Abstract
Coronaviruses may cause respiratory, enteric and central nervous system diseases in many species, including humans. Until recently, the relatively low burden of disease in humans caused by few of these viruses hampered development of coronavirus specific therapeutics. However, the emergence of severe acute respiratory syndrome coronavirus (SARS-CoV) has prompted the discovery of such drugs. Subsequent studies in animal models demonstrated the efficacy of SARS-CoV specific monoclonal antibodies, pegylated-interferon-α and siRNAs against SARS-CoV. Furthermore, several antivirals shown to be effective against other viruses were tested in vitro. Because of availability and shown efficacy, the use of interferons may be considered should SARS-CoV or a related coronavirus (re)-emerge. The more recent design of wide-spectrum inhibitors targeting the coronavirus main proteases may lead to the discovery of new antivirals against multiple coronavirus induced diseases.
Keywords: Coronaviruses, SARS, Antivirals, Therapy
1. Introduction
Coronaviruses are positive stranded RNA viruses named after their “corona” like morphological appearance (Weiss and Navas-Martin, 2005). Their genome is the largest RNA genome to date and packaged together with the nucleocapsid protein, several membrane proteins (M, E and sometimes a hemagglutinin esterase protein) and the spike protein. Translation of the replicase gene produces two large poly-proteins with diverse enzymatic activities needed for efficient replication. A phylogenetic analysis of the replicase gene, using a distantly related torovirus as an outgroup, demonstrated that three groups of coronaviruses can be distinguished and that despite a number of unique features, severe acute respiratory syndrome coronavirus (SARS-CoV) is most closely related to group 2 coronaviruses (Snijder et al., 2003). Viruses that belong to group1 include transmissible gastroenteritis virus of pigs, feline infectious peritonitis virus of cats and the human coronaviruses 229E and HCoV-NL63 (Fig. 1 ). In group 2, viruses such as murine hepatitis virus, bovine coronavirus, human coronavirus OC43, HKU1 and SARS-CoV are classified. Group 3 constitutes only avian coronaviruses, such as infectious bronchitis virus and turkey coronavirus. Each of the three groups of viruses classified thus far encodes a set of unique small proteins with unknown functions. Coronaviruses cause acute and chronic respiratory, enteric and or central nervous system diseases in many species, including humans (Weiss and Navas-Martin, 2005). Until recently, the need to develop antiviral drugs was limited because human coronaviruses like 229E and OC43 only cause a mild disease in humans. The burden of disease for HCoV-NL63 and HKU1 are not known at present. However, the emergence of SARS changed this picture. In this review we discuss the latest developments in antiviral drug testing specifically with regard to SARS-CoV and whenever applicable we broaden the scope to other coronaviruses. A range of compounds may interfere with the lifecycle of SARS-CoV as shown in Fig. 2 and discussed below.
Fig. 1.
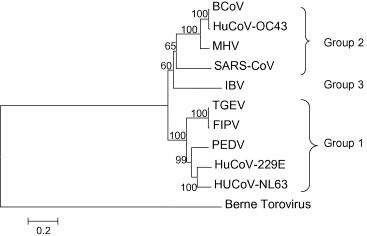
Phylogenetic tree based on deduced amino acid sequences of the coronavirus replicase ORF1b gene for bovine coronavirus (BCoV), human coronavirus 22E (HuCoV-OC43), mouse hepatitis virus (MHV), SARS-CoV, infectious bronchitis virus (IBV), transmissible gastroenteritis virus (TGEV), feline infectious peritonitis virus (FIPV), porcine epidemic diarrhea virus (PEDV), human coronavirus 229E (HuCoV-229E), human coronavirus NL63 (HuCoV-NL63) and Berne Torovirus (used as an outgroup).
Fig. 2.
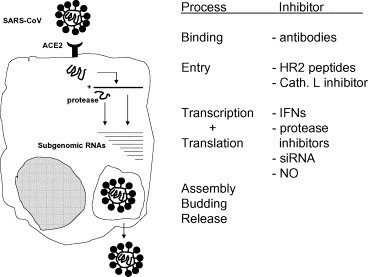
Schematic overview of the SARS-CoV lifecycle and inhibitors of viral replication.
2. SARS
The 2002–2003 SARS outbreak affected more than 8000 individuals worldwide and caused 774 fatalities (Poon et al., 2004). The lung pathology of fatal SARS showed bronchial epithelial denudation, diffuse alveolar damage and type-2 pneumocyte hyperplasia. In patients who died late in the course of the disease, also syncytial cells were seen in the alveoli. Subsequently, three laboratories independently reported the isolation of a novel coronavirus from clinical specimens of SARS patients (Drosten et al., 2003, Peiris et al., 2003, Ksiazek et al., 2003). The virus was visualized by electron microscopy and identification of the virus was accomplished through sequencing of different fragments of the replicase gene, obtained by random-priming RT-PCR and coronavirus consensus primers. Most importantly, SARS-CoV was detected in lung biopsies and bronchoalveolar lavage of SARS patients using virus culture, RT-PCR, and electron microscopy, whereas viral antigen was detected in alveolar epithelial cells and macrophages by immunohistology (Nicholls et al., 2006). To further establish SARS-CoV as the cause of the disease, Koch's postulates were fulfilled by reproduction of the disease in a relevant animal model. Infection of cynomolgous macaques with SARS-CoV led to disease that was pathologically similar to that seen in human patients with SARS, with epithelial necrosis, serosanguinous alveolar exudates, hyaline membranes, type-2 pneumocyte hyperplasia, diffuse alveolar damage and the presence of syncytia (Fouchier et al., 2003, Kuiken et al., 2003). These experiments also led to the development of an animal model to test the efficacy of candidate vaccines and antivirals.
