Abstract
Four truncated porcine aminopeptidase N (pAPN, a cellular receptor for porcine coronaviruses) proteins were expressed in prokaryotic cells. The recognizing of a specific serum against pAPN to these proteins was investigated by enzyme-linked immunosorbent assay (ELISA) and immunoblotting. The binding ability of the proteins to transmissible gastroenteritis virus (TGEV), a porcine coronavirus, was analyzed by ELISA. The inhibitory effect of these proteins to cell infection by TGEV was analyzed using plaque assays. Our data indicate that three truncated pAPNs positively reacted with the specific antiserum and the major binding regions of pAPN were limited in regions 36aa–223aa, 349aa–591aa and 592–963aa. The proteins showed discrepant binding activity to either pAPN antibody or TGE virions. Moreover, the truncated proteins blocked the infection of cells by TGEV to different extent. The results suggest that the major antibody-binding domains of pAPN may associate with the receptor-binding determinants. The role of APN is discussed in the context of virus receptor usage.
Keywords: pAPN, Receptor, Binding, Coronavirus
1. Introduction
Aminopeptidase N (APN) is a kind of membrane-bound metallopeptidase (Delmas et al., 1994). It is a type II glycoprotein of about 150–160 kDa, in molecular weight and its large extracellular carboxy-terminal domain contains a pentapeptide catalytic sequence (His-Glu-X-X-His), characteristic of zinc metalloprotease (Hooper, 1994). APN is distributed on the surface of diverse cell lines, for example, it is expressed on the plasma membranes of granulocytes, lymphocytes, monocytes, fibroblasts and synaptic membrane in the central nervous system (Miguel et al., 2002). It is highly expressed on the surface of the brush border membrane of the enterocytes, where it participates in the final steps of digestion by cleaving peptides preferentially after N-terminal neutral amino acids. It has been suggested that APN is involved in modulating signals of bioactive peptides in the brain (Delmas et al., 1994).
APN acts as a cellular receptor for several coronaviruses. It is documented that human coronavirus 229E (HCoV-229E) and feline infectious peritonitis virus (FIPV) uses human APN (hAPN, also called CD13) and feline APN (fAPN) as their respective receptor. Transmissible gastroenteritis virus (TGEV) is a porcine coronavirus, which causes severe diarrhea and high mortality rate in seronegative piglets (Ren et al., 2008). The enveloped virus consists of a positive single-strand RNA approximately 28.5-kb in size. TGEV has four major structural proteins: the spike (S), the integral membrane (M) protein, the nucleocapsid (N) protein and a small envelope protein (sM) (Spaan et al., 1988, Laude et al., 1993, Penzes et al., 2001).
TGEV S protein plays important roles in initiating cell infection by interacting with porcine APN, pAPN (Gebauer et al., 1991, Enjuanes et al., 1992, Schwegmann-Wessels et al., 2003, Ren et al., 2008) and sialic acid residues (Krempl et al., 1997, Krempl et al., 2000, Krempl and Herrler, 2001). The pAPN has been identified as a functional receptor for TGEV, although this virus can also use fAPN as another receptor (Delmas et al., 1992, Tresnan and Holmes, 1998). At present, the role of APN in coronavirus infection is not fully understood.
In the current study, four truncated pAPN proteins were expressed in Escherichia coli (E. coli). Their reactivity with anti-pAPN antibody was analyzed by ELISA and Western blot. We found that one of the truncated pAPN proteins had the lowest reactivity with an anti-pAPN antibody compared with other truncated pAPN proteins and whole mature pAPN. At the same time, it was not recognized by the antibody in Western blot. Interestingly, the tentative antibody-binding determinants showed a discrepant binding to TGE virions. Moreover, the dissimilar binding ability was also related with their inhibitory effect on cell infection by TGEV.
2. Materials and methods
2.1. Cells, virus and other reagents
Swine testis (ST) cells were grown in Eagle's Minimum Essential Medium (EMEM) containing 10% newborn bovine serum (NBS, Excell Bio., China) at 37 °C in a CO2 incubator. TGEV strain PUR46-MAD (a generous gift from Dr. L. Enjuanes of CSIC-UAM Canto Blanco, Madrid, Spain) was propagated in the ST cells and passaged twice a week. The plasmid purification kit was purchased from (KeyGen Biotech, Nanjing, China). Other reagents were molecular biology grade products.
