Abstract
A novel coronavirus (CoV) has been described in association with cases of severe acute respiratory syndrome (SARS). The virus, SARS-CoV, differs from the previously described human coronaviruses, 229E and OC43. 229E was previously shown to productively infect human monocytes/macrophages, whereas OC43 poorly infected the cells. In this study, we examined whether SARS-CoV could productively infect purified monocytes/macrophages (PM) derived from human donor cells. Unlike 229E-infected cells, which produced viral titers of 103.5 to 106 TCID50/ml, SARS-CoV replicated poorly in PM, producing titers of 101.75 to 102 TCID50/ml. This finding was similar to results reported for OC43-infected cells, with titers ranging from 101.2 to 102.7 TCID50/ml. Of interest, SARS-CoV proteins were detected only in PM that did not produce significant amounts of interferon (IFN)-α, and in one such case, preliminary electron microscope studies demonstrated that SARS-CoV-like particles could enter the cells, possibly via phagocytosis. These results suggest that SARS-CoV, like human CoV OC43, poorly infects human PM, and production of IFN-α by these cells further limits the infection. Given the importance of monocytes/macrophages to the immune response, it is possible that their infection by SARS-CoV and alteration of this infection by IFN-α may be important to the course of the infection in humans.
Keywords: SARS-CoV, Monocytes/macrophages, Interferon
1. Introduction
SARS-coronavirus (SARS-CoV), which was discovered in association with cases of severe acute respiratory syndrome (SARS), is a newly identified member of a diverse group of large, enveloped, positive-strand RNA viruses of the order Nidovirales in the family Coronaviridae (Holmes, 2003, Berger et al., 2004). A number of coronaviruses have been described. They include the human coronaviruses 229E and OC43 and a number of animal coronaviruses, such as mouse hepatitis virus (Stadler et al., 2003, Holmes, 2003). The structural proteins of SARS-CoV include the large type I membrane glycoprotein spike (S) (∼200 kDa), which binds to the receptor, the nucleoprotein (N) (∼45.9 kDa), the membrane glycoprotein or matrix protein (M) (∼25 kDa), and the small envelope protein (E) (∼14 kDa) (Stadler et al., 2003, Ying et al., 2004). The angiotensin-converting enzyme 2, or ACE-2 receptor, assumed to be absent from monocyte/macrophage surfaces (Harmer et al., 2002), was recently identified as a receptor for SARS-CoV entry into the permissive cell line Vero E6 (Li et al., 2003b).
Studies in Vero E6 cells have helped describe several characteristics of SARS-CoV infection (Ksiazek et al., 2003, Kuiken et al., 2003; Ng et al., 2003a, Ng et al., 2003b; Goldsmith et al., 2004). The virus can bind, enter, and uncoat in cells within 30 min (Ng et al., 2003a). Ultrastructural characterization studies demonstrate replication in the cytoplasm of infected cells by virions, which bud upon the membranes of the endoplasmic reticulum and migrate to the cell surface while within the cisternae of the endoplasmic reticulum, where they will then be extruded to the extracellular space (Ng et al., 2003a, Ng et al., 2003b; Goldsmith et al., 2004). Extracellular virus particles can be seen accumulating in large quantities along the plasma membranes of the infected cells (Ng et al., 2003b, Goldsmith et al., 2004).
SARS-CoV appears highly susceptible to the type I interferons (IFNs), as these studies reveal (Cinatl et al., 2003, Hensley et al., 2004, Spiegel et al., 2004, Ströher et al., 2004). It is well known that by producing IFN-α/β, monocytes/macrophages participate in inducing resistance to viral replication. The cytokines activate cellular genes that destroy viral mRNA and inhibit the translation of proteins. In addition, IFN-α/β induce major histocompatibility complex class I protein expression in infected cells, increasing the level of protein presentation to CD8+ T cells and the ability of uninfected cells to resist killing by natural killer cells (Goodbourn et al., 2000).
In a previous study, monocytes/macrophages were infected with human CoVs, 229E and OC43 (Collins, 2002). Macrophages express human peptidase N (Yeager et al., 1992), which has been identified as the receptor for 229E; the receptor for OC43 is still unknown (Collins, 2002). The study by Collins showed that 229E efficiently infected monocytes and induced apoptosis, as determined by DNA changes and annexin V staining, but OC43 poorly infected the cells. The apoptosis correlated with differential release of infectious virus.
We therefore determined whether monocyte/macrophage donor cells could be infected with SARS-CoV by examining their ability to produce infectious virus. Since we had previously shown that measles virus induced IFN-α production in purified monocyte/macrophage (PM) cultures (Yilla et al., 2003), we also asked whether similar cultures produced IFN-α in response to SARS-CoV infection. SARS-CoV (Urbani strain) was used to infect PM derived from healthy adult donors. Results from the study suggest that SARS-CoV infection of the PM cultures yields little productive virus and that IFN-α might curb SARS-CoV infection of these cells.
