi: 10.1007/BF00450633. [DOI] [PubMed] [Google Scholar]
- Enjuanes L., Brian D., Cavanagh D., Holmes K., Lai M.M.C., Laude H., Masters P.S., Rottier P., Siddell S., Spaan W., Taguchi F., Talbot P. Family Coronaviridae. In: van Regenmortel M.H.V., Fauquet C.M., Bishop D.H.L., Carstens E.B., Estes M.K., Lemon S.M., Maniloff J., Mayo M.A., McGeoch D.J., Pringle C.R., Wickner R.B., editors. Virus Taxonomy, 7th Report of the International Committee on Taxonomy of Viruses. Academic Press; San Diego: 2000. pp. 835–849. [Google Scholar]
- Escors D., Camafeita E., Ortego J., Laude H., Enjuanes L. Organization of two transmissible gastroenteritis coronavirus membrane protein topologies within the virion and core. J. Virol. 2001;75:12228–12240. doi: 10.1128/JVI.75.24.12228-12240.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escors D., Ortego J., Laude H., Enjuanes L. The membrane M protein carboxy terminus binds to transmissible gastroenteritis coronavirus core and contributes to core stability. J. Virol. 2001;75:1312–1324. doi: 10.1128/JVI.75.3.1312-1324.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauquet C.M., Mayo M.A., Maniloff J., Desselberger U., Ball L.A. Elsevier, Academic Press; Amsterdam: 2005. Virus Taxonomy, Eighth Report of the International Committee on Taxonomy of Viruses. [Google Scholar]
- Feder M., Pas J., Wyrwicz L.S., Bujnicki J.M. Molecular phylogenetics of the RrmJ/fibrillarin superfamily of ribose 2′-O-methyltransferases. Gene. 2003;302:129–138. doi: 10.1016/s0378-1119(02)01097-1. [DOI] [PubMed] [Google Scholar]
- Ferron F., Longhi S., Henrissat B., Canard B. Viral RNA-polymerases—a predicted 2′-O-ribose methyltransferase domain shared by all Mononegavirales. Trends Biochem. Sci. 2002;27:222–224. doi: 10.1016/s0968-0004(02)02091-1. [DOI] [PubMed] [Google Scholar]
- Fischer F., Peng D., Hingley S.T., Weiss S.R., Masters P.S. The internal open reading frame within the nucleocapsid gene of mouse hepatitis virus encodes a structural protein that is not essential for viral replication. J. Virol. 1997;71:996–1003. doi: 10.1128/jvi.71.2.996-1003.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer F., Stegen C.F., Masters P.S., Samsonoff W.A. Analysis of constructed E gene mutants of mouse hepatitis virus confirms a pivotal role for E protein in coronavirus assembly. J. Virol. 1998;72:7885–7894. doi: 10.1128/jvi.72.10.7885-7894.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouchier R.A.M., Hartwig N.G., Bestebroer T.M., Niemeyer B., de Jong J.C., Simon J.H., Osterhaus A.D.M.E. A previously undescribed coronavirus associated with respiratory disease in humans. Proc. Natl. Acad. Sci. U.S.A. 2004;101:6212–6216. doi: 10.1073/pnas.0400762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gioia U., Laneve P., Dlakic M., Arceci M., Bozzoni I., Caffarelli E. Functional characterization of XendoU, the endoribonuclease involved in small nucleolar RNA biosynthesis. J. Biol. Chem. 2005;280:18996–19002. doi: 10.1074/jbc.M501160200. [DOI] [PubMed] [Google Scholar]
- Godeny E.K., de Vries A.A., Wang X.C., Smith S.L., de Groot R.J. Identification of the leader-body junctions for the viral subgenomic mRNAs and organization of the simian hemorrhagic fever virus genome: evidence for gene duplication during arterivirus evolution. J. Virol. 1998;72:862–867. doi: 10.1128/jvi.72.1.862-867.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldbach R.W. Molecular evolution of plant RNA viruses. Ann. Rev. Phytopathol. 1986;24:289–310. [Google Scholar]
- González J.M., Gomez-Puertas P., Cavanagh D., Gorbalenya A.E., Enjuanes L. A comparative sequence analysis to revise the current taxonomy of the family Coronaviridae. Arch. Virol. 2003;148:2207–2235. doi: 10.1007/s00705-003-0162-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez M.E., Carrasco L. Viroporins. FEBS Lett. 2003;552:28–34. doi: 10.1016/s0014-5793(03)00780-4. [DOI] [PubMed] [Google Scholar]
- Gorbalenya A.E. Big nidovirus genome: when count and order of domains matter. Adv. Exp. Med. Biol. 2001;494:1–17. [PubMed] [Google Scholar]
- Gorbalenya A.E., Koonin E.V. Viral-proteins containing the purine ntp-binding sequence pattern. Nucleic Acids Res. 1989;17:8413–8440. doi: 10.1093/nar/17.21.8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E., Koonin E.V. Comparative analysis of the amino acid sequences of the key enzymes of the replication and expression of positive-strand RNA viruses. Validity of the approach and functional and evolutionary implications. Sov. Sci. Rev. D Physicochem. Biol. 1993;11:1–84. [Google Scholar]
- Gorbalenya A.E., Koonin E.V. Helicases: amino acid sequence comparisons and structure-function relationships. Curr. Opin. Struct. Biol. 1993;3:419–429. [Google Scholar]
