Abstract
The recombinant 3C-like protease of Chiba virus, a Norovirus, expressed in Escherichia coli cells was purified and characterized as to effects of pH, temperature, salt contents, and SH reagents on its proteolytic activity. The optimal pH and temperature of the 3C-like protease for the proteolytic activity were 8.6 and 37 °C, respectively. Increased concentration (∼100 mM) of monovalent cations such as Na+ and K+ was inhibitory to the activity. Hg2+ and Zn2+ remarkably inhibited the protease activity, while Mg2+ and Ca2+ had no virtual effect. Several sulfhydryl reagents such as p-chloromercuribenzoic acid, methyl methanethiosulfonate, N-ethylmaleimide and N-phenylmaleimide also blocked the activity, confirming the previous result that cysteine residue(s) were responsible for the proteolysis.
Keywords: 3C-like protease, Chiba virus, Norovirus, SH reagents, Proteolytic activity
1. Introduction
Viral proteases play a central role in the maturation of functional viral proteins, and hence in its genome replication and the formation of virus particles (Dougherty and Semler, 1993). The 3C and 3C-like proteases belonging to the chymotrypsin-like protease superfamily are found in several animal, insect, and plant virus families such as Picornaviridae, Comoviridae, Caliciviridae, Coronaviridaee, and Arteriviridae (Dougherty and Semler, 1993, Gorbalenya et al., 1989). Their three-dimensional structures have been determined by X-ray crystallography in poliovirus (Mosimann et al., 1997), rhinovirus (Matthews et al., 1994), hepatitis A virus (Bergmann et al., 1997), and coronavirus (Anand et al., 2002, Anand et al., 2003, Yang et al., 2003), tobacco etch virus (Phan et al., 2002) and equine arteritis virus (Barrette-Ng et al., 2002). These studies allowed us to identify the active-site amino acid residues with a combination of site-directed mutagenesis studies (Boniotti et al., 1994, Cheah et al., 1990, Gosert et al., 1997, Hammerle et al., 1991). Several 3C and 3C-like proteases were purified and their biochemical properties were well characterized (Baum et al., 1991, Chisholm et al., 2001, Davis et al., 1997, Ziebuhr et al., 1997). They are characterized by their proteolytic activities being inhibited by both cysteine and serine protease inhibitors. As a result of these studies, therapeutic drugs, especially for rhinovirus, are being developed (Hammerle et al., 1991, McKinlay, 2001, Turner, 2001).
Norovirus (formerly Norwalk-like virus) is a genus of the family Caliciviridae and is a major causative agent of non-bacterial acute gastroenteritis in humans (Clarke et al., 1998, Estes et al., 1997). Norovirus cannot be propagated in cell cultures, and animal models have not been developed, which hampers the progress of biochemical and molecular biological studies of Norovirus. We cloned and sequenced the whole genome of a Chiba strain of Norovirus (Chiba virus), and its 3C-like protease has been heterologously expressed in Escherichia coli in an active form (Someya et al., 2000). Furthermore, we identified the active-site amino acid residues and concluded that Norovirus 3C-like protease had a catalytic dyad consisting of His30 and Cys139 (Someya et al., 2002). This is a unique feature since most of the 3C and 3C-like proteases have a catalytic triad consisting of His, Asp/Glu, and Cys/Ser, similar to chymotrypsin (Dougherty and Semler, 1993, Gorbalenya et al., 1989). More recently, it was shown that the active site of Coronavirus 3C-like protease was also composed of two residues, His and Cys (Anand et al., 2002).
To obtain information regarding the biochemical properties of norovirus 3C-like protease, we purified the bacterially expressed recombinant 3C-like protease from Chiba virus and characterized as to effects of pH, temperature, salt contents, and SH reagents on its activity.