2.2. Construction of recombinant plasmids
The recombinant plasmid, pcDNA-APN encoding full-length pAPN was provided by Dr. Georg Herrler (Institute for Virology, University of Veterinary Medicine, Hannover, Germany) and used as a template for subsequent PCR amplification. The sequence of pAPN gene has been deposited in the GenBank database of NCBI and was assigned an accession no. NM_214277. Five pairs of primers were used to amplify five gene fragments encoding a signal peptide sequence-deleted pAPN and four truncated pAPN (Table 1 ). The PCR profile included 95 °C for 5 min, 30 cycles of 95 °C for 5 min, 63.2 °C for 30 s, 72 °C for 1.5 min and followed by a final extension of 72 °C for 10 min. The amplicons were inserted into the multiple cloning sites BamHI and XhoI of pET-30a(+) vector (Novagen, Germany) using standard molecular cloning techniques.
Table 1.
Primer information used in the study.
| Primer pairs | PCR product in length (bp) |
|---|---|
| Sense 5′-GGGGGGATCCGAGAAGAACAAGAATGCC-3′ | T/pAPN1 |
| Antisense 5′-CCCCCTCGAGTGGCACCGGCATTGAAGTC-3′ | 939 |
| Sense 5′-GGGGGGATCCATGAAGGCCACGTTCAAC-3′ | T/pAPN2 |
| Antisense 5′-CCCCCTCGAGTGGCACCGGCATTGAAGTC-3′ | 375 |
| Sense 5′-GGGGGGATCCATGGAGAACTGGGGGCTG-3′ | T/pAPN3 |
| Antisense 5′-CCCCCTCGAGTCACACCATTTTTAATAGA-3′ | 729 |
| Sense 5′-GGGGGGATCCATGCAGGATCACTACTGG-3′ | T/pAPN4 |
| Antisense 5′-CCCCCTCGAGTGCTGTGCTCTATGAACCA-3′ | 1116 |
| Sense 5′-GGGGGGATCCGAGAAGAACAAGAATGCC-3′ | pAPN |
| Antisense 5′-CCCCCTCGAGTGCTGTGCTCTATGAACCA-3′ | 2784 |
Primers used for amplification of pAPNs.
2.3. Expression and purification of pAPN in E. coli
The above-mentioned recombinant plasmids were transformed into E. coli BL21(DE3)pLysS (Novagen, Germany) and the transformed cells were cultured in Luria bertani (LB) medium containing Ampicillin (50 μg/ml) at 37 °C with shaking until the optical density (OD) of the culture at 600 nm reached 0.5. Isopropyl β-d-thiogalactoside (IPTG) was then added into the culture to a final concentration of 1 mM to induce pAPN expression at 37 °C for 6 h. The empty vector-transformed culture was used as control.
The purification and renaturation of inclusion bodies were performed as previously described (Liu et al., 2009). Briefly, the bacteria were pelleted at 8000 × g, at 4 °C for 5 min. The pellets were re-suspended in buffer I (50 mM Tris and 1 mM EDTA, pH 8.0) followed by digestion with lysozyme (100 μg/l) at room temperature for 30 min. The cell suspension was sonicated on ice five times, each for 30 s with 30 s intervals. After the lysate was centrifuged at 5000 rpm for 10 min, the supernatant was discarded and the pellet was re-suspended in buffer II (50 mM Tris, 0.5 mM EDTA, and 1% Triton X-100, pH 8.0). The sample was centrifuged at 10,000 rpm at 4 °C for 10 min. The precipitated inclusion bodies were washed with PBS for three times and then were re-suspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer, boiled for 10 min and centrifuged at 12,000 rpm for 5 min. The supernatant was isolated by 10% SDS-PAGE. The protein of interest was collected by gel-cutting and homogenized gel was dissolved in PBS. After freezing and thawing with liquid nitrogen twice, the samples were centrifuged at 5000 rpm for 10 min and the supernatant was collected. For protein renaturation, the purified protein-containing dialysis bag was put into 1 l renaturation solution (20 mM Tris–HCl pH 8.0, 0.1 mM glutathione of oxidized form, and 0.9 mM glutathione of reduced form) at 4 °C for 2 h, then put into 1 l new renaturation solution for 4 h, and then put into 1 l TE buffer (10 mM Tris–HCl, pH 7.5, 1 mM EDTA) for 4 h. The protein was concentrated using 50% PEG8000 at 4 °C. The concentration of purified proteins was determined using Ultrospec 3000 spectrophotometer (Pharmacia Biotech, USA) according to the manufacturer's instructions.