- Gorbalenya A.E., Koonin E.V., Donchenko A.P., Blinov V.M. A novel superfamily of nucleoside triphosphate-binding motif containing proteins which are probably involved in duplex unwinding in DNA and RNA replication and recombination. FEBS Lett. 1988;235:16–24. doi: 10.1016/0014-5793(88)81226-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E., Koonin E.V., Donchenko A.P., Blinov V.M. Coronavirus genome: prediction of putative functional domains in the non-structural polyprotein by comparative amino acid sequence analysis. Nucleic Acids Res. 1989;17:4847–4861. doi: 10.1093/nar/17.12.4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E., Koonin E.V., Lai M.M.C. Putative papain-related thiol proteases of positive-strand RNA viruses. FEBS Lett. 1991;288:201–205. doi: 10.1016/0014-5793(91)81034-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E., Pringle F.M., Zeddam J.L., Luke B.T., Cameron C.E., Kalmakoff J., Hanzlik T.N., Gordon K.H., Ward V.K. The palm subdomain-based active site is internally permuted in viral RNA-dependent RNA polymerases of an ancient lineage. J. Mol. Biol. 2002;324:47–62. doi: 10.1016/S0022-2836(02)01033-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E., Snijder E.J. Viral cysteine proteinases. Perspect. Drug Discov. Des. 1996;6:64–86. doi: 10.1007/BF02174046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E., Snijder E.J., Spaan W.J. Severe acute respiratory syndrome coronavirus phylogeny: toward consensus. J. Virol. 2004;78:7863–7866. doi: 10.1128/JVI.78.15.7863-7866.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowda S., Satyanarayana T., Ayllon M.A., Albiach-Marti M.R., Mawassi M., Rabindran S., Garnsey S.M., Dawson W.O. Characterization of the cis-acting elements controlling subgenomic mRNAs of Citrus tristeza virus: production of positive- and negative-stranded 3′-terminal and positive-stranded 5′-terminal RNAs. Virology. 2001;286:134–151. doi: 10.1006/viro.2001.0987. [DOI] [PubMed] [Google Scholar]
- Graham R.L., Sims A.C., Brockway S.M., Baric R.S., Denison M.R. The nsp2 replicase proteins of murine hepatitis virus and severe acute respiratory syndrome coronavirus are dispensable for viral replication. J. Virol. 2005;79:13399–13411. doi: 10.1128/JVI.79.21.13399-13411.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarino L.A., Bhardwaj K., Dong W., Sun J.C., Holzenburg A., Kao C. Mutational analysis of the SARS virus Nsp15 endoribonuclease: identification of residues affecting hexamer formation. J. Mol. Biol. 2005;353:1106–1117. doi: 10.1016/j.jmb.2005.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haijema B.J., Volders H., Rottier P.J.M. Live, attenuated coronavirus vaccines through the directed deletion of group-specific genes provide protection against feline infectious peritonitis. J. Virol. 2004;78:3863–3871. doi: 10.1128/JVI.78.8.3863-3871.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J.F., Peng G.W., Min J., Yu D.W., Liang W.J., Zhang S.Y., Xu R.H., Zheng H.Y., Wu X.W., Xu J., Wang Z.H., Fang L., Zhang X., Li H., Yan X.G., Lu J.H., Hu Z.H., Huang J.C., Wan Z.Y., Hou J.L., Lin J.Y., Song H.D., Wang S.Y., Zhou X.J., Zhang G.W., Gu B.W., Zheng H.J., Zhang X.L., He M., Zheng K., Wang B.F., Fu G., Wang X.N., Chen S.J., Chen Z., Hao P., Tang H., Ren S.X., Zhong Y., Guo Z.M., Liu Q., Miao Y.G., Kong X.Y., He W.Z., Li Y.X., Wu C.I., Zhao G.P., Chiu R.W.K., Chim S.S.C., Tong Y.K., Chan P.K.S., Tam J.S., Lo Y.M.D. Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science. 2004;303:1666–1669. doi: 10.1126/science.1092002. [DOI] [PubMed] [Google Scholar]
- Hegyi A., Ziebuhr J. Conservation of substrate specificities among coronavirus main proteases. J. Gen. Virol. 2002;83:595–599. doi: 10.1099/0022-1317-83-3-595. [DOI] [PubMed] [Google Scholar]
- Herold J., Siddell S.G. An ’elaborated’ pseudoknot is required for high frequency frameshifting during translation of HCV 229E polymerase mRNA. Nucleic Acids Res. 1993;21:5838–5842. doi: 10.1093/nar/21.25.5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold, J., Siddell, S.G., Gorbalenya, A.E., 1999. A human RNA viral cysteine proteinase that depends upon a unique Zn2+-binding finger connecting the two domains of a papain-like fold (published erratum appears in J Biol Chem 1999;274(30):21490). J. Biol. Chem. 274, 14918–14925. [DOI] [PMC free article] [PubMed]
- Herrewegh A.A.P.M., Smeenk I., Horzinek M.C., Rottier P.J.M., de Groot R.J. Feline coronavirus type II strains 79-1683 and 79-1146 originate from a double recombination between feline coronavirus type I and canine coronavirus. J. Virol. 1998;72:4508–4514. doi: 10.1128/jvi.72.5.4508-4514.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heusipp G., Harms U., Siddell S.G., Ziebuhr J. Identification of an ATPase activity associated with a 71-kilodalton polypeptide encoded in gene 1 of the human coronavirus 229E. J. Virol. 1997;71:5631–5634. doi: 10.1128/jvi.71.7.5631-5634.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson T., Britton P., Cavanagh D. Neither the RNA nor the proteins of open reading frames 3a and 3b of the coronavirus infectious bronchitis virus are essential for replication. J. Virol. 2006;80:296–305. doi: 10.1128/JVI.80.1.296-305.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q.L., Yu L.P., Petros A.M., Gunasekera A., Liu Z.H., Xu N., Hajduk P., Mack J., Fesik S.W., Olejniczak E.T. Structure of the N-terminal RNA-binding domain of the SARS CoV nucleocapsid protein. Biochemistry. 2004;43:6059–6063. doi: 10.1021/bi036155b. [DOI] [PubMed] [Google Scholar]