2. Materials and methods
2.1. Construction of plasmids
The DNA fragment encoding all residues (Ala1 to Glu181) of the Chiba virus 3C-like protease was amplified by PCR, in that NdeI and Aor51HI restriction sites were introduced at the 5′ and 3′ ends, respectively. The NdeI–Aor51HI fragment was placed downstream of the T7 promoter with a linker encoding a sequence containing six consecutive His residues. The resultant plasmid, pT7ProHis, encodes a 3C-like protease with a Met residue in the N-terminal and with a SAGHHHHHHG sequence in the C-terminal. The NdeI–SphI fragment from pUCHis3CD (Someya et al., 2000) encoding the 3CD region was placed downstream of the T7 promoter to construct pT7His3CD. Next, the C139A mutation of the 3C-like protease was introduced and designated pT7His3CD-C139A. The DNA fragment encoding glutathione S-transferase (GST) was amplified by PCR, in that ApaI and SphI restriction sites were introduced at the 5′ and 3′ ends, respectively. The ApaI–SphI fragment encoding GST was ligated to the ApaI–SphI fragment of pT7His3CD-C139A. The resultant plasmid was designated pT7His3Cd-GST. This plasmid encodes the N-terminal His-tag sequence (MGGHHHHHHGASA), followed by the whole sequence of the C139A mutant 3C-like protease and Gly1 to Pro19 of 3D RNA-dependent RNA polymerase, and then followed by GST.
2.2. Purification of proteins
E. coli BL21-CodonPlus (DE3)-RIL cells (Stratagene, La Jolla, CA) were transformed with pT7ProHis or pT7His3Cd-GST. Cells were grown on medium A (McMurry et al., 1980) supplemented with 0.2% glucose and 0.1% casamino acids at 37 °C. When the absorbance at 530 nm reached 0.5, IPTG was added to a final concentration of 1 mM for induction of gene expression. After 2 h, cells were harvested, then washed and resuspended with 20 mM Tris–HCl (pH 8.0) buffer containing 0.1 M NaCl. Cells were disrupted by sonication. After the removal of cell debris, the supernatant was subjected to ultracentrifugation (150,000 × g, 1 h). The resultant supernatant was mixed with TALON Metal Affinity Resin (CLONTECH, Palo Alto, CA) equilibrated with 20 mM Tris–HCl (pH 8.0) buffer containing 0.1 M NaCl. The resin was washed with 20 mM Tris–HCl (pH 8.0) buffer containing 0.1 M NaCl and 0.1 M imidazole for removal of non-specifically bound proteins. The proteins of interest were eluted with 20 mM Tris–HCl (pH 8.0) buffer containing 0.1 M NaCl and 0.5 M imidazole and concentrated using Ultrafree-15 unit with Biomax-10 membrane (MILLIPORE, Bedford, MA). Proteins were determined by the method of Bradford (Bradford, 1976).
2.3. Proteolytic reaction
Usually, 5 μM enzyme and 5 μM substrate were reacted in 20 μl of the mixture containing 50 mM bis–tris–propane–HCl buffer (pH 8.62) for 16 h. Details of reaction mixtures were described in the respective sections. The reaction was stopped by the addition of an equal volume of 2× sample buffer, and the proteins were then analyzed by SDS-polyacrylamide (13%) gel electrophoresis. Gels were stained with Coomassie Brilliant Blue R-250. In order to estimate the efficiency of the proteolytic cleavage, density of the respective stained bands was scanned and calculated using NIH Image for Macintosh version 1.62.