2. Materials and methods
2.1. Cell culture
Blood from healthy adult donors, labeled A–H, was collected into heparinized tubes, and peripheral blood mononuclear cells (PBMC) were isolated from buffy coats generated by lymphocyte separation medium (ICN/Cappel). Cells were collected and processed as described previously (Yilla et al., 2003). Monocytes were isolated by using CD14 microbeads (Mitenyi Biotec) according to the manufacturer's protocol. Purity was assessed by flow cytometry (CD14, CD11b, CD3, HLA-A, -B, -C, HLA-DR) and was >99%. Viability by Trypan blue exclusion was >95%. The same donor cells could not be used in every experiment because their numbers were limited. To minimize inter-donor variability, PBMC were obtained in bulk and frozen for use in replicate experiments. Vero E6 cells (a gift from Anthony Sanchez, Centers for Disease Control and Prevention [CDC], Atlanta) were cultured in Dulbecco's modified Eagle medium (DMEM; Invitrogen Life Technologies) supplemented with 5% fetal bovine serum. [35S]-methionine (specific radioactivity, >1000 Ci/mmol) was obtained from ICN, and fetal calf serum, RPMI-1640, penicillin/streptomycin (100×), and methionine-free medium were from Invitrogen Life Technologies.
2.2. Antibodies
The human convalescent serum (HCS) that was used for protein detection was obtained from a SARS-CoV-positive patient (see Ksiazek et al., 2003), 18 days after infection. The anti-Spike (anti-S) monoclonal antibody (mAb; a gift from Ralph Tripp, CDC, Atlanta) used in these experiments neutralizes SARS-CoV. Both antibodies were used to identify SARS-CoV proteins in infected PM and Vero E6 cells.
2.3. Virus preparation
SARS-CoV (Urbani strain) was isolated from a SARS-infected patient described in Ksiazek et al. (2003) and Rota et al. (2003) and propagated in Vero E6 cells. The titer of the Urbani SARS-CoV as determined on a Vero E6 monolayer was 106 50% tissue culture-infective dose (TCID50)/ml. In brief, TCID50 was determined by adding serial dilutions of the virus to a Vero E6 monolayer in a 96-well plate. The wells were inspected daily for virus-induced cytopathic effects. Four days after infection, the supernatant was removed and the cells were washed twice with PBS and stained with 2 ml of crystal violet solution for 1 h. After being stained, the cells were washed twice with PBS and allowed to air dry. Titers were subsequently recorded as Log10 TCID50 units, using the Karber method (Gray, 1999).
2.4. In vitro assays
PM (1 × 106) were plated in 24-well plates (Corning Inc.) and infected with 1 × 105 TCID50 of SARS-CoV Vero E6 supernatant, contained in a volume of 100 μl, for 1 h at 37 °C in serum-free medium while rocking. Cells were subsequently cultured in the presence of serum for 0, 24, 48, or 72 h (Fig. 1 ). Control cells were mock-infected with uninfected Vero E6 cell supernatant from equivalent cell numbers. Approximately 12–16 h after infection, the monocytes displayed characteristics typical of macrophages.
Fig. 1.
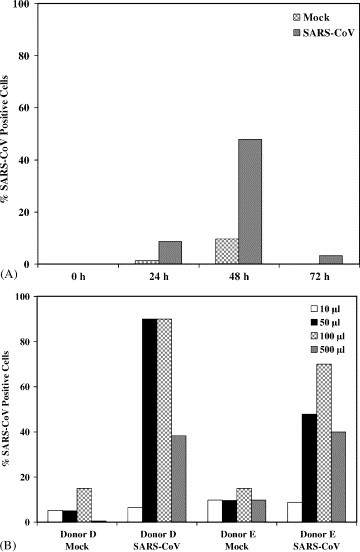
(A) PM from donor E was either mock infected with Vero E6 supernatant or infected with 1 × 105 TCID50 of SARS-CoV for 0, 24, 48, or 72 h. To detect SARS-CoV proteins by flow cytometric analysis, cells were permeabilized and stained with human convalescent serum that recognizes SARS-CoV proteins. The percentages of protein-positive cells (y-axis) are recorded as a function of time (x-axis). (B) PM from donors D and E were either mock infected or infected with SARS-CoV with the indicated amounts of cell supernatant for 48 h. Cells were analyzed as described for Fig. 1A. Ten thousand events were collected in this and subsequent experiments.
2.5. Cytokine assays
An indirect ELISA that reacts with all isotypes was used to quantitate the levels of IFN-α in experimental samples. Human IFN-α (PBL Biomedical Labs) was used as the assay standard to estimate the concentration of IFN induced in the experiment. Cytokine controls and experiment samples were added to anti-IFN antibody-coated microtiter wells (PBL Biomedical Labs) and assayed according to the manufacturer's protocol. One unit of IFN-α was equivalent to 4 pg. In antibody neutralization experiments, PM (1 × 106) were treated 30 min prior to infection with >1000 neutralizing U/ml anti-IFN-α antibody (Chemicon International). PM were cultured 48 h after infection and then analyzed for the presence of SARS-CoV proteins. To test if infectious virus was produced from donors A, H, and I, the innoculum was washed off after 1 h and cells were re-incubated for 48 h with fresh medium containing anti-IFN-α antibody prior to processing for SARS antigen.