2.4. Western blot
Purified pAPN proteins were subjected to SDS-PAGE and transferred to a nitrocellulose (NC) membrane. The NC membrane was blocked using 5% non-fat dry milk in PBS-0.05% Tween 20 (PBST) at 4 °C overnight, then incubated with a polyclonal antibody against pAPN (1:1000 diluted in PBS) at room temperature for 2 h. After the membrane was washed three times with PBST, it was incubated with horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG at room temperature for 1 h. The protein bands were visualized using o-phenylenediamine (OPD) substrate.
2.5. Interaction between the truncated pAPNs and antibody
Recently, a polyclonal antibody against the mature pAPN protein has been prepared in our laboratory (Liu et al., 2009). In this study, the reaction between the pAPN proteins and the antibody was analyzed by indirect ELISA. Briefly, ELISA plates were coated with the purified pAPN proteins at a final concentration of 100 μg/ml (100 μl/well) at 4 °C overnight in carbonate–bicarbonate buffer (15 mM Na2CO3, 35 mM NaHCO3, pH 9.6). The wells were incubated with 5% non-fat dry milk in PBST at 37 °C for 2 h. After three times washing with PBST, the wells were incubated with the anti-pAPN antibody (1:1000 dilution in PBS) at 37 °C for 1 h. After four times washing with PBST, the plates were incubated with HRP-conjugated goat anti-rabbit IgG at 37 °C for 1 h. OPD substrate (100 μl/well) was added and incubated for 15 min after washing with PBST. After stop buffer (2 M H2SO4) was added to each well (50 μl/well), the OD490 was read using an ELISA reader.
2.6. Virus binding assay
To analyze the binding activity of virus and the pAPN proteins, TGEV Purdue 46-MAD was included in indirect ELISA. The procedure for the ELISA was performed as above, in addition to using purified TGEV particles (2 μg/well) as coating antigen followed by the successive adding of the pAPN proteins (10 μg/well), anti-pAPN antibody as well as HRP-conjugated secondary antibody. For ELISA, a negative control rabbit serum was included. The rabbit was immunized with infectious bronchitis virus (IBV) S1 protein. The protein was expressed, purified and renaturated in the same way as the pAPN proteins. The ELISA results were judged by comparing the value between the positive OD490 value of the sample (P)/negative OD490 value of control (N). The P/N value > 2 was regarded as positive.
2.7. Blockade of virus infection by the pAPNs
The blocking activity of the proteins to TGEV infection was evaluated. TGEV (1 × 106 pfu/ml) was incubated with the pAPN proteins diluted in serum-free medium at 37 °C for 1 h, and then the treated viruses at an multiplicity of infection (MOI) of 10 were infected ST cells seeded in 6-well plates at 37 °C. Virus infected cells, mock-infected cells, and viruses treated with IBV S1 protein expressed in the same expression system were included as controls. The wells were subjected to plaque assays as previously described with minor modifications (Li et al., 2009, Sui et al., 2010). Briefly, the inoculums were replaced with 1% methylcellulose in DMEM, after 1 h absorption of the viruses. Then, the cells were cultured for 48–72 h. After the overlay medium was removed, the cells were washed three times with PBS and fixed with 3% paraformaldehyde in PBS for 30 min at room temperature. Then, the cells were stained with 1% crystal violet (v/v) diluted in 5% ethanol for 20 min at room temperature. The clear plaque number was counted post-washing. All the experiments were performed in triplicate.
3. Results
3.1. Cloning of truncated pAPN genes
The recombinant plasmids encoding five domains in the pAPN were constructed by conventional DNA recombination technique. The correctness of the inserts was confirmed by DNA sequencing. The identified recombinant plasmids encoding the fragments (106–2889 nt), (106–1044 nt), (670–1044 nt), (1045–1773 nt), and (1774–2889 nt) in the full-length pAPN gene were designated as pAPN, truncated (T)/pAPN1, T/pAPN2, T/pAPN3 and T/pAPN4, respectively. The schematic drawing of the pAPN proteins was shown in Fig. 1 .