3. Results and discussion
3.1. Choice of the substrate protein
The viral non-structural proteins and the proteolytic cleavage sites within the ORF1 polyprotein are depicted in Fig. 1A. In order to purify the Chiba virus 3C-like protease, we constructed pT7ProHis with the gene fragment encoding the protease with His-tag in its C-terminus (Fig. 1B). The ProHis protein (calculated molecular mass of 20.6 kDa) expressed from pT7ProHis was used as an enzyme in this study. In order to observe proteolysis at the cleavage site between the 3C and 3D, we at first used the His-3CD-C139A mutant protein expressed from pT7His3CD-C139A as a substrate, which was the N-terminal His-tagged 3CD fragment containing the Ala mutation of active-site Cys139 of the 3C-like protease (Fig. 1B). However, this protein tended to aggregate at higher pH after prolonged incubation. Other parts from the native Chiba virus ORF1 polyprotein could not be effectively produced in E. coli cells. Therefore, the His3Cd-GST protein (calculated molecular mass of 48.0 kDa) was used as a substrate, which retains the cleavage site between the 3C and 3D. The region covering the entire 3C and the N-terminal 19 amino acids of 3D was placed under the control of T7 promoter with the N-terminal His-tag and the C-terminal glutathione S-transferase (GST), the resultant plasmid being designated pT7His3Cd-GST (Fig. 1B). Although this protein itself has the 3C-like protease moiety, it never undergoes autocatalytic cleavage because the C139A mutation is introduced. If His3Cd-GST is cleaved at the 3C/3D junction (LE/GG) by the active ProHis, the His3C-C139A moiety of 20.7 kDa and the 3D′-GST moiety of 27.3 kDa are produced. Enzymatic reaction was initiated by mixing enzyme (ProHis) and substrate (His3Cd-GST). We could not obtain clear results when the reaction was performed in a short period (2–4 h) (data not shown). However, even if a substrate was mixed with an enzyme for a long period (more than 24 h), complete cleavage could not be achieved. One possibility for the incomplete cleavage is that the cleavage site of the substrate (His3Cd-GST) could not be accessible by the protease because the substrate was not properly folded. Alternatively, part of the cleavage site in the substrate might be masked by its own catalytic site or the catalytic site of other substrate molecule, so that the enzyme might have competed with the substrate for the cleavage site.
Fig. 1.
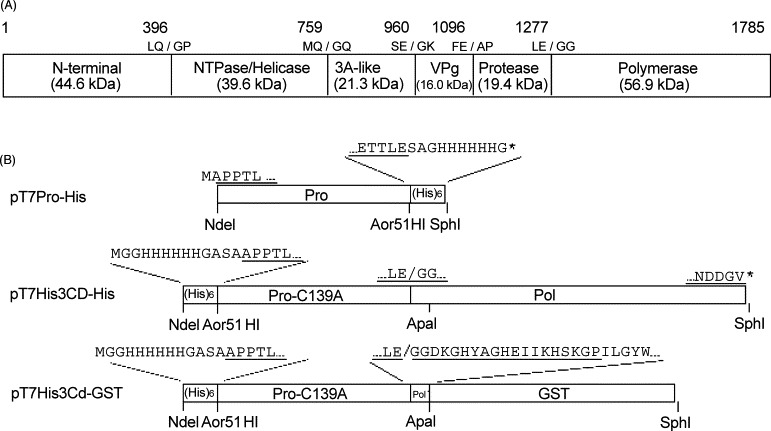
Construction of plasmids. (A) Map of the proteolytic cleavage sites within the ORF1 polyprotein. The numbers indicate the positions of the amino acids. Slashes indicate the cleavage sites recognized by the protease. The calculated molecular masses of the cleavage products are indicated. (B) Construction of the expression plasmids. Details are described in Section 2. Amino acids derived from the Chiba virus sequence are underlined.
Very recently, Belliot et al. (2003) reported trans cleavage activity of the 3C-like protease from the genogroup II MD145 Norovirus (Belliot et al., 2003). They used bacterially expressed 3C-like protease with its C-terminus His-tagged as an enzyme and the entire ORF1 protein or 3CD fragment containing the active-site Cys mutation as a substrate which was expressed in the in vitro transcription/translation reaction. Cleavage reaction was performed in the pH 7.4 buffer at 30 °C. Data showed that the 3C-like protease cleaved not only the 3C/3D junction but also other proposed junctions, and that substrate protein appeared to be cleaved completely after 24-h incubation. However, consistent with our experiment, a long time incubation was required for the result being obvious.