2.6. Biochemical analysis and gel electrophoresis
Vero E6 cells (2.5 × 105) were starved in 1 ml of methionine-free DMEM for 20 min at 37 °C prior to labeling with 0.1 mCi/ml [35S]-methionine. Incorporation of label was terminated by the addition of 1 mM cold methionine after 1 h. Cells were collected by centrifugation and lysed for 20 min in lysis buffer (0.1% SDS; 1.0% TX-100; 1% NaDOC, 150 mM NaCl, 10 mM Tris–HCl, pH 7.2 and 5 mM EDTA) containing 1 mM PMSF and 100 μM Pefablock (Boehringer Mannheim). All immunoprecipitations of total lysates were preceded by preclearing the cell lysate with normal mouse serum. Immune complexes were then removed by adsorption to protein G-sepharose beads. Precleared lysates were immunoprecipitated with the different antisera in parallel (Fig. 2 ), and the immune complexes, which were adsorbed to protein G-sepharose beads, were collected by centrifugation. Immune complexes were washed and prepared for SDS-polyacrylamide gel electrophoresis as previously described (Burke et al., 1984).
Fig. 2.
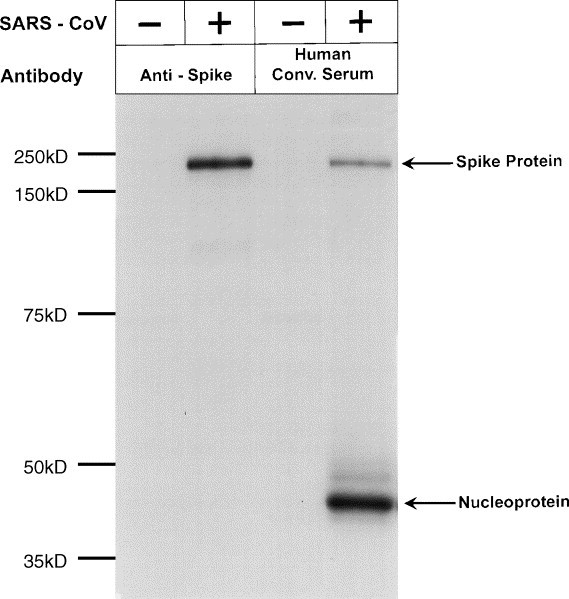
Vero E6 monolayers were either mock infected with Vero E6 supernatant or infected with 105 TCID50 of SARS-CoV. Cells were radiolabeled with [35S]-methionine for 1 h. The cells were harvested and lysed as described in Section 2. Subsequently, the clarified cell lysates were immunoprecipitated with anti-S mAb or with human convalescent serum (Human Conv. Serum) and immune complexes were collected by adsorption to protein G-sepharose beads. Proteins were identified following separation on 12.5% SDS-PAGE gel.
2.7. Flow cytometry
In brief, PM were washed in PBS containing 0.5% bovine serum albumin and 0.05% sodium azide (FACS buffer) and resuspended in FACS buffer containing appropriately diluted antibody. Cells were stained for SARS proteins, using either HCS (a gift from Tom Ksiazek, CDC) or anti-S mAb (a gift from Ralph Tripp, CDC). Anti-HLA-A, -B, and -C locus products (clone G46-2.6; BD Bioscience Pharmingen), anti-HLA-DR (clone Tu36; BD Bioscience Pharmingen), anti-CD14 (clone RMO52; Beckman Coulter), anti-CD11b (clone Bear 1; Beckman Coulter), and anti-CD3 (clone UCHT1; Beckman Coulter) were used to detect the presence or absence of the respective proteins on the PM surface. Cells were surface-stained with antibodies for 20 min at 4 °C, washed in FACS buffer, and then fixed with 4% paraformaldehyde for 20 min at 4 °C. Cells were then permeabilized in FACS buffer containing 0.1% Saponin (permeabilization buffer) for 10 min at room temperature to test for the intracellular presence of SARS proteins. Internal staining for SARS-CoV was carried out for 30 min at room temperature with either HCS or anti-S mAb as indicated in the figure legend (Fig. 1, Fig. 3, Fig. 4, Fig. 5). Reactive antibodies were detected by indirect staining, using either goat anti-mouse or goat anti-human secondary antibodies conjugated to AlexaFluor 488 (Molecular Probes, Inc.). Stained cells were analyzed by four-color flow cytometry (Beckman Coulter EPICS-XL) with Beckman Coulter EXPO32 ADC software. Side scatter versus forward scatter parameters were used to gate cells by size. Side scatter versus HLA-DR (a marker for monocytes and macrophages) was used to analyze monocyte/macrophage cell populations. Quadrant analysis using FL1 (fluorescein isothiocyanate) versus FL3 (phycoerythrin-texas red) was used to differentiate infected CD14 cells from uninfected cells. Mean fluorescence intensity measurements from the generated plots were displayed as graphs in the figures.