Fig. 1.
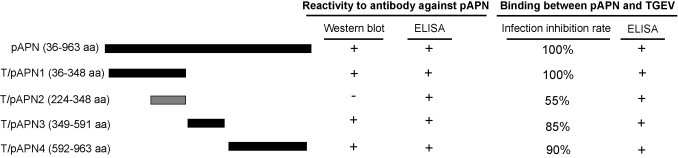
Schematic drawing of the truncated pAPN constructs and functional activities. Five recombinant plasmids bearing either the signal peptide sequence-deleted pAPN (pAPN) or four truncated pAPN (T/pAPNs) were constructed using conventional molecular cloning techniques. The name and insert length of the constructs are indicated. Their ability in binding to either an anti-pAPN antibody or TGE virions are indicated inferred from the results of Western blot, plaque asssays or ELISA. “+” means positive reaction or effect; “−” means negative result. The infection inhibition rate shows the maximum inhibition ability of the pAPN proteins to cell infection by TGEV. It should be noted that the sizes of the lines are not proportional to the length of the amino acid chain. The details on the inhibition ability of the pAPN proteins to TGEV infection or binding to the antibody or virions are shown in other figures.
3.2. Immunoblotting analysis of expressed pAPNs
The expression of the pAPN has been reported (Liu et al., 2009). Four truncated pAPN proteins were expressed in the form of inclusion bodies in E. coli. The expressed proteins were isolated in SDS-PAGE, purified by gel-purification and re-natured. Subsequently, they were transferred onto the NC membrane and identified by specific antibody against pAPN. As shown in Fig. 2 , in addition to T/pAPN2, other truncated pAPN proteins were detectable in the Western blot analysis.
Fig. 2.
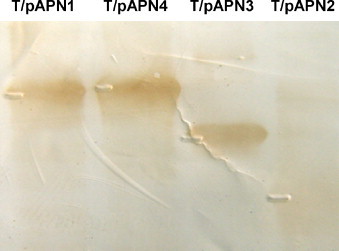
Immunoblotting analysis of the pAPN proteins. After the truncated pAPN proteins were transferred onto a nitrocellulose membrane, the membrane was incubated with the anti-pAPN antibody and followed by the incubation of HRP-conjugated secondary antibody. The blot result is shown.
3.3. Protein-based ELISA
To confirm the results from Western blot, indirect ELISA was performed using the recombinant proteins as detected antigens, followed by the addition of the primary antibody and HRP-labeled secondary antibody. The results showed that truncated T/pAPN1, 3 and 4 proteins had stronger reactivity with the specific antiserum than T/pAPN2; however, the pAPN had the highest reaction ability with the antibody (Fig. 3 ).
Fig. 3.
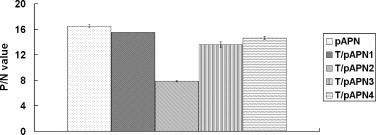
Binding of the pAPNs to anti-pAPN antibody. The pAPN proteins were used as coating antigen and then they were incubated with anti-pAPN antibody (positive sample) or anti-IBV S1 antibody (negative control) followed by addition of HRP-conjucated secondary antibody. The OD490 was read. The P/N value = the postive sample value/negative control value, which is indicated in the y-axis.
3.4. Binding of the truncated pAPN proteins to TGEV
Using indirect ELISA, the binding activity between the truncated pAPN proteins and TGEV was analyzed. The results showed that all the truncated proteins reacted with TGEV particles; however, the binding activity between T/pAPN2 was the lowest among the pAPN proteins tested in this study (Fig. 4 ).
Fig. 4.
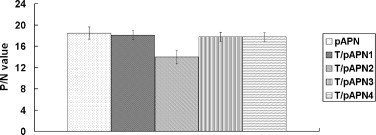
Binding of the pAPNs to TGE virions. The purified TGEV particles were coated as antigens and then incubated with the pAPN proteins followed by indirect ELISA. The anti-pAPN serum (positive sample) or anti-IBV S1 antibody (negative control) as primary antibody followed by addition of HRP-conjugated secondary antibody. The The P/N value measured at OD490 is indicated in y-axis.
