Highlights
► We established a novel in vitro magnetic pull down assay to measure Ebola full length recombinant VP35–dsRNA binding. ► We determined the full length rVP35 Kd value for dsRNA to be 2.8 nM. ► We measured the IFN-inhibiting defective R305A, K309A and R312A rVP35 mutants dsRNA binding Kd values. ► We identified ATA as the first rVP35–dsRNA binding inhibitor.
Keywords: Ebola virus, VP35, dsRNA binding, Biochemical screening assay
Abstract
The Ebola viruses (EBOVs) VP35 protein is a multifunctional major virulence factor involved in EBOVs replication and evasion of the host immune system. EBOV VP35 is an essential component of the viral RNA polymerase, it is a key participant of the nucleocapsid assembly and it inhibits the innate immune response by antagonizing RIG-I like receptors through its dsRNA binding function and, hence, by suppressing the host type I interferon (IFN) production. Insights into the VP35 dsRNA recognition have been recently revealed by structural and functional analysis performed on its C-terminus protein. We report the biochemical characterization of the Zaire ebolavirus (ZEBOV) full-length recombinant VP35 (rVP35)–dsRNA binding function. We established a novel in vitro magnetic dsRNA binding pull down assay, determined the rVP35 optimal dsRNA binding parameters, measured the rVP35 equilibrium dissociation constant for heterologous in vitro transcribed dsRNA of different length and short synthetic dsRNA of 8 bp, and validated the assay for compound screening by assessing the inhibitory ability of auryntricarboxylic acid (IC50 value of 50 μg/mL). Furthermore, we compared the dsRNA binding properties of full length wt rVP35 with those of R305A, K309A and R312A rVP35 mutants, which were previously reported to be defective in dsRNA binding-mediated IFN inhibition, showing that the latter have measurably increased Kd values for dsRNA binding and modified migration patterns in mobility shift assays with respect to wt rVP35. Overall, these results provide the first characterization of the full-length wt and mutants VP35–dsRNA binding functions.
1. Introduction
Ebolaviruses (EBOVs) constitute a group of five species of filamentous, enveloped, non-segmented single- and negative-stranded RNA viruses belonging to the family of Filoviridae, order of Mononegavirales (Barrette et al., in press, Kuhn et al., 2010). EBOVs are causative agents of highly lethal hemorrhagic fevers in humans and non-human primates and they are classified as category A pathogens, for which treatments and vaccination are currently lacking (Falzarano et al., 2011, Feldmann and Geisbert, 2011). Although human outbreaks are rare and generally confined to sub-Saharan African regions, the risk of infection due to travel-imported cases or their misuse as bioterrorism agents place EBOVs among the highest public health threats (Hartman et al., 2010, Leroy et al., 2011). In 2008, the discovery in the Philippines of the presence of viruses belonging to the Reston ebolavirus (REBOV) species in infected domestic swine, gave rise to worldwide concern due to EBOVs theoretical capability of entering the human food chain (Barrette et al., 2009). More recently, susceptibility of pigs to EBOVs infection was assessed, showing that the virus is able to replicate with high titers in their respiratory tract and to shed to naïve hosts from the oronasal mucosa (Kobinger et al., 2011). Furthermore, the EBOVs capability of inducing disease following aerosol inhalation exposure has been recently demonstrated for three species of non-human primates (Reed et al., 2011).
In humans, EBOV hemorrhagic fever often results in very high mortality rates, reaching 90% for the most lethal species such as Zaire ebolavirus (ZEBOV). As recently observed in ZEBOV human fatal cases (Wauquier et al., 2010), as well as in previously described studies with experimentally infected animal models, death is associated with a markedly impaired innate immunity reaction, a strong production of pro-inflammatory cytokines and a profound immunosuppression, resulting in peripheral T lymphocyte apoptosis and lack of adaptive immunity (Mahanty and Bray, 2004, Mohamadzadeh et al., 2007, Zampieri et al., 2007). By contrast, survivors to EBOVs infection seem to develop an effective immune response (Becquart et al., 2010, Wauquier et al., 2009). This disparity suggests that events early in the EBOV infection may influence the patients’ ability to activate an effective immune response (Basler and Amarasinghe, 2009, Mohamadzadeh, 2009).
In general, pathogen associated molecular patterns (PAMPs) are recognized, early in infection, by PAMP recognition receptors (PRRs) (Randall and Goodbourn, 2008) which activate the host innate immune response (Koyama et al., 2008). Among PAMPs, dsRNA is a unique viral product, very effectively detected by cellular PRRs such as retinoic acid-inducible gene I (RIG-I) and melanoma differentiation-associated gene 5 (MDA-5) proteins (Loo and Gale, 2011, Yoneyama and Fujita, 2010) which, once recognized dsRNAs, initiate signaling cascades that activate interferon regulatory factor 3 (IRF-3), leading to production of interferon α/β (IFN-α/β) which, in turn, activates the antiviral responses (Sadler and Williams, 2008). To circumvent viral PAMPs recognition by cellular PRRs, viruses evolved different strategies to block IFN-α/β activation (Bowie and Unterholzner, 2008, Conzelmann, 2005, Katze et al., 2008). In particular, EBOVs code for two viral proteins, VP24 and VP35, which display innate immune antagonism (Basler and Amarasinghe, 2009).
VP35 is a multifunctional protein that is indispensable for EBOVs replication. In fact, VP35 is an essential cofactor of the EBOV RNA polymerase complex (Boehmann et al., 2005, Mühlberger et al., 1998, Mühlberger et al., 1999), it serves as a viral assembly factor (Huang et al., 2002, Johnson et al., 2006), it is a RNAi suppressor in mammalian cells (Haasnoot et al., 2007, Fabozzi et al., 2011) and, moreover, it counteracts the host innate immune response by blocking, at many different steps, the production of IFNs-α/β. In this respect, it has been reported that VP35 (i) binds to dsRNA, suppressing RIG-I helicase signaling cascade triggered by dsRNA recognition (Cárdenas et al., 2006); (ii) impairs IKK-ε and TBK-1 kinases functionality (Prins et al., 2009); (iii) enhances PIAS1-mediated SUMOylation of IRF-7, decreasing IFN induction (Chang et al., 2009); (iv) abrogates IRF-3 phosphorylation–dimerization and nuclear translocation (Basler et al., 2000, Basler et al., 2003, Hartman et al., 2006, Hartman et al., 2008a) and (v) inhibits dsRNA-induced activation of PKR (Feng et al., 2007, Schümann et al., 2009). In particular, among VP35 functions, dsRNA binding seems to be the most important in the context of EBOVs pathogenesis, since mutations in its dsRNA binding domain (RBD) (i) abolish its IFN-antagonism properties (Cárdenas et al., 2006, Hartman et al., 2004); (ii) result in greatly attenuated viral growth rate and virulence loss (Hartman et al., 2006, Hartman et al., 2008a, Prins et al., 2010); (iii) abrogate VP35 capability to suppress RNA silencing (Haasnoot et al., 2007).
VP35 contains an N-terminal coiled-coil domain that is important for its homo-oligomerization (Möller et al., 2005, Reid et al., 2005) and a C-terminal RBD (Cárdenas et al., 2006, Hartman et al., 2004, Leung et al., 2009, Leung et al., 2010b, Leung et al., 2010c). The coiled-coil domain is required for several VP35-mediated functions such as viral replication (Möller et al., 2005, Reid et al., 2005) nucleocapsid formation (Huang et al., 2002, Johnson et al., 2006, Shi et al., 2008) and immune suppression (Feng et al., 2007, Jin et al., 2010, Reid et al., 2005), while the VP35 RBD is involved in dsRNA binding, mainly linked to immune suppression (in fact, at our best knowledge, the VP35–dsRNA binding properties seems to be not involved in the replication complex function), even though, when the sole VP35 RDB is expressed into cells, it is not able to suppress the type I IFN activation at the same level than the full length protein (Basler and Amarasinghe, 2009, Cárdenas et al., 2006, Feng et al., 2007, Reid et al., 2005). Currently, the complete tertiary structure of the full length EBOV VP35 is not yet available. However, the crystallographic resolution of the RBD C-terminal portion (amino acid residues 221–340) has been recently solved for the ZEBOV and REBOV species, either alone, or bound to short dsRNA molecules (Kimberlin et al., 2010, Leung et al., 2009, Leung et al., 2010c, Leung et al., 2010d). EBOV VP35 RBD presents a unique fold among the dsRNA binding proteins, different from the αβββα fold of canonical cellular dsRNA binding proteins (Leung et al., 2009, Leung et al., 2010c) as well as from that of other viral multifunctional proteins which also inhibit IFN-α/β activation, such as the influenza virus NS1 protein (Kimberlin et al., 2010, Leung et al., 2009). The structural uniqueness of the VP35 RDB, the importance of the VP35 inhibition of the IFN-α/β activation following EBOV infection for viral pathogenesis and the essential VP35 role in EBOV replication clearly indicate that the VP35 RBD is a promising therapeutic target for viral inhibition.
With the aim of characterizing the VP35–dsRNA binding function, we have previously expressed in bacterial systems and purified the full-length ZEBOV recombinant VP35 (rVP35), demonstrating its ability to bind dsRNA and its suitability for biochemical studies (Zinzula et al., 2009). Since all the biochemical data currently available on the VP35 binding to dsRNA had been obtained using the truncated version of its isolated RBD, which encompasses only one third of the VP35 protein, we report the first biochemical characterization of the full length rVP35–dsRNA binding. In order to perform such study, we established a new in vitro magnetic pull down assay and characterized the optimal parameters for rVP35–dsRNA binding. We determined the VP35-dsRNA equilibrium dissociation constant (K d) obtained with substrates of different length, diverse end-capping characteristics and at different temperatures. Furthermore, we validated this assay as a tool for compound screening by assessing the ability of the auryntricarboxylic acid (ATA) to inhibit rVP35–dsRNA interaction. Finally, since studies performed on the isolated VP35 RBD domain have suggested that single mutations in a RBD central basic patch are sufficient to completely loose the dsRNA binding ability (Leung et al., 2009, Leung et al., 2010c, Leung et al., 2010d) we characterized the VP35–dsRNA interaction of the full length R305A, K309A and R312A rVP35 mutants.