SARS was characterised by a rapidly progressive atypical pneumonia refractory to conventional antibiotic therapy. Next efforts to treat SARS patients mainly were based on the use of ribavirin and corticosteroids. Ribavirin, which targets IMP dehydrogenase, has been known a long time as a broad-spectrum antiviral agent. However, it is efficacy in SARS patients remains questionable and in vitro studies also did not show significant antiviral activity (Cinatl et al., 2003a). More recent studies demonstrated that ribavirin even enhanced the infectivity of SARS-CoV in mice (Barnard et al., 2006). Collectively these data do not support the use of ribavirin for treating SARS in humans. The use of steroids in SARS patients was mainly based on the observation that a clinical deterioration was observed in most patients late during the disease when SARS-CoV became undetectable. In most cases the concomitant use of other drugs confounded the results and the fact that most studies did not contain a control group made it quite difficult to determine whether steroids exerted a beneficial effect. Although efforts were made to control the infection through use of antivirals and corticosteroids, isolation and quarantine measures eventually proved instrumental in control of the epidemic at that time.
3. Inhibition of SARS-CoV entry
Antibodies to the SARS-CoV spike protein have been shown to block entry (Sui et al., 2004). In SARS patients that recover, high levels of spike glycoprotein-specific antibody responses are observed, suggesting that antibody responses play a role in determining the ultimate disease outcome of SARS-CoV-infected patients (Zhang et al., 2006). Although attempts have been made to test the efficacy of serum preparations from seroconvalescent SARS patients in the acute phase of SARS, no conclusive evidence has been obtained regarding their efficacy (Wong et al., 2003, Cheng et al., 2005), although convalescent immunoglobulines exerted antiviral activity when given to macaques prior SARS-CoV challenge (Haagmans et al., unpublished observations).
The SARS-CoV spike protein binds to angiotensin-converting enzyme 2 (ACE2), which acts as the functional receptor for SARS-CoV (Li et al., 2003) and soluble forms of ACE2 and ACE2 antibodies block this interaction and inhibit SARS-CoV infection. The S1 region within the spike protein and more specifically a 193-amino acid fragment of the S protein (corresponding to residues 318–510) has been identified as the region which interacts with ACE2 (Wong et al., 2004). Monoclonal antibodies directed against the S1 domain of the SARS-CoV spike protein were shown to block association with ACE2 (Sui et al., 2004). Because other epitopes may be hidden as a result of extensive glycosylation, Balzarini (2005) hypothesized that drugs directed against these carbohydrate components may select for mutant virus strains that progressively gain deletions in these glycosylation sites uncovering previously hidden epitopes, resulting in increased immunological neutralization.
The protective efficacy of SARS-CoV neutralizing antibodies was demonstrated convincingly in several animal models. Mice efficiently controlled SARS-CoV infection upon passive transfer of serum from mice that recovered from SARS-CoV infection (Subbarao et al., 2004). The concept that antibodies protect against SARS has been further explored through the generation of human monoclonal antibodies against SARS-CoV. Prophylactic administration of a monoclonal antibody at 10 mg/kg reduced replication of SARS coronavirus in the lungs of infected ferrets by 3 logs, completely prevented the development of SARS coronavirus-induced macroscopic lung pathology, and abolished shedding of virus in pharyngeal secretions (ter Meulen et al., 2004). Other monoclonal antibodies were evaluated for their efficacy in mouse- and hamster-models (Sui et al., 2005, Roberts et al., 2006). When given prophylactically to mice at doses therapeutically achievable in humans, viral replication was reduced by more than 4 orders of magnitude to below assay limits. Post exposure treatment also alleviated the viral burden and associated pathological findings in a golden Syrian hamster model of SARS-CoV infection. After hamsters were treated with the antibody their viral burden was reduced by 102.4 to 103.9 50% tissue-culture infectious doses per gram of tissue, and the severity of associated pathological findings, including interstitial pneumonitis and consolidation, was also remarkably reduced (Roberts et al., 2006).
Traggiai et al. (2004) developed an efficient method to make human monoclonal SARS-CoV neutralizing antibodies from memory B cells. These results further proved the feasibility of immunoprophylaxis using human monoclonal Abs and provided evidence that vaccines able to elicit neutralizing antibodies are likely to be effective against SARS. However, as has been demonstrated for feline coronavirus, antibodies directed against the coronavirus spike protein sometimes enhance infection. Macrophages are able to take up feline coronavirus-antibody complexes more efficiently causing the virus to replicate to higher titers. Passive and active immunization against feline coronavirus thus may enhance feline coronavirus infection in vivo and cause early death (Vennema et al., 1990). The question remains whether such a scenario also holds for SARS-CoV. Although enhanced entry of certain SARS-CoV pseudotyped viruses in Vero cells was observed in the presence of antibodies directed against the spike protein (Yang et al., 2005a), its relevance in vivo still awaits further investigation. Thus, caution has to be taken into account by developing strategies to block SARS-CoV entry by antibodies.