3.2. Optimal pH of the proteolytic activity
Five micromolar enzyme and 5 μM substrate were mixed in 50 mM bis–tris–propane–HCl buffer of various pH containing 25 mM NaCl, and reacted at 37 °C for 16 h. Fig. 2 shows that maximum cleavage was obtained at pH 8.62, although the proteolytic cleavage occurred over a wide range of pH. Faint bands of 80 and 60 kDa were thought to be dimers of the substrate and its cleaved product (3D′-GST), respectively. When the efficiency of cleavage at pH 8.62 is 100%, those at pH 7.22, 7.66, 8.12, and 9.08 were estimated as 70, 86, 89, and 79%, respectively. A slope of relative activity was gentle at acidic pH below 8.62, while it was sharp at alkaline pH above 8.62. In the subsequent experiments, a buffer of pH 8.62 was used.
Fig. 2.
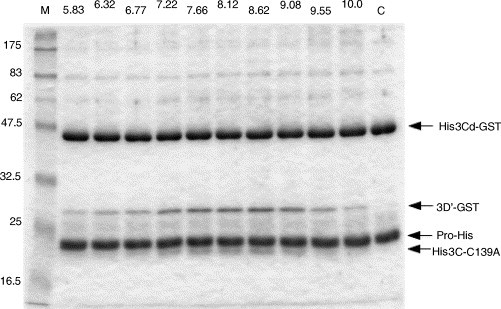
Effects of pH on protease activity. Five micromolar enzyme (ProHis) and 5 μM substrate (His3Cd-GST) were reacted in the 50 mM bis–tris–propane–HCl buffer of various pH (indicated at the top of the respective lanes) containing 25 mM NaCl at 37 °C for 16 h. M and C indicate molecular weight marker and reaction mixture at 0 time, respectively. His3Cd-GST was cleaved into 3D′-GST and His3C-C139A by ProHis.
3.3. Optimal temperature of the proteolytic activity
Five micromolar enzyme and 5 μM substrate were mixed in 50 mM bis–tris–propane–HCl (pH 8.62) containing 25 mM NaCl, and reacted for 16 h at 4, 15, 26, 37, or 48 °C. As shown in Fig. 3 , optimal temperature for the activity was 37 °C. The activity at 4, 15, 26, and 48 °C was estimated as 23, 31, 55, and 80% of that at 37 °C, respectively.
Fig. 3.
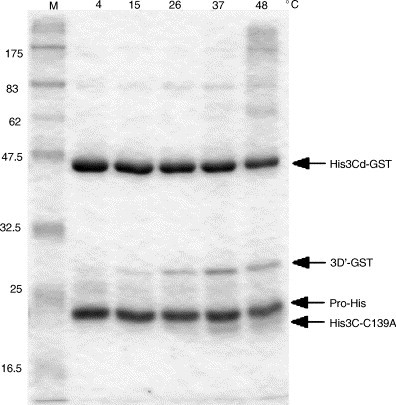
Effects of temperature on protease activity. Five micromolar enzyme (ProHis) and 5 μM substrate (His3Cd-GST) were reacted in the 50 mM bis–tris–propane–HCl (pH 8.62) buffer containing 25 mM NaCl for 16 h at the indicated temperature.