2. Material and methods
2.1. ZEBOV rVP35 wt and mutant plasmids
Cloning of the ZEBOV VP35 gene into pET45b(+) vector (Novagen) to obtain the pET45b-ZEBOV-VP35 plasmid was previously described (Zinzula et al., 2009). Site-directed mutagenesis was performed using the QuickChange II Site-Directed Mutagenesis kit (Agilent technologies) to introduce the single point mutations R305A, K309A and R312A in the VP35 gene of the mutant plasmids pET45b–ZEBOV–VP35/R305A, pET45b–ZEBOV–VP35/K309A and pET45b–ZEBOV–VP35/R312A, respectively.
2.2. Expression and purification of full-length wt and mutants ZEBOV rVP35
The full-length, wt and mutants, bacterially-expressed rVP35s were obtained as previously described (Zinzula et al., 2009). Briefly, protein expression was carried out, using the above described plasmids, in BL21AI Escherichia coli cells, cultured in LB media at 37 °C and induced at an optical density of 0.6 at 600 nm with 0.4% l-arabinose. rVP35s were purified with the IMAC resin Ni-Sepharose High Performance (GE Healthcare) using a BioLogic FPLC system (Biorad) and dialyzed in desalting buffer (50 mM sodium phosphate, 300 mM NaCl, 10% glycerol, 0.014% β-mercaptoethanol). As previously published (Zinzula et al., 2009), rVP35 full-length protein was purified at ≈95% homogeneity, its integrity was assessed by PAGE analysis and its concentration was calculated using the Protein Quantification kit-Rapid (Fluka).
2.3. dsRNA in vitro transcription and dsRNA labeling
Heterologous 500 bp dsRNA was produced by in vitro transcription using the T7 MEGAscript RNAi kit (Ambion) from the linearized DNA provided with the kit, according to manufacturer’s instructions. The 500 bp labeled dsRNA was generated by supplementing the in vitro transcription reaction with 0.15 μCi of 3H-GTP (35.5 Ci/mmol) (Perkin–Elmer). The DNA control template provided by Ambion was used to generate, by standard PCR protocols, other two linearized DNA templates to in vitro transcribe the 150 and 50 bp dsRNA as above. All in vitro transcribed (IVT) dsRNA molecules were purified from transcription reaction with the MEGAclear kit (Ambion) or with Quick Spin G25 columns (Roche), and quantified by spectrophotometry. dsRNA oligos of 8 bp in length (sequence 5′-GCGUACGC-3′) bearing 5′ phosphate and 5′ hydroxyl ends were purchased from Metabion International AG (Germany). The integrity of DNA templates, IVT dsRNAs and synthetic dsRNA molecules was assessed by agarose-gel electrophoresis.
2.4. Electrophoresis mobility shift assay (EMSA)
Purified IVT dsRNA molecules of different length were incubated at a concentration of 40 nM with 0.3 μg rVP35 in a reaction volume of 50 μL containing 50 mM sodium phosphate pH 7.5, 100 mM NaCl, 20 mM MgCl2 and 1 μg tRNA for 30 min at 23 °C under gentle rotating agitation (20 rpm). Five microliters of 6× non-denaturing gel loading buffer (37% glycerol, 0.025% bromophenol blue, 0.025% xylene cyanol, 20 mM Tris–HCl pH 8.0) were then added to the reaction and the samples were loaded on a 1.7% non-denaturing agarose gel. Electrophoresis was allowed to proceed in 1X TBE running buffer at 4 °C for 2.5 h at a constant voltage (5.5 V/cm). Gels were stained with 1X SYBR Green II RNA gel stain (Invitrogen) and migration profiles of reaction products were visualized on a UV transilluminator.
2.5. Magnetic pull down assay
The rVP35–dsRNA complex formation was assessed exploiting the properties of the TALON paramagnetic Dynabeads (Invitrogen). Firstly, 1 μg of rVP35 was conjugated to 50 μL TALON beads in a volume of 700 μL of binding buffer (50 mM sodium phosphate pH 7.5, 150 mM NaCl, 0.05% Tween-20) for 15 min at 23 °C under gentle rotating agitation (20 rpm). Unconjugated rVP35 was removed by magnetic field application, supernatant removal and further washing with binding buffer. Pellets with conjugated rVP35 were re-suspended in a 100 μL volume binding buffer containing 20 mM MgCl2 and 1.5 nM 500 bp 3H-dsRNA (0.1 Ci/mmoles) and incubated for 60 min at 37 °C (20 rpm). Unbound 3H-dsRNA was separated by the conjugated rVP35–dsRNA complex by magnetic field application and supernatant removal. A further washing step was performed to completely remove unbound 3H-dsRNA. 3H-dsRNA elution was performed by incubation of the pellets in 300 μL elution buffer (binding buffer plus 1 M imidazole pH 7.5) for 10 min at 23 °C (20 rpm), subsequent magnetic field application and supernatant removal. The supernatant was transferred to vials and its radioactivity determined with a Beckman LS 6500 beta-counter.
2.6. Determination of biochemical parameters for optimal dsRNA binding
The determination of the equilibrium dissociation constants (K d), was performed by competition binding studies using the homologous competition approach for the 500 bp dsRNA and the heterologous competition for the 150 and 50 bp dsRNAs, as described (Bylund and Murrin, 2000, Motulsky and Neubig, 2010). In both cases, increasing amounts of unlabeled dsRNA were titrated against a fixed amount of labeled 500 bp 3H-dsRNA (0.1 Ci/mmoles). Raw data from at least three independent binding experiments were used to calculate K d value utilizing non-linear regression fitting model provided by Prism software (GraphPad).
2.7. Thermodynamic Van’t Hoff analysis
The effect of temperature on rVP35 binding affinity for dsRNA was evaluated using a comparative thermodynamic Van’t Hoff analysis, as described (Chung et al., 2010). Briefly, triplicate 500 bp dsRNA homologous-competition binding experiments were conducted at 23 °C, 30 and 37 °C and titration curves were generated for K d determination. The K d values obtained at each temperature were plotted as ln (K d) versus 1/temperature (T; expressed in Kelvin), and the simplified Van’t Hoff equation ln (K d) = [(ΔH/R) − (1/T)] − ΔS/R was used to estimate enthalpy and entropy, by assuming that these two parameters are constant within the temperature range assayed. ln (K d) is the natural log of the equilibrium dissociation constant, ΔH and ΔS are the enthalpy and entropy and R is the molar gas constant (8.314 kJ−1 mol−1).
3. Results
3.1. rVP35 shifts the electrophoretic migration of an heterologous 500 bp IVT dsRNA
It has been previously shown that full length EBOV VP35 expressed in mammalian cells can bind to poly-IC, poly-AU as well as IVT dsRNA molecules ranging from 200 to 1000 bp (Cárdenas et al., 2006). When truncated, E. coli-expressed VP35 proteins were tested, different dsRNA binding abilities were observed. In fact, while a VP35 truncated version containing the amino acid residues 169–340 was still able to bind poly-IC dsRNA (Cárdenas et al., 2006), the protein containing only the 201–340 amino acid residues was not able to co-precipitate with the poly-IC substrate (Feng et al., 2007). However, more recently, a shorter VP35 truncated protein containing the amino acid residues 221–340 was found to bind dsRNA as short as 8 bp in length (Kimberlin et al., 2010, Leung et al., 2009, Leung et al., 2010a, Leung et al., 2010b, Prins et al., 2009). Within this context, we previously reported the expression and purification of a full-length rVP35 protein and demonstrated that it co-precipitates with agarose-beaded poly-CG in a standard pull down assay, thereby assessing the rVP35 ability to bind dsRNA, similarly to the native VP35 (Zinzula et al., 2009). Therefore, in order to further characterize the full-length rVP35–dsRNA binding ability, we first asked whether the rVP35 protein could effectively bind to heterologous IVT dsRNA molecules obtained using a T7-promoter-driven DNA–plasmid. To answer this question, we performed an EMSA using IMAC purified ZEBOV rVP35 (Fig. 1 A) and IVT dsRNAs, showing that rVP35 binds to the 500 bp dsRNA and leads to a shift in its electrophoretic migration (Fig. 1B, compare lanes 3 to 4). In addition to the shifted dsRNA signal into the gel (second band from the top of the gel), a further fluorescent signal could also be observed stuck in the well of the gel, which was possibly ascribed to higher molecular mass rVP35–dsRNA specific aggregates (Fig. 1B, lanes 4 first band from the top). In this respect, it is important to note that (i) the migration of rVP35 alone was unable to produce any UV fluorescing signal in the gel (Fig. 1B, lane 2), therefore excluding the possible presence of bacterial RNA bound to rVP35; (ii) the addition of an excess of tRNA in the mixture did not alter the rVP35-dsRNA bands pattern (Fig. 1B, lane 5); (iii) heat-denatured rVP35 did not display any form of rVP35–dsRNA aggregates (Fig. 1B, lane 7). Moreover, EMSA performed using dsRNA substrates of 150 and 50 bp in length led to similar results (data not shown), in agreement with the length-independent modality proposed for VP35 binding to dsRNA (Kimberlin et al., 2010, Leung et al., 2010c). Altogether, EMSA experiments showed that bacterially expressed rVP35 is able to modify the migration profile of IVT dsRNAs of 50–500 bp in length. Noteworthy, even though the two distinct bands observed in the gel are probably due to specific rVP35–dsRNA complexes of different weight, their functional significance is still not determined. Overall, these data demonstrate the suitability of the heterologous IVT dsRNA molecules as effective substrates for quantitative studies on VP35–dsRNA binding.