- Ingallinella P., Bianchi E., Finotto M., Cantoni G., Eckert D.M., Supekar V.M., Bruckmann C., Carfi A., Pessi A. Structural characterization of the fusion-active complex of severe acute respiratory syndrome (SARS) coronavirus. Proc. Natl. Acad. Sci. U.S.A. 2004;101:8709–8714. doi: 10.1073/pnas.0402753101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito N., Mossel E.C., Narayanan K., Popov V.L., Huang C., Inoue T., Peters C.J., Makino S. Severe acute respiratory syndrome coronavirus 3a protein is a viral structural protein. J. Virol. 2005;79:3182–3186. doi: 10.1128/JVI.79.5.3182-3186.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov K.A., Thiel V., Dobbe J.C., van der Meer Y., Snijder E.J., Ziebuhr J. Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J. Virol. 2004;78:5619–5632. doi: 10.1128/JVI.78.11.5619-5632.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov K.A., Hertzig T., Rozanov M., Bayer S., Thiel V., Gorbalenya A.E., Ziebuhr J. Major genetic marker of nidoviruses encodes a replicative endoribonuclease. Proc. Natl. Acad. Sci. U.S.A. 2004;101:12694–12699. doi: 10.1073/pnas.0403127101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov K.A., Ziebuhr J. Human coronavirus 229E nonstructural protein 13: characterization of duplex-unwinding, nucleoside triphosphatase, and RNA 5′-triphosphatase activities. J. Virol. 2004;78:7833–7838. doi: 10.1128/JVI.78.14.7833-7838.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang B.M., Monroe S.S., Koonin E.V., Stine S.E., Glass R.I. RNA sequence of astrovirus—distinctive genomic organization and a putative retrovirus-like ribosomal frameshifting signal that directs the viral replicase synthesis. Proc. Natl. Acad. Sci. U.S.A. 1993;90:10539–10543. doi: 10.1073/pnas.90.22.10539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadare G., Haenni A.-L. Virus-encoded RNA helicases. J. Virol. 1997;71:2583–2590. doi: 10.1128/jvi.71.4.2583-2590.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamer G., Argos P. Primary structural comparison of RNA-dependent polymerases from plant, animal and bacterial viruses. Nucleic Acids Res. 1984;12:7269–7282. doi: 10.1093/nar/12.18.7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazi L., Lissenberg A., Watson R., de Groot R.J., Weiss S.R. Expression of hemagglutinin esterase protein from recombinant mouse hepatitis virus enhances neurovirulence. J. Virol. 2005;79:15064–15073. doi: 10.1128/JVI.79.24.15064-15073.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E.V. The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses. J. Gen. Virol. 1991;72:2197–2206. doi: 10.1099/0022-1317-72-9-2197. [DOI] [PubMed] [Google Scholar]
- Koonin E.V. Computer-assisted identification of a putative methyltransferase domain in NS5 protein of flaviviruses and λ2 protein of reovirus. J. Gen. Virol. 1993;74:733–740. doi: 10.1099/0022-1317-74-4-733. [DOI] [PubMed] [Google Scholar]
- Koonin E.V., Gorbalenya A.E. Evolution of RNA genomes—does the high mutation-rate necessitate high-rate of evolution of viral-proteins. J. Mol. Evol. 1989;28:524–527. doi: 10.1007/BF02602932. [DOI] [PubMed] [Google Scholar]
- Koonin E.V., Gorbalenya A.E., Purdy M.A., Rozanov M.N., Reyes G.R., Bradley D.W. Computer-assisted assignment of functional domains in the nonstructural polyprotein of hepatitis E virus: delineation of an additional group of positive-strand RNA plant and animal viruses. Proc. Natl. Acad. Sci. U.S.A. 1992;89:8259–8263. doi: 10.1073/pnas.89.17.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroneman A., Cornelissen L.A.H.M., Horzinek M.C., de Groot R.J., Egberink H.F. Identification and characterization of a porcine Torovirus. J. Virol. 1998;72:3507–3511. doi: 10.1128/jvi.72.5.3507-3511.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T.A., Bebenek R. DNA replication fidelity. Ann. Rev. Biochem. 2000;69:497–529. doi: 10.1146/annurev.biochem.69.1.497. [DOI] [PubMed] [Google Scholar]
- Kunkel T.A., Erie D.A. DNA mismatch repair. Ann. Rev. Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- Kuo L.L., Masters P.S. The small envelope protein E is not essential for murine coronavirus replication. J. Virol. 2003;77:4597–4608. doi: 10.1128/JVI.77.8.4597-4608.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M.M., Baric R.S., Brayton P.R., Stohlman S.A. Characterization of leader RNA sequences on the virion and mRNAs of mouse hepatitis virus, a cytoplasmic RNA virus. Proc. Natl. Acad. Sci. U.S.A. 1984;81:3626–3630. doi: 10.1073/pnas.81.12.3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M.M.C. RNA recombination in animal and plant-viruses. Microbiol. Rev. 1992;56:61–79. doi: 10.1128/mr.56.1.61-79.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M.M.C. Recombination in large RNA viruses: coronaviruses. Semin. Virol. 1996;7:381–388. doi: 10.1006/smvy.1996.0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M.M.C., Cavanagh D. The molecular biology of coronaviruses. Adv. Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M.M.C., Holmes K.V. Coronaviruses. In: Knipe D.M., Howley P.M., editors. Fields Virology. Lippincott, Williams & Wilkins; Philadelphia, PA: 2001. pp. 1163–1185. [Google Scholar]
- Laneve P., Altieri F., Fiori M.E., Scaloni A., Bozzoni I., Caffarelli E. Purification, cloning, and characterization of XendoU, a novel endoribonuclease involved in processing of intron-encoded small nucleolar RNAs in Xenopus laevis. J. Biol. Chem. 2003;278:13026–13032. doi: 10.1074/jbc.M211937200. [DOI] [PubMed] [Google Scholar]