4. Inhibition of SARS-CoV fusion
After engagement with ACE2, SARS-CoV fuses with host cell membranes by a fusion mechanism similar to that exerted by class I fusion proteins. The conformational changes of the two heptad regions located in the S2 region, HR-1 and HR-2, cause the formation of an oligomeric structure, leading to fusion between the viral and target-cell membranes. Therefore, blocking the HR1–HR2 interaction may reveal novel strategies to inhibit entry of SARS-CoV. Indeed, synthetic peptides derived from the HR2 region that bind with high affinity to a peptide from the HR1 region interfere with conformational changes leading to the 6-helix bundle formation and subsequently block SARS-CoV infection (Zhu et al., 2004, Liu et al., 2004, Bosch et al., 2004, Ni et al., 2005). However, their relatively low efficacy may hamper their development as SARS-CoV antivirals at this moment.
More recent observations have demonstrated that besides conformational changes in the HR1 and HR2 interaction, lysosomal cysteine proteases are required for productive infection. SARS-CoV but not HCoV-NL63 utilizes cathepsin L to infect ACE2-expressing cells. Inhibitors of cathepsin L, like calpain inhibitor III or VI block SARS-CoV infection (Simmons et al., 2005, Huang et al., 2006). At this moment the exact mechanism of action of these enzymes is not clear.
5. Inhibition of SARS-CoV replication by interferons
After SARS-CoV has entered the cell and has fused with the cellular membranes, the positive stranded coronavirus genome is released and viral replication is initiated. However, the subsequent formation of double stranded RNA intermediates is a strong trigger of innate immune responses. Through interaction with toll-like receptors, production of type 1 interferons (IFNs) is initiated to limit viral replication. Interestingly, several studies have demonstrated that IFN production by SARS-CoV infected cells may be limited; macrophages as well as 293 cells infected with SARS-CoV do not show induction of IFN-beta gene expression (Cheung et al., 2005, Spiegel et al., 2005). Therefore, SARS-CoV most likely encodes viral proteins that interfere with one of the signaling cascades leading to the production of type 1 IFNs in these cells. On the other hand, SARS-CoV remains sensitive to the action of exogenous IFNs. Several studies noted that IFNs inhibited the replication of SARS-CoV in cell culture in vitro, IFN-β being more potent than either IFN-α or -γ (Cinatl et al., 2003a, Haagmans et al., 2004, Hensley et al., 2004, Stroher et al., 2004). IFN-β in conjunction with IFN-γ synergistically inhibit the replication of SARS-CoV (Sainz et al., 2004). Similarly, IFNs inhibit replication of other animal and human coronaviruses (Turner et al., 1986, Smith et al., 1987, Pei et al., 2001).
Viruses causing lysis of their target cell are most effectively inhibited by IFNs through their antiviral activity in non-infected cells. Therefore, IFNs have their highest utility in the prophylaxis or early post-exposure management of SARS. Pegylated IFN-α has been shown to effectively reduce viral replication and excretion, viral antigen expression by type 1 pneumocytes and the pulmonary damage in cynomolgous macaques that were infected experimentally with SARS-CoV (Haagmans et al., 2004). Evidence of a protective effect of IFN-alpha also has been obtained in a preliminary study during the SARS outbreak with interferon alfacon-1 in combination with corticosteroids (Loutfy et al., 2003). Because IFNs are commercially available for the treatment of hepatitis B and C, these drugs could be considered for treatment of SARS should it re-emerge.
6. Inhibition of SARS-CoV replication by RNA interference
Because RNA interference (RNAi) provides effective antiviral defence in plants and other organisms, several studies have focused on harnessing RNAi to inhibit viral infection (Leonard and Schaffer, 2006). To explore the possibility of interrupting SARS-coronavirus replication with siRNAs, specific siRNAs targeting the spike gene in SARS coronavirus have been synthesised. These siRNAs effectively and specifically inhibited gene expression of the spike protein in SARS-coronavirus-infected cells (Zhang et al., 2004). Another study assessed the in vitro efficiacy of six siRNA molecules targeting different sites of the replicase 1A region of the SARS-coronavirus genome (He et al., 2003) Judged by morphological changes, three of the molecules markedly inhibited the cytopathic effects caused by viral infection and replication. The three siRNAs also inhibited the infection and replication of different strains of SARS coronavirus, indicating that siRNAs targeting the replicase 1A region may be an option for future clinical use.
Potent siRNA inhibitors of SARS-CoV in vitro, targeting the SARS-CoV genome at S protein- and nsp12-coding regions, were further evaluated for their efficacy in a rhesus macaque SARS model (Li et al., 2005), and found to provide relief from SARS-CoV infection-induced fever, diminish SARS-CoV levels and reduce acute diffuse alveolar damage.