Fig. 1.
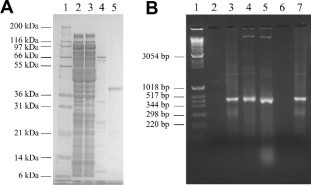
rVP35 shifts the electrophoretic migration of heterologous IVT dsRNAs. (A) Coomassie blue-stained 12% SDS–PAGE showing IMAC-purified, full length ZEBOV rVP35. Lane 1, molecular weight marker; lane 2, E. coli crude extract cell lysate; lane 3, IMAC unbound flow through proteins; lane 4, unspecific washed proteins; lane 5, eluted rVP35. (B) 1.7% non-denaturing agarose EMSA shows that rVP35 is able to bind dsRNA of 500 bp in length, retarding its migration on a gel. Lane 1, molecular weight marker; lane 2, rVP35 alone; lane 3, IVT 500 bp dsRNA; lane 4, IVT 500 bp dsRNA + rVP35; lane 5, IVT 500 bp dsRNA + tRNA + rVP35; lane 6, rVP35 alone (heat-denatured); lane 7, IVT 500 bp + rVP35 (heat-denatured).
3.2. rVP35 binds to a 3H-labeled 500 bp dsRNA in a magnetic pull down assay
With the aim of establishing a quantitative method to measure the rVP35 binding to IVT dsRNA we exploited the biochemical properties of the TALON Dynabeads which are paramagnetic beads that specifically bind His-tagged recombinant proteins. Purified rVP35 was first coated to Dynabeads through its N-terminal His-tag to form stable complexes of conjugated beaded-proteins. The use of excess rVP35 assured that all beads were conjugated with rVP35 and the application of magnetic field to the solution allowed the washing out of the non-conjugated rVP35 (data not shown). The conjugated rVP35 was incubated with a 500 bp IVT 3H-dsRNA, allowing the binding to dsRNA. The unbound 3H-dsRNA was washed out by magnetic field application while the rVP35–3H-dsRNA complex was subsequently eluted with imidazole and quantified. Data showed that dsRNA was specifically retained by conjugated rVP35, in fact, the 3H-dsRNA binding to the empty beads (background signal) was very low as compared to the rVP35–3H-dsRNA binding (data not shown). Optimal binding conditions were determined to be at pH 7.5, 100 mM NaCl and 20 mM MgCl2 (Fig. 2 A–C, respectively) and the quantification of the dsRNA bound to rVP35 was shown to be linearly dependent on the conjugated-rVP35 concentration (Fig. 2D). Overall, these results validated the magnetic pull down technique as a tool to measure the rVP35–dsRNA binding.
Fig. 2.
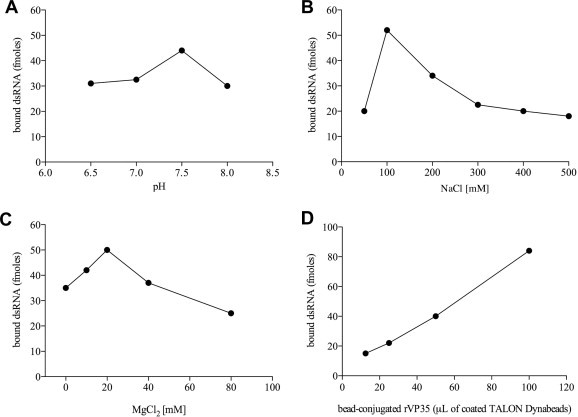
Characterization of optimal binding conditions in the magnetic pull down assay. The formation of a stable complex between VP35 and IVT dsRNA is dependent to pH, ionic strength and concentration of the divalent cation Mg2+. Optimal binding was observed at pH 7.5 (A), in the presence of 100 mM NaCl (B) and 20 mM MgCl2 (C). rVP35 binding to IVT dsRNA was shown to be correlated to the amount of conjugated protein to TALON Dynabeads: a rVP35 dose–response binding curve shows the linear dependence between VP35–dsRNA binding and rVP35 concentration (D).
3.3. Full length rVP35 binds dsRNAs with high affinity
It has been reported that, as measured by isothermal titration calorimetry (ITC), the RBD of VP35 (aminoacids 221–340) binds to a synthetic 8 bp dsRNA with a K d value of 500 nM, while it binds to an IVT 8 bp dsRNA with higher affinity, showing a K d value of 30 nM (Leung et al., 2010c, Leung et al., 2010d). Hence, we wanted to estimate the full length rVP35 equilibrium dissociation constant using more natural and longer substrates utilizing the magnetic pull down assay. Homologous competition binding experiments, in which the IVT 500 bp dsRNA, 3H-labeled and unlabeled, were used as ligand and competitor, respectively, were firstly performed (Fig. 3 A). Data showed that the full-length rVP35 K d value for IVT 500 bp dsRNA binding at 37 °C temperature was 2.8 ± 0.1 nM.
Fig. 3.
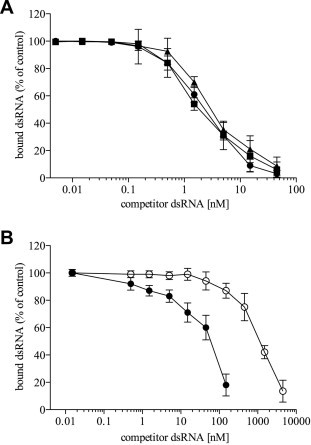
rVP35 binds to IVT dsRNAs of different length with a very high affinity. Competition binding experiments, performed with the magnetic pull down assay at 37 °C at optimal biochemical conditions, show that: (A) rVP35 binds IVT dsRNA molecules of 50–500 bp in a length-independent manner. Unlabeled dsRNA of 500 bp (full circle), 150 bp (full square) and 50 bp (full triangle) in length are titrated to compete against a fixed amount of the 500 bp 3H–dsRNA ligand. Comparable values of the apparent equilibrium dissociation constant were observed: 2.8 ± 0.1 nM (500 bp dsRNA), 2.4 ± 0.3 nM (150 bp dsRNA) and 3.2 ± 0.5 nM (50 bp dsRNA); (B) rVP35 binding to synthetic short dsRNAs with different 5′ ends. Unlabeled 5′-phosphate dsRNA (full circle) and 5′-hydroxyl dsRNA (empty circle) of 8 bp in length are titrated to compete against a fixed amount of the 500 bp 3H–dsRNA ligand. Different equilibrium dissociation constant were observed: of 64 ± 9 nM (5′-phosphate) and 1.1 ± 0.2 μM (5′-hydroxyl). Concentration of dsRNA competitor is plotted versus the bound percentage of the radiolabeled ligand, each experimental point represents the mean ± SD of specific bound dsRNA from at least three independent experiments.
Since our EMSA studies performed with IVT dsRNA of different length suggested that rVP35 could bind to IVT dsRNA of 500, 150 and 50 bp in length with a similar mode, we wanted to determine whether, despite this similarity, any difference would exist in the rVP35 binding affinity constant for IVT 500, 150 and 50 bp dsRNAs. Heterologous competition binding curves were performed at 37 °C temperature using 500 bp IVT 3H-dsRNA as ligand and unlabeled IVT 150 and 50 bp dsRNA molecules as competitors, and the K d values were calculated to be 2.4 ± 0.3 and 3.2 ± 0.5 nM, respectively (Fig. 3A). Furthermore, given that it was reported that the VP35 RBD is able to bind with high affinity dsRNA molecules as short as 8 bp in length, we also tested the full-length rVP35–dsRNA binding function towards such short substrates. To this end, heterologous competition binding curves were performed using 500 bp IVT 3H-dsRNA as ligand and two synthetic 8 bp dsRNA as unlabeled competitors. One of the 8 bp dsRNA competitors terminated with a 5′-phosphate, the other with a 5′-hydroxyl group. Data showed that rVP35 bound to the 5′-phosphate 8 bp dsRNA with high affinity, showing a K d value of 64 ± 9 nM, while it bound to the 5′-hydroxyl 8 bp dsRNA with a lower affinity, showing a K d value of 1.1 ± 0.2 μM. Overall, these data show that rVP35 binding affinity for IVT dsRNA in the magnetic pull down assay is independent from dsRNA length, at least for substrates ranging from 50 to 500 bp. In addition, rVP35 demonstrates a lower affinity for a 5′-phosphate 8 bp dsRNA, and a dramatic affinity reduction for a 8 bp dsRNA molecule lacking the 5′ phosphate group.
In the process of the magnetic pull down assay optimization, we observed that rVP35 binding ability to dsRNA was slightly dependent on temperature (data not shown). Therefore, we wanted to precisely determine the effect of the temperature on the rVP35 binding by carrying out homologous competition curves with IVT 500 bp dsRNA also at 30 and 23 °C (Fig. 4 A). Results showed that, as temperature decreases, the rVP35 binding affinity for dsRNA increases. In fact, the rVP35 K d values for the binding to IVT 500 bp dsRNA were equal to 1.5 ± 0.5 and 1.1 ± 0.1 nM, at 30 and 23 °C, respectively, indicating that the rVP35–dsRNA binding at these temperatures does not change significantly. The effect of temperature on the rVP35–dsRNA binding affinity was also examined using the Van’t Hoff analysis which allows to determine ΔG, ΔH and ΔS values (Chung et al., 2010). The final Van’t Hoff plot was linear (Fig. 4B) and the thermodynamic parameters were calculated to be ΔH = −14.9 kcal/mol, ΔS = −0009 eu and ΔG = −12.12 kcal/(mol K) at 37 °C. These values are in agreement with the thermodynamic parameters calculated for other dsRNA binding proteins such as the protein kinase PKR, which were reported to be from −41.7 to −26.3 kcal/(mol K) for ΔG and from −21.1 to −7.3 kcal/(mol K) for ΔH (Zheng and Bevilacqua, 2000).