- Lau S.K.P., Woo P.C.Y., Li K.S.M., Huang Y., Tsoi H.W., Wong B.H.L., Wong S.S.Y., Leung S.Y., Chan K.H., Yuen K.Y. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. U.S.A. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W.D., Shi Z.L., Yu M., Ren W.Z., Smith C., Epstein J.H., Wang H.Z., Crameri G., Hu Z.H., Zhang H.J., Zhang J.H., McEachern J., Field H., Daszak P., Eaton B.T., Zhang S.Y., Wang L.F. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- Lindner H.A., Fotouhi-Ardakani N., Lytvyn V., Lachance P., Sulea T., Menard R. The papain-like protease from the severe acute respiratory syndrome coronavirus is a deubiquitinating enzyme. J. Virol. 2005;79:15199–15208. doi: 10.1128/JVI.79.24.15199-15208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lissenberg A., Vrolijk M.M., van Vliet A.L.W., Langereis M.A., Groot-Mijnes J.D.F., Rottier P.J.M., de Groot R.J. Luxury at a cost? Recombinant mouse hepatitis viruses expressing the accessory hemagglutinin esterase protein display reduced fitness in vitro. J. Virol. 2005;79:15054–15063. doi: 10.1128/JVI.79.24.15054-15063.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S.W., Xiao G.F., Chen Y.B., He Y.X., Niu J.K., Escalante C.R., Xiong H.B., Farmar J., Debnath A.K., Tien P., Jiang S.B. Interaction between heptad repeat 1 and 2 regions in spike protein of SARS-associated coronavirus: implications for virus fusogenic mechanism and identification of fusion inhibitors. Lancet. 2004;363:938–947. doi: 10.1016/S0140-6736(04)15788-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luytjes W., Bredenbeek P.J., Noten A.F.H., Horzinek M.C., Spaan W.J.M. Sequence of mouse hepatitis virus A59 messenger RNA-2—indications for RNA recombination between coronaviruses and influenza-C virus. Virology. 1988;166:415–422. doi: 10.1016/0042-6822(88)90512-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Keck J.G., Stohlman S.T., Lai M.M.C. High-frequency RNA recombination of murine coronaviruses. J. Virol. 1986;57:729–737. doi: 10.1128/jvi.57.3.729-737.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra M.A., Jones S.J.M., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S.N., Khattra J., Asano J.K., Barber S.A., Chan S.Y., Cloutier A., Coughlin S.M., Freeman D., Girn N., Griffin O.L., Leach S.R., Mayo M., McDonald H., Montgomery S.B., Pandoh P.K., Petrescu A.S., Robertson A.G., Schein J.E., Siddiqui A., Smailus D.E., Stott J.E., Yang G.S., Plummer F., Andonov A., Artsob H., Bastien N., Bernard K., Booth T.F., Bowness D., Czub M., Drebot M., Fernando L., Flick R., Garbutt M., Gray M., Grolla A., Jones S., Feldmann H., Meyers A., Kabani A., Li Y., Normand S., Stroher U., Tipples G.A., Tyler S., Vogrig R., Ward D., Watson B., Brunham R.C., Krajden M., Petric M., Skowronski D.M., Upton C., Roper R.L. The genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- Martzen M.R., McCraith S.M., Spinelli S.L., Torres F.M., Fields S., Grayhack E.J., Phizicky E.M. A biochemical genomics approach for identifying genes by the activity of their products. Science. 1999;286:1153–1155. doi: 10.1126/science.286.5442.1153. [DOI] [PubMed] [Google Scholar]
- Mazumder R., Iyer L.M., Vasudevan S., Aravind L. Detection of novel members, structure-function analysis and evolutionary classification of the 2H phosphoesterase superfamily. Nucleic Acids Res. 2002;30:5229–5243. doi: 10.1093/nar/gkf645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller W.A., Brown C.M., Wang S.P. New punctuation for the genetic code: luteovirus gene expression. Semin. Virol. 1997;8:3–13. [Google Scholar]
- Miller W.A., Koev G. Synthesis of subgenomic RNAs by positive-strand RNA viruses. Virology. 2000;273:1–8. doi: 10.1006/viro.2000.0421. [DOI] [PubMed] [Google Scholar]
- Minskaia, E., Hertzig, T., Gorbalenya, A.E., Campanacci, V., Cambillau, C., Canard, B., Ziebuhr, J. Discovery of an RNA virus 3′ → 5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc. Natl. Acad. Sci. U.S.A. 103, in press. [DOI] [PMC free article] [PubMed]
- Molenkamp R., van Tol H., Rozier B.C., van der Meer Y., Spaan W.J., Snijder E.J. The arterivirus replicase is the only viral protein required for genome replication and subgenomic mRNA transcription. J. Gen. Virol. 2000;81:2491–2496. doi: 10.1099/0022-1317-81-10-2491. [DOI] [PubMed] [Google Scholar]
- Moser M.J., Holley W.R., Chatterjee A., Mian I.S. The proofreading domain of Escherichia coli DNA polymerase I and other DNA and/or RNA exonuclease domains. Nucleic Acids Res. 1997;25:5110–5118. doi: 10.1093/nar/25.24.5110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moya A., Holmes E.C., Gonzalez-Candelas F. The population genetics and evolutionary epidemiology of RNA viruses. Nat. Rev. Microbiol. 2004;2:279–288. doi: 10.1038/nrmicro863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson C.A., Pekosz A., Lee C.A., Diamond M.S., Fremont D.H. Structure and intracellular targeting of the SARS-coronavirus Orf7a accessory protein. Structure. 2005;13:75–85. doi: 10.1016/j.str.2004.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortego J., Escors D., Laude H., Enjuanes L. Generation of a replication-competent, propagation-deficient virus vector based on the transmissible gastroenteritis coronavirus genome. J. Virol. 2002;76:11518–11529. doi: 10.1128/JVI.76.22.11518-11529.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortego J., Sola I., Almazan F., Ceriani J.E., Riquelme C., Balasch M., Plana J., Enjuanes L. Transmissible gastroenteritis coronavirus gene 7 is not essential but influences in vivo virus replication and virulence. Virology. 2003;308:13–22. doi: 10.1016/S0042-6822(02)00096-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasternak A.O., van den Born E., Spaan W.J.M., Snijder E.J. Sequence requirements for RNA strand transfer during nidovirus discontinuous subgenomic RNA synthesis. EMBO J. 2001;20:7220–7228. doi: 10.1093/emboj/20.24.7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pehrson J.R., Fuji R.N. Evolutionary conservation of histone macroH2A subtypes and domains. Nucleic Acids Res. 1998;26:2837–2842. doi: 10.1093/nar/26.12.2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng C.W., Peremyslov V.V., Snijder E.J., Dolja V.V. A replication-competent chimera of plant and animal viruses. Virology. 2002;294:75–84. doi: 10.1006/viro.2001.1306. [DOI] [PubMed] [Google Scholar]
