Highlights
-
•
The prevalence of PDCoV was high in Henan pigs and the novel virus can infect sows.
-
•
The complete genomes of 4 representative PDCoV Henan strains were sequenced.
-
•
The PDCoV Henan strains were grouped within the Chinese clade.
-
•
The PDCoV strains from Henan might evolve from the latest Chinese epidemic strains.
Keywords: Porcine deltacoronavirus, Prevalence, Complete genome, Phylogenetic analysis, Evolution
Abstract
Porcine deltacoronavirus (PDCoV) is a novel porcine enteric coronavirus that causes diarrhea, vomiting and dehydration in piglets. This newly virus has spread rapidly and has caused serious economic losses for pig industry since the outbreak in USA in 2014. In this study, 430 faecal and intestinal samples (143 faecal samples and 287 intestinal samples) were collected from individual pigs with diarrhea and 211 serum samples were also collected from the sows with mild diarrhea in 17 regions in Henan province, China from April 2015 to March 2018. The RT-PCR detection indicated that the infection of PDCoV was high up to 23.49% (101/430), and co-infection with PEDV were common (60.40%, 61/101) in Henan pigs. The prevalence of PDCoV in suckling piglets was the highest (36.43%, 94/258). We also found that PDCoV could be detected in sows faeces and sera while the sows showed mild, self-limited diarrhea in clinic. The complete genomes of 4 PDCoV Henan strains (CH-01, HNZK-02, HNZK-04, HNZK-06) were sequenced and analyzed. Phylogenetic analysis based on the complete genome, spike and nucleocapsid gene sequences revealed that the PDCoV Henan strains were closely related to other PDCoV reference strains that located in the Chinese clade. Furthermore, the phylogenetic analysis showed PDCoV CH-01 strain was closely related to CHN-HB-2014 strain and HKU15-44 strain, while the other PDCoV Henan strains were more related to PDCoV CHJXNI2 and CH-SXD1-2015 strains, indicating that the ancestor of these sequenced strains may different. These results would support the understanding of the prevalence and evolution characteristics of PDCoV in China.
1. Introduction
Coronaviruses are single-stranded positive-sense, enveloped RNA viruses that can be classified into four genera, Alphacoronavirus, Betacoronavirus, Gammacoronavirus and Deltacoronavirus (Ma et al., 2015). Coronaviruses from different genera can infect diverse host species, mainly including mammals and birds (Woo et al., 2010; Chan et al., 2013). Deltacoronavirus, a novel genus, was first found in Asian leopard cat and Chinese ferret badgers in Hong Kong and then identified in a variety of avian species (Dong et al., 2007). In 2012, the porcine deltacoronavirus (PDCoV) was first discovered in pigs in Hong Kong (Woo et al., 2012). To date, six coronaviruses have been identified from pigs, including the Alphacoronavirus porcine epidemic diarrhea virus (PEDV), transmissible gastroenteritis virus (TGEV), porcine respiratory coronavirus (PRCV) and swine acute diarrhea syndrome coronavirus (SADS-CoV), the Betacoronavirus genus porcine haemagglutinating encephalomyelitis coronavirus (PHEV), and the Deltacoronavirus genus PDCoV (Jung et al., 2016b; Gong et al., 2017; Pan et al., 2017; Hu et al., 2018; Zhou et al., 2018). The clinical symptom caused by PDCoV in piglets is similar to that of the PEDV and TGEV infection, including severe diarrhea, vomiting and dehydration (Hu et al., 2016; Jung et al., 2016a). However, the clinical symptom and mortality caused by PDCoV are milder than PEDV infection in piglets (Jung et al., 2016a). Furthermore, PDCoV commonly co-infected with PEDV and TGEV in natural infections and has seriously harmed the development of the pig industry (Hu et al., 2015). Currently, there are no effective reagents and vaccines to control PDCoV infection.