Fig. 4.
Effect of temperature on rVP35 binding to IVT 500 bp dsRNA. Analysis of the rVP35-binding affinity to IVT 500 bp dsRNA at different temperatures. (A) Homologous-competition binding curves performed with the magnetic pull down assay at optimal binding conditions at different temperatures (full circle, 37 °C; full triangle, 30 °C; full square, 23 °C). Concentration of IVT 500 bp unlabeled dsRNA is plotted versus the bound percentage of the same IVT 500 bp 3H–dsRNA molecule, each experimental point represents the mean ± SD of specific bound dsRNA from three independent experiments. (B) Van’t Hoff plot describing the effect of temperature change upon rVP35 binding affinity to 500 bp dsRNA. The natural log of the equilibrium dissociation constant is plotted versus the reciprocal value (in Kelvin) of the incubation temperature.
3.4. rVP35–dsRNA interaction is inhibited by ATA
The EBOV VP35 RBD has a unique fold among the dsRNA binding proteins, which is different from the αβββα-fold of the canonical cellular dsRNA binding proteins (Kimberlin et al., 2010, Leung et al., 2009, Leung et al., 2010c). Thus, given the important role of the VP35 inhibition of the IFN-α/β activation following EBOV infection for viral pathogenesis (Leung et al., 2010a), the VP35–dsRNA binding ability represents a promising therapeutic target for viral inhibition. Therefore, we wanted to validate our newly established magnetic pull down assay also as a method for the screening of molecules that may inhibit the interaction between dsRNA and VP35. In the absence of a specific VP35–dsRNA binding inhibitor, we used a known inhibitor of nucleic acid–protein interactions such as the ATA (Ghosh et al., 2009, Gonzáles et al., 1980) (Fig. 5 ). Results showed that ATA inhibited rVP35–dsRNA binding in a dose–response manner with an IC50 of 50 μg/mL. However, to be suitable for the identification of antivirals by screening compound libraries, a biochemical assay is required to be up-scalable to a medium or high-throughput system. To satisfy such requirement, we also tested the magnetic pull down assay in a 96-wells format, which is cost-effective and amenable of automation. In this scale-up, ATA was found to inhibit the rVP35–dsRNA binding with an IC50 of 61 μg/mL (data not shown), confirming that the rVP35 magnetic pull down assay is an effective in vitro biochemical assay for the screening of potential antiviral agents.
Fig. 5.
Inhibition of rVP35–dsRNA interaction by ATA. Dose–response curve of the rVP35 binding to IVT 500 bp dsRNA inhibition by ATA. The reactions were performed at 37 °C at optimal biochemical conditions and titrated with compound concentrations ranging from 0.1 to 300 μg/mL. Results are shown as the percentage of specific bound dsRNA and represent the average of three independent determinations.
3.5. R305A, K309A and R312A rVP35 mutants display modified migration profiles on EMSA and bind to IVT 500 bp dsRNA with lower affinity with respect to wt rVP35
It has been reported that some amino acid residues in the RBD are important for dsRNA binding. In particular, the R312A mutation severely impaired the VP35–dsRNA binding ability and its IFN antagonist activity in cell culture, without affecting the VP35 function as part of the viral polymerase complex (Cárdenas et al., 2006, Hartman et al., 2004, Hartman et al., 2008a, Hartman et al., 2008b, Leung et al., 2009). More recently, the 221–340 amino acid R312A VP35 truncated protein was shown to lose the dsRNA binding ability, as measured by EMSA (Leung et al., 2009) and ITC analysis (Leung et al., 2010c). Similarly, the K309A mutation has also been reported to greatly impair the VP35–dsRNA binding ability and its IFN antagonist activity in cell culture, without altering the VP35 function in the viral polymerase complex (Cárdenas et al., 2006, Hartman et al., 2004, Leung et al., 2009) and the 221–340 amino acid K309A VP35 mutant was unable to bind dsRNA, as measured by EMSA analysis (Leung et al., 2009). In addition, the R305A mutation was recently reported to reduce by 3-fold the 221–340 amino acid VP35 truncated protein K d value for dsRNA without affecting the viral replication (Leung et al., 2010c). Within this picture, we were interested in assessing the impact of these mutations on the full-length rVP35–dsRNA binding ability. Therefore, we performed in vitro site-directed mutagenesis of the wt plasmid construct to express the R305A, K309A and R312A rVP35s in E. coli and purified these mutants as full length, recombinant, His-tagged proteins. Next, we assessed the ability of the R305A, K309A and R312A rVP35 mutants to bind to heterologous dsRNAs in EMSA studies. Using the IVT 500 bp dsRNA, the rVP35 mutants, when compared to wt rVP35, displayed a diminished ability to shift the dsRNA into the gel, even though the shifted band was only scarcely visible (Fig. 6 A and B). Differently, all mutants maintained their ability to form the higher order aggregates, which were retained into the wells. Similar results were obtained using dsRNA substrates of 150 and 50 bp in length (data not shown).
Fig. 6.
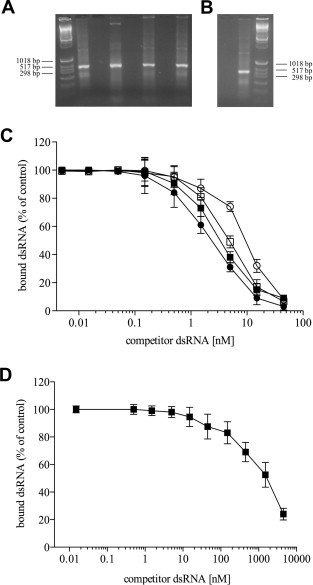
R305A, K309A and R312A rVP35 mutants display diminished dsRNA binding on native gel shift and decreased affinity for dsRNA as compared with wt rVP35. Non-denaturing agarose EMSA shows that R305A, K309A and R312A rVP35 mutants are not able to shift the IVT dsRNA of 500 bp in length migration in the gel as wt rVP35, even though they retain a certain ability to bind IVT dsRNA. (A) Lane 1, molecular weight marker; lane 2, IVT 500 bp dsRNA alone; lane 3, wt rVP35 alone; lane 4, wt rVP35 + IVT 500 bp dsRNA; lane 5, R305A rVP35 alone; lane 6, R305A rVP35 + IVT 500 bp dsRNA; lane 7, K309A rVP35 alone; lane 8, K309A rVP35 + IVT 500 bp dsRNA. (B) Lane 1, R312A rVP35 alone; lane 2, R312A rVP35 + IVT 500 bp dsRNA; lane 3, molecular weight marker. (C) Homologous-competition binding, performed with the magnetic pull down assay at 37 °C at optimal biochemical conditions, showed that R305A rVP35 (full square, Kd = 3.85 ± 0.6 nM), K309A rVP35 (empty square, Kd = 4.99 ± 1.03 nM) and R312A rVP35 (empty circle, Kd = 10.6 ± 1.7 nM) bind to 500 bp IVT dsRNA with lower affinity with respect to wt rVP35 (full circle, Kd = 2.8 ± 0.1 nM). (D) Heterologous competition binding experiment, performed with the magnetic pull down assay at 37 °C at optimal biochemical conditions, showed that R312A rVP35 (full square, Kd value of 1.76 ± 0.2 μM) binds to 8 bp 5′-phosphate dsRNA with significantly lower affinity with respect to wt rVP35. Concentration of unlabeled competitor dsRNA is plotted versus the bound percentage of the radiolabeled dsRNA ligand, each experimental point is the mean ± SD of specific bound dsRNA from three independent experiments.
Subsequently, given that the EMSA analysis was merely qualitative, we wanted to quantify the differences in dsRNA binding observed for the different rVP35 mutants using the magnetic pull down assay. Therefore, we performed homologous competition binding curve using the IVT 500 bp dsRNA as ligand at 37 °C temperature for the three rVP35 mutants (Fig. 6C). Results showed that all three rVP35 mutants were able to bind the IVT 500 bp dsRNA but showed measurable increased K d values with respect to wt rVP35. In particular, the R305A rVP35–dsRNA binding affinity appeared to be only slightly lower than wt rVP35 affinity, showing a K d value dsRNA of 3.85 ± 0.6 nM (with a p value equal to 0.0418 with respect to wt rVP35). Interestingly, the K d value for R309A rVP35 was 4.99 ± 1.0 nM (with a p value equal to 0.0219 with respect to wt rVP35) and the K d value for R312A rVP35 was 10.60 ± 1.7 nM (with a p value equal to 0.0015 with respect to wt rVP35). In addition, given that K d value for wt rVP35 binding to the 5′-phosphate 8 bp dsRNA was roughly 25-fold higher than the K d values observed for 50–500 bp dsRNAs, we wanted to assess whether a comparable decrease in dsRNA binding affinity could be observed for the R312A–rVP35 binding. As shown by heterologous competition binding curve (Fig. 6D), the K d value calculated for the R312A–rVP35 binding to the 5′-phosphate 8 bp dsRNA was 1.76 ± 0.2 μM, around 166-fold higher than the value shown for the 500 bp dsRNA.
