Highlights
-
•
Peptide ligands to TGEV M protein was identified.
-
•
A phage-based ELISA was established.
-
•
TGEV M peptide (pepTGEV-M7; HALTPIKYIPPG) had antiviral potential.
Keywords: TGEV, Membrane protein, Peptide, Antivirals
Abstract
The membrane (M) protein is one of the major structural proteins of coronavirus particles. In this study, the M protein of transmissible gastroenteritis virus (TGEV) was used to biopan a 12-mer phage display random peptide library. Three phages expressing TGEV-M-binding peptides were identified and characterized in more depth. A phage-based immunosorbent assay (phage-ELISA) capable of differentiating TGEV from other coronaviruses was developed using one phage, phTGEV-M7, as antigen. When the phage-ELISA was compared to conventional antibody-based ELISA for detecting infections, phage-ELISA exhibited greater sensitivity. A chemically synthesized, TGEV-M7 peptide (pepTGEV-M7; HALTPIKYIPPG) was evaluated for antiviral activity. Plaque-reduction assays revealed that pepTGEV-M7 was able to prevent TGEV infection in vitro (p < 0.01) following pretreatment of the virus with the peptide. Indirect immunofluorescence and real-time RT-PCR confirmed the inhibitory effects of the peptide. These results indicate that pepTGEV-M7 might be utilized for virus-specific diagnostics and treatment.
1. Introduction
Transmissible gastroenteritis (TGE) is a highly contagious disease of swine characterized by up to 100% mortality in suckling piglets (Laude et al., 1993). The clinical symptoms include acute diarrhea, vomiting and dehydration. Pigs of all ages and categories are susceptible. In seronegative herds, TGE can cause devastating economic losses (Saif and Wesley, 1999; Ren et al., 2010a, Ren et al., 2010b; Yin et al., 2010). The virus responsible for TGE (TGEV) has a glycoprotein surface envelope and positive-sense RNA genome of approximately 28.5 kb (Ortego et al., 2002). The virus consists of four structural proteins: spike (S), small membrane (sM or E), membrane (M), and nucleocapsid (N) proteins (Spaan et al., 1988, Saif and Wesley, 1999, Penzes et al., 2001). The S protein is a large transmembrane surface glycoprotein that induces virus-neutralizing (VN) antibodies (Jiménez et al., 1986, Laude et al., 1987, Suñé et al., 1990); the N protein, together with genomic RNA, forms the viral nucleocapsid (Suñé et al., 1990, Cavanagh, 1994); and the sM protein regulates virion assembly and release (Laude et al., 1990). The M protein is the most abundant component of the coronavirus particle (Rottier, 1995). Roughly one-third of the M protein assumes a topology where part of the endo domain constitutes the fourth transmembrane segment, thereby positioning the carboxy terminus on the exterior portion of the virion (Risco et al., 1995, Masters, 2006). It has been suggested that the M protein induces innate immunity including interferon production (Charley and Laude, 1988, Laude et al., 1992).
Phage display is a powerful technology that has been applied to antibody engineering (Hayden et al., 1997, Cyranka-Czaja and Otlewski, 2012), drug discovery and manufacturing (Kay et al., 1998, Harper et al., 2011), ligand identification (Ehrlich and Bailon, 2001, Ladner and Ley, 2001, Yi et al., 2003, Beer and Liu, 2012), and development of new diagnostics (Ren et al., 2010a, Ren et al., 2010b; Gazarian et al., 2012) and vaccines (Lesinski and Westerink, 2001, Samoylova et al., 2012). Phage display yields billions of heterologous fusion peptides that are expressed on the surfaces of filamentous bacteriophage (Scott and Smith, 1990). Inasmuch as each phage expresses only one fusion peptide, the technology permits high throughput panning of a phage library with the intent of identifying single phage particles with the capacity to bind a target protein. This permits epitope mapping and easy purification at marginal costs. In the current study, three TGEV M-binding phage and the concomitant peptides were identified from a phage display library. Results indicate the ability of these peptides to function as TGEV-specific diagnostic reagents. One peptide was further studied for its potential as an inhibitor of TGEV infectivity.
2. Materials and methods
2.1. Cells and virus
Swine Testis (ST) cells were grown in Dulbecco’s MEM with 10% fetal bovine serum at 37 °C, 5% CO2. TGEV (PUR46-MAD strain) (Sánchez et al., 1990) was propagated in ST cells in the absence of serum, followed by gradient ultracentrifugation purification (Krempl and Herrler, 2001).
2.2. Virus titration
Cytopathic effect (CPE) assays were performed as described (Ren et al., 2011b). Briefly, ST cells seeded onto 96-well tissue culture plates (Costar, USA) were inoculated with 10-fold serially-diluted TGEV. After incubation at 37 °C for 48 h, CPE was recorded at 100× magnification using the BDS200 microscope (OPTEC, China).
2.3. Cloning and expression of the TGEV-M gene
Total RNA was isolated from TGEV-infected ST cells using a commercial protocol (220010, Fastgene, China). Sense (P1: 5′-GGGGGGATCCATGCGCTATTGTGCTATG) and antisense (P2: 5′- CCCCGAATTCTTATACCATATGTAATAA) primers were used in RT-PCR. The amplified M gene was inserted into the BamHI-EcoRI site of a prokaryotic expression vector, pGEX (Novagen, Germany), resulting in the recombinant plasmid, pGEX-TGEV-M. After pGEX-TGEV-M was transformed into Rosette Escherichia coli bacteria by heat shock (Bowyer, 2001), expression of the glutathione S-transferase (GST) tagged-TGEV-M fusion protein was induced with isopropyl-β-d-thiogalactoside (IPTG) and visualized by 12% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE). The recombinant protein of interest (rTGEV-M) was gel purified (Ren et al., 2011a). In parallel, rGST from pGEX, was expressed and purified as well. The concentration of rTGEV-M was measured spectrophotometrically using a NANODROP 2000 spectrophotometer according to manufacturer’s instructions.