3.5. Effect of pAPN proteins on TGEV infection in vitro
The inhibitory effect of the truncated pAPN proteins to cell infection by TGEV was further investigated. Virus plaque-reduction assays indicated that all the proteins blocked the virus infection in a dose-dependent manner and the truncated T/pAPN1, 3 and 4 proteins showed more than 85% inhibition rate (Fig. 5 ). In contrast, the maximal inhibition rate of T/pAPN2 was approximately 55%.
Fig. 5.
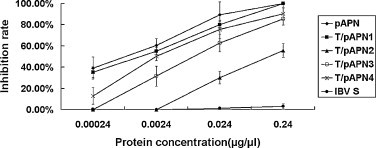
Blockade of TGEV infection by the pAPNs. TGE virons were treated with pAPN proteins at various concentrations and then the viruses were used to infect ST cells. A control protein (IBV S1) produced in the same system as pAPN was used as control to treat viruses. The protein concentration is shown in x-axis and the inhibition rate of the pAPNs determined by plaque assays is indicatd in y-axis. The 100% infectivity value corresponds to an average plaque number of approx. 200.
4. Discussion
The purpose of the current study allows the identification of the functional domains of pAPN. In an earlier study, we expressed the full-length pAPN without the signal peptide and the protein has been used to raise specific polyclonal antiserum (Liu et al., 2009). In this study, we expressed four truncated proteins covering different regions of the whole mature pAPN. The recognizing of the anti-pAPN antibody to the proteins indicated that three regions, 36aa–223aa, 349aa–591aa and 592–963aa were the major antibody-binding domains. This was firstly identified by Western blot and then confirmed by ELISA. The bacterial expression system is very optimal for protein expression due to its advantages consisting of low cost, convenience and high fermentation potential and numerous heterologous proteins have been expressed in this system (Kataoka et al., 2004, Hamann and Lange, 2006, Yin et al., 2007, Hino et al., 2008, Cai et al., 2009, Ren et al., 2010a, Ren et al., 2010b, Li et al., 2010, Meng et al., 2010). In this study, the specificity of the polyclonal antibody raised against the bacterially expressed pAPN was clarified by including a control rabbit antibody generated by immunizing IBV S1 protein expressed in the same way as the pAPN proteins. It is known that pAPN serves as a cellular receptor for several group I coronaviruses including HCoV-229E, TGEV, FIPV and canine coronavirus (CCV) (Delmas et al., 1992, Yeager et al., 1992, Tresnan et al., 1996). Therefore, in addition to investigation on the binding between the truncated proteins with anti-pAPN antibody, the binding ability between TGE virions and the proteins was compared. Our results indicated that other three truncated pAPN proteins had stronger binding activity than T/pAPN2, suggesting the binding domains of pAPN identified in this study are important functional sites for recognizing antibody and interacting with TGEV. Since the truncated pAPN proteins react with the pAPN antibody to different extent, the identified domains in the pAPN may represent different epitopes in the pAPN.
The TGEV S protein is responsible for initiating the cell infection by interacting with the cellular receptor, pAPN. In this study, we analyzed the neutralizing effect of the truncated pAPN on cell infection by TGEV. Our data showed that the different amino acid regions had discrepant inhibition abilities. Among the four truncated pAPNs, the region (T/pAPN1), amino acids 36–348 showed the best inhibitory effect on TGEV infection, and this region also had the strongest binding activity with the anti-pAPN serum and viruses. Therefore, we suppose that this region should contain the major receptor sites for TGEV and neutralization determinants. Because this region encompasses the T/pAPN2 that showed a poor binding activity, the determinant should be localized in the amino acids 36–223. It has been shown that, analogously to the human APN protein, a region within the amino-terminal part of the feline APN protein (encompassing amino acids 132–295) is essential for its receptor function of human CoV (Kolb et al., 1997). There are some overlaid regions between the identified determinants among pAPN, hAPN and fAPN. At the same time, the APN sequences from different species share a high homologous identity (70–80% amino acid identity) (Tusell et al., 2007). Therefore, the binding determinant domains of pAPN may be also important in the counterpart of hAPN and fAPN. As far as the ability of the pAPN proteins to neutralize TGEV infection in vitro is concerned, the current results, together with our previous finding indicated that both full-length pAPN and the amino-terminal part of pAPN were able to neutralize TGEV completely (Liu et al., 2009). However, the binding activity of the truncated pAPNs to TGEV or pAPN-antiserum was lower than matured pAPN protein in our ELISA analysis. This result gave rise to another possibility that the complementation of other domains in the pAPN may be required in virus neutralization and antibody binding. In addition, it has been proved that four major antigenic sites of TGEV S protein is in the half of its amino terminus (S1), it would be therefore interesting to analyze the interaction between TGEV S1 and the truncated pAPN proteins in the future.