3.4. Effects of monovalent and divalent cations on proteolytic activity
We next examined the effects of monovalent and divalent cations on the proteolytic activity. The activity was compared with that under the condition where 50 mM bis–tris–propane–HCl (pH 8.62) containing 5 mM EDTA without any cations was used. The addition of 25, 50, or 100 mM NaCl decreased the proteolytic activity to 83, 52, or 36%, respectively (Fig. 4A). KCl also had an inhibitory effect on the activity (data not shown). As for the divalent cations, 5 mM MgCl2 or 5 mM CaCl2 did not affect the activity (108 and 105%) (Fig. 4B). In contrast, 5 mM ZnCl2 or 5 mM HgCl2 completely inhibited the activity (Fig. 4B). This result was consistent with Cys being a critical residue of the active site (Someya et al., 2002), and inhibition by Zn2+ indicated that Norovirus 3C-like protease was different from metalloprotease. When Hg2+ was used, two faint bands of 30 and 15 kDa slightly appeared (lane HgCl2, Fig. 4B). It was possible that high concentrations of Hg2+ allowed the enzyme to alternatively cleave the substrate and/or the enzyme itself. With the addition of 5 mM MnCl2, part of the proteins used for reaction appeared to be aggregated since Coomassie-stained materials stayed in the well (Fig. 4B). Because Mn2+ oxidized the proteins, the part may have remained in the well and the other part may be flown from the gel after degradation into small molecular weight peptides.
Fig. 4.
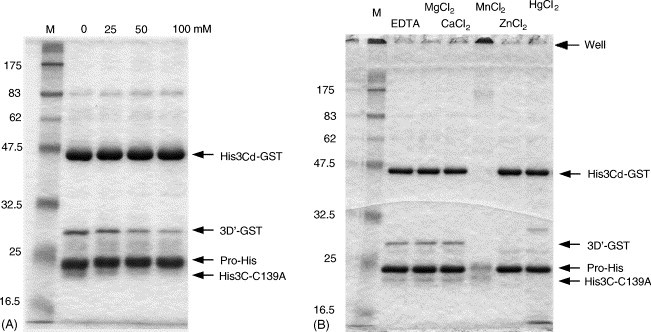
Effects of NaCl (A) and various divalent cations (B) on protease activity. Five micromolar enzyme (ProHis) and 5 μM substrate (His3Cd-GST) were reacted in the 50 mM bis–tris–propane–HCl (pH 8.62) buffer containing the indicated components at 37 °C for 16 h. (A) Reaction mixture contained 5 mM EDTA and the indicated concentration of NaCl. Essentially the same result was obtained when KCl was added instead of NaCl (data not shown). (B) Reaction mixture contained the indicated reagent of 5 mM.
3.5. Effects of sulfhydryl reagents on the proteolytic activity
Most of the 3C and 3C-like proteases have a cysteine residue as a nucleophile of the active site (Dougherty and Semler, 1993, Gorbalenya et al., 1989). These proteases should therefore be sensitive to sulfhydryl reagents. In fact, as shown in Fig. 4B, high concentrations (5 mM) of Hg2+ and Zn2+ blocked the activity of the 3C-like protease. In Fig. 5 , the effects of several sulfhydryl reagents were tested. Five micromolar substrate was reacted with 5 μM enzyme in the presence of 10 μM sulfhydryl reagents. Ten micromolar Hg2+ or Zn2+ almost completely inhibited the activity. p-Chloromercuribenzoate (PCMB), which includes an Hg(II) within the molecule, was also a good inhibitor of the 3C-like protease (82% inhibition). In contrast, p-chloromercuribenzenesulfonate (PCMBS) slightly inhibited the activity (27% inhibition), despite PCMBS also having an Hg(II) similarly to PCMB. Methyl methanethiosulfonate (MMTS), N-ethylmaleimide (NEM), and N-phenylmaleimide (NPM), which form a covalent bond with a sulfur atom of the cysteine residue, almost completely inhibited the activity. The reaction with NEM and NPM slightly altered the mobility of the enzyme molecules, possibly because a bulkier modified side chain of the active-site cysteine residue affected the structure of the enzyme, or because more than one molecule of the inhibitors attached to an enzyme. Neither 10 μM iodoacetate (IAA) nor iodoacetamide (IAM) inhibited the activity. These two reagents may be less sensitive to an SH group than other modifying reagents. Phenyl methylsulfonylfluoride (PMSF) is well known as an irreversible inhibitor of serine proteases. Because the 3C and 3C-like proteases have a structural architecture similar to chymotrypsin, and with essentially the same proteolysis mechanism (Dougherty and Semler, 1993, Gorbalenya et al., 1989), PMSF may inhibit the activity of the 3C and 3C-like proteases. Ten micromolar PMSF was less effective but significantly inhibited the activity of the Chiba virus 3C-like protease (20% inhibition). The effect of β-mercaptoethanol (β-ME) was also examined here. The residual activity of the 3C-like protease was approximately 80% of the control in the presence of 10 μM β-ME (Fig. 5). Even in the presence of much higher concentrations (5 mM) of β-ME, approximately 60% activity remained (data not shown). These results suggest that a disulfide bond is not essential to the activity. However, we cannot exclude the possibility that a disulfide bond is buried inside the folded protein and cannot be reduced by high concentrations of β-ME.