2.4. Biopanning
Biopanning of the phage was performed using the Ph.D™-12 Phage Display Peptide Library Kit according to manufacturer’s instructions (E8110SC, New England Biolabs, USA) with minor modifications (Ren et al., 2010a, Ren et al., 2010b, Ren et al., 2011c). During the first round of binding the phage library to the rTGEV-M, 15 μg/well of rTGEV-M was used. In the second round, rTGEV-M was replaced by rGST as a negative control to remove non-specific binding to GST. The 3rd-6th rounds were performed under the similar conditions except for gradually decreasing the concentration of rTGEV-M (10, 7.5, 5, 2.5 μg/well) and increasing the concentration of Tween 20 from 0.1% to 0.5% to increase the stringency of binding. The phage remaining after the last round of biopanning were titered and 10 were selected for phage amplification and further study.
2.5. Analysis of binding activity of individual phages with rTGEV-M
Phages interacting positively with rTGEV-M were identified by indirect ELISA. The 96-well plates (FEP201896, Jet Bio, China) were coated either with rTGEV-M in 0.1 M NaHCO3 (pH 8.6) or with purified TGEV in DMEM at 15 μg/well at 4 °C. Plates were blocked with 1% BSA in TBS buffer (50 mM Tris–HCl, pH 7.5, 150 mM NaCl) for 2 h at 37 °C then washed 6X with TBS-Tween. The 10 selected phages derived from the last round of biopanning were added separately to the wells at concentrations of 1.5 × 1011 plaque forming units (pfu)/ml in 0.1 M NaHCO3 (pH 8.6) and incubated at 37 °C for 1 h. The remaining steps were performed as described (Ren et al., 2011a). Plates were read at OD450 with an ELX808 (BioTek, USA) ELISA reader.
2.6. Virus detection using ELISA
Ten TGEV-positive phage clones were amplified using an E. coli expression system (36). The DNAs of interest were extracted, purified (51106, Qiagen, Germany), PCR amplified and sequenced to corroborate the presence of TGEV-M sequences (3). Deduced amino acid sequences were determined (Wu et al., 2011). Phage ELISA and antibody-ELISA were compared for their abilities to detect the presence of the virus as described (Wu et al., 2011). For phage-ELISA, TGEV serially-diluted in DMEM was coated onto ELISA plates overnight at 4 °C. Then either the specific phage clone or a non-specific phage complex (negative control) from the phage display library was diluted in PBS (1.5 × 1011 pfu/100 μl) and added to the wells. Next, the wells were incubated with commercially-available rabbit anti-M13 antibody (1:1000 in PBS) followed by horseradish peroxidase-conjugated goat anti-rabbit antibody (GARP) (1:5000 in PBS). For antibody-mediated ELISA, rabbits were immunized once with 2 mg rTGEV-M in Freund’s complete adjuvant followed by three additional immunizations at 1 week intervals with the same amount of antigen in Freund’s incomplete adjuvant. Animals were bled 5 days after the final boost. TGEV was coated onto ELISA plates to which was added rabbit anti-TGEV-M followed by GARP secondary antibody (1:5000). In all experiments, the concentrations of TGEV were determined experimentally such that ELISA values could be measured and where [OD450 phage (P)/OD450 negative control (N)]>2 was considered positive.
2.7. Specificity of TGEV-reactive phages
The specificity of TGEV-reactive phages for TGEV infection was evaluated by phage-ELISA. A panel of selected viruses prepared in our laboratory consisting of TGEV (Ren et al., 2008), porcine epidemic diarrhea virus (PEDV) strain HLJBY (Ren et al., 2010a, Ren et al., 2010b), porcine reproductive and respiratory syndrome virus (PRRSV) strain JilinTN1 (Gao et al., 2012), porcine rotavirus (PRV) (Ren et al., 2011c), porcine pseudorabies virus (PrV) strain Kaplan (Sui et al., 2010), porcine parvovirus (PPV) isolate PPV2010 (Cui et al., 2012), and porcine circovirus type II (PCV2) strain PCV2-LJR (Zhu and Ren, 2012) were assembled for comparative studies. These viruses were diluted in DMEM to final concentrations of 5 μg/well and coated onto ELISA plates at 4 °C for 4 h. ELISA was performed according to standard protocols (Wu et al., 2011). The unpanned Ph.D™-12 Phage Display Peptide Library (phage L) was used as a negative control. Statistical significance of the OD450 values among all viral antigens was evaluated where [OD450 virus /OD450 phage L]>2 was judged as positive and subsequently analyzed using the t-test.
The amino acid sequence corresponding to the phage with the highest binding activity to TGEV (pepTGEV-M7) was commercially synthesized (BOSHI, China). The peptide was diluted in sterile water to a final concentration of 1 mg/ml. Purified rTGEV-M or control protein rTGEV S-AD (Meng et al., 2011) was coated onto ELISA plates (15 μg/well) at 37 °C. The 50 μl of serially-diluted pepTGEV-M7 (500, 100, 20 or 4 μg/ml) were mixed with 50 μl of phTGEV-M7 or 50 μl of phTGEV-S-AD (1.5 × 1011 pfu/ml) and incubated at 37 °C for 1 h. After washing, the wells were incubated with rabbit anti-M13 antibody (1:1000 dilution; Abcam, China) for 1 h followed by a 1:5000 dilution of GARP for 50 min; the OD450 values were recorded.