Most APN proteins serve as a species-specific receptor for coronaviruses, for example, human APN only mediates the infection of human coronavirus 229E, and pAPN exclusively mediates TGEV infection (Hegyi and Kolb, 1998). This principle is not always true for several APN proteins in terms of their roles as species-specific receptors for coronaviruses. Transfection of fAPN into cells renders the infection of TGEV, it is therefore suggestive that fAPN is another receptor for TGEV (Tresnan et al., 1996, Kolb et al., 1997). It has been reported that aa 704–831 of fAPN was required for entry of TGEV, feline coronavirus (FCoV), and CCV, particularly, mutational analysis of APN indicated that the amino acid region 732–746 in fAPN is indispensable for TGEV entry (Tusell et al., 2007). In our study, the homologous region in pAPN was identified as one of the major binding regions (T/pAPN4) in ELISA, and it blocked TGEV infection in vitro, confirming the importance of the functional domain. Previously, fAPN has been identified as a receptor for an avian coronavirus, namely, infectious bronchitis virus (IBV). The assertion was based on the observation that the hamster kidney fibroblasts became permissive to IBV strain Ark 99 after transfection with a fAPN cDNA (Miguel et al., 2002). Interestingly, both transient transfection and stable expression of fAPN on the same cell line (BHK cells) rescued FIPV and TGEV infection in non-permissive BHK cells; however, fAPN expression did not rescue infection by the prototype IBV strain Mass41 (Chu et al., 2007). In another report, TGEV was found to use a human/bovine APN chimera as a receptor, although it is unable to replicate in bovine cells (Benbacer et al., 1997). These paradoxical results complicate the delineation of APN as a cellular receptor for virus infection. Additionally, there are amino acid differences in the APN proteins from different species, therefore, more virus strains, cell lines as well as related receptor genes should be analyzed to completely elucidate the mechanism of coronavirus infection in the context of receptor usage.
Acknowledgements
National Natural Science Foundation of China (30700590; 30972195), Cultivation Fund of the Key Scientific and Technical Innovation Project, Ministry of Education of China (NO706019), Funding supported by Program for New Century Excellent Talents in Heilongjiang Provincial University (1155-NCET-005), Heilongjiang Provincial Science and Technology Department, China (ZJN0702-01) and Northeast Agricultural University, China (CXZ008-1) are acknowledged.
References
- Benbacer L., Kut E., Besnardeau L., Laude H., Delmas B. Interspecies aminopeptidase-N chimeras reveal species-specific receptor recognition by canine coronavirus, feline infectious peritonitis virus, and transmissible gastroenteritis virus. J. Virol. 1997;71:734–737. doi: 10.1128/jvi.71.1.734-737.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L., Chen H., Miao Q., Wu S., Shang Y., Zhen Y. Binding capability of the enediyne-associated apoprotein to human tumors and constitution of a ligand oligopeptide-integrated protein. J. Biotechnol. 2009;144:142–150. doi: 10.1016/j.jbiotec.2009.09.001. [DOI] [PubMed] [Google Scholar]
- Chu V.C., McElroy L.J., Aronson J.M., Oura T.J., Harbison C.E., Bauman B.E., Whittaker G.R. Feline aminopeptidase N is not a functional receptor for avian infectious bronchitis virus. Virol. J. 2007;4:20. doi: 10.1186/1743-422X-4-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmas B., Gelfi J., Kut E., Sjostrom H., Noren O., Laude H. Determinants essential for the transmissible gastroenteritis virus-receptor interaction reside within a domain of aminopeptidase-N that is distinct from the enzymatic site. J. Virol. 1994;68:5216–5224. doi: 10.1128/jvi.68.8.5216-5224.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmas B., Gelfi J., L’Haridon R., Vogel L.K., Sjöström H., Norén O., Laude H. Aminopeptidase N is a major receptor for the entero-pathogenic coronavirus TGEV. Nature. 