Fig. 3.
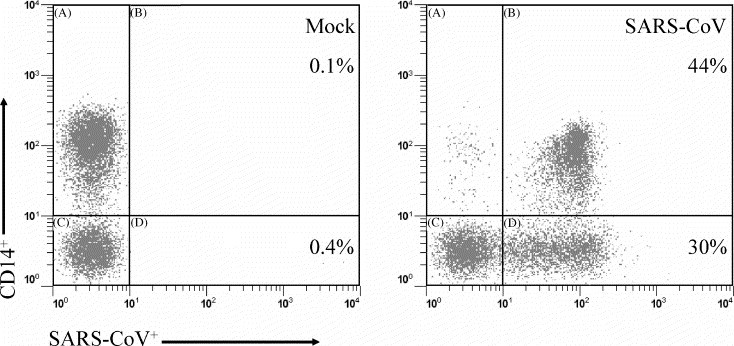
A representative flow cytometry plot displaying reactivity against SARS-CoV antigens is shown. All cells shown were HLA-DR positive. CD14+ cells (y-axis) versus SARS-CoV-positive cells (x-axis) (revealed with the human convalescent serum) are shown. The percentage of HLA-DR+ cells that express both CD14 and SARS-CoV S protein are shown in the upper quadrant.
Fig. 4.

Donor PM (A–I) were either mock infected with Vero E6 supernatant or infected with 105 TCID50 of SARS-CoV for 48 h. To detect SARS-CoV proteins by flow cytometric analysis, cells were permeabilized and stained with human convalescent serum that recognizes SARS-CoV proteins. The percentages of protein-positive cells (y-axis) are recorded for each donor tested. (Inset) PM from donors B, D, H, and I were either mock infected with Vero E6 supernatant or infected with 1 × 105 TCID50 of SARS-CoV for 48 h. SARS-CoV S protein was detected by flow cytometric analysis, using anti-S mAb.
Fig. 5.

PM from two donors were either mock infected or infected with 105 TCID50 of SARS-CoV for 48 h in the presence (top panel) or absence (bottom panel) of 1000 U/ml neutralizing anti-IFN-α. Mock-infected (dashed line) or SARS-CoV-infected (solid line) cells were prepared for flow cytometry. Cells were permeabilized and stained for S protein with anti-S mAb. The histogram plots show the amount of S protein detected, expressed as mean fluorescence intensity (MFI) values along the x-axis. The peak height shown on the y-axis is a measure of the number of events collected.
2.8. Electron microscopy
Cells were prepared for electron microscopy according to published protocols (Goldsmith et al., 2003). In brief, PM were infected with SARS-CoV for 15 h and then fixed in 2.5% buffered glutaraldehyde. Specimens were processed for embedding in epoxy resins and then sectioned.
3. Results
3.1. Analysis of SARS-CoV infection of peripheral monocytes/macrophages
We examined PM for the presence of SARS-CoV proteins following infection with SARS-CoV or mock infection with Vero E6 supernatant. A time-course experiment (0–72 h) was carried out in order to determine whether SARS-CoV could replicate in PM (Fig. 1A). Protein was detected by using an HCS that recognizes the SARS-CoV S (∼200 kDa) and N (∼50 kDa) proteins from 35[S]-methionine-labeled, infected Vero E6 cells (Fig. 2). Radiolabeling with cysteine is required to reveal additional SARS-CoV proteins (unpublished observations). Peak protein levels were detected in PM 48 h after infection. Fig. 1B shows results from titrations done on cells from two donors. Since the highest amount of protein was detected following addition of 105 TCID50 of virus, all subsequent experiments were carried out at this concentration for 48 h. At higher inoculum of virus, the Vero E6 cell supernatant was toxic to the cells and resulted in fewer infected cells (Fig. 1B). In these experiments, the level of background staining was usually low as shown in Fig. 3 .
We next tested PM from the nine donors (A–I; Fig. 4 ), using HCS: cells from four of the nine donors were positive for SARS-CoV protein, which was detected internally after the cells were permeabilized. No reactivity to the serum was detected on the cell surface (data not shown). When anti-S mAb became available, we used it to test mock-infected and SARS-CoV-infected PM from four donors (B, D, H, and I) for SARS S protein (Fig. 4 inset). This mAb reacts with a single ∼200 kDa protein from SARS-CoV-infected Vero E6 cells (Fig. 2). We found that donor PM that were reactive to the HCS were also reactive with the mAb to S protein. Similarly, the absence of S protein in the PM of infected donors H and I was consistent with the absence of other SARS-CoV proteins that could be detected by the HCS. Thus, infection of PM with SARS-CoV was successful in some but not all PM studied. We tested donors C and F for the production of infectious virus. Very low virus titers (102 TCID50/ml) were obtained from donor F, and no infectious virus was secreted by infected PM from donor C. This implied that despite the detection of SARS-CoV protein, high-titered virus was not being secreted from the infected PM.