2.8. Antiviral activity of pepTGEV-M
Three experimental approaches were used to evaluate the antiviral activity of pepTGEV-M7 (Miyazaki et al., 2010, Ren et al., 2011c). First, to assess the ability of the peptide to bind TGEV in vitro, 100 TCID50 of TGEV was pre-incubated with pepTGEV-M7 at 500, 250, 125, 62.5 or 31.25 μg/ml before infecting ST cells. Second, to determine any impact of pepTGEV-M7 on ST cells, cells were treated with serially-diluted peptide at 37 °C for 1 h prior to TGEV infection. Finally, to determine the effect of pepTGEV–M7 on ST cells after TGEV infection, cells were first infected with TGEV (100TCID50) at 37 °C for 1 h then washed and incubated with serially-diluted pepTGEV-M7 at 37 °C for 1 h. All the cells were grown to 100% confluency in 24-well plates overlaid with 1% methylcellulose then cultured for 48–72 h to maximize plaque development. To evaluate the effects of pepTGEV-M7 on TGEV replication in vitro, indirect immunofluorescence assay (IFA) was performed. Briefly, confluent ST cells were incubated with peptide-treated TGEV [15 μl of serially diluted (2−1–2−5) peptide was incubated with 15 μl of TGEV (100TCID50)] at 37 °C for 48 h then processed for IFA as previously described (Meng et al., 2011); an unrelated peptide that binds the PEDV-S1 protein, pepPEDV-S1 (diluted 2−1) was used as a negative control.
The impact of pepTGEV-M7 on virus replication was evaluated by real time RT-PCR targeting a portion of the TGEV-S gene. The TGEV was treated with serially-diluted peptide ranging from 500 to 31.25 μg/ml at 37 °C for 1 h; 500 μg/ml of pepPEDV-S1 was used as negative control. After washing the ST cells with PBS, confluent monolayers were infected with peptide treated TGEV (100TCID50) at 37 °C for 36 h. Then the virus-containing culture was frozen and thawed three times followed by the addition of an equal volume of 20% PGE-8000 at room temperature for 30 min. The samples were centrifuged at 12,000 rpm for 5 min and the pellets were suspended in RNase-free water. Total RNA was extracted according to the manufacturer’s instructions (Fastgene, China) and real-time RT-PCR was performed on cDNA as described (Ren et al., 2011b). It was anticipated that a detrimental effect of pepTGEV-M7 on viral infectivity would be reflected in significantly reduced numbers of TGEV S gene copies.
3. Results
3.1. Expression of rTGEV-M and biopanning
Using the previously described techniques (Ren et al., 2010a, Ren et al., 2010b; Ren et al., 2011a), the rTGEV-M protein was expressed in E. coli (Fig. 1 ) for use in biopanning the phage display library to identify peptides that bind the M protein of TGEV. During the isolation process it was determined that rTGEV-M was present within inclusion bodies. Peak expression was observed at 5 h post-IPTG induction. The mass of the rTGEV-M was ∼55 kDa (Fig. 1).
Fig. 1.
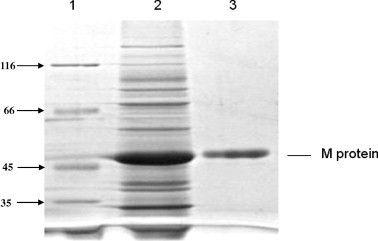
Expression and purification of TGEV M protein. Protein expression was induced in the recombinant E. coli harboring the TGEV M gene and bacterial proteins were separated by SDS–PAGE. Lane 1, marker; lane 2, cell lysate; lane 3, purified rTGEV-M.
Five rounds of biopanning were performed using the rTGEV-M as the target and one additional round with GST as target. The amount of eluted phages increased between rounds 1 and 5 from 2.9 × 1011 to 8.4 × 1011, respectively. Ten different TGEV-M protein-reactive phages were identified by ELISA, when rTGEV-M was used as the coating antigen (Fig. 2 a). All ten phages exhibited better binding to rTGEV-M than to other phage-expressed protein controls (p < 0.05, Fig. 2a). Phages M1, M3 and M5-M9 had higher binding affinity (p < 0.01) with TGEV than with the other phages (Fig. 2b). Among all ten reactive phages that bound rTGEV-M with high affinity, phTGEV-M7 exhibited the highest binding to TGEV (p < 0.01, Fig. 2b) followed by phTGEV-M5.and phTGEV-M6.
Fig. 2.
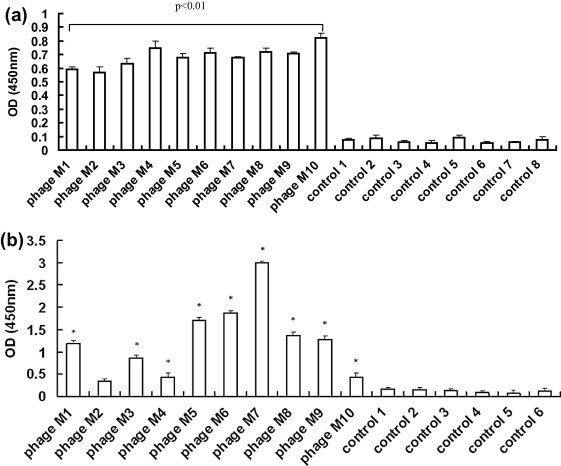
Binding analysis of the selected phages to TGEV M protein in ELISA. Ten selected phages were incubated with rTGEV-M or TGEV in ELISA plates to test their binding activities as measured by OD450 values. Individual phage are labeled M1–M10. The controls (1–8) contain; (1) phage library; (2) PRRSV GP5; (3) rTGEV N; (4) no primary antibody; (5) no phage; (6) blocking buffer; (7) protein dilution buffer; and (8) elution buffer. The OD450 readings were performed in triplicates. (a) Binding antigen was rTGEV-M. (b) Binding antigen was TGEV. Statistical significance is noted by “∗” (p < 0.01) compared to control groups.