- Plant E.P., Perez-Alvarado G.C., Jacobs J.L., Mukhopadhyay B., Hennig M., Dinman J.D. A three-stemmed mRNA pseudoknot in the SARS coronavirus frameshift signal. PLoS Biol. 2005;3:1012–1023. doi: 10.1371/journal.pbio.0030172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon L.L.M., Chu D.K.W., Chan K.H., Wong O.K., Ellis T.M., Leung Y.H.C., Lau S.K.P., Woo P.C.Y., Suen K.Y., Yuen K.Y., Guan Y., Peiris J.S.M. Identification of a novel coronavirus in bats. J. Virol. 2005;79:2001–2009. doi: 10.1128/JVI.79.4.2001-2009.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posthuma C.C., Nedialkova D.D., Zevenhoven-Dobbe J.C., Blokhuis J.H., Gorbalenya A.E., Snijder E.J. Site-directed mutagenesis of the nidovirus replicative endoribonuclease NendoU exerts pleiotropic effects on the arterivirus life cycle. J. Virol. 2006;80:1653–1661. doi: 10.1128/JVI.80.4.1653-1661.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentice E., McAuliffe J., Lu X., Subbarao K., Denison M.R. Identification and characterization of severe acute respiratory syndrome coronavirus replicase proteins. J. Virol. 2004;78:9977–9986. doi: 10.1128/JVI.78.18.9977-9986.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putics A., Filipowicz W., Hall J., Gorbalenya A.E., Ziebuhr J. ADP-ribose-1″-monophosphatase: that is dispensable for viral a conserved coronavirus enzyme replication in tissue culture. J. Virol. 2005;79:12721–12731. doi: 10.1128/JVI.79.20.12721-12731.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putics A., Gorbalenya A.E., Ziebuhr J. Identification of protease and ADP-ribose 1″-monophosphatase activities associated with transmissible gastroenteritis virus non-structural protein 3. J. Gen. Virol. 2006;87:651–656. doi: 10.1099/vir.0.81596-0. [DOI] [PubMed] [Google Scholar]
- Putics, A., Slaby, J., Filipowicz, W., Gorbalenya, A.E., Ziebuhr, J., in press. ADP-ribose-1″-phosphatase activities of the human coronavirus 229E and SARS coronavirus X domains. Adv. Exp. Med. Biol. [DOI] [PMC free article] [PubMed]
- Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.H., Tong S.X., Tamin A., Lowe L., Frace M., Derisi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C.T., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., Gunther S., Osterhaus A.D.M.E., Drosten C., Pallansch M.A., Anderson L.J., Bellini W.J. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- Saikatendu K.S., Joseph J.S., Subramanian V., Clayton T., Griffith M., Moy K., Velasquez J., Neuman B.W., Buchmeier M.J., Stevens R.C., Kuhn P. Structural basis of severe acute respiratory syndrome coronavirus ADP-ribose-1″-phosphate dephosphorylation by a conserved domain of nsP3. Structure. 2005;13:1665–1675. doi: 10.1016/j.str.2005.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salemi M., Fitch W.M., Ciccozzi M., Ruiz-Alvarez M.J., Rezza G., Lewis M.J. Severe acute respiratory syndrome coronavirus sequence characteristics and evolutionary rate estimate from maximum likelihood analysis. J. Virol. 2004;78:1602–1603. doi: 10.1128/JVI.78.3.1602-1603.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki D.L., Wang T., Sawicki S.G. The RNA structures engaged in replication and transcription of the A59 strain of mouse hepatitis virus. J. Gen. Virol. 2001;82:385–396. doi: 10.1099/0022-1317-82-2-385. [DOI] [PubMed] [Google Scholar]
- Sawicki S.G., Sawicki D.L. Coronaviruses use discontinuous extension for synthesis of subgenome-length negative strands. Adv. Exp. Med. Biol. 1995;380:499–506. doi: 10.1007/978-1-4615-1899-0_79. [DOI] [PubMed] [Google Scholar]
- Sawicki S.G., Sawicki D.L. Coronavirus transcription: a perspective. Curr. Top. Microbiol. Immunol. 2005;287:31–55. doi: 10.1007/3-540-26765-4_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki S.G., Sawicki D.L., Younker D., Meyer Y., Thiel V., Stokes H., Siddell S.G. Functional and genetic analysis of coronavirus replicase-transcriptase proteins. PLoS Pathog. 2005;1:e39. doi: 10.1371/journal.ppat.0010039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schelle B., Karl N., Ludewig B., Siddell S.G., Thiel V. Selective replication of coronavirus genomes that express nucleocapsid protein. J. Virol. 2005;79:6620–6630. doi: 10.1128/JVI.79.11.6620-6630.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz B., Routledge E., Siddell S.G. Murine coronavirus nonstructural protein ns2 is not essential for virus replication in transformed cells. J. Virol. 1990;64:4784–4791. doi: 10.1128/jvi.64.10.4784-4791.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seybert A., Hegyi A., Siddell S.G., Ziebuhr J. The human coronavirus 229E superfamily 1 helicase has RNA and DNA duplex-unwinding activities with 5′-to-3′ polarity. RNA. 2000;6:1056–1068. doi: 10.1017/s1355838200000728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seybert A., van Dinten L.C., Snijder E.J., Ziebuhr J. Biochemical characterization of the equine arteritis virus helicase suggests a close functional relationship between arterivirus and coronavirus helicases. J. Virol. 2000;74:9586–9593. doi: 10.1128/jvi.74.20.9586-9593.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seybert A., Posthuma C.C., van Dinten L.C., Snijder E.J., Gorbalenya A.E., Ziebuhr J. A complex zinc finger controls the enzymatic activities of nidovirus helicases. J. Virol. 2005;79:696–704. doi: 10.1128/JVI.79.2.696-704.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddell S.G., Ziebuhr J., Snijder E.J. Coronaviruses, toroviruses and arteriviruses. In: Mahy B.W.J., ter Meulen V., editors. Topley & Wilson's Microbiology and Microbial Infections, Virology. Hodder Arnold; 2005. pp. 823–856. [Google Scholar]