3.2. Interferon-α assay
PM have been shown to release IFNs after viral infection (Yilla et al., 2003). Therefore, we asked whether SARS-CoV also induced IFN-α in PM. Supernatants from SARS-CoV-infected PM or mock-infected PM were tested for IFN-α production 48 h after infection (Table 1 ). In cells from three of the nine donors studied (donors A, H, and I), significant levels of IFN-α (∼60–260 U/ml) were measured following SARS-CoV infection. Furthermore, SARS-CoV protein was not detected in these donor PM. In the remaining six donors, B–G, absent or low levels of IFN-α (≤8 U/ml) were detected following infection. SARS-CoV proteins could be observed in cells from four of these six donors (B–E) (Table 1). We examined the age, race, and sex of the donors to determine whether any of these parameters influenced the response to SARS-CoV (Table 1). Due to the small number of donors, we were unable to convincingly show that any of these factors influenced the results we obtained. Despite secreting low levels of IFN-α, SARS-CoV proteins failed to be detected in Donor G. We were unable to identify a reason for this variation from the pattern seen with other donors. It does suggest that multiple factors may affect the ability of donor cells to support SARS-CoV infection.
Table 1.
Donor profiles and PM response to SARS-CoV infection
| Donor | Age (years) | Race | Sex | IFN-α (U/ml) | SARS proteins |
|---|---|---|---|---|---|
| A | 33 | White | F | 73.75 ± 6.72 | − |
| B | 23 | White | F | ≤0.00 | + |
| C | 50 | White | F | 2.11 ± 0.22 | + |
| D | 54 | Black | F | 0.36 ± 0.06 | + |
| E | 55 | White | M | 8.55 ± 0.03 | + |
| F | 55 | White | M | 8.82 ± 0.25 | − |
| G | 40 | White | M | ≤0.00 | − |
| H | 29 | Asian | F | 266.10 ± 15.47 | − |
| I | 45 | Asian | M | 60.0 ± 3.44 | − |
Donor profiles, reactivity to SARS antibodies and the results from IFN-α assays are shown. IFN-α levels in supernatants were measured by ELISA. Media from PM (106) were evaluated. Mean values ± standard deviations from two independent experiments done on donor cells are shown (P < 0.01). Vero E6 cells do not produce IFN (data not shown; Diaz et al., 1988). + = positive for SARS-CoV proteins; − = negative for SARS-CoV.
3.3. IFN-α neutralization experiments
Since SARS-CoV infection, as specified by the presence of protein, was associated with low levels or the absence of IFN-α, we looked at the effect of IFN-α-neutralizing antibodies on SARS-CoV infection. After treating cells from two IFN-α-producing donors (H and I) with IFN-α-neutralizing antibody, we analyzed SARS-CoV protein expression in the cells (Fig. 5 ). Addition of IFN-α-neutralizing antibodies led to detection of SARS-CoV proteins in both previously negative donor cells. The percentage of positive cells was higher in donor I (56%) than donor H (34%) possibly because donor I cells produced lower levels of IFN (i.e., levels that could be more easily neutralized). Addition of anti-IFN-β and IFN-γ failed to improve detection of S protein (data not shown). Furthermore, in the presence of IFN-α-neutralizing antibody, infected cells produced low levels of infectious virus (∼101.75 TCID50/ml).
3.4. Ultrastructural analysis of infected cells
Electron microscopy was used to further characterize SARS-CoV infection in cells (from donor B) that did not produce IFN-α following infection. Clusters of virus-like particles could be seen engulfed by pseudopod-like extensions of the macrophage in preparation for entry into the cell, possibly by phagocytosis (Fig. 6 A). Within the cytoplasm, SARS-CoV virions were seen aggregated in enlarged membrane-bound structures presumed to be phagolysosomes (Fig. 6B), although the phagocytic nature of these structures has yet to be confirmed. None of the morphologic features indicative of viral replication were detected in the PM culture from donor B, including nascent viral particles forming on cytoplasmic membranes, double-membrane vesicles, the proposed replication complex for coronaviruses, or the adherence of virions to the cell surface.
Fig. 6.

PM from SARS-CoV-infected cells from donor B were examined by electron microscopy. (A) Evidence of possible phagocytic entry of SARS-CoV viral particles into PM from donor B (arrow). (B) Intracellular vesicles, presumed to be phagolysosomes, contain SARS-CoV particles (arrows). Bars: 100 nm.
4. Discussion
This study examined whether human monocytes/macrophages can support SARS-CoV replication and, as such, participate in the progression of SARS disease. We found evidence of virus entry, limited replication, and minimal production of infectious virus. Viral titers from monocyte/macrophage cultures inoculated with SARS-CoV did not exceed 102 TCID50/ml, while titers in excess of 106 were seen in Vero E6 cells. Electron microscope studies suggest that SARS-CoV might enter a non-IFN-α-producing PM by phagocytosis rather than by adhesion to the cell surface and membrane fusion as seen in more permissive cells, but further examination would be necessary to determine the actual mode of viral entry into monocytes and macrophages. Dendritic cells were also recently shown to take up SARS-CoV (Yang et al., 2004).