RNA samples isolated from TGEV-M-reactive and control phages were amplified by RT-PCR. All samples yielded products approximately 250 bp in length (data not shown). All deduced peptides (12 aa in length) were unique (Table 1 ). Phages bearing the peptides LTFPVTTTPPAV, MTHNMHGPNSEP and HALTPIKYIPPG were selected for their high-affinity binding with rTGEV-M (Fig. 2a) and TGEV (Fig. 2b). These three peptides were named pepTGEV-M5, pepTGEV-M6 and pepTGEV-M7, respectively.
Table 1.
Deduced amino acid sequences of phage clones.
| Phage clones (number) | Phage displayed peptide sequence |
|---|---|
| PhageM1 | MQLPKADARHPH |
| PhageM2 | YQITLPYRYEMP |
| PhageM3 | TFSWEFTGWWGQ |
| PhageM4 | Failed |
| PhageM5 | LTFPVTTTPPAV |
| PhageM6 | MTHNMHGPNSEP |
| PhageM7 | HALTPIKYIPPG |
| PhageM8 | SSAYYYHYEYFH |
| PhageM9 | LSISPQHALVFA |
| PhageM10 | AMYHGHYTITRW |
Ten selected phages (phages M1–M10) were subjected to phage DNA extraction and PCR. The deduced amino acid sequences are presented.
3.2. TGEV-specific ELISA
The binding efficiencies of the selected TGEV-M-reactive phages were examined by phage-ELISA. Virus concentrations between 0.1–10 μg/well all exceeded the signal limitations of the assay. As illustrated in Fig. 3 a, titratable virus concentrations fell in the range <0.1 ug/well. The lowest concentrations of virus detectable with phTGEV-M5, phTGEV-M6 and phTGEV-M7 where P/N ⩾ 2 were 0.07, 0.03, and 0.02 μg/well, respectively.
Fig. 3.
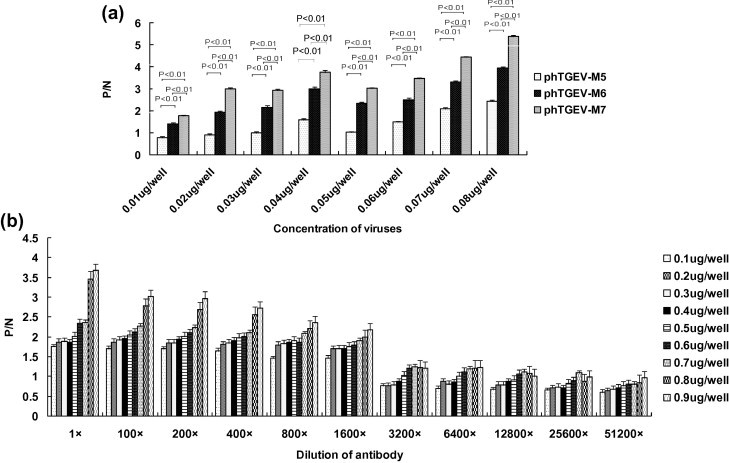
Detection limits of TGEV. (a) Phage-mediated ELISA. Serially-diluted TGEV was used as coating antigen followed by successive incubation with phTGEV-M5, phTGEV-M6 or phTGEV-M7, and antibody detection. OD450 ratios where P/N (sample/negative control) > 2 were considered positive. The experiment and determination of OD values were derived from three independent assays. The concentration of the virus is indicated on the x axis. (b) Antibody-based ELISA. Serially-diluted TGEV in DMEM was coated into ELISA plates followed by incubation with serially-diluted rabbit against TGEV serum and antibody binding using GARP; normal rabbit serum was used as negative control. OD450 ratios where P/N > 2 were considered positive. The experiment and determination of OD values were derived from three independent assays. The concentration of the virus is indicated on the x axis. Antibody and virus dilutions are as indicated.
For antibody-based ELISA, the rabbit antibody titers against rTGEV-M where P/N ⩾ 2 were determined to be 1:2.6 × 105. When assaying the antibody binding to TGEV, at virus concentrations >1.0 μg/ml, P/N values began to exceed the limits of detection. Virus (0.8–1.0 μg/well) were reproducibly detected (P/N ⩾ 2) with antibody dilutions less than 1/400 (Fig. 3b).
The binding specificities of phTGEV-M5, phTGEV-M6, phTGEV-M7 for non-TGE viruses were examined (Fig. 4 ). In all cases P/N > 2 for TGEV only. Demonstrable binding was not observed with any other viruses tested. Further, our results showed that pepTGEV-M7 bound to rTGEV-M and did not bind other proteins (Fig. 5 ). Increasing the concentration of pepTGEV-M7 effectively competed with binding of phTGEV-M7 for rTGEV-M as evidenced by decreasing OD450 values; however, pepTGEV-M7 did not compete with phage binding when a non-specific protein, rTGEV-S-AD was used as the coating antigen (Fig. 5).
Fig. 4.
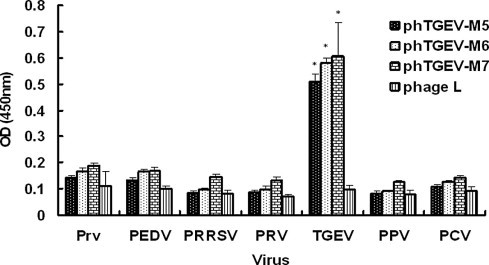
Specificity of phage- mediated ELISA. Three TGEV-selected phages, phTGEV-M5, phTGEV-M6, and phTGEV-M7, were tested for specificity of binding to TGEV, PEDV, PRRSV, PRV, PrV, PPV and PCV. All viruses were coated onto ELISA plates at final concentrations of 5 μg/well. The phage complex from the phage library (phage L) was used as a negative control. Statistical significance is indicated by ‘‘∗” (p < 0.01). All experiments were performed in triplicate.