4. Discussion
At nearly 40 years from their identification, EBOVs are still highly infectious pathogens without available therapy and, despite the vast knowledge achieved in the last two decades on their biology and pathogenesis, the molecular basis for their extreme lethality remain to a large extent unraveled (Feldmann and Geisbert, 2011). However, the impairment of the innate immunity, and particularly the suppression of the IFN-α/β response, has been proved to be critical in supporting an efficient EBOV replication (Basler and Amarasinghe, 2009). Recently, the multifunctional virally-coded VP35 has been proposed to be the key determinant of the EBOVs virulence (Jin et al., 2010), and its ability to antagonize the IFN-α/β response has been ascribed mainly to its capacity to bind and sequestrate dsRNA, thereby suppressing RIG-I signaling cascade triggered by this viral nucleic acid (Cárdenas et al., 2006, Hartman et al., 2004, Hartman et al., 2006). In fact, EBOVs bearing mutations in the VP35 dsRNA RBD, which reduce or suppress VP35–dsRNA binding, display greatly attenuate viral growth rate or are avirulent in animal models (Hartman et al., 2008b, Prins et al., 2010). Until now, all the in vitro information on the VP35–dsRNA binding were referred to data obtained using a truncated version of the VP35 protein, including only the RBD, and dsRNA molecules as short ad 8–18 bp (Kimberlin et al., 2010, Leung et al., 2009, Leung et al., 2010c, Leung et al., 2010d). Therefore, it was of interest to perform the first characterization of the full-length rVP35 binding to heterologous IVT dsRNA as long as 50–500 bp.
Data from the EMSA study we performed to demonstrate that the IVT dsRNA could be used for subsequent quantitative studies, clearly indicate that full length rVP35 binds to 50–500 bp IVT dsRNAs. rVP35 determined a shift in their mobility, forming two main rVP35–dsRNA complexes regardless of the dsRNA length, which seem to be both specific. The precise composition of the two complexes are not known yet, however, it is worth to note that gel filtration studies performed on the full length VP35 suggest that VP35 forms dimers, tri-tetramers and higher order aggregates (Zinzula et al., 2009, Reid et al., 2005, Möller et al., 2005, Leung et al., 2010c). Therefore, it is possible to hypothesize that the VP35 dimers or tri-tetramers may bind dsRNA forming the low order complex that can be seen into the gel, while the VP35 high order aggregates are still able to bind IVT dsRNAs but are too big to enter the gel. The specificity of such interactions is also supported by the fact that a C-terminal His-tagged rVP35, when tested in the same EMSA, totally fails to shift the IVT dsRNA, forming neither low nor high order complexes (data not shown). Given that rVP35 C-terminal residues such as I340 and R339 have been reputed critical for stabilization of the RBD (Leung et al., 2010a, Leung et al., 2010c), the presence of an His-tag at the C-terminus could be sufficient to create some hurdle in the interaction between VP35 and dsRNA. The functional role of such low and high order VP35–dsRNA complexes is not clear, however, the fact that the R305A, R309A and R312A rVP35 mutants, which reduced or abrogated the EBOV IFN-α/β inhibition (Leung et al., 2010c), display a diminished ability to form low order rVP35–dsRNA aggregates, while they still formed high order aggregates, may suggest that the low order VP35–dsRNA complex may have an important functional role in the VP35 IFN-antagonistic function.
The fact that the disruption of IFN-α/β production is essential for the efficient EBOV propagation in vivo and that the loss of VP35–dsRNA binding capability strongly correlates with the loss of EBOV virulence, highlights the importance of VP35–dsRNA binding as an attractive and promising target for antiviral development. Therefore, we established a novel, straightforward and reproducible biochemical in vitro assay to measure the VP35–dsRNA binding, we used it to quantitatively characterize this interaction and validated it as a tool for compound screening. Firstly, we showed that full-length rVP35 can be stably conjugated to paramagnetic beads and that these conjugates specifically retain a radiolabeled dsRNA probe in a linear dose-dependent manner. Secondly, we determined the optimal conditions for the in vitro reaction at 37 °C, describing the VP35–dsRNA binding dependency by pH, ionic strength, Mg2+ concentration, temperature, time of incubation and amount of beaded-protein. Interestingly, such conditions were comparable to those observed in a different biochemical assay developed to target the dsRNA binding function of the influenza virus NS1 protein (Maroto et al., 2008), a known viral IFN-antagonist that shares several functions with EBOVs VP35. Thirdly, we showed that the full-length rVP35 binds to IVT dsRNA of 50–500 bp in length with high affinity, since the K d values ranged from 1 to 3 nM according to the temperature used. These K d values were comparable, although determined with different techniques, with those calculated for other RNA binding proteins, such as EBOV VP30 (K d = 61 nM) (John et al., 2007), HIV-1 Tat (K d = 10 nM) (Slice et al., 1992), Hantaan virus N (K d = 14 nM) (Severson et al., 2005), influenza virus NS1 (K d = 1 μM) (Chien et al., 2004, Maroto et al., 2008) and the cellular antiviral protein PKR (K d = 15 nM at one binding site) (Lemaire et al., 2008). Noteworthy, the EBOV rVP35 K d value for dsRNA was lower than the K d values of all these proteins. In particular, it is interesting to compare the affinity for dsRNA between VP35 and the cellular protein RIG-I. In fact, inhibition of the IFN-α/β response due to viral antagonism primarily occurs through the inhibition of the RIG-I-dependent signaling, since this cellular helicase is a crucial sensor of infection through the recognition of viral dsRNA. In this regard, RIG-I stimulation and overexpression has been demonstrated to attenuate EBOVs replication in cell cultures (Spiropoulou et al., 2009) and it is likely that an effective suppression of the IFN-α/β response by EBOVs would require an efficient sequestration of dsRNA by VP35. Determination by ITC of the RIG-I equilibrium dissociation constant for a 12 bp substrate with a 5′-triphosphate (5′-ppp) terminus showed K d values from 30 to 214 nM, while the K d value was 1 μM for the same dsRNA with 5′-OH substrate (Wang et al., 2010). Other studies performed by surface plasmon resonance at 25 °C with a 14 bp dsRNA with a 5′-ppp terminus yielded a K d value of 0.3 nM, while the K d value was 5 nM for the same dsRNA with 5′-OH ends (Lu et al., 2010). Using an 8 bp dsRNA we determined that rVP35 K d values was around 70 nM and 1 μM for 5′-p and 5′-OH oligos, respectively. Therefore, our results show that VP35 could effectively compete with RIG-I for dsRNA binding in the infected cell. Furthermore, the full length rVP35 demonstrated a comparable affinity for dsRNA with respect to its truncated C-terminal RBD. In fact, when the VP35 RDB has been studied by ITC, it showed K d values for dsRNA molecules of 8 bp in length of 30 and 500 nM for dsRNAs with 5′-ppp and 5′-OH ends, respectively (Leung et al., 2010c, Leung et al., 2010d), while when it has been studied by dual filter dot blot binding assay it showed a K d value for IVT 18 bp dsRNA of 890 nM (Kimberlin et al., 2010). Both reported values are in substantial agreement with those obtained in our magnetic pull down assay using similar short dsRNA substrates. However, it is worth to note that full-length rVP35 showed a 25-fold increase in dsRNA binding affinity for slightly longer IVT dsRNA (50 bp in length). Hence, it is possible to speculate that VP35 domains other than RDB may efficiently contribute to the establishment of a stronger or more stable dsRNA–protein complex with longer dsRNAs, leading to an higher binding affinity. In addition, it is important to note that, while a bimodal strategy has been recently proposed for VP35–RBD–dsRNA binding in which two RBD monomers cooperatively undertake a dimeric assembly to bind dsRNA (Kimberlin et al., 2010, Leung et al., 2010c), our data analysis used a single site binding approach because the rVP35 that conjugates to the magnetic beads, and binds to the IVT dsRNA, is a tri-tetrameric homo oligomer (Zinzula et al., 2009 and data not shown). Therefore, at least under our experimental conditions, the oligomerization of the full length rVP35 proteins takes place independently of the dsRNA presence and rVP35 probably binds to dsRNA as a tri-tetrameric homo oligomer.
Two recently solved crystallographic structures of the VP35 RBD bound to short RNA duplexes describe a binding mode to dsRNA where a patch of conserved basic residues interacts with the end-proximal phosphate backbone and a pocket of hydrophobic residues end-caps the dsRNA blunt ends (Kimberlin et al., 2010, Leung et al., 2010c). According to this model, the simultaneous recognition of both strands of the RNA double helix leads to the formation of a VP35-RBD dimer that mimics the strategies employed by PRRs to detect viral dsRNA (Kimberlin et al., 2010, Leung et al., 2010c). Our results agree with a model for which dsRNA binding is not dependent on the dsRNA sequence or length. In fact, the full-length rVP35 shows no significant differences in dsRNA binding affinity for heterologous IVT molecules ranging from 500 to 50 bp. Moreover, they also agree with an end-capping binding modality. In fact, the full-length rVP35 binding affinity is affected by the nature of dsRNA ends, since a 14-fold lower K d value is observed for 5′-phosphorylated versus 5′-unphosphorylated 8 bp dsRNAs. In addition, the fact that, in the EMSA studies, the intensity of the low order wt VP35–dsRNA complexes obtained with the three IVT dsRNAs is comparable, is also in agreement with this model.
It has been shown that VP35 RBD has a relatively thermal stability when tested in ThermoFluor assays, yielding Tm values of 57 and 63 °C for the ZEBOV and REBOV proteins, respectively (Leung et al., 2010b). As indicated by Van’t Hoff analysis, at increasing temperatures the rVP35 affinity to dsRNA diminishes linearly with a relatively shallow slope, suggesting that the binding reduction is likely due to changes in the rVP35–dsRNA complex stability following the thermodynamics of the interaction rather than in any loss in rVP35 binding ability. Noteworthy, even though higher affinities were shown to be 23 and 30 °C, we selected the more biologically relevant 37 °C for all our studies.