- Smits S.L., Lavazza A., Matiz K., Horzinek M.C., Koopmans M.P., de Groot R.J. Phylogenetic and evolutionary relationship among torovirus field variants: evidence for multiple intertypic recombination events. J. Virol. 2003;77:9567–9577. doi: 10.1128/JVI.77.17.9567-9577.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits, S.L., Snijder, E.J., de Groot, R.J. Characterization of a torovirus main proteinase. J. Virol. 80, in press. [DOI] [PMC free article] [PubMed]
- Snijder E.J., Bredenbeek P.J., Dobbe J.C., Thiel V., Ziebuhr J., Poon L.L.M., Guan Y., Rozanov M., Spaan W.J.M., Gorbalenya A.E. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 2003;331:991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Brinton M.A., Faaberg K.S., Godeny E.K., Gorbalenya A.E., MacLachlan N.J., Mengeling W.L., Plagemann P.G. Family Arteriviridae. In: Fauquet C.M., Mayo M.A., Maniloff J., Desselberger U., Ball L.A., editors. Virus Taxonomy, Eighth Report of the International Committee on Taxonomy of Viruses. Elsevier, Academic Press; Amsterdam: 2005. pp. 965–974. [Google Scholar]
- Snijder E.J., den Boon J.A., Bredenbeek P.J., Horzinek M.C., Rijnbrand R., Spaan W.J. The carboxyl-terminal part of the putative Berne virus polymerase is expressed by ribosomal frameshifting and contains sequence motifs which indicate that toro- and coronaviruses are evolutionarily related. Nucleic Acids Res. 1990;18:4535–4542. doi: 10.1093/nar/18.15.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., den Boon J.A., Horzinek M.C., Spaan W.J. Comparison of the genome organization of toro- and coronaviruses: evidence for two nonhomologous RNA recombination events during Berne virus evolution. Virology. 1991;180:448–452. doi: 10.1016/0042-6822(91)90056-H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., den Boon J.A., Spaan W.J., Weiss M., Horzinek M.C. Primary structure and post-translational processing of the Berne virus peplomer protein. Virology. 1990;178:355–363. doi: 10.1016/0042-6822(90)90332-L. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Horzinek M.C. Toroviruses: replication, evolution and comparison with other members of the coronavirus-like superfamily. J. Gen. Virol. 1993;74:2305–2316. doi: 10.1099/0022-1317-74-11-2305. [DOI] [PubMed] [Google Scholar]
- Snijder E.J., Horzinek M.C., Spaan W.J. A 3′-coterminal nested set of independently transcribed mRNAs is generated during Berne virus replication. J. Virol. 1990;64:331–338. doi: 10.1128/jvi.64.1.331-338.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Horzinek M.C., Spaan W.J. The coronaviruslike superfamily. Adv. Exp. Med. Biol. 1993;342:235–244. doi: 10.1007/978-1-4615-2996-5_37. [DOI] [PubMed] [Google Scholar]
- Snijder E.J., Meulenberg J.J. The molecular biology of arteriviruses. J. Gen. Virol. 1998;79:961–979. doi: 10.1099/0022-1317-79-5-961. [DOI] [PubMed] [Google Scholar]
- Snijder E.J., Siddell S.G., Gorbalenya A.E. The order Nidovirales. In: Mahy B.W.J., ter Meulen V., editors. Topley & Wilson's Microbiology and Microbial Infections, Virology. Hodder Arnold; 2005. pp. 390–404. [Google Scholar]
- Snijder E.J., Wassenaar A.L., Spaan W.J., Gorbalenya A.E. The arterivirus Nsp2 protease. An unusual cysteine protease with primary structure similarities to both papain-like and chymotrypsin-like proteases. J. Biol. Chem. 1995;270:16671–16676. doi: 10.1074/jbc.270.28.16671. [DOI] [PubMed] [Google Scholar]
- Snijder E.J., Wassenaar A.L., van Dinten L.C., Spaan W.J., Gorbalenya A.E. The arterivirus nsp4 protease is the prototype of a novel group of chymotrypsin-like enzymes, the 3C-like serine proteases. J. Biol. Chem. 1996;271:4864–4871. doi: 10.1074/jbc.271.9.4864. [DOI] [PubMed] [Google Scholar]
- Snijder E.J., Wassenaar A.L.M., Spaan W.J.M. The 5′ end of the equine arteritis virus replicase gene encodes a papain-like protease. J. Virol. 1992;66:7040–7048. doi: 10.1128/jvi.66.12.7040-7048.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H.D., Tu C.C., Zhang G.W., Wang S.Y., Zheng K., Lei L.C., Chen Q.X., Gao Y.W., Zhou H.Q., Xiang H., Zheng H.J., Chern S.W.W., Cheng F., Pan C.M., Xuan H., Chen S.J., Luo H.M., Zhou D.H., Liu Y.F., He J.F., Qin P.Z., Li L.H., Ren Y.Q., Liang W.J., Yu Y.D., Anderson L., Wang M., Xu R.H., Wu X.W., Zheng H.Y., Chen J.D., Liang G., Gao Y., Liao M., Fang L., Jiang L.Y., Li H., Chen F., Di B., He L.J., Lin J.Y., Tong S., Kong X., Du L., Hao P., Tang H., Bernini A., Yu X.J., Spiga O., Guo Z.M., Pan H.Y., He W.Z., Manuguerra J.C., Fontanet A., Danchin A., Niccolai N., Li Y.X., Wu C.I., Zhao G.P. Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc. Natl. Acad. Sci. U.S.A. 2005;102:2430–2435. doi: 10.1073/pnas.0409608102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spann K.M., Vickers J.E., Lester R.J.G. Lymphoid organ virus of Penaeus-Monodon from Australia. Dis. Aquat. Organ. 1995;23:127–134. [Google Scholar]
- Spaan W., Delius H., Skinner M., Armstrong J., Rottier P., Smeekens S., van der Zeijst B.A., Siddell S.G. Coronavirus mRNA synthesis involves fusion of non-contiguous sequences. EMBO J. 1983;2:1839–1844. doi: 10.1002/j.1460-2075.1983.tb01667.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaan W.J.M., Brian D., Cavanagh D., de Groot R.J., Enjuanes L., Gorbalenya A.E., Holmes K.V., Masters P.S., Rottier P., Taguchi F., Talbot P. Family Coronaviridae. In: Fauquet C.M., Mayo M.A., Maniloff J., Desselberger U., Ball L.A., editors. Virus Taxonomy, Eighth Report of the International Committee on Taxonomy of Viruses. Elsevier, Academic Press; Amsterdam: 2005. pp. 947–964. [Google Scholar]