7. Inhibition of SARS-CoV replication by targeting viral proteases
The pivotal roles played by coronavirus main proteases in controlling viral replication and transcription through extensive processing of replicase polyproteins, together with the absence of closely related cellular homologues, identified these proteins as a potentially important target for antiviral drug design (Anand et al., 2003). The SARS-CoV encoded main protease cleaves the replicase polyproteins at 11 conserved sites, in order to generate the functional proteins necessary for virus replication. The crystal structures of the SARS-CoV main protease complexed with various substrate analogues have been determined (Yang et al., 2003). Recently, Yang et al. (2005b) reported the discovery of a highly conserved region based on four crystal structures and one homology model of main protease representing all three genetic clusters of the genus Coronavirus. Furthermore they revealed a uniform inhibition mechanism from the structures of main protease-inhibitor complexes from SARS-CoV and TGEV. A structure-assisted optimization program yielded compounds with fast in vitro inactivation of multiple CoV main proteases, potent antiviral activity, and extremely low cellular toxicity in cell-based assays. Further modification could rapidly lead to the discovery of a single agent with clinical potential against existing and possible future emerging CoV-associated diseases. Also some other main protease inhibitors were shown to inhibit SARS-CoV replication in vitro. Cinanserin, niclosamide anilide and a Phe–Phe dipeptide inhibitor were found to inhibit the main protease and inhibited virus replication in Vero cells (Shie et al., 2005, Chen et al., 2005).
The lopinavir/ritonavir combination was first considered a potentially useful treatment after in vitro studies showed it had antiviral activity against SARS-CoV. Chan et al. (2003) compared outcomes in people who received lopinavir/ritonavir as initial treatment, and as rescue therapy, with matched controls; all patients were given ribavirin and steroids according to a standardised protocol. The addition of lopinavir/ritonavir as initial treatment was associated with a statistically significant reduction in the overall death rate and intubation rate compared with matched controls. Chu et al. (2004) also assessed treatment with lopinavir/ritonavir compared with historic controls; all patients were also treated with ribavirin and steroids in a similar protocol to that of Chan and colleagues. Adverse events (development of acute respiratory distress syndrome [ARDS] or death within 21 days) were significantly lower in the lopinavir/ritonavir group than in the historic controls. In addition, a significant reduction in the need for rescue pulsed steroids for severe respiratory deterioration and significantly lower nosocomial infections were also noted in those treated with lopinavir/ritonavir, compared with controls. By multivariate analysis, it was demonstrated that the lack of treatment with lopinavir/ritonavir, age 60 years old or greater, and positive hepatitis B carrier status were independent predictors of an adverse outcome including death or the development of ARDS requiring intensive care within 21 days of onset of illness. Based on these studies, lopinavir/ritonavir appears to be a promising anti-SARS-CoV agent. Other protease inhibitors have been studied in vitro for potential antiviral effects in SARS. For example, Yamamoto et al. (2004) screened a set of compounds that included antiviral drugs already widely used, and found that nelfinavir strongly inhibited SARS-CoV replication. In addition, calpain inhibitor VI (Val-Leu-CHO) and calpain inhibitor III (Z-Val-Phe-Ala-CHO) inhibited SARS-CoV (Barnard et al., 2004), suggesting that other protease inhibitors may also be useful in the treatment of SARS.
8. Inhibition of SARS-CoV replication by other compounds
A large number of compounds that inhibit SARS-CoV replication with unknown mechanism of action have been identified. Glycyrrhizin, which consists of one molecule glycyrrhetinic acid linked to two molecules of glucuronic acid, is known to exert antiviral activity against a range of viruses. It also inhibits SARS-CoV replication, although relatively high concentrations are needed (Cinatl et al., 2003b). The mechanism of glycyrrhizin-induced inhibition of viral replication – and specifically SARS-CoV replication – is unclear, but possibly involves inhibition of replication through an antiviral effect of nitric oxide (NO). SARS-CoV replication is inhibited when DETA NONOate – a NO donor compound – is added to the culture medium (Cinatl et al., 2003b). Inhibition of SARS-CoV infection in vitro has also been reported for a nitric oxide generating compound, S-nitroso-N-acetylpenicillamine (SNAP), but only at high concentrations (Keyaerts et al., 2004b).
A wide variety of pyridine N-oxide derivatives have been found to be inhibitory against feline coronavirus (FIPV strain) and human SARS-CoV (Frankfurt strain-1) in CRFK and simian kidney (Vero) cell cultures, respectively (Balzarini et al., 2006).
Bananin shown to inhibit both the ATPase and helicase activity of the SARS-CoV inhibits SARS-CoV replication in vitro (Tanner et al., 2005). The 4-aminoquinoline chloroquine, best known for its antimalarial effects, has been recommended for its potential use, preferably in combination with other antivirals, in the treatment of SARS (Keyaerts et al., 2004a). Chloroquine inhibits SARS-CoV replication in Vero cells at an EC50 of 8.8 μM which approximates the plasma concentrations of chloroquine reached during treatment of acute malaria.