The three basic residues R305, K309 and R312 lie in the VP35 central basic patch that was originally proposed to be required for dsRNA binding on the basis of its sequence homology with the RBD of the influenza virus NS1 protein (Hartman et al., 2004). Alanine substitution of these residues was demonstrated to be critical for dsRNA binding and to impair VP35 suppression of IFN-α/β production, even though on a differential extent (Cárdenas et al., 2006, Hartman et al., 2006, Hartman et al., 2008a, Hartman et al., 2008b). In particular, in cell culture, R305A and K309A VP35s suppressed IFN-β activation slightly less efficiency than wt VP35 (Leung et al., 2010c), and K309A VP35 failed to bind pIC beads in a pull down assay (Cárdenas et al., 2006). In addition, while in silico modeling predictions have attributed to R305 only limited interactions with dsRNA, in vitro the truncated (comprising only the RBD) R305A VP35 mutant K d value measured by ITC was 3- to 5-fold higher than wt VP35 (Leung et al., 2010c). Furthermore, the R312A VP35 was shown to be severely impaired in its IFN-inhibition capability in cell culture (viruses bearing such mutation were attenuated and avirulent) (Hartman et al., 2008a, Hartman et al., 2008b), the structural data on the VP35 RBD indicated that R312 is critical for VP35–dsRNA binding (Kimberlin et al., 2010, Leung et al., 2010a, Leung et al., 2010c, Leung et al., 2010d) and the truncated VP35 R312A mutant showed a total loss of dsRNA binding by ITC, using an 8 bp IVT dsRNA (Leung et al., 2010c). Our data on full length rVP35 mutants are in substantial agreement with these observations. However, our data on full length rVP35 mutants describe a somehow more subtle gradient in 500 bp dsRNA binding ability. In fact, all mutants retained a major dsRNA binding capacity, even though their K d values were significantly higher than the one observed for wt rVP35 (a 4-fold difference was observed between wt and R312A–rVP35 binding affinities). Importantly however, when compared to wt rVP35, the full length rVP35 R312A showed a 25-fold reduction in binding affinity for the 5′-phosphorylated 8 bp dsRNA. Furthermore, the comparison of the full length rVP35 R312A K d values for the 500 bp IVT dsRNA and for the 5′-phosphorylated 8 bp dsRNA shows a 166-fold difference, indicating that the length of the dsRNA severely affects the mutant binding ability.
Taking together these data, it is possible to hypothesize that, on the one side, when other VP35 domains are present in addition to the sole RBD, the VP35 interaction with dsRNA takes place in a more complex, and/or stable, modality than the one described so far. In this hypothesis, the contribution of the N-terminal domain may represent a stabilizing factor. On the other side, it is possible that these single VP35 mutations may affect only one of the existing functional modes through which VP35 binds dsRNA. Given that other VP35 residues have been suggested to be critically implicated in dsRNA binding-mediated IFN inhibition (Leung et al., 2010a, Leung et al., 2010c, Prins et al., 2010), further mutagenesis studies on the full-length rVP35 might help to elucidate these aspects.
Among the strategies adopted to interfere with EBOVs propagation in animal models, two successful attempts based on RNA-interference approach and the use of phosphorodiamidate morpholino antisense oligomers have been recently reported (Enterlein et al., 2006, Geisbert et al., 2010, Warfield et al., 2006). Interestingly, both studies included the silencing of VP35 gene to knockdown virus replication. According to this vision, the most suitable EBOVs countermeasure would be to impair viral replication and to allow the innate and adaptive immune responses to clear the infection (Feldmann and Geisbert, 2011). This strategy further validates the choice of targeting the VP35–dsRNA interaction. With the aim of finding a molecule that would inhibit this interaction and would validate the magnetic pull down reliability for compound screening, we assayed ATA, which was originally recognized as a non-specific inhibitor of protein interactions with nucleic acids (Gonzáles et al., 1980), including the HIV-1 integrase–dsDNA interaction (Fabozzi et al., 2011, Cushman and Sherman, 1992, Tramontano et al., 1998). More recently, ATA has been identified to be a potent and selective inhibitor of viral SARS-CoV and HCV RNA polymerases (Chen et al., 2009), and it has been shown to be 10–100 times more potent than IFN-α/β in inhibiting SARS-CoV replication in infected cells (He et al., 2004, Yap et al., 2005). In our in vitro assay, ATA displayed a dose dependent inhibitory effect on the formation of rVP35–dsRNA complexes, with an IC50 value equal to 50 μg/mL, which is in agreement with similar results obtained for other RNA binding proteins. Importantly, the presently described magnetic pull down assay can be easily scaled up to a high-throughput format and, in fact, studies in this directions are currently underway.
5. Conclusions
In summary, in the present study we performed the first characterization of the full length rVP35 wt and mutants dsRNA binding function that brought novel insights into the VP35–dsRNA interaction. Moreover, we validated a novel and straightforward biochemical in vitro assay for the screening of antiviral agents targeted to the interaction between rVP35 and dsRNA. Finally, given that EBOVs VP35 is a paradigm protein for potent viral dsRNA binding-dependent IFN-suppression, the described method can be further applied to other viral proteins that are implicated in the IFN-antagonist inhibition of the innate immunity by dsRNA binding.
Acknowledgements
Luca Zinzula and Francesca Esposito were supported by the research Grants CRP2_682 and CRP2_683, respectively, held by Regione Autonoma della Sardegna (RAS, PO Sardegna FSE 2007-2013, L.R. 7/2007 “Promozione della ricerca scientifica e dell’innovazione tecnologica in Sardegna”). Authors thank Prof. Osvaldo Giorgi for helpful discussions.
References
- Barrette R.W., Metwally S.A., Rowland J.M., Xu L., Zaki S.R., Nichol S.T., Rollin P.E., Towner J.S., Shieh W.-J., Batten B., Sealy T.K., Carrillo C., Moran K.E., Bracht A.J., Mayr G.A., Sirios-Cruz M., Cathagan D.P., Lautner E.A., Ksiazek T.G., White W.R., McIntosh M.T. Discovery of Swine as a host for the Reston ebolavirus. Science. 2009;325:204–206. doi: 10.1126/science.1172705. [DOI] [PubMed] [Google Scholar]
- Barrette, R.W., Xu, L., Rowland, J.M., McIntosh, M.T., in press. Current perspectives on the phylogeny of Filoviridae. Infect. Genet. Evol. doi: 10.1016/j.meegid.2011.06.017 [DOI] [PMC free article] [PubMed]
- Basler C.F., Wuang X., Mühlberger E., Volchkov V., Paragas J., Klenk H.-D., Garcia-Sastre A., Palese P. The Ebola virus VP35 protein functions as a type I IFN antagonist. Proc. Natl. Acad. Sci. USA. 2000;97:12289–12294. doi: 10.1073/pnas.220398297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basler C.F., Mikulasova A., Martinez-Sobrido L., Paragas J., Mühlberger E., Bray M., Klenk H.-D., Palese P., Garcia-Sastre A. The Ebola virus VP35 protein inhibits activation of interferon regulatory factor 3. J. Virol. 2003;77:7945–7956. doi: 10.1128/JVI.77.14.7945-7956.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basler C.F., Amarasinghe G.K. Evasion of interferon responses by Ebola and Marburg viruses. J. Interferon Cytokine Res. 2009;29:511–520. doi: 10.1089/jir.2009.0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becquart P., Wauquier N., Mahlakõv T., Nkoghe D., Padilla C., Souris M., Ollomo B., Gonzales J.-P., De Lamballerie X., Kazanji M., Leroy E.M. High prevalence of both humoral and cellular immunity to Zaire ebolavirus among rural populations in Gabon. PLoS One. 2010;5:e9126. doi: 10.1371/journal.pone.0009126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehmann Y., Enterlein S., Randolf A., Mühlberger E. A reconstituted replication and transcription system for Ebola virus Reston and comparison with Ebola virus Zaire. Virology. 2005;332:406–417. doi: 10.1016/j.virol.2004.11.018. [DOI] [PubMed] [Google Scholar]
- Bowie A.G., Unterholzner L. Viral evasion and subversion of pattern-recognition receptor signaling. Nat. Rev. Immunol. 2008;8:911–922. doi: 10.1038/nri2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bylund D.B., Murrin L.C. Radioligand saturation binding experiments over large concentration ranges. Life Sci. 2000;67:2897–2911. doi: 10.1016/s0024-3205(00)00877-8. [DOI] [PubMed] [Google Scholar]
- Cárdenas W.B., Loo Y.M., Gale M., Jr., Hartman A.L., Kimberlin C.R., Martinez-Sobrido L., Saphire E.O., Basler C.F. Ebola virus VP35 protein binds double-stranded RNA and inhibits alpha/beta interferon production induced by RIG-I signaling. J. Virol. 2006;80:5168–5178. doi: 10.1128/JVI.02199-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang T.H., Kubota T., Matsuoka M., Jones S., Bradfute S.B., Bray M., Ozato K. Ebola Zaire virus blocks type I interferon production by exploiting the host SUMO modification machinery. PLoS Pathog. 2009;5:e1000493. doi: 10.1371/journal.ppat.1000493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Bopda-Waffo A., Basu A., Krishnan R., Silberstein E., Taylor D.R., Talele T.T., Arora P., Kaushik-Basu N. Characterization of aurintricarboxylic acid as a potent hepatitis C virus replicase inhibitor. Antivir. Chem. Chemother. 2009;20:19–36. doi: 10.3851/IMP1286. [DOI] [PubMed] [Google Scholar]