Fig. 5.
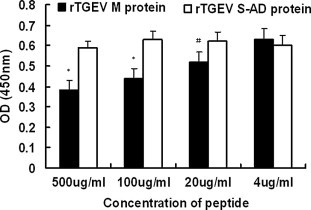
Binding of the peptides to rTGEV-M in ELISA. To confirm the specific binding of pepTGEV-M7 to rTGEV M protein and not to other phage components, rTGEV-M was coated onto ELISA plates and incubated with mixtures of pepTGEV-M7 and phTGEV-M7. The amount of phTGEV-M7 (1.5 × 1011 pfu) was kept constant amidst decreasing concentrations (500 μg/ml – 4.0 μg/ml) of pepTGEV-M7. Binding of phTGEV-M7 to rTGEV M protein was monitored using anti-M13 antibody and GARP secondary antibody which target phTGEV-M7 only. In parallel, rTGEV S-AD was screened as a negative control. Concentrations of pepTGEV-M7 are shown on the y axis. Statistical significance is indicated by ‘‘∗” (p < 0.01) or, ‘‘#” (0.01 < p < 0.05) relative to controls. All experiments were performed in triplicate.
3.3. Antiviral effects of pepTGEV-M1 in vitro
To determine the impact of pepTGEV-M7 on TGEV infection, three separate approaches were used: (1) TGEV was treated with peptide before inoculation of ST cells; (2) ST cells were treated with peptide prior to TGEV inoculation, and; (3) ST cells were infected with TGEV before being treated with peptide.
As expected, the results showed no impact of pepTGEV-M7 on ST cells already infected with TGEV or on ST cells pretreated with pepTGEV-M7 prior to incubation with the virus (data not shown). However, when TGEV was pre-incubated with pepTGEV-M7, its infectivity decreased in a dose-dependent manner: At 100 μg/ml of pepTGEV-M7, the drop in infectivity was nearly 55% whereas at 500 μg/ml a 66% drop in virus infectivity was observed (Table 2 ) suggesting peak inhibition was reached. These data indicate that pepTGEV-M7 is able to interfere with the ability of the virus to infect ST cells. The antiviral effects of pepTGEV-M7-treated virus were further investigated by IFA. Results demonstrated a dose-dependent reduction in the infection of ST cells in the presence of pepTGEV-M7 whereas the unrelated peptide, pepPEDV-S1, had no effect on virus uptake (Fig. 6 ).
Table 2.
Inhibition of peptide M to plaque production on ST cells.
| Concentration of peptide (μg/ml) | Virus (average plaque number) | Virus + peptide (average plaque number) | Inhibition rate (%) |
|---|---|---|---|
| 500 | 196 | 66 | 66.2 |
| 100 | 188 | 85 | 54.8 |
| 20 | 202 | 190 | 5.9 |
| 4 | 190 | 187 | 1.6 |
Fig. 6.
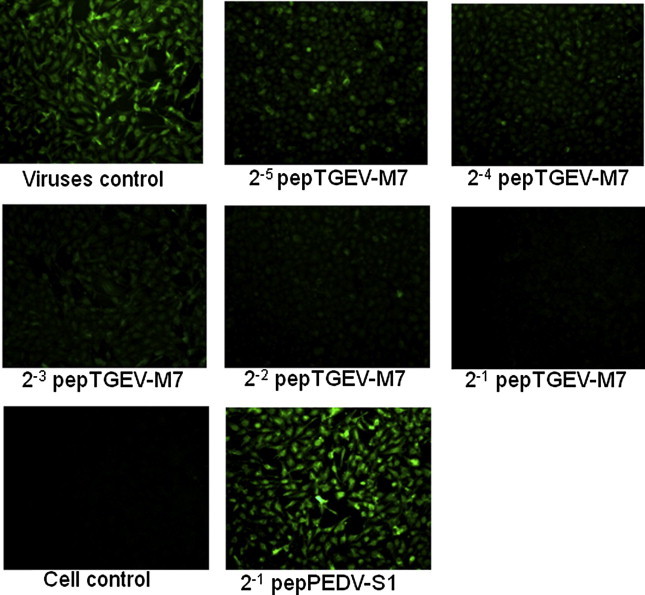
Inhibition of virus infectivity monitored by IFA. 100TCID50 TGEV virus was pre-incubated with 2-fold, serially-diluted (500–31.25 μg/ml) peptide (pepTGEV-M7) then added to ST cells for 48 h. Loss of fluorescence intensity coincides with loss of viral infectivity; an un-related peptide pepPEDV-S1 was used as control.
Real-time RT-PCR was used to confirm and quantify the TGEV inhibitory effects of pepTGEV-M7 (Fig. 7 ). Results showed that the amount of viral RNA was lower (p < 0.01) when TGEV was pretreated with pepTGEV-M7 than in peptide-untreated controls. At the highest concentrations (500 μg/ml), virus RNA levels dropped 93.45%; however, significant (p < 0.01) and reproducible reductions (70%) were observed when 62.5 ug/ml of pepTGEV-M7 were used. These data corroborated the IFA studies.
Fig. 7.