PDCoV has the smallest genome size among coronaviruses and the full-length genome is approximately 25.4 kb with a 5′ cap and a 3′ polyadenylated tail (Fang et al., 2016). The two-thirds of PDCoV genome at the 5′ end encodes genome replication proteins (ORF1ab) and one-third at the 3′ end encodes four mainly structural proteins, spike glycoprotein (S), envelope protein (E), membrane protein (M), and nucleocapsid protein (N), and three accessory proteins, including nonstructural protein 6 (NS6), NS7, and NS7a (Woo et al., 2012; Jang et al., 2017). The PDCoV S protein is a glycoprotein on the viral surface and plays an essential function in mediating viral entry into host cells by binding with host receptors (Graham and Baric, 2010; Shang et al., 2017). In addition, PDCoV S gene encodes the most antigenic protein and frequently used for studying the genetic relationships among different PDCoV strains and the epidemiological investigation (Shang et al., 2017). The highly conserved coronaviruses N protein produces largely in infected cells and plays a vital function in viral replication and pathogenesis. The recent study showed that the multifunctional role of PDCoV N protein is similar to other coronaviruses (Lee and Lee, 2015).
In February 2014, PDCoV was first detected in swine herds with diarrhea in Ohio, USA (Wang et al., 2014a). This novel coronavirus was then further detected in more than 20 US states (Marthaler et al., 2014; Wang et al., 2014b; Sinha et al., 2015). Subsequently, PDCoVs were detected in pigs in Canada, South Korea, mainland China, Thailand and Laos (Lee and Lee, 2014; Janetanakit et al., 2016; Saeng-Chuto et al., 2017; Ajayi et al., 2018). In China, PDCoV infection in pig farms with diarrhea occurred in some provinces and regions since 2014 (Wang et al., 2015; Dong et al., 2016; Zhai et al., 2016). Previous study also showed that PDCoV was detected in diarrheic pig samples collected as early as 2004 in China (Dong et al., 2015). Sequence analyses suggested that all the known PDCoVs strains have a high nucleotide similarity with the earliest discovered strains, HKU15-44 and HKU15-155 (Jung et al., 2016b), and all reported China PDCoV strains are closely related with each other and located in the same clade (Song et al., 2015; Liu et al., 2018; Mai et al., 2018; Wang et al., 2018). Henan province is a major pig-raising region in China. According to the data of the National Bureau of Statistics of China, the number of pigs in stock in the first half year of 2018 in Henan province was about 42 million with the percentage of 10.2% in the whole stock pigs in China. However, little is reported regarding the prevalence and epidemiology of PDCoV in Henan province.
In this study, we aimed to investigate the PDCoV prevalence in the diarrheal pigs and full-length genome sequence properties of PDCoV in Henan province. 430 faecal and intestinal samples were collected from pigs with diarrhea and vomiting in 17 regions for PDCoV testing. 211 serum samples were also collected from the sows with mild diarrhea to detect PDCoV infection. The characteristic of PDCoV prevalence in the diarrheal pigs in different regions of Henan province was analyzed. Then the complete genomes of 4 representative PDCoV Henan strains were sequenced, and the phylogenetic and evolutionary characterization of these four PDCoV strains were further analyzed. Our study will provide more information about prevalence and genetic evolution of PDCoV.
2. Materials and methods
2.1. Clinical sample collection
From April 2015 to March 2018, 430 faecal and intestinal samples (143 faecal samples and 287 intestinal samples) were collected from pigs with diarrhea outbreaks that diagnosed by pig farm veterinarian in 17 regions in Henan province, China. The number of sampled farms and pigs in different years and regions was as shown in Table 1 . 211 serum samples were also collected from the sows with mild diarrhea in this period. The swine faeces and intestinal contents were diluted 10-fold with phosphate-buffered saline (PBS). The suspensions were vortexed and centrifuged at 1847 g at 4 °C for 10 min. The clarified supernatants were collected and stored at −80 °C for RNA extraction. The research protocol for animal experiments of live pigs in this study was approved by the Animal Care and Use Committee of Henan Agricultural University (Zhengzhou, China) and was performed in accordance with the “Guidelines for Experimental Animals” of the Ministry of Science and Technology (Beijing, China).