Finally, Wu et al. (2004) tested >10,000 agents against SARS-CoV in Vero cells and selected approximately 50 active compounds. Valinomycin, a peptidic insecticide which acts as a potassium ion transporter, was the most potent inhibitor.
References
- Anand K., Ziebuhr J., Wadhwani P., Mesters J.R., Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003;300:1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- Balzarini J. Targeting the glycans of gp120: a novel approach aimed at the Achilles heel of HIV. Lancet Infect Dis. 2005;5(11):726–731. doi: 10.1016/S1473-3099(05)70271-1. [DOI] [PubMed] [Google Scholar]
- Balzarini J., Keyaerts E., Vijgen L., Vandermeer F., Stevens M., De Clercq E., Egberink H.J., Van Ranst M. Pyridine N-oxide derivatives are inhibitory to the human SARS and feline infectious peritonitis coronavirus in cell culture. J. Ant. Chemother. 2006;57:472–481. doi: 10.1093/jac/dki481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnard D.L., Hubbard V.D., Burton J., Smee D.F., Morrey J.D., Otto M.J., Sidwell R.W. Inhibition of severe acute respiratory syndrome-associated coronavirus (SARSCoV) by calpain inhibitors and beta-d-N4-hydroxycytidine. Antiviral Chem. Chemother. 2004;15:15–22. doi: 10.1177/095632020401500102. [DOI] [PubMed] [Google Scholar]
- Barnard D.L., Day C.W., Bailey K., Heiner M., Montgomery R., Lauridsen L., Winslow S., Hoopes J., Li J.K., Lee J., Carson D.A., Cottam H.B., Sidwell R.W. Enhancement of the infectivity of SARS-CoV in BALB/c mice by IMP dehydrogenase inhibitors, including ribavirin. Antiviral Res. 2006 doi: 10.1016/j.antiviral.2006.03.001. (Epub) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch B.J., Martina B.E.E., Van der Zee R., Lepault J., Haijema B.J., Versluis C., Heck A.J.R., De Groot R., Osterhaus A.D.M.E., Rottier P.J.M. Severe acute respiratory syndrome coronavirus (SARS-CoV) infection inhibition using spike protein heptad repeat-derived peptides. Proc. Natl. Acad. Sci. U.S.A. 2004;101:8455–8460. doi: 10.1073/pnas.0400576101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K.S., Lai S.T., Chu C.M., Tsui E., Tam C.Y., Wong M.M., Tse M.W., Que T.L., Peiris J.S., Sung J., Wong V.C., Yuen K.Y. Treatment of severe acute respiratory syndrome with lopinavir/ritonavir: a multicentre retrospective matched cohort study. Hong Kong Med. J. 2003;9:399–406. [PubMed] [Google Scholar]
- Chen L., Gui C., Luo X., Yang Q., Günther S., Scandella E., Drosten C., Bai D., He X., Ludewig B., Chen J., Luo H., Yang Y., Yang Y., Zou J., Thiel V., Chen K., Shen J., Shen X., Jiang H. Cinanserin is an inhibitor of the 3C-like proteinase of severe acute respiratory syndrome coronavirus and strongly reduces virus replication in vitro. J. Virol. 2005;79:7095–7103. doi: 10.1128/JVI.79.11.7095-7103.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., Wong R., Soo Y.O., Wong W.S., Lee C.K., Ng M.H., Chan P., Wong K.C., Leung C.B., Cheng G. Use of convalescent plasma therapy in SARS patients in Hong Kong. Eur. J. Clin. Microbiol. Infect. Dis. 2005;24:44–46. doi: 10.1007/s10096-004-1271-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C.M., Cheng V.C., Hung I.F., Wong M.M., Chan K.H., Chan K.S., Kao R.Y., Poon L.L., Wong C.L., Guan Y., Peiris J.S., Yuen K.Y. HKU/UCH SARS Study Group: role of lopinavir/ritonavir in the treatment of SARS: initial virological and clinical findings. Thorax. 2004;59:252–256. doi: 10.1136/thorax.2003.012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung C.Y., Poon L.L.M., Ng I.H.Y., Luk W., Sia S.F., Wu M.H.S., Chan K.H., Yuen K.Y., Gordon S., Guan Y., Peiris J.S.M. Cytokine responses in severe acute respiratory syndrome coronavirus-infected macrophages in vitro: possible relevance to pathogenesis. J. Virol. 2005;79:7819–7826. doi: 10.1128/JVI.79.12.7819-7826.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinatl J., Morgenstern B., Bauer G., Chandra P., Rabenau H., Doerr H.W. Treatment of SARS with human interferons. Lancet. 2003;362:293–294. doi: 10.1016/S0140-6736(03)13973-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinatl J., Morgenstern B., Bauer G., Chandra P., Rabenau H., Doerr H.W. Glycyrrhizin, an active component of liquorice roots, and replication of SARS-associated coronavirus. Lancet. 2003;361:2045–2046. doi: 10.1016/S0140-6736(03)13615-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., Berger A., Burguiere A.M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.C., Muller S., Rickerts V., Sturmer M., Vieth S., Klenk H.D., Osterhaus A.D., Schmitz H., Doerr H.W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- Fouchier R.A., Kuiken T., Schutten M., van Amerongen G., van Doornum G.J., van den Hoogen B.G., Peiris M., Lim W., Stohr K., Osterhaus A.D. Aetiology: Koch's postulates fulfilled for SARS virus. Nature. 