In PM, virus is detected in phagolysosomes but not on the cell surface, and virus is not associated with cell membrane fusion. Flow cytometric studies of SARS-CoV-infected cells detected no S protein on the cell surface, thus further suggesting that PM may lack a specific receptor for SARS entry. Electron microscope studies suggested differences in the replication process between PM and the more permissive Vero E6 cells. Replicating centers seen in SARS-CoV-infected Vero E6 cells were absent in the infected PM that were examined. Most significantly, SARS-CoV infection of PM appeared to be inhibited by the presence of IFN-α. SARS-CoV antigens were not detected in PM that produced significant levels of IFN-α (>10 U/ml); however, they were detected in PM producing <10 U/ml IFN-α (the PM were from four of six donors) and in cells producing significant levels of IFN-α after IFN-α-neutralizing antibody was added to the culture system. Taken together these data suggest human monocytes/macrophages can support low-level productive infection by SARS-CoV and that the level of infection can be modified by IFN-α.
Other coronaviruses, 229E and OC43, were previously shown to infect monocyte/macrophage cultures, but with very different outcomes (Collins, 2002). 229E infected monocytes well, inducing apoptosis and releasing higher titers of infectious virus into the culture media. In contrast, OC43 infected monocytes poorly, failed to induce increased apoptosis, and released low amounts of infectious virus (Collins, 2002). Our studies of SARS-CoV-infected PM suggest that like OC43, SARS-CoV also infects monocytes/macrophages poorly. Both viruses may lack a viable receptor for entry into monocytes. PBMC of SARS patients were also examined for evidence of infection and replication of SARS-CoV (Li et al., 2003a). In this report, minus-strand RNA copies of the SARS-CoV genome were observed in PBMC and used as indicators of replicating virus. However such intermediates were not long lasting and could not be detected 6 days after infection (Li et al., 2003a).
Sensitivity of SARS-CoV to IFN-α/β has been shown in monkeys and monkey cell lines. For example, treatment of Vero E6 cells with IFN resulted in inhibition of SARS-CoV replication (Cinatl et al., 2003, Hensley et al., 2004, Spiegel et al., 2004, Ströher et al., 2004). Expression of SARS-CoV proteins in type I pneumocytes of infected macaques was also severely reduced after prophylactic treatment of the SARS-CoV-infected monkeys with IFN-α (Haagmans et al., 2004). A reduction in viral replication and excretion, in protein expression in pneumocytes, and in pulmonary damage were all noted in infected macaques who were treated compared with infected macaques who were not treated (Haagmans et al., 2004). Our results showing that SARS-CoV infection of monocytes/macrophages can be modulated by IFN-α are consistent with these findings.
The role that SARS-CoV infection of monocytes/macrophages plays in the course of the disease in humans is yet to be determined. The role could be more important than would otherwise be indicated by the low level of productive infection, given the importance of these cells in guiding the immune response to infection through antigen presentation and cytokine production. Our data suggest that differences in levels of IFN-α production with infection may occur, as indicated by infection in monocytes/macrophages, and such differences could contribute to the disease course. For example, co-infection with a paramyxovirus, human metapneumovirus, has been noted in some SARS-infected patients, and this infection has been hypothesized to render some persons more susceptible to SARS-CoV disease (Chan et al., 2003, Stephenson, 2003). Although human metapneumovirus infections have not been described previously as interfering with IFN-mediated antiviral responses, other paramyxoviruses have been shown to interfere with such responses (Gotoh et al., 2001, Bossert et al., 2003). Thus, it is possible that such interference could facilitate SARS-CoV infection. Another example where variability in IFN-α production could explain differences in disease outcome is the association between increasing severity of disease with increasing age. Young children appear to be the least susceptible and the elderly the most susceptible to serious complications of infection (Wong et al., 2003). It is known that IFN-α levels are highest in the young and decrease with age (Katschinski et al., 1994, Shodell and Siegal, 2002). Perhaps age-associated variations in IFN-α production may help explain age-associated differences in SARS-CoV disease.
Yang and colleagues recently demonstrated that myeloid dendritic cells can interact with SARS-CoV S protein and bind virus (Yang et al., 2004). Although these dendritic cells could not be infected by SARS-CoV, the authors suggest they could serve as vehicles to disseminate the virus. This potential of monocytes/macrophages and dendritic cells to take up SARS-CoV may be relevant to SARS pathogenesis in vivo.
Acknowledgements
We thank Tom Ksiazek and Ralph Tripp for generously providing the seed virus and antibodies, respectively, used for this study; Jim Gathany for photographic assistance, and Claudia Chesley for editing. MM received funding from the McKing Consulting Corporation.