- Chien C., Xu Y., Xiao R., Aramini J.M., Sahasrabudhe P.V., Krug R.M., Montelione G.T. Biophysical characterization of the complex between double-stranded RNA and the N-terminal domain of the NS1 protein from influenza A virus: evidence for a novel RNA-binding mode. Biochemistry. 2004;43:1950–1962. doi: 10.1021/bi030176o. [DOI] [PubMed] [Google Scholar]
- Chung S., Wendeler M., Rausch J.W., Beilhartz G., Gotte M., O’Keefe B.R., Bermingham A., Beutler J.A., Liu S., Zhuang X., Le Grice S.F. Structure–activity analysis of vinylogous urea inhibitors of human immunodeficiency virus-encoded ribonuclease H. Antimicrob. Agents Chemother. 2010;54:3913–3921. doi: 10.1128/AAC.00434-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conzelmann K.K. Transcriptional activation of alpha/beta interferon genes: interference by non segmented negative-stranded RNA viruses. J. Virol. 2005;79:5241–5248. doi: 10.1128/JVI.79.9.5241-5248.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cushman M., Sherman P. Inhibition of HIV-1 integration protein by aurintricarboxylic acid monomers, monomer analogs, and polymer fractions. Biochim. Biophys. Res. Commun. 1992;185:85–90. doi: 10.1016/s0006-291x(05)80958-1. [DOI] [PubMed] [Google Scholar]
- Enterlein S., Warfield K.L., Swenson D.L., Stein D.A., Smith J.L., Gamble C.S., Kroeker A.D., Iversen P.L., Bavari S., Mühlberger E. VP35 knockdown inhibits Ebola virus amplification and protects against lethal infection in mice. Antimicrob. Agents Chemother. 2006;50:984–993. doi: 10.1128/AAC.50.3.984-993.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabozzi G., Nabel C.S., Dolan M.A., Sullivan N.J. Ebolavirus proteins suppress the effects of small interfering RNA by direct interaction with the mammalian RNA interference pathway. J. Virol. 2011;85:2512–2523. doi: 10.1128/JVI.01160-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falzarano D., Geisbert T.W., Feldmann H. Progress in filovirus vaccine development: evaluating the potential for clinical use. Expert. Rev. Vaccines. 2011;10:63–77. doi: 10.1586/erv.10.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann H., Geisbert T.W. Ebola hemorrhagic fever. Lancet. 2011;377:849–862. doi: 10.1016/S0140-6736(10)60667-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z., Cerveny M., Yan Z., He B. The VP35 protein of Ebola virus inhibits the antiviral effect mediated by double-stranded RNA-dependent protein kinase PKR. J. Virol. 2007;81:182–192. doi: 10.1128/JVI.01006-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisbert T.W., Lee A.C.H., Robbins M., Geisbert J.B., Honko A.N., Sood V., Johnson J.C., de Jong S., Tavakoli I., Judge A., Hensley L.E., Mac Lachlan I. Post exposure protection of non-human primates against a lethal Ebola virus challenge with RNA interference: a proof of concept study. Lancet. 2010;375:1896–1905. doi: 10.1016/S0140-6736(10)60357-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh U., Giri K., Bhattacharyya N.P. Interaction of auryntricarboxylic acid (ATA) with four nucleic acid proteins DNase I, RNase A, reverse transcriptase and Taq polymerase. Spectrochim. Acta, Part A. 2009;74:1145–1151. doi: 10.1016/j.saa.2009.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzáles R.G., Haxo R.S., Schleich T. Mechanism of action of polymeric auryntricarboxylic acid, a potent inhibitor of protein–nucleic acid interactions. Biochemistry. 1980;19:4299–4303. doi: 10.1021/bi00559a023. [DOI] [PubMed] [Google Scholar]
- Haasnoot J., de Vries W., Geutjes E.-J., Prins M., de Haan P., Berkhout P. The Ebola virus VP35 protein is a suppressor of RNA silencing. PLoS Pathog. 2007;3:e86. doi: 10.1371/journal.ppat.0030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman A.L., Towner J.S., Nichol S.T. A C-terminal basic amino acid motif of Zaire ebolavirus VP35 is essential for type I interferon antagonism and displays high identity with the RNA-binding domain of another interferon antagonist, the NS1 protein of influenza A virus. Virology. 2004;328:177–184. doi: 10.1016/j.virol.2004.07.006. [DOI] [PubMed] [Google Scholar]
- Hartman A.L., Dover J.E., Towner J.S., Nichol S.T. Reverse genetic generation of recombinant Zaire Ebola Viruses containing disrupted IRF-3 inhibitory domains results in attenuated virus growth in vitro and higher levels of IRF-3 activation without inhibiting viral transcription or replication. J. Virol. 2006;80:6430–6440. doi: 10.1128/JVI.00044-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman A.L., Bird B.H., Towner J.S., Antoniadou Z.-A., Zaki S.R., Nichol S.T. Inhibition of IRF-3 activation by VP35 is critical for the high level of virulence of Ebola virus. J. Virol. 2008;82:2699–2704. doi: 10.1128/JVI.02344-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman A.L., Ling L., Nichol S.T., Hibberd M.L. Whole-genome expression profiling reveals that inhibition of host innate immune response pathways by Ebola virus can be reversed by a single amino acid change in the VP35 protein. J. Virol. 2008;82:5348–5358. doi: 10.1128/JVI.00215-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman A.L., Towner J.S., Nichol S.T. Ebola and Marburg hemorrhagic fever. Clin. Lab. Med. 2010;30:161–177. doi: 10.1016/j.cll.2009.12.001. [DOI] [PubMed] [Google Scholar]
- He R., Adonov A., Traykova-Adonova M., Cao J., Cutts T., Grudesky E., Deschambaul Y., Berry J., Drebot M., Li X. Potent and selective inhibition of SARS coronavirus replication by aurintricarboxylic acid. Biochem. Biophys. Res. Commun. 2004;320:1199–1203. doi: 10.1016/j.bbrc.2004.06.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Xu L., Sun Y., Nabel G.J. The assembly of Ebola virus nucleocapsid requires virion-associated proteins 35 and 24 and posttranslational modification of nucleoprotein. Mol. Cell. 2002;10:307–316. doi: 10.1016/s1097-2765(02)00588-9. [DOI] [PubMed] [Google Scholar]
- Jin H., Yan Z., Prabakhar B.S., Feng Z., Ma Y., Verpooten D., Ganesh B., He B. The VP35 protein of Ebola virus impairs dendritic cell maturation induced by virus and lipopolysaccharide. J. Gen. Virol. 2010;91:352–361. doi: 10.1099/vir.0.017343-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John S.P., Wang T., Steffen S., Longhi S., Schmaljohn C.S., Jonsson C.B. VP30 is an RNA binding protein. J. Virol. 2007;81:8967–8976. doi: 10.1128/JVI.02523-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R.F., McCarthy S.E., Godlewski P.J., Harty R.N. Ebola virus VP35-VP40 interaction is sufficient for packaging 3E–5E minigenome RNA into virus-like particles. J. Virol. 2006;80:5135–5144. doi: 10.1128/JVI.01857-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katze M.G., Fornek J.L., Palermo R.E., Walters K.-A., Korth M.J. Innate immune modulation by RNA viruses: emerging insights from functional genomics. Nat. Rev. Immunol. 2008;8:644–654. doi: 10.1038/nri2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimberlin C.R., Bornholdt Z.A., Li S., Woods V.L., Jr., MacRae I.J., Saphire E.O. Ebolavirus VP35 uses a bimodal strategy to bind dsRNA for innate immune suppression. Proc. Natl. Acad. Sci. USA. 2010;107:314–319. doi: 10.1073/pnas.0910547107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobinger G.P., Leung A., Neufeld J., Richardson J.S., Falzarano D., Smith G., Tierney K., Patel A., Weingartl H.M. Replication, pathogenicity, shedding and transmission of Zaire ebolavirus in pigs. J. Infect. Dis. 2011;204:200–208. doi: 10.1093/infdis/jir077. [DOI] [PubMed] [Google Scholar]
- Koyama S., Ishii K.J., Coban C., Akira S. Innate immune response to viral infection. Cytokine. 2008;43:336–341. doi: 10.1016/j.cyto.2008.07.009. [DOI] [PubMed] [Google Scholar]
- Kuhn J.H., Becker S., Ebihara H., Geisbert T.W., Johnson K.M., Kawaoka Y., Lipkin W.I., Negredo A.I., Netesov S.V., Nichol S.T., Palacios G., Peters C.J., Tenorio A., Volchkov V.E., Jahrling P.B. Proposal for a revised taxonomy of the family Filoviridae: classification, names of taxa and viruses, and virus abbreviations. Arch. Virol. 2010;155:2083–2103. doi: 10.1007/s00705-010-0814-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire P.A., Anderson E., Lary J., Cole J.L. Mechanism of PKR activation by dsRNA. J. Mol. Biol. 2008;381:351–360. doi: 10.1016/j.jmb.2008.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy E.M., Gonzales J.-P., Baize S. Ebola and Marburg hemorrhagic fever viruses: major scientific advances, but a relatively minor public health threat for Africa. Clin. Microbiol. Infect. 2011;17:964–976. doi: 10.1111/j.1469-0691.2011.03535.x. [DOI] [PubMed] [Google Scholar]