Fig. 5.
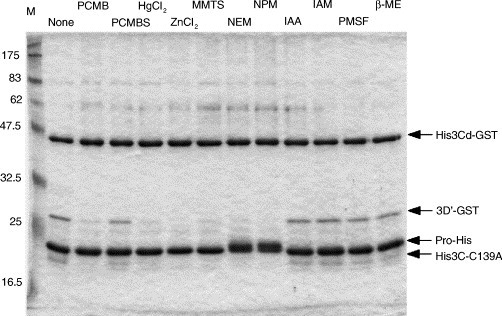
Effects of various sulfhydryl reagents on protease activity. Five micromolar enzyme (ProHis) and 5 μM substrate (His3Cd-GST) were reacted in the 50 mM bis–tris–propane–HCl (pH 8.62) buffer containing the indicated sulfhydryl reagent of 10 μM at 37 °C for 16 h. PCMB, p-chloromercuribenzoate; PCMBS, p-chloromercuribenzenesulfonate; MMTS, methyl methanethiosulfonate; NEM, N-ethylmaleimide; NPM, N-phenylmaleimide; IAA, iodoacetate; IAM, iodoacetamide; PMSF, phenylmethylsulfonylfluoride; β-ME, β-mercaptoethanol.
The optimal pH for the proteolytic reaction by the Chiba virus 3C-like protease was around 8.6, which is much more alkaline than physiological pH (Fig. 2). The 3C-like protease was sensitive to increased concentrations of Na+ (Fig. 4A). In addition, high concentrations of K+, which is abundant inside the cell in a normal state, were also inhibitory to the activity (data not shown). The presence of Mg2+ or Ca2+ did not affect the activity at all (Fig. 4B). These features might suggest that the 3C-like protease is localized and works in a limited environment inside the infected cells, rather than spreading throughout the cells. More information is necessary to elucidate when and where the 3C-like protease does work in the infected cell. We have previously shown that the active site of the Chiba virus 3C-like protease is composed of Cys139 and His30 (Someya et al., 2002). As expected, sulfhydryl reagents as well as Hg2+ and Zn2+ inhibited the activity of the 3C-like protease, although the degree of inhibition varied dependent on the sulfhydryl reagents used. Small (Hg2+, Zn2+, and MMTS) and hydrophobic (NEM and NPM) reagents tended to inhibit the activity more strongly than hydrophilic reagents (PCMB and PCMBS) (Fig. 5). This tendency may reflect the environment of the active site. The present results provide the basic biochemical features of the Norovirus 3C-like protease.
Acknowledgments
We thank T. Mizoguchi for her secretarial work. This work was supported in part by a grant for Research on Emerging and Re-emerging Infectious Diseases from the Ministry of Health, Labor, and Welfare.