References
- Berger A., Drosten Ch., Doerr H.W., Sturmer M., Preiser W. Severe acute respiratory syndrome (SARS)—paradigm of an emerging viral infection. J. Clin. Virol. 2004;29:13–22. doi: 10.1016/j.jcv.2003.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossert B., Marozin S., Conzelmann K.K. Nonstructural proteins NS1 and NS2 of bovine respiratory syncytial virus block activation of interferon regulatory factor 3. J. Virol. 2003;77:8661–8668. doi: 10.1128/JVI.77.16.8661-8668.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke B., Matlin K., Bause E., Legler G., Peyrieras N., Ploegh H.L. Inhibition of N-linked oligosaccharide trimming does not interfere with surface expression of certain integral membrane proteins. EMBO J. 1984;3:551–556. doi: 10.1002/j.1460-2075.1984.tb01845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan P.K., Tam J.S., Lam C.W., Chan E., Wu A., Li C.K., Buckley T.A., Ng K.C., Joynt G.M., Cheng F.W., To K.F., Lee N., Hui D.S., Cheung J.L., Chu I., Liu E., Chung S.S., Sung J.J. Human metapneumovirus detection in patients with severe acute respiratory syndrome. Emerg. Infect. Dis. 2003;9:1058–1063. doi: 10.3201/eid0909.030304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinatl J., Morgenstern B., Bauer G., Chandra P., Rabenau H., Doerr H.W. Treatment of SARS with human interferons. Lancet. 2003;362:293–294. doi: 10.1016/S0140-6736(03)13973-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins A.R. In vitro detection of apoptosis in monocytes/macrophages infected with human coronavirus. Clin. Diagn. Lab. Immunol. 2002;9:1392–1395. doi: 10.1128/CDLI.9.6.1392-1395.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz M.O., Ziemin S., Le Beau M.M., Pitha P., Smith S.D., Chilcote R.R., Rowley J.D. Homozygous deletion of the alpha- and beta 1-interferon genes in human leukemia and derived cell lines. Proc. Natl. Acad. Sci. U.S.A. 1988;85:5259–5263. doi: 10.1073/pnas.85.14.5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsmith C.S., Tatti K.M., Ksiazek T.G., Rollin P.E., Comer J.A., Lee W.W., Rota P.A., Bankamp B., Bellini W.J., Zaki S.R. Ultrastructural characterization of SARS coronavirus. Emerg. Infect. Dis. 2004;10:320–326. doi: 10.3201/eid1002.030913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsmith C.S., Whistler T., Rollin P.E., Ksiazek T.G., Rota P.A., Bellini W.J., Daszak P., Wong K.T., Shieh W.J., Zaki S.R. Elucidation of Nipah virus morphogenesis and replication using ultrastructural and molecular approaches. Virus Res. 2003;92:89–98. doi: 10.1016/s0168-1702(02)00323-4. [DOI] [PubMed] [Google Scholar]
- Goodbourn S., Didcock L., Randall R.E. Interferons: cell signalling, immune modulation, antiviral response and virus countermeasures. J. Gen. Virol. 2000;81:2341–2364. doi: 10.1099/0022-1317-81-10-2341. [DOI] [PubMed] [Google Scholar]
- Gotoh B., Komatsu T., Takeuchi K., Yokoo J. Paramyxovirus accessory proteins as interferon antagonists. Microbiol. Immunol. 2001;45:787–800. doi: 10.1111/j.1348-0421.2001.tb01315.x. [DOI] [PubMed] [Google Scholar]
- Gray J. Assays for Virus Infection. In: Cann A.J., editor. Virus Culture: A Practical Approach. Oxford University Press; New York: 1999. 84 pp. [Google Scholar]
- Haagmans B.L., Kuiken T., Martina B.E., Fouchier R.A., Rimmelzwaan G.F., van Amerongen G., van Riel D., de Jong T., Itamura S., Chan K.H., Tashiro M., Osterhaus A.D. Pegylated interferon-alpha protects type 1 pneumocytes against SARS coronavirus infection in macaques. Nat. Med. 2004;10:290–293. doi: 10.1038/nm1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer D., Gilbert M., Borman R., Clark K.L. Quantitative mRNA expression profiling of ACE 2, a novel homologue of angiotensin converting enzyme. FEBS Lett. 2002;532:107–110. doi: 10.1016/s0014-5793(02)03640-2. [DOI] [PubMed] [Google Scholar]
- Hensley L.E., Fritz E.A., Jahrling P.B., Karp C., Huggins J.W., Geisbert T.W. Interferon-beta 1a and SARS coronavirus replication. Emerg. Infect. Dis. 2004;10:317–319. doi: 10.3201/eid1002.030482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes K.V. SARS coronavirus: a new challenge for prevention and therapy. J. Clin. Invest. 2003;111:1605–1609. doi: 10.1172/JCI18819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katschinski D.M., Neustock P., Kluter H., Kirchner H. Influence of various factors on interferon-alpha production in cultures of human leukocytes. J. Interferon Res. 1994;14:105–110. doi: 10.1089/jir.1994.14.105. [DOI] [PubMed] [Google Scholar]