- Spaan W.J.M., Cavanagh D., de Groot R.J., Enjuanes L., Gorbalenya A.E., Snijder E.J., Walker P.J. Order Nidovirales. In: Fauquet C.M., Mayo M.A., Maniloff J., Desselberger U., Ball L.A., editors. Virus Taxonomy, Eighth Report of the International Committee on Taxonomy of Viruses. Elsevier, Academic Press; Amsterdam: 2005. pp. 937–945. [Google Scholar]
- Sperry S.M., Kazi L., Graham R.L., Baric R.S., Weiss S.R., Denison M.R. Single-amino-acid substitutions in open reading frame (ORF) 1b-nsp14 and ORF 2a proteins of the coronavirus mouse hepatitis virus are attenuating in mice. J. Virol. 2005;79:3391–3400. doi: 10.1128/JVI.79.6.3391-3400.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephensen C.B., Casebolt D.B., Gangopadhyay N.N. Phylogenetic analysis of a highly conserved region of the polymerase gene from 11 coronaviruses and development of a consensus polymerase chain reaction assay. Virus Res. 1999;60:181–189. doi: 10.1016/S0168-1702(99)00017-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss J.H., Strauss E.G. Evolution of RNA viruses. Ann. Rev. Microbiol. 1988;42:657–683. doi: 10.1146/annurev.mi.42.100188.003301. [DOI] [PubMed] [Google Scholar]
- Sulea T., Lindner H.A., Purisima E.O., Menard R. Deubiquitination, a new function of the severe acute respiratory syndrome coronavirus papain-like protease? J. Virol. 2005;79:4550–4551. doi: 10.1128/JVI.79.7.4550-4551.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supekar V.M., Bruckmann C., Ingallinella P., Bianchi E., Pessi A., Carfi A. Structure of a proteolytically resistant core from the severe acute respiratory syndrome coronavirus S2 fusion protein. Proc. Natl. Acad. Sci. U.S.A. 2004;101:17958–17963. doi: 10.1073/pnas.0406128102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton G., Fry E., Carter L., Sainsbury S., Walter T., Nettleship J., Berrow N., Owens R., Gilbert R., Davidson A., Siddell S., Poon L.L.M., Diprose J., Alderton D., Walsh M., Grimes J.M., Stuart D.I. The nsp9 replicase protein of SARS-coronavirus, structure and functional insights. Structure. 2004;12:341–353. doi: 10.1016/j.str.2004.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y.J., Lim S.G., Hong W.J. Characterization of viral proteins encoded by the SARS-coronavirus genome. Antivir. Res. 2005;65:69–78. doi: 10.1016/j.antiviral.2004.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner J.A., Watt R.M., Chai Y.B., Lu L.Y., Lin M.C., Peiris J.S., Poon L.L., Kung H.F., Huang J.D. The severe acute respiratory syndrome (SARS) coronavirus NTPase/helicase belongs to a distinct class of 5′ to 3′ viral helicases. J. Biol. Chem. 2003;278:39578–39582. doi: 10.1074/jbc.C300328200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijms M.A., van Dinten L.C., Gorbalenya A.E., Snijder E.J. A zinc finger-containing papain-like protease couples subgenomic mRNA synthesis to genome translation in a positive-stranded RNA virus. Proc. Natl. Acad. Sci. U.S.A. 2001;98:1889–1894. doi: 10.1073/pnas.041390398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoek L., Pyrc K., Jebbink M.F., Vermeulen-Oost W., Berkhout R.J.M., Wolthers K.C., Wertheim-van Dillen P.M.E., Kaandorp J., Spaargaren J., Berkhout B. Identification of a new human coronavirus. Nat. Med. 2004;10:368–373. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Most R.G., Spaan W.J.M. Coronavirus replication, transcription, and RNA recombination. In: Siddell S.G., editor. The Coronaviridae. Plenum Press; New York: 1995. pp. 11–31. [Google Scholar]
- van Dinten L.C., den Boon J.A., Wassenaar A.L., Spaan W.J., Snijder E.J. An infectious arterivirus cDNA clone: identification of a replicase point mutation that abolishes discontinuous mRNA transcription. Proc. Natl. Acad. Sci. U.S.A. 1997;94:991–996. doi: 10.1073/pnas.94.3.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dinten L.C., van Tol H., Gorbalenya A.E., Snijder E.J. The predicted metal-binding region of the arterivirus helicase protein is involved in subgenomic mRNA synthesis, genome replication, and virion biogenesis. J. Virol. 2000;74:5213–5223. doi: 10.1128/jvi.74.11.5213-5223.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Marle G., Dobbe J.C., Gultyaev A.P., Luytjes W., Spaan W.J., Snijder E.J. Arterivirus discontinuous mRNA transcription is guided by base pairing between sense and antisense transcription-regulating sequences. Proc. Natl. Acad. Sci. U.S.A. 1999;96:12056–12061. doi: 10.1073/pnas.96.21.12056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Marle G., van Dinten L.C., Spaan W.J., Luytjes W., Snijder E.J. Characterization of an equine arteritis virus replicase mutant defective in subgenomic mRNA synthesis. J. Virol. 1999;73:5274–5281. doi: 10.1128/jvi.73.7.5274-5281.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Vliet A.L.W., Smits S.L., Rottier P.J.M., de Groot R.J. Discontinuous and non-discontinuous subgenornic RNA transcription in a nidovirus. EMBO J. 2002;21:6571–6580. doi: 10.1093/emboj/cdf635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijgen L., Lemey P., Keyaerts E., Van Ranst M. Genetic variability of human respiratory coronavirus OC43. J. Virol. 2005;79:3223–3224. doi: 10.1128/JVI.79.5.3223-3225.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Grotthuss M., Wyrwicz L.S., Rychlewski L. mRNA cap-1 methyltransferase in the SARS genome. Cell. 2003;113:701–702. doi: 10.1016/S0092-8674(03)00424-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker P.J., Bonami J.R., Boonsaeng V., Chang P.S., Cowley J.A., Enjuanes L., Flegel T.W., Lightner D.V., Loh P.C., Snijder E.J., Tang K. Family Roniviridae. In: Fauquet C.M., Mayo M.A., Maniloff J., Desselberger U., Ball L.A., editors. Virus Taxonomy, Eighth Report of the International Committee on Taxonomy of Viruses. Elsevier, Academic Press; Amsterdam: 2005. pp. 975–979. [Google Scholar]