2003;423:240. doi: 10.1038/423240a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haagmans B.L., Kuiken T., Martina B.E., Fouchier R.A., Rimmelzwaan G.F., van Amerongen G., van Riel D., de Jong T., Itamura S., Chan K.H., Tashiro M., Osterhaus A.D. Pegylated interferon-alpha protects type 1 pneumocytes against SARS coronavirus infection in macaques. Nat. Med. 2004;10:290–293. doi: 10.1038/nm1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M.L., Zheng B., Peng Y., Peiris J.S., Poon L.L., Yuen K.Y., Lin M.C., Kung H.F., Guan Y. Inhibition of SARS-associated coronavirus infection and replication by RNA interference. JAMA. 2003;290:2665–2666. doi: 10.1001/jama.290.20.2665. [DOI] [PubMed] [Google Scholar]
- Hensley L.E., Fritz L.E., Jahrling P.B., Karp C.L., Huggins J.W., Geisbert T.W. Interferon-beta 1a and SARS coronavirus replication. Emerg. Infect. Dis. 2004;10:317–319. doi: 10.3201/eid1002.030482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang I.C., Bosch B.J., Li F., Li W., Lee K.H., Ghiran S., Vasilieva N., Dermody T.S., Harrison S.C., Dormitzer P.R., Farzan M., Rottier P.J.M., Choe H. SARS coronavirus, but not human coronavirus NL63, utilizes cathepsin L to infect ACE2-expressing cells J. Biol. Chem. 2006;281:3198–3203. doi: 10.1074/jbc.M508381200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyaerts E., Vijgen L., Maes P., Neyts J., Van Ranst M. In vitro inhibition of severe acute respiratory syndrome coronavirus by chloroquine. Biochem. Biophys. Res. Commun. 2004;323:264–268. doi: 10.1016/j.bbrc.2004.08.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyaerts E., Vijgen L., Chen L., Maes P., Hedenstierna G., Van Ranst M. Inhibition of SARS-coronavirus infection in vitro by S-nitroso-N-acetylpenicillamine, a nitric oxide donor compound. Int. J. Infect. Dis. 2004;8:223–226. doi: 10.1016/j.ijid.2004.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A.E., Humphrey C.D., Shieh W.J., Guarner J., Paddock C.D., Rota P., Fields B., DeRisi J., Yang J.Y., Cox N., Hughes J.M., LeDuc J.W., Bellini W.J., Anderson L.J. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- Kuiken T., Fouchier R.A., Schutten M., Rimmelzwaan G.F., van Amerongen G., van Riel D., Laman J.D., de Jong T., van Doornum G., Lim W., Ling A.E., Chan P.K., Tam J.S., Zambon M.C., Gopal R., Drosten C., van der Werf S., Escriou N., Manuguerra J.C., Stohr K., Peiris J.S., Osterhaus A.D. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet. 2003;362:263–270. doi: 10.1016/S0140-6736(03)13967-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard J.N., Schaffer D.V. Antiviral RNAi therapy: emerging approaches for hitting a moving target. Gene Ther. 2006;13:532–540. doi: 10.1038/sj.gt.3302645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B.J., Tang Q., Cheng D., Qin C., Xie F.Y., Wei Q., Xu J., Liu Y., Zheng B.J., Woodle M.C., Zhong N., Lu P.Y. Using siRNA in prophylactic and therapeutic regimens against SARS coronavirus in rhesus macaque. Nat. Med. 2005;11:944–951. doi: 10.1038/nm1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Xiao G., Chen Y., He Y., Niu J., Escalante C.R., Xiong H., Farmar J., Debnath A.K., Tien P., Jiang S. Interaction between heptad repeat 1 and 2 regions in spike protein of SARS associated coronavirus: implications for virus fusogenic mechanism and identification of fusion inhibitors. Lancet. 2004;363:938–947. doi: 10.1016/S0140-6736(04)15788-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loutfy M.R., Blatt L.M., Siminovitch K.A., Ward S., Wolff B., Lho H., Pham D.H., Deif H., LaMere E.A., Chang M., Kain K.C., Farcas G.A., Ferguson P., Latchford M., Levy G., Dennis J.W., Lai E.K., Fish E.N. Interferon alfacon-1 plus corticosteroids in severe acute respiratory syndrome: a preliminary study. JAMA. 2003;290:3222–3228. doi: 10.1001/jama.290.24.3222. [DOI] [PubMed] [Google Scholar]
- Ni L., Zhu J., Zhang J., Yan M., Gao G.F., Tien P. Design of recombinant protein-based SARS-CoV entry inhibitors targeting the heptad-repeat regions of the spike protein S2 domain. Biochem. Biophys. Res. Commun. 2005;330:39–45. doi: 10.1016/j.bbrc.2005.02.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls J.M., Butany J., Poon L.L.M., Chan K.H., Beh S.I., Poutanen S., Peiris J.S.M., Wong M. Time course and cellular localization of SARS-CoV nucleoprotein and RNA in lungs from fatal cases of SARS. PloS Med. 2006;3:e27. doi: 10.1371/journal.pmed.0030027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei J., Sekellick M.J., Marcus P.I., Choi I.S., Collison E.W. Chicken interferon type I inhibits infectious bronchitis virus replication and associated respiratory ilness. J. Interferon. Cytokine Res. 2001;21:1071–1077. doi: 10.1089/107999001317205204. [DOI] [PubMed] [Google Scholar]