1992;357:417–420. doi: 10.1038/357417a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enjuanes L., Suñé C., Gebauer F., Smerdou C., Camacho A., Antón I.M., González S., Talamillo A., Méndez A., Ballesteros M.L., Sánchez C. Antigen selection and presentation to protect against transmissible gastroenteritis coronavirus. Vet. Microbiol. 1992;33:249–262. doi: 10.1016/0378-1135(92)90053-V. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebauer F., Posthumus W.P., Correa I., Suñé C., Smerdou C., Sánchez C.M., Lenstra J.A., Meloen R.H., Enjuanes L. Residues involved in the antigenic sites of transmissible gastroenteritis coronavirus S glycoprotein. Virology. 1991;183:225–238. doi: 10.1016/0042-6822(91)90135-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamann T., Lange L. Discovery, cloning and heterologous expression of secreted potato proteins reveal erroneous pre-mRNA splicing in Aspergillus oryzae. J. Biotechnol. 2006;126:265–276. doi: 10.1016/j.jbiotec.2006.04.026. [DOI] [PubMed] [Google Scholar]
- Hegyi A., Kolb A.F. Characterization of determinants involved in the feline infectious peritonitis virus receptor function of feline aminopeptidase N. J. Gen. Virol. 1998;79:1387–1391. doi: 10.1099/0022-1317-79-6-1387. [DOI] [PubMed] [Google Scholar]
- Hino M., Kataoka M., Kajimoto K., Yamamoto T., Kido J., Shinohara Y., Baba Y. Efficiency of cell-free protein synthesis based on a crude cell extract from Escherichia coli, wheat germ, and rabbit reticulocytes. J. Biotechnol. 2008;133:183–189. doi: 10.1016/j.jbiotec.2007.08.008. [DOI] [PubMed] [Google Scholar]
- Hooper N.M. Families of zinc metalloproteases. FEBS Lett. 1994;354:1–6. doi: 10.1016/0014-5793(94)01079-x. [DOI] [PubMed] [Google Scholar]
- Kataoka M., Kotaka A., Thiwthong R., Wada M., Nakamori S., Shimizu S. Cloning and overexpression of the old yellow enzyme gene of Candida macedoniensis, and its application to the production of a chiral compound. J. Biotechnol. 2004;114:1–9. doi: 10.1016/j.jbiotec.2004.04.033. [DOI] [PubMed] [Google Scholar]
- Kolb A.F., Hegyi A., Siddell S.G. Identification of residues critical for the human coronavirus 229E receptor function of human aminopeptidase N. J. Gen. Virol. 1997;78:2795–2802. doi: 10.1099/0022-1317-78-11-2795. [DOI] [PubMed] [Google Scholar]
- Krempl C., Ballesteros M.L., Zimmer G., Enjuanes L., Klenk H.D., Herrler G. Characterization of the sialic acid binding activity of transmissible gastroenteritis coronavirus by analysis of haemagglutination-deficient mutants. J. Gen. Virol. 2000;81:489–496. doi: 10.1099/0022-1317-81-2-489. [DOI] [PubMed] [Google Scholar]
- Krempl C., Herrler G. Sialic acid binding activity of transmissible gastroenteritis coronavirus affects sedimentation behavior of virions and solubilized glycoproteins. J. Virol. 2001;75:844–849. doi: 10.1128/JVI.75.2.844-849.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krempl C., Schultze B., Laude H., Herrler G. Point mutations in the S protein connect the sialic acid binding activity with the enteropathogenicity of transmissible gastroenteritis coronavirus. J. Virol. 1997;71:3285–3287. doi: 10.1128/jvi.71.4.3285-3287.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laude H., Reeth K.V., Pensaert K.A. Porcine respiratory coronavirus: molecular features and virus-host interactions. Vet. Res. 1993;24:125–150. [PubMed] [Google Scholar]
- Li G., Zeng Y., Yin J., Lillehoj H.S., Ren X. Cloning, prokaryotic expression, and biological analysis of recombinant chicken IFN-gamma. Hybridoma (Larchmt) 2010;29:1–6. doi: 10.1089/hyb.2009.0053. [DOI] [PubMed] [Google Scholar]
- Li J., Yin J., Sui X., Li G., Ren X. Comparative analysis on the effect of glycyrrhizin diammonium and lithium chloride on infectious bronchitis virus infection in vitro. Avian Pathol. 2009;38:215–221. doi: 10.1080/03079450902912184. [DOI] [PubMed] [Google Scholar]