- Wassenaar A.L., Spaan W.J., Gorbalenya A.E., Snijder E.J. Alternative proteolytic processing of the arterivirus replicase ORF1a polyprotein: evidence that NSP2 acts as a cofactor for the NSP4 serine protease. J. Virol. 1997;71:9313–9322. doi: 10.1128/jvi.71.12.9313-9322.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss M., Steck F., Horzinek M.C. Purification and partial characterization of a new enveloped RNA virus (Berne virus) J. Gen. Virol. 1983;64:1849–1858. doi: 10.1099/0022-1317-64-9-1849. [DOI] [PubMed] [Google Scholar]
- Wilson L., Mckinlay C., Gage P., Ewart G. SARS coronavirus E protein forms cation-selective ion channels. Virology. 2004;330:322–331. doi: 10.1016/j.virol.2004.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C.Y., Lau S.K.P., Chu C.M., Chan K.H., Tsoi H.W., Huang Y., Wong B.H.L., Poon R.W.S., Cai J.J., Luk W.K., Poon L.L.M., Wong S.S.Y., Guan Y., Peiris J.S.M., Yuen K.Y. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 2005;79:884–895. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worobey M., Holmes E.C. Evolutionary aspects of recombination in RNA viruses. J. Gen. Virol. 1999;80:2535–2543. doi: 10.1099/0022-1317-80-10-2535. [DOI] [PubMed] [Google Scholar]
- Xu Y.H., Liu Y.W., Lou Z.Y., Qin L., Li X., Bai Z.H., Pang H., Tien P., Gao G.F., Rao Z. Structural basis for coronavirus-mediated membrane fusion—crystal structure of mouse hepatitis virus spike protein fusion core. J. Biol. Chem. 2004;279:30514–30522. doi: 10.1074/jbc.M403760200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Yang M., Ding Y., Liu Y., Lou Z., Zhou Z., Sun L., Mo L., Ye S., Pang H., Gao G.F., Anand K., Bartlam M., Hilgenfeld R., Rao Z. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc. Natl. Acad. Sci. U.S.A. 2003;100:13190–13195. doi: 10.1073/pnas.1835675100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yount B., Roberts R.S., Sims A.C., Deming D., Frieman M.B., Sparks J., Denison M.R., Davis N., Baric R.S. Severe acute respiratory syndrome coronavirus group-specific open reading frames encode nonessential functions for replication in cell cultures and mice. J. Virol. 2005;79:14909–14922. doi: 10.1128/JVI.79.23.14909-14922.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanotto P.M., Gibbs M.J., Gould E.A., Holmes E.C. A reevaluation of the higher taxonomy of viruses based on RNA polymerases. J. Virol. 1996;70:6083–6096. doi: 10.1128/jvi.70.9.6083-6096.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Y.J., Sun F., Li X.M., Pang H., Xu X.L., Bartlam M., Rao Z.H. Insights into SARS-CoV transcription and replication from the structure of the nsp7-nsp8 hexadecamer. Nat. Struct. Mol. Biol. 2005;12:980–986. doi: 10.1038/nsmb999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr J. Molecular biology of severe acute respiratory syndrome coronavirus. Curr. Opin. Microbiol. 2004;7:412–419. doi: 10.1016/j.mib.2004.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr J. The coronavirus replicase. Curr. Top. Microbiol. Immunol. 2005;287:57–94. doi: 10.1007/3-540-26765-4_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr J., Bayer S., Cowley J.A., Gorbalenya A.E. The 3C-like proteinase of an invertebrate nidovirus links coronavirus and potyvirus homologs. J. Virol. 2003;77:1415–1426. doi: 10.1128/JVI.77.2.1415-1426.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr J., Heusipp G., Siddell S.G. Biosynthesis, purification, and characterization of the human coronavirus 229E 3C-like proteinase. J. Virol. 1997;71:3992–3997. doi: 10.1128/jvi.71.5.3992-3997.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr J., Siddell S.G. Processing of the human coronavirus 229E replicase polyproteins by the virus-encoded 3C-like proteinase: identification of proteolytic products and cleavage sites common to pp1a and pp1ab. J. Virol. 1999;73:177–185. doi: 10.1128/jvi.73.1.177-185.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr J., Snijder E.J., Gorbalenya A.E. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 2000;81:853–879. doi: 10.1099/0022-1317-81-4-853. [DOI] [PubMed] [Google Scholar]
- Ziebuhr J., Thiel V., Gorbalenya A.E. The autocatalytic release of a putative RNA virus transcription factor from its polyprotein precursor involves two paralogous papain-like proteases that cleave the same peptide bond. J. Biol. Chem. 2001;276:33220–33232. doi: 10.1074/jbc.M104097200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuniga S., Sola I., Alonso S., Enjuanes L. Sequence motifs involved in the regulation of discontinuous coronavirus subgenomic RNA synthesis. J. Virol. 2004;78:980–994. doi: 10.1128/JVI.78.2.980-994.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Y.H., Deutscher M.P. Exoribonuclease superfamilies: structural analysis and phylogenetic distribution. Nucleic Acids Res. 2001;29:1017–1026. doi: 10.1093/nar/29.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]