- Peiris J.S. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon L., Guan Y., Nicholls J., Yuen K., Peiris J. The aetiology, origins, and diagnosis of severe acute respiratory syndrome. Lancet Infect. Dis. 2004;4:663–671. doi: 10.1016/S1473-3099(04)01172-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A., Thomas W.D., Guarner J., Lamirande E.W., Babcock G.J., Greenough T.C., Vogel L., Hayes N., Sullivan J.L., Zaki S., Subbarao K., Ambrosino D.M. Therapy with a severe acute respiratory syndrome–associated coronavirus–neutralizing human monoclonal antibody reduces disease severity and viral burden in golden syrian hamsters. J. Infect. Dis. 2006;193:685–692. doi: 10.1086/500143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainz B., Jr., Mossel E.C., Peters C.J., Garry R.F. Interferon-beta and interferon-gamma synergistically inhibit the replication of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) Virology. 2004;329:11–17. doi: 10.1016/j.virol.2004.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shie J.J., Fang J.M., Kuo T.H., Kuo C.J., Liang P.H., Huang H.J., Wu Y.T., Jan J.T., Cheng Y.S.E., Wong C.H. Inhibition of the severe acute respiratory syndrome 3CL protease by peptidomimetic α,β-unsaturated esters. Bioorg. Med. Chem. 2005;13:5240–5252. doi: 10.1016/j.bmc.2005.05.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons G., Gosalia D.N., Rennekamp A.J., Reeves J.D., Diamond S.L., Bates P. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. U.S.A. 2005;102:11876–11881. doi: 10.1073/pnas.0505577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A.L., Barthold S.W., Beck D.S. Intranasally administered alpha/beta interferon prevents extension of mouse hepatitis virus, strain JHM, into the brains of BALB/cByJ mice. Ant. Res. 1987;8:239–245. doi: 10.1016/S0166-3542(87)80002-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Bredenbeek P.J., Dobbe J.C., Thiel V., Ziebuhr J., Poon L.L., Guan Y., Rozanov M., Spaan W.J., Gorbalenya Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 2003;331:991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegel M., Pilchmair A., Martinez-Sobrido L., Cros J., Garcìa-Sastre A., Haller O., Weber F. Inhibition of beta interferon induction by severe acute respiratory syndrome coronavirus suggests a two-step model for activation of interferon regulatory factor 3. J. Virol. 2005;79:2079–2086. doi: 10.1128/JVI.79.4.2079-2086.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroher U., DiCaro A., Li Y., Strong J.E., Aoki F., Plummer F., Jones S.M., Feldmann H. Severe acute respiratory syndromerelated coronavirus is inhibited by interferon-alpha. J. Infect. Dis. 2004;189:1164–1167. doi: 10.1086/382597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbarao K., McAuliffe J., Vogel L., Fahle G., Fischer S., Tatti K., Packard M., Shieh W.J., Zaki S., Murphy B. Prior infection and passive transfer of neutralizing antibody prevent replication of severe acute respiratory syndrome coronavirus in the respiratory tract of mice. J. Virol. 2004;78:3572–3577. doi: 10.1128/JVI.78.7.3572-3577.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui J., Li W., Murakami A., Tamin A., Matthews L.J., Wong S.K., Moore M.J., Tallarico A.S., Olurinde M., Choe H., Anderson L.J., Bellini W.J., Farzan M., Marasco W.A. Potent neutralization of severe acute respiratory syndrome (SARS) coronavirus by a human mAb to S1 protein that blocks receptor association. Proc. Natl. Acad. Sci. U.S.A. 2004;101:2536–2541. doi: 10.1073/pnas.0307140101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui J., Li W., Roberts A., Matthews L.J., Murakami A., Vogel L., Wong S.K., Subbarao K., Farzan M., Marasco W.A. Evaluation of human monoclonal antibody 80R for immunoprophylaxis of severe acute respiratory syndrome by an animal study, epitope mapping, and analysis of spike variants. J. Virol. 2005;79:5900–5906. doi: 10.1128/JVI.79.10.5900-5906.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner J.A., Zheng B.J., Zhou J., Watt R.M., Jiang J.Q., Wong K.L., Lin Y.P., Lu L.Y., He M.L., Kung H.F., Kesel A.J., Huang J.D. The adamantane-derived bananins are potent inhibitors of the helicase activities and replication of SARS coronavirus. Chem. Biol. 2005;12:303–311. doi: 10.1016/j.chembiol.2005.