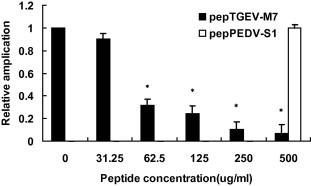
Inhibition of virus infectivity monitored by RT-PCR. Real time RT-PCR was used to quantify TGEV RNA in ST cells and therefore virus infectivity. PCR was performed on the TGEV S-AD gene. 100TCID50 TGEV virus was pre-incubated with 2-fold, serially-diluted (500–31.25 μg/ml) peptide (pepTGEV-M7) then added to ST cells for 48 h. An un-related peptide pepPEDV-S1 at the maximal concentration was used as control. Virus-derived cDNA was amplified and ΔCt values were measured in triplicate. The relative amplification of TGEV S-AD in TGEV-infected cells was normalized to beta-actin and calculated using 2−△△Ct method. Statistical significance is indicated by ‘‘∗” (p < 0.01) relative to 0 μg/ml peptide.
4. Discussion
Despite the availability of commercial vaccines, TGE still poses a regional threat to the swine industry due to the high morbidity and mortality that TGEV causes in suckling piglets under 2 weeks of age (Miyazaki et al., 2010). Although passive, lactogenic immunity induced by virulent or attenuated TGEV vaccines can provide effective protection, sufficient risk remains. Albeit safer, inactivated vaccines provide less protection. As such, accurate diagnostic tests are needed to better effect TGE prevention and therapy. Phage display and biopanning are powerful methods for identifying key ligands within large target antigens that may be involved in protection and/or diagnosis (Wu et al., 2011). This technology has been used for identifying receptor binding domains (RBDs) in TGEV that may interact with its pAPN cellular receptor (Ren et al., 2011a). It has also been used for identifying immunogenic proteins in pig sera from convalescent animals with a history of Salmonella typhimurium infection (Meyer et al., 2012), and for targeting tissues like bone-marrow dendritic cells and kidney, liver, lung, spleen and visceral adipose tissues (Jung et al., 2012).
Since the M protein is one of the three major TGEV structural proteins, it was used as a target in biopanning a 12-mer phage display peptide library to identify peptides that ultimately inhibit virus binding. To this end, the TGEV-M protein was first expressed in E. coli as a GST fusion protein. Given the possibility of identifying GST-binding phage, we included additional panning steps using purified recombinant GST as the target to remove phage that did not specifically target the rTGEV-M protein. Also, the stringency was systematically increased by panning with decreasing concentrations of rTGEV-M and increasing concentrations of Tween-20.
Three of 10 TGEV-M phages identified in this study were evaluated by ELISA for detecting TGEV infection. Results showed that the widely used TGEV-M antibody-ELISA and phage-ELISA both could detect the virus successfully; however, phage-ELSIA is more cost-effective because it does not involve animals and phages are in unlimited supply. Further, even using serum from animals boosted 3X with rTGEV-M, the phage-ELISA appeared more sensitive. Clearly, targeting the M protein rather than the whole virus could sacrifice sensitivity. Although we demonstrated specificity in phage and peptide binding to rTGEV-M, we have not determined how well-exposed the binding region is to the surface of the virus. As such, using the M protein as the biopanning agent is probably not as effective as using whole virus. Also, this may have contributed to the lower than expected signal strength in the polyclonal Ab-ELISA especially when the Ab titer using rTGEV-M protein as antigen was so high.
The use of antivirals represents one approach to treat coronavirus infections (Ren et al., 2011b). In this study, a peptide encoded by phTGEV-M7 that interacted with the rTGEV-M protein was synthesized. The interaction was deemed specific in that it was titratable and similar results were not observed with interactions between pepTGEV-M7 and another TGEV structural protein, rTGEV S-AD (Meng et al., 2011). Plaque-reduction assays showed that TGEV infectivity decreased only when virus was pre-treated with the peptide. In contrast, when the pepTGEV-M7 was incubated with ST cells alone or with TGEV-infected ST cells, no inhibitory effect was observed. This is an expected outcome given that rTGEV-M was used to pan for bioreactive phage and this protein is not present on the surface of or within ST cells. Collectively the data presented here are consistent with pepTGEV-M7 binding to the surface of the virus and either interfering with the ability of the virus to invade the cell or generate progeny virus. Neither incubating the peptide with ST cells prior to or after adding virus had a demonstrable effect on replication. The virus-specific effects of pepTGEV-M7 were confirmed by IFA and RT-PCR.
5. Conclusions
In summary, peptide sequences that recognize the TGEV-M protein were identified in our study using phage display technology. Phages bearing these peptides may be utilized in serology-based diagnosis of TGE, pepTGEV-M7 had direct inhibitory effects on TGEV infectivity in vitro. In future research, peptides identified in this study may be subjected to antiviral testing.
Acknowledgements
The research work of the authors was supported by the Program for New Century Excellent Talents at the Heilongjiang Provincial University (1155–NCET–005).