References
- Anand K., Palm G.J., Mesters J.R., Siddell S.G., Ziebuhr J., Hilgenfeld R. Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra alpha-helical domain. EMBO J. 2002;21(13):3213–3224. doi: 10.1093/emboj/cdf327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand K., Ziebuhr J., Wadhwani P., Mesters J.R., Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003;300(5626):1763–1767. doi: 10.1126/science.1085658. Epub 13 May 2003. [DOI] [PubMed] [Google Scholar]
- Barrette-Ng I.H., Ng K.K., Mark B.L., Van Aken D., Cherney M.M., Garen C., Kolodenko Y., Gorbalenya A.E., Snijder E.J., James M.N. Structure of arterivirus nsp4. The smallest chymotrypsin-like proteinase with an alpha/beta C-terminal extension and alternate conformations of the oxyanion hole. J. Biol. Chem. 2002;277(42):39960–39966. doi: 10.1074/jbc.M206978200. Epub 5 August 2002. [DOI] [PubMed] [Google Scholar]
- Baum E.Z., Bebernitz G.A., Palant O., Mueller T., Plotch S.J. Purification, properties, and mutagenesis of poliovirus 3C protease. Virology. 1991;185(1):140–150. doi: 10.1016/0042-6822(91)90762-z. [DOI] [PubMed] [Google Scholar]
- Belliot G., Sosnovtsev S.V., Mitra T., Hammer C., Garfield M., Green K.Y. In vitro proteolytic processing of the MD145 norovirus ORF1 non-structural polyprotein yields stable precursors and products similar to those detected in calicivirus-infected cells. J. Virol. 2003;77(20):10957–10974. doi: 10.1128/JVI.77.20.10957-10974.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann E.M., Mosimann S.C., Chernaia M.M., Malcolm B.A., James M.N. The refined crystal structure of the 3C gene product from hepatitis A virus: specific proteinase activity and RNA recognition. J. Virol. 1997;71(3):2436–2448. doi: 10.1128/jvi.71.3.2436-2448.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boniotti B., Wirblich C., Sibilia M., Meyers G., Thiel H.J., Rossi C. Identification and characterization of a 3C-like protease from rabbit hemorrhagic disease virus, a calicivirus. J. Virol. 1994;68(10):6487–6495. doi: 10.1128/jvi.68.10.6487-6495.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Cheah K.C., Leong L.E., Porter A.G. Site-directed mutagenesis suggests close functional relationship between a human rhinovirus 3C cysteine protease and cellular trypsin-like serine proteases. J. Biol. Chem. 1990;265(13):7180–7187. [PubMed] [Google Scholar]
- Chisholm J., Wieczorek A., Sanfacon H. Expression and partial purification of recombinant tomato ringspot nepovirus 3C-like proteinase: comparison of the activity of the mature proteinase and the VPg-proteinase precursor. Virus Res. 2001;79(1–2):153–164. doi: 10.1016/S0168-1702(01)00344-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke I.N., Lambden P.R., Caul E.O. Human enteric RNA viruses: caliciviruses and astroviruses. In: Mahy B.W., Collier L., editors. ninth ed. vol. 1. Arnold; London: 1998. pp. 511–535. (Topley and Wilson's Microbiology and Microbial Infections). [Google Scholar]
- Davis G.J., Wang Q.M., Cox G.A., Johnson R.B., Wakulchik M., Dotson C.A., Villarreal E.C. Expression and purification of recombinant rhinovirus 14 3CD proteinase and its comparison to the 3C proteinase. Arch. Biochem. Biophys. 1997;346(1):125–130. doi: 10.1006/abbi.1997.0291. [DOI] [PubMed] [Google Scholar]
- Dougherty W.G., Semler B.L. Expression of virus-encoded proteinases: functional and structural similarities with cellular enzymes. Microbiol. Rev. 1993;57(4):781–822. doi: 10.1128/mr.57.4.781-822.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estes M.K., Atmar R.L., Hardy M.E. Norwalk and related diarrhea viruses. In: Richmann D.D., Whitley R.J., Hayden F.G., editors. Clinical Virology. Churchill Livingstone Inc.; New York: 1997. pp. 1073–1095. [Google Scholar]