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Meulen J., Bakker A.B., van den Brink E.N., Weverling G.J., Martina B.E., Haagmans B.L., Kuiken T., de Kruif J., Preiser W., Spaan W., Gelderblom H.R., Goudsmit J., Osterhaus A.D. Human monoclonal antibody as prophylaxis for SARS coronavirus infection in ferrets. Lancet. 2004;363:2139–2141. doi: 10.1016/S0140-6736(04)16506-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traggiai E., Becker S., Subbarao K., Kolesnikova L., Uematsu Y., Gismondo M.R., Murphy B.R., Rappuoli R., Lanzavecchia A. An efficient method to make human monoclonal antibodies from memory B cells: potent neutralization of SARS coronavirus. Nat. Med. 2004;10:871–875. doi: 10.1038/nm1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner R.B., Felton A., Kosak K., Kelsey D.K., Mescievitz C.K. Prevention of experimental coronavirus colds with intranasal alpha-2b interferon. J. Infect. Dis. 1986;154:443–447. doi: 10.1093/infdis/154.3.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vennema H., De Groot R.J., Harbour D.A., Dalderup M., Gruffydd-Jones T., Horzinek M.C., Spaan W.J. Early death after feline infectious peritonitis virus challenge due to recombinant vaccinia virus immunization. J. Virol. 1990;64:1407–1409. doi: 10.1128/jvi.64.3.1407-1409.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss S.R., Navas-Martin S. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol. Mol. Biol. Rev. 2005;69:635–664. doi: 10.1128/MMBR.69.4.635-664.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong V.W., Dai D., Wu A.K., Sung J.J. Treatment of severe acute respiratory syndrome with convalescent plasma. Hong Kong Med. J. 2003;9:199–201. [PubMed] [Google Scholar]
- Wong S.K., Li W., Moore M.J., Choe H., Farzan M. A 193-amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. J. Biol. Chem. 2004;279:3197–3201. doi: 10.1074/jbc.C300520200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.Y., Jan J.T., Ma S.H., Kuo C.J., Juan H.F., Cheng Y.S., Hsu H.H., Huang H.C., Wu D., Brik A., Liang F.S., Liu R.S., Fang J.M., Chen S.T., Liang P.H., Wong C.H. Small molecules targeting severe acute respiratory syndrome human coronavirus. Proc. Natl. Acad. Sci. U.S.A. 2004;101:10012–10017. doi: 10.1073/pnas.0403596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto N., Yang R., Yoshinaka Y., Amari S., Nakano T., Cinatl J., Rabenau H., Doerr H.W., Hunsmann G., Otaka A., Tamamura H., Fujii N., Yamamoto N. HIV protease inhibitor nelfinavir inhibits replication of SARS-associated coronavirus. Biochem. Biophys. Res. Commun. 2004;318:719–725. doi: 10.1016/j.bbrc.2004.04.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Yang M., Ding Y., Liu Y., Lou Z., Zhou Z., Sun L., Mo L., Ye S., Pang H., Gao G.F., Anand K., Bartlam M., Hilgenfeld R., Rao Z. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc. Natl. Acad. Sci. U.S.A. 2003;100:13190–13195. doi: 10.1073/pnas.1835675100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z.Y., Werner H.C., Kong W.P., Leung K., Traggiai E., Lanzavecchia A., Nabel G.J. Evasion of antibody neutralization in emerging severe acute respiratory syndrome coronaviruses. Proc. Natl. Acad. Sci. U.S.A. 2005;102:797–801. doi: 10.1073/pnas.0409065102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Xie W., Xue X., Yang K., Ma J., Liang W., Zhao Q., Zhou Z., Pei D., Ziebuhr J., Hilgenfeld R., Yuen K.Y., Wong L., Gao G., Chen S., Chen Z., Ma D., Bartlam M., Rao Z. Design of wide-spectrum inhibitors targeting coronavirus main proteases. PLoS Biol. 2005;3:e324. doi: 10.1371/journal.pbio.0030324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Li T., Fu L., Yu C., Li Y., Xu X., Wang Y., Ning H., Zhang S., Chen W., Babiuk L.A., Chang Z. Silencing SARS-CoV Spike protein expression in cultured cells by RNA interference. FEBS Lett. 2004;560:141–146. doi: 10.1016/S0014-5793(04)00087-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Zhang F., Yu W., He T., Yu J., Yi C.E., Ba L., Li W., Farzan M., Chen Z., Yuen K.Y., Ho D. Antibody responses against SARS coronavirus are correlated with disease outcome of infected individuals. J. Med. Virol. 2006;78:1–8. doi: 10.1002/jmv.20499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Xiao G., Xu Y., Yuan F., Zheng C., Liu Y., Yan H., Cole D.K., Bell J.D., Rao Z., Tien P., Gao G.F. Following the rule: formation of the 6-helix bundle of the fusion core from severe acute respiratory syndrome coronavirus spike protein and identification of potent peptide inhibitors. Biochem. Biophys. Res. Commun. 2004;319:283–288. doi: 10.1016/j.bbrc.2004.04.141. [DOI] [PMC free article] [PubMed] [Google Scholar]