References
- Beer M., Liu C.Q. Panning of a phage display library against a synthetic capsule for peptide ligands that bind to the native capsule of Bacillus anthracis. Plos One. 2012;7:e45472. doi: 10.1371/journal.pone.0045472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowyer P. DNA-mediated transformation of fungi. In: Talbot N., editor. Molecular and Cellular Biology of Filamentous Fungi. Oxford Univ. Press; Oxford: 2001. pp. 33–46. [Google Scholar]
- Cavanagh D. The coronaviridae study group of the International committee on taxonomy of viruses, revision of the taxonomy of the coronavirus, Torovirus, and Arterivirus genera. Arch. Virol. 1994;135:226–237. doi: 10.1007/BF01309782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charley B., Laude H. Induction of interferon alpha by transmissible gastroenteritis coronavirus: role of transmembrane glycoprotein E1. J. Virol. 1988;62:8–11. doi: 10.1128/jvi.62.1.8-11.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., Wang X., Ren Y., Cui S., Li G., Ren X. Genome sequence of Chinese porcine parvovirus strain PPV2010. J. Virol. 2012;86:2379. doi: 10.1128/JVI.06852-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyranka-Czaja A., Otlewski J. A novel, stable, helical scaffold as an alternative binder – construction of phage display libraries. Acta Biochim. Pol. 2012;59:383–390. [PubMed] [Google Scholar]
- Ehrlich G.K., Bailon P. Identification of model peptides as affinity ligands for the purification of humanized monoclonal antibodies by means of phage display. J. Biochem. Biophys. Methods. 2001;49:443–454. doi: 10.1016/s0165-022x(01)00212-3. [DOI] [PubMed] [Google Scholar]
- Gao M., Cui J., Ren Y., Suo S., Li G., Sun X., Su D., Opriessnig T., Ren X. Development and evaluation of a novel reverse transcription loop-mediated isothermal amplification (RT-LAMP) assay for detection of type II porcine reproductive and respiratory syndrome virus. J. Virol. Methods. 2012;185:18–23. doi: 10.1016/j.jviromet.2012.05.016. [DOI] [PubMed] [Google Scholar]
- Gazarian K., Rowlay M., Gazarian T., Vazquez Buchelli J.E., Hernández Gonzáles M. Mimotope peptides selected from phage display combinatorial library by serum antibodies of pigs experimentally infected with Taenia solium as leads to developing diagnostic antigens for human neurocysticercosis. Peptides. 2012;38:381–388. doi: 10.1016/j.peptides.2012.09.021. [DOI] [PubMed] [Google Scholar]
- Harper D.R., Anderson J., Enright M.C. Phage therapy: delivering on the promise. Ther. Deliv. 2011;2:935–947. doi: 10.4155/tde.11.64. [DOI] [PubMed] [Google Scholar]
- Hayden M.S., Gillilandt L.K., Ledbetter J.A. Antibody engineering. Curr. Opin. Immunol. 1997;9:201–212. doi: 10.1016/s0952-7915(97)80136-7. [DOI] [PubMed] [Google Scholar]
- Jiménez G., Correa I., Melgosa M.P., Bullido M.J., Enjuanes L. Critical epitopes in transmissible gastroenteritis virus neutralization. J. Virol. 1986;60:131–139. doi: 10.1128/jvi.60.1.131-139.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung E., Lee N.K., Kang S.K., Choi S.H., Kim D., Park K., Choi K., Choi Y.J., Jung D.H. Identification of tissue-specific targeting peptide. J. Comput. Aided Mol. Des. 2012;26:1267–1275. doi: 10.1007/s10822-012-9614-6. [DOI] [PubMed] [Google Scholar]
- Kay B.K., Kurakin A.V., Hyde-DeRuyscher R. From peptides to drugs via phage display. Drug Discov. Today. 1998;3:370–378. [Google Scholar]
- Krempl C., Herrler G. Sialic acid binding activity of transmissible gastroenteritis coronavirus affects sedimentation behavior of virions and solubilized glycoproteins. J. Virol. 2001;75:844–849. doi: 10.1128/JVI.75.2.844-849.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladner R.C., Ley A.C. Novel frameworks as a source of high-affinity ligands. Curr. Opin. Biotechnol. 2001;12:406–410. doi: 10.1016/s0958-1669(00)00235-4. [DOI] [PubMed] [Google Scholar]
- Laude H., Rasschaert D., Huet J.C. Sequence and N-terminal processing of the transmembrane protein El of the coronavirus transmissible gastroenteritis virus. J. Gen. Virol. 1987;68:1687–1693. doi: 10.1099/0022-1317-68-6-1687. [DOI] [PubMed] [Google Scholar]
- Laude H., Rasschaert D., Delmas B., Godet M., Gelfi J., Charley B. Molecular biology of transmissible gastroenteritis virus. Vet. Microbiol. 1990;23:147–154. doi: 10.1016/0378-1135(90)90144-K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laude H., Gelfi J., Lavenant L., Charley B. Single amino acid changes in the viral glycoprotein M affect induction of alpha interferon by the coronavirus transmissible gastroenteritis virus. J. Virol. 1992;66:743–749. doi: 10.1128/jvi.66.2.743-749.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laude H., Reeth K.V., Pensaert M. Porcine respiratory coronavirus: molecular features and virus-host interactions. Vet. Res. 1993;24:125–150. [PubMed] [Google Scholar]
- Lesinski G.B., Westerink J. Novel vaccine strategies to T-independent antigens. J. Microbiol. Methods. 2001;47:135–149. doi: 10.1016/s0167-7012(01)00290-1. [DOI] [PubMed] [Google Scholar]
- Masters P.S. The molecular biology of coronaviruses. Adv. Virus Res. 2006;66:193–292. doi: 10.1016/S0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng F., Zhao Z., Li G., Suo S., Shi N., Yin J., Zarlenga D., Ren X. Bacterial expression of antigenic sites A and D in the spike protein of transmissible gastroenteritis virus and evaluation of their inhibitory effects on viral infection. Virus Genes. 2011;43:335–341. doi: 10.1007/s11262-011-0637-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer T., Svhirrmann T., Frenzel A., Miethe S., Stratmann-Selke J., Gerlach G.F., Strutzberg-Minder K., Dubel S., Hust M. Identification of immunogenic proteins and generation of antibodies against Salmonella Typhimurium using phage display. BMC Biotechnol. 2012;12:9. doi: 10.1186/1472-6750-12-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki A., Fukuda M., Kuga K., Takagi M., Tsunemitsu H. Prevalence of antibodies against transmissible gastroenteritis virus and porcine respiratory coronavirus among pigs in six regions in Japan. J. Vet. Med. Sci. 2010;72(7):943–946. doi: 10.1292/jvms.09-0377. [DOI] [PubMed] [Google Scholar]