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A.E., Humphrey C.D., Shieh W.J., Guarner J., Paddock C.D., Rota P., Fields B., DeRisi J., Yang J.Y., Cox N., Hughes J.M., LeDuc J.W., Bellini W.J., Anderson L.J., and SARS Working Group A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- Kuiken T., Fouchier R.A., Schutten M., Rimmelzwaan G.F., van Amerongen G., van Riel D., Laman J.D., de Jong T., van Doornum G., Lim W., Ling A.E., Chan P.K., Tam J.S., Zambon M.C., Gopal R., Drosten C., van der Werf S., Escriou N., Manuguerra J.C., Stohr K., Peiris J.S., Osterhaus A.D. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet. 2003;362:263–270. doi: 10.1016/S0140-6736(03)13967-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Wo J., Shao J., Zhu H., Wu N., Li M., Yao H., Hu M., Dennin R.H. SARS-coronavirus replicates in mononuclear cells of peripheral blood (PBMCs) from SARS patients. J. Clin. Virol. 2003;28:239–244. doi: 10.1016/S1386-6532(03)00195-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng M.L., Tan S.H., See E.E., Ooi E.E., Ling A.E. Early events of SARS coronavirus infection in Vero cells. J. Med. Virol. 2003;71:323–331. doi: 10.1002/jmv.10499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng M.L., Tan S.H., See E.E., Ooi E.E., Ling A.E. Proliferative growth of SARS coronavirus in Vero E6 cells. J. Gen. Virol. 2003;84:3291–3303. doi: 10.1099/vir.0.19505-0. [DOI] [PubMed] [Google Scholar]
- Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.H., Tong S., Tamin A., Lowe L., Frace M., DeRisi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., Gunther S., Osterhaus A.D., Drosten C., Pallansch M.A., Anderson L.J., Bellini W.J. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- Shodell M., Siegal F.P. Circulating, interferon-producing plasmacytoid dendritic cells decline during human ageing. Scand. J. Immunol. 2002;56:518–521. doi: 10.1046/j.1365-3083.2002.01148.x. [DOI] [PubMed] [Google Scholar]
- Spiegel M., Pichlmair A., Muhlberger E., Haller O., Weber F. The antiviral effect of interferon-beta against SARS-coronavirus is not mediated by MxA protein. J. Clin. Virol. 2004;30:211–213. doi: 10.1016/j.jcv.2003.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler K., Masignani V., Eickmann M., Becker S., Abrignani S., Klenk H.D., Rappuoli R. SARS—beginning to understand a new virus. Nat. Rev. Microbiol. 2003;1:209–218. doi: 10.1038/nrmicro775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson J. Studies explore impact of new pathogens: investigators report on metapneumovirus, SARS. JAMA. 2003;290:2112–2115. doi: 10.1001/jama.290.16.2112. [DOI] [PubMed] [Google Scholar]
- Ströher U., DiCaro A., Li Y., Strong J.E., Aoki F., Plummer F., Jones S.M., Feldmann H. Severe acute respiratory syndrome-related coronavirus is inhibited by interferon-alpha. J. Infect. Dis. 2004;189:1164–1167. doi: 10.1086/382597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong G.W., Li A.M., Ng P.C., Fok T.F. Severe acute respiratory syndrome in children. Pediatr. Pulmonol. 2003;36:261–266. doi: 10.1002/ppul.10367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z.Y., Huang Y., Ganesh L., Leung K., Kong W.P., Schwartz O., Subbarao K., Nabel G.J. pH-dependent entry of severe acute respiratory syndrome coronavirus is mediated by the spike glycoprotein and enhanced by dendritic cell transfer through DC-SIGN. J. Virol. 2004;78:5642–5650. doi: 10.1128/JVI.78.11.5642-5650.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeager C.L., Ashmun R.A., Williams R.K., Cardellichio C.B., Shapiro L.H., Look A.T., Holmes K.V. Human aminopeptidase N is a receptor for human coronavirus 229E. Nature. 1992;357:420–422. doi: 10.1038/357420a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilla M., Hickman C., McGrew M., Meade E., Bellini W.J. Edmonston measles virus prevents increased cell surface expression of peptide-loaded major histocompatibility complex class II proteins in human peripheral monocytes. J. Virol. 2003;77:9412–9421. doi: 10.1128/JVI.77.17.9412-9421.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying W., Hao Y., Zhang Y., Peng W., Qin E., Cai Y., Wei K., Wang J., Chang G., Sun W., Dai S., Li X., Zhu Y., Li J., Wu S., Guo L., Dai J., Wang J., Wan P., Chen T., Du C., Li D., Wan J., Kuai X., Li W., Shi R., Wei H., Cao C., Yu M., Liu H., Dong F., Wang D., Zhang X., Qian X., Zhu Q., He F. Proteomic analysis on structural proteins of severe acute respiratory syndrome coronavirus. Proteomics. 2004;4:492–504. doi: 10.1002/pmic.200300676. [DOI] [PMC free article] [PubMed] [Google Scholar]