- Leung D.W., Ginder N.D., Fulton D.B., Nix J., Basler C.F., Honzatko R.B., Amarasinghe G.K. Structure of the Ebola VP35 interferon inhibitory domain. Proc. Natl. Acad. Sci. USA. 2009;106:441–446. doi: 10.1073/pnas.0807854106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung D.W., Borek D., Farahbakhsh M., Ramanan P., Nix J.C., Wang T., Prins K.C., Otwinowski Z., Honzatko R.B., Helgeson L.A., Basler C.F., Amarasinghe G.K. Crystallization and preliminary X-ray analysis of Ebola VP35 interferon inhibitory domain mutant proteins. Acta Crystallogr. Sect. F: Struct. Biol. Cryst. Commun. 2010;66:689–692. doi: 10.1107/S1744309110013266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung D.W., Prins K.C., Basler C.F., Amarasinghe G.K. Ebolavirus VP35 is a multifunctional virulence factor. Virulence. 2010;6:526–531. doi: 10.4161/viru.1.6.12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung D.W., Prins K.C., Borek D.M., Farahbakhsh M., Tufariello J.M., Ramaman P., Nix J.C., Helgeson L.A., Otwinowski Z., Honzatko R.B., Basler C.F., Amarasinghe G.K. Structural basis for dsRNA recognition and interferon antagonism by Ebola VP35. Nat. Struct. Mol. Biol. 2010;17:165–172. doi: 10.1038/nsmb.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung D.W., Shabman R.S., Farahbakhsh M., Prins K.C., Borek D.M., Wang T., Mühlberger E., Basler C.F., Amarasinghe G.K. Structural and functional characterization of Reston Ebola virus VP35 interferon inhibitory domain. J. Mol. Biol. 2010;3:347–357. doi: 10.1016/j.jmb.2010.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo Y.-M., Gale M., Jr. Immune signaling by RIG-I-like receptors. Immunity. 2011;34:680–692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C., Ranjith-Kumar C.T., Hao L., ChengKao C., Li P. Crystal structure of RIG-I C-terminal domain bound to blunt-ended double-strand RNA without 5′triphosphate. Nucleic Acids Res. 2010;39:1565–1575. doi: 10.1093/nar/gkq974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahanty S., Bray M. Pathogenesis of filoviral hemorrhagic fevers. Lancet Infect. Dis. 2004;4:487–498. doi: 10.1016/S1473-3099(04)01103-X. [DOI] [PubMed] [Google Scholar]
- Maroto M., Fernandez Y., Ortin J., Pelaez F., Cabello M.A. Development of an HTS assay for the search of anti-influenza agents targeting the interaction of viral RNA with the NS1 protein. J. Biomol. Screen. 2008;13:581–590. doi: 10.1177/1087057108318754. [DOI] [PubMed] [Google Scholar]
- Mohamadzadeh M., Chen L., Schmaljohn A.L. How Ebola and Marburg viruses battle the immune system. Nat. Rev. Immunol. 2007;7:556–567. doi: 10.1038/nri2098. [DOI] [PubMed] [Google Scholar]
- Mohamadzadeh M. Potential factors induced by filoviruses that lead to immune suppression. Curr. Mol. Med. 2009;9:174–185. doi: 10.2174/156652409787581628. [DOI] [PubMed] [Google Scholar]
- Möller P., Pariente N., Klenk H.-D., Becker S. Homo-oligomerization of Marburgvirus VP35 is essential for its function in replication and transcription. J. Virol. 2005;79:14876–14886. doi: 10.1128/JVI.79.23.14876-14886.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motulsky H.J., Neubig R.R. Analyzing binding data. Curr. Protoc. Neurosci. 2010;52:751–7565. doi: 10.1002/0471142301.ns0705s52. [DOI] [PubMed] [Google Scholar]
- Mühlberger E., Löftering B., Klenk H.-D., Becker S. Three of the four nucleocapsid proteins of Marburg virus, NP, VP35, and L, are sufficient to mediate replication and transcription of Marburg virus-specific monocistronic minigenomes. J. Virol. 1998;72:8756–8764. doi: 10.1128/jvi.72.11.8756-8764.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlberger E., Weik M., Volchkov V.E., Klenk H.-D., Becker S. Comparison of the transcription and replication strategies of Marburg virus and Ebola virus by using artificial replication systems. J. Virol. 1999;73:2333–2342. doi: 10.1128/jvi.73.3.2333-2342.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prins K.C., Cárdenas W.B., Basler C.F. Ebola virus protein VP35 impairs the function of interferon regulatory factor-activating kinases IKK epsilon and TBK-1. J. Virol. 2009;83:3069–3077. doi: 10.1128/JVI.01875-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prins K.C., Delpeut S., Leung D.W., Reynard O., Volchkova V.A., Reid S.P., Ramanan P., Cárdenas W.B., Amarasinghe G.K., Volchkov V.E., Basler C.F. Mutations abrogating VP35 interactions with double-stranded RNA render Ebola virus avirulent in guinea pigs. J. Virol. 2010;84:3004–3015. doi: 10.1128/JVI.02459-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall R.E., Goodbourn S. Interferons and viruses: an interplay between induction, signaling, antiviral responses and virus countermeasures. J. Gen. Virol. 2008;89:1–47. doi: 10.1099/vir.0.83391-0. [DOI] [PubMed] [Google Scholar]
- Reed D.S., Lackemeyer M.G., Garza N.L., Sullivan L.J., Nichols D.K. Aerosol exposure to Zaire ebola virus in three non human primate species: differences in disease course and clinical pathology. Microbes Infect. 2011;13:930–936. doi: 10.1016/j.micinf.2011.05.002. [DOI] [PubMed] [Google Scholar]
- Reid S.P., Cárdenas W.B., Basler C.F. Homo-oligomerization facilitates the interferon-antagonist activity of the ebolavirus VP35 protein. Virology. 2005;341:179–189. doi: 10.1016/j.virol.2005.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler A.J., Williams B.R. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 2008;8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schümann M., Gantke T., Mühlberger E. Ebola virus VP35 antagonizes PKR activity through its C-terminal interferon inhibitory domain. J. Virol. 2009;83:8993–8997. doi: 10.1128/JVI.00523-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severson W., Xu X., Kuhn M., Senutovitch N., Thokala M., Ferron F., Longhi S., Canard B., Jonsson C.B. Essential amino acids of the Hantaan virus N protein in its interaction with RNA. J. Virol. 2005;79:10032–10039. doi: 10.1128/JVI.79.15.10032-10039.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W., Huang Y., Sutton-Smith M., Tissot B., Panico M., Morris H.R., Dell A., Haslam S.M., Boyington J., Graham B.S., Yang Z.-Y., Nabel G.J. A filovirus-unique region of Ebola virus nucleoprotein confers aberrant migration and mediates its incorporation into virions. J. Virol. 2008;82:6190–6199. doi: 10.1128/JVI.02731-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slice L.W., Codner E., Antelman D., Holly M., Wegrzynski B., Wang J., Toome V., Hsu M.C., Nalin C.M. Characterization of recombinant HIV-1 Tat and its interaction with TAR RNA. Biochemistry. 1992;31:12062–12068. doi: 10.1021/bi00163a014. [DOI] [PubMed] [Google Scholar]
- Spiropoulou C.F., Ranjan P., Pearce M.B., Sealy T.K., Albariño C.G., Gangappa S., Fujita T., Rollin P.E., Nichol S.T., Ksiazek T.G., Sambhara S. RIG-I activation inhibits ebola virus replication. Virology. 2009;392:11–15. doi: 10.1016/j.virol.2009.06.032. [DOI] [PubMed] [Google Scholar]
- Tramontano E., La Colla P., Cheng Y.-C. Biochemical characterization of the HIV-1 integrase 3′-processing activity and its inhibition by phosphorothioate oligonucleotides. Biochemistry. 1998;37:7237–7243. doi: 10.1021/bi972792o. [DOI] [PubMed] [Google Scholar]
- Wang Y., Ludwig J., Schuberth C., Goldeck M., Schlee M., Li H., Juranek S., Sheng G., Micura R., Tuschl T., Hartmannand G., Patel D.J. Structural and functional insights into 5′-ppp RNA pattern recognition by the innate immune receptor RIG-I. Nat. Struct. Mol. Biol. 2010;17:781–787. doi: 10.1038/nsmb.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warfield K.L., Swenson D.L., Olinger G.G., Nichols D.K., Pratt W.D., Blouch R., Stein D.A., Aman M.J., Iversen P.I., Bavari S. Gene-specific countermeasures against Ebola virus based on antisense phosphorodiamidate morpholino oligomers. PLoS Pathog. 2006;2:e1. doi: 10.1371/journal.ppat.0020001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wauquier N., Becquart P., Gasquet C., Leroy E.M. Immunoglobulin G in Ebola outbreak survivors. Gabon. Emerg. Infect. Dis. 2009;15:1136–1137. doi: 10.3201/eid1507.090402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wauquier N., Becquart P., Padilla C., Baize S., Leroy E.M. Human fatal Zaire Ebola virus infection is associated with an aberrant innate immunity and with massive lymphocyte apoptosis. PLoS Negl. Trop. Dis. 2010;5:e837. doi: 10.1371/journal.pntd.0000837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap Y., Zhang X., Andonov A., He R. Structural analysis of inhibition mechanisms of aurintricarboxylic acid on SARS-CoV polymerase and other proteins. Comput. Biol. Chem. 2005;29:212–219. doi: 10.1016/j.compbiolchem.2005.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama M., Fujita T. Recognition of viral nucleic acids in innate immunity. Rev. Med. Virol. 2010;20:4–22. doi: 10.1002/rmv.633. [DOI] [PubMed] [Google Scholar]
- Zampieri C.A., Sullivan N.J., Nabel G.J. Immunopathology of highly virulent pathogens: insights from Ebola virus. Nat. Immunol. 2007;8:1159–1164. doi: 10.1038/ni1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Bevilacqua P.C. Straightening of bulged RNA by the double-stranded RNA-binding domain from the protein kinase PKR. Proc. Natl. Acad. Sci. USA. 2000;97:14162–14167. doi: 10.1073/pnas.011355798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinzula L., Esposito F., Mühlberger E., Trunschke M., Conrad D., Piano D., Tramontano E. Purification and functional characterization of the full length recombinant Ebola virus VP35 protein expressed in E. coli. Protein Expr. Purif. 2009;66:113–119. doi: 10.1016/j.pep.2009.02.008. [DOI] [PubMed] [Google Scholar]