- Ortego J., Escoes D., Laude H., Enjuanes L. Generation of a replication-competent, propagation-deficient virus vector based on the transmissible gastroenteritis coronavirus genome. J. Virol. 2002;76:11518–11529. doi: 10.1128/JVI.76.22.11518-11529.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penzes Z., Gonzalez J.M., Calvo E., Izeta A., Smerdou C., Méndez A., Sanchez C.M., Sola I., Almazan F., Enjuanes L. Complete genome sequence of transmissible gastroenteritis coronavirus PUR46-MAD clone and evolution of the Purdue virus cluster. Virus Genes. 2001;23:105–118. doi: 10.1023/A:1011147832586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X., Glende J., Yin J., Schwegmann-Wessels C., Herrler G. Importance of cholesterol for infection of cells by transmissible gastroenteritis virus. Virus Res. 2008;137:220–224. doi: 10.1016/j.virusres.2008.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X., Wang M., Yin J., Li G. Phages harboring specific peptides to N protein of PRRSV distinguish the virus from other viruses. J. Clin. Microbiol. 2010;48:1875–1881. doi: 10.1128/JCM.01707-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X., Wang M., Yin J., Ren Y., Li G. Heterologous expression of fused genes encoding the glycoprotein 5 from PRRSV: a way for producing functional protein in prokaryotic microorganism. J. Biotechnol. 2010;147:130–135. doi: 10.1016/j.jbiotec.2010.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X., Liu B., Yin J., Zhang H., Li G. Phage displayed peptides recognizing porcine aminopeptidase N inhibit transmissible gastroenteritis coronavirus infection in vitro. Virology. 2011;410:299–306. doi: 10.1016/j.virol.2010.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X., Meng F., Yin J., Li G., Li X., Wang C., Herrler G. Action mechanisms of lithium chloride on cell infection by transmissible gastroenteritis coronavirus. PloS One. 2011;6:e18669. doi: 10.1371/journal.pone.0018669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X., Yin J., Li G., Herrler G. Cholesterol dependence of pseudorabies herpesvirus entry. Curr. Microbiol. 2011;62:261–266. doi: 10.1007/s00284-010-9700-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risco C., Antón I.M., Suñé C., Pedregosa A.M., Martín-Alonso J.M., Parra F., Carrascosa J.L., Enjuanes L. Membrane protein molecules of transmissible gastroenteritis coronavirus also expose the carboxy-terminal region on the external surface of the virion. J. Virol. 1995;69:5269–5277. doi: 10.1128/jvi.69.9.5269-5277.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottier P.J.M. The coronavirusmembrane glycoprotein. In: Siddell S.G., editor. The Coronaviridae. Plenum Press; New York, N.Y: 1995. pp. 115–139. [Google Scholar]
- Saif L.J., Wesley R.D. Transmissible gastroenteritis and porcine respiratory coronavirus. In: Straw B.E., D’Allaire S., Mengeling W.L., Taylor D.J., editors. Diseases of Swine. 8th edition. Iowa State University Press; Ames: 1999. pp. 295–325. [Google Scholar]
- Samoylova T.I., Cochran A.M., Samoylov B., Breiteneicher A.H., Ditchkoff S.S., Peternko V.A., Cox N.R. Phage display allows identification of zona pellucida-binding peptides with species-specific properties: novel approach for development of contraceptive vaccines for wildlife. J. Biotechnol. 2012;162:311–318. doi: 10.1016/j.jbiotec.2012.10.006. [DOI] [PubMed] [Google Scholar]
- Sánchez C.M., Jiménez G., Laviada M.D., Correa I., Suñé C., Bullido M., Gebauer F., Smerdou C., Callebaut P., Escribano J.M. Antigenic homology among coronaviruses related to transmissible gastroenteritis virus. Virology. 1990;174:410–417. doi: 10.1016/0042-6822(90)90094-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott J.K., Smith G.P. Searching for peptide ligands with an epitope library. Science. 1990;249:386–390. doi: 10.1126/science.1696028. [DOI] [PubMed] [Google Scholar]
- Spaan W., Cavanagh D., Horzinek C. Coronaviruses: structure and genome expression. J. Gen. Virol. 1988;69:2939–2952. doi: 10.1099/0022-1317-69-12-2939. [DOI] [PubMed] [Google Scholar]
- Sui X., Yin J., Ren X. Antiviral effect of diammonium glycyrrhizinate and lithium chloride on cell infection by pseudorabies herpesvirus. Antiviral Res. 2010;85:346–353. doi: 10.1016/j.antiviral.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suñé C., Jiménez G., Correa I., Bullido M.J., Gebauer F., Smerdou C., Enjuanes L. Mechanisms of transmissible gastroenteritis coronavirus neutralization. Virology. 1990;177:559–569. doi: 10.1016/0042-6822(90)90521-R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D., Li G., Qin C., Ren X. Phage displayed peptides to avian H5N1 virus distinguished the virus from other viruses. PloS One. 2011;6:e23058. doi: 10.1371/journal.pone.0023058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi G., Qian J., Wang Z., Qi Y. A phage-displayed peptide can inhibit infection by white spot syndrome virus of shrimp. J. Gen. Virol. 2003;84:2545–2553. doi: 10.1099/vir.0.19001-0. [DOI] [PubMed] [Google Scholar]
- Yin J., Glende J., Schwegmann-Wessels C., Enjuanes L., Herrler G., Ren X. Cholesterol is important for a post-adsorption step in the entry process of transmissible gastroenteritis virus. Antiviral Res. 2010;88:311–316. doi: 10.1016/j.antiviral.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W., Ren X. Isolation, genome phylogenetic analysis and in vitro rescue of a newly emerging porcine circovirus type 2. Pak. Vet. J. 2012;32:165–170. [Google Scholar]