- Gorbalenya A.E., Donchenko A.P., Blinov V.M., Koonin E.V. Cysteine proteases of positive strand RNA viruses and chymotrypsin-like serine proteases A distinct protein superfamily with a common structural fold. FEBS Lett. 1989;243(2):103–114. doi: 10.1016/0014-5793(89)80109-7. [DOI] [PubMed] [Google Scholar]
- Gosert R., Dollenmaier G., Weitz M. Identification of active-site residues in protease 3C of hepatitis A virus by site-directed mutagenesis. J. Virol. 1997;71(4):3062–3068. doi: 10.1128/jvi.71.4.3062-3068.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerle T., Hellen C.U., Wimmer E. Site-directed mutagenesis of the putative catalytic triad of poliovirus 3C proteinase. J. Biol. Chem. 1991;266(9):5412–5416. [PubMed] [Google Scholar]
- Matthews D.A., Smith W.W., Ferre R.A., Condon B., Budahazi G., Sisson W., Villafranca J.E., Janson C.A., McElroy H.E., Gribskov C.L. Structure of human rhinovirus 3C protease reveals a trypsin-like polypeptide fold, RNA-binding site, and means for cleaving precursor polyprotein. Cell. 1994;77(5):761–771. doi: 10.1016/0092-8674(94)90059-0. [DOI] [PubMed] [Google Scholar]
- McKinlay M.A. Recent advances in the treatment of rhinovirus infections. Curr. Opin. Pharmacol. 2001;1(5):477–481. doi: 10.1016/s1471-4892(01)00083-2. [DOI] [PubMed] [Google Scholar]
- McMurry L., Petrucci R.E., Jr., Levy S.B. Active efflux of tetracycline encoded by four genetically different tetracycline resistance determinants in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 1980;77(7):3974–3977. doi: 10.1073/pnas.77.7.3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosimann S.C., Cherney M.M., Sia S., Plotch S., James M.N. Refined X-ray crystallographic structure of the poliovirus 3C gene product. J. Mol. Biol. 1997;273(5):1032–1047. doi: 10.1006/jmbi.1997.1306. [DOI] [PubMed] [Google Scholar]
- Phan J., Zdanov A., Evdokimov A.G., Tropea J.E., Peters H.K., III, Kapust R.B., Li M., Wlodawer A., Waugh D.S. Structural basis for the substrate specificity of tobacco etch virus protease. J. Biol. Chem. 2002;277(52):50564–50572. doi: 10.1074/jbc.M207224200. Epub 10 October 2002. [DOI] [PubMed] [Google Scholar]
- Someya Y., Takeda N., Miyamura T. Complete nucleotide sequence of the chiba virus genome and functional expression of the 3C-like protease in Escherichia coli. Virology. 2000;278(2):490–500. doi: 10.1006/viro.2000.0672. [DOI] [PubMed] [Google Scholar]
- Someya Y., Takeda N., Miyamura T. Identification of active-site amino acid residues in the Chiba virus 3C-like protease. J. Virol. 2002;76(12):5949–5958. doi: 10.1128/JVI.76.12.5949-5958.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner R.B. The treatment of rhinovirus infections: progress and potential. Antiviral. Res. 2001;49(1):1–14. doi: 10.1016/S0166-3542(00)00135-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Yang M., Ding Y., Liu Y., Lou Z., Zhou Z., Sun L., Mo L., Ye S., Pang H., Gao G.F., Anand K., Bartlam M., Hilgenfeld R., Rao Z. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc. Natl. Acad. Sci. U.S.A. 2003;100(23):13190–13195. doi: 10.1073/pnas.1835675100. Epub 29 October 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr J., Heusipp G., Siddell S.G. Biosynthesis, purification, and characterization of the human coronavirus 229E 3C-like proteinase. J. Virol. 1997;71(5):3992–3997. doi: 10.1128/jvi.71.5.3992-3997.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]