- Liu B., Li G., Sui X., Yin J., Wang H., Ren X. Expression and functional analysis of porcine aminopeptidase N produced in prokaryotic expression system. J. Biotechnol. 2009;141:91–96. doi: 10.1016/j.jbiotec.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng F., Yin J., Li X., Yang W., Li G., Ren X. Production and characterization of a monoclonal antibody against spike protein of transmissible gastroenteritis virus. Hybridoma (Larchmt) 2010;29(4) doi: 10.1089/hyb.2010.0009. doi:10.1089/hyb.2010.0009. [DOI] [PubMed] [Google Scholar]
- Miguel B., Pharr G.T., Wang C. The role of feline aminopeptidase N as a receptor for infectious bronchitis virus. Arch. Virol. 2002;147:2047–2056. doi: 10.1007/s00705-002-0888-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penzes Z., González J.M., Calvo E., Izeta A., Smerdou C., Mendez A., Sánchez C.M., Sola I., Almazán F., Enjuanes L. Complete genome sequence of transmissible gastroenteritis coronavirus PUR46-MAD clone and evolution of the Purdue virus cluster. Virus Genes. 2001;23:105–118. doi: 10.1023/A:1011147832586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X., Wang M., Yin J., Ren Y., Li G. Importance of cholesterol for infection of cells by transmissible gastroenteritis virus. Virus Res. 2008;137:220–224. doi: 10.1016/j.virusres.2008.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X., Wang M., Yin J., Ren Y., Li G. Heterologous expression of fused genes encoding the glycoprotein 5 from PRRSV: a way for producing functional protein in prokaryotic microorganism. J. Biotechnol. 2010;147:130–135. doi: 10.1016/j.jbiotec.2010.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X., Wang M., Yin J., Ren Y., Li G. Phages harboring specific peptides to N protein of PRRSV distinguish the virus from other viruses. J. Clin. Microbiol. 2010;48:1875–1881. doi: 10.1128/JCM.01707-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwegmann-Wessels C., Zimmer G., Schröder B., Breves G., Herrler G. Binding of transmissible gastroenteritis coronavirus to brush border membrane sialoglycoproteins. J. Virol. 2003;77:11846–11848. doi: 10.1128/JVI.77.21.11846-11848.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaan W., Cavanagh D., Horzinek C. Coronaviruses: structure and genome expression. J. Gen. Virol. 1988;69:2939–2952. doi: 10.1099/0022-1317-69-12-2939. [DOI] [PubMed] [Google Scholar]
- Sui X., Yin J., Ren X. Antiviral effect of diammonium glycyrrhizinate and lithium chloride on cell infection by pseudorabies herpesvirus. Antiviral Res. 2010;85:346–353. doi: 10.1016/j.antiviral.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tresnan D.B., Holmes K.V. Feline aminopeptidase N is a receptor for all group I coronaviruses. Adv. Exp. Med. Biol. 1998;440:69–75. doi: 10.1007/978-1-4615-5331-1_9. [DOI] [PubMed] [Google Scholar]
- Tresnan D.B., Levis R., Holmes K.V. Feline aminopeptidase Nserves as a receptor for feline, canine, porcine, and human coronaviruses in serogroup I. J. Virol. 1996;70:8669–8674. doi: 10.1128/jvi.70.12.8669-8674.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusell S.M., Schittone S.A., Holmes K.V. Mutational analysis of aminopeptidase N, a receptor for several group 1 coronaviruses, identifies key determinants of viral host range. J. Virol. 2007;81:1261–1273. doi: 10.1128/JVI.01510-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeager C.L., Ashmun R.A., Williams R.K., Cardellichio C.B., Shapiro L.H., Look A.T., Holmes K.V. Human aminopeptidase N is a receptor for human coronavirus 229E. Nature. 1992;357:420–422. doi: 10.1038/357420a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J., Li G., Ren X., Herrler G. Select what you need: a comparative evaluation of the advantages and limitations of frequently used expression systems for foreign genes. J. Biotechnol. 2007;127:335–347. doi: 10.1016/j.jbiotec.2006.07.012. [DOI] [PubMed] [Google Scholar]