Table 1.
The number of sampled farms and pigs in different years and regions in Henan province, China.
| Regions | The number of sampled farms |
The number of sampled pigs |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 2015 | 2016 | 2017 | 2018 | Total | 2015 | 2016 | 2017 | 2018 | Total | |
| An yang | 0 | 1 | 1 | 0 | 2 | 0 | 2 | 4 | 0 | 6 |
| He Bi | 1 | 4 | 3 | 1 | 9 | 3 | 8 | 12 | 3 | 26 |
| Ji Yuan | 0 | 0 | 2 | 0 | 2 | 0 | 0 | 6 | 0 | 6 |
| Kai Feng | 4 | 2 | 5 | 1 | 12 | 15 | 6 | 14 | 2 | 37 |
| Luo Yang | 1 | 5 | 1 | 0 | 7 | 1 | 17 | 6 | 0 | 24 |
| Luo He | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 3 | 0 | 3 |
| Nan Yang | 3 | 3 | 4 | 1 | 11 | 4 | 20 | 17 | 2 | 43 |
| Ping Dingshan | 3 | 1 | 3 | 1 | 8 | 7 | 6 | 18 | 6 | 37 |
| Pu Yang | 2 | 2 | 6 | 1 | 11 | 10 | 16 | 51 | 2 | 79 |
| San Menxia | 0 | 1 | 3 | 0 | 4 | 0 | 1 | 9 | 0 | 10 |
| Zheng Zhou | 1 | 0 | 4 | 1 | 6 | 2 | 0 | 10 | 3 | 15 |
| Xin Xiang | 1 | 3 | 1 | 0 | 5 | 2 | 7 | 5 | 0 | 14 |
| Xin yang | 0 | 0 | 3 | 2 | 5 | 0 | 0 | 3 | 3 | 6 |
| Zhou Kou | 2 | 3 | 4 | 2 | 11 | 5 | 8 | 27 | 31 | 71 |
| Zhu Madian | 1 | 0 | 5 | 0 | 6 | 7 | 0 | 21 | 0 | 28 |
| Shang Qiu | 1 | 1 | 4 | 1 | 7 | 3 | 1 | 17 | 1 | 22 |
| Xu Chang | 0 | 0 | 1 | 1 | 2 | 0 | 0 | 1 | 2 | 3 |
| Total | 20 | 26 | 51 | 12 | 109 | 59 | 92 | 224 | 55 | 430 |
2.2. RNA extraction and PDCoV detection
Total RNAs were extracted from samples using Trizol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. Synthesis of cDNA from the extracted RNA was carried out by using the Vazyme Reverse Transcription Kit (Nanjing, China). To detect PDCoV from the samples’ RNA, a pair of primers targeting the S gene of PDCoV (5′- GACCCTAAATCTGCCGTTAGAG-3′ and 5′-TGTTGGAGAGGTGAATGCTATG-3′) were designed based on the sequence of HKU15-155 strain (GenBank accession no. JQ065043), and the expected size of the amplicon is 543 bp. PCR was performed in a 25 μL volume containing 1 μL cDNA, 12.5 μL 2×Rapid Taq Master Mix, and 0.5 μL of each primer (25 μmol/L). The reactions were subjected to 95℃ for 5 min, followed by 35 cycles of 95℃ for 50 s, 58℃ for 45 s, and 72℃ for 30 s, with a final extension step at 72℃ for 10 min. The PCR products were purified using the SanPrep Column DNA Gel Extraction Kit (Sangon Biotech, Shanghai, China). Both strands of the PCR products were sequenced twice with an ABI Prism 3700 DNA analyzer (Sangon Biotech, Shanghai, China) by using the forward and reverse PCR primers. The sequences of the PCR products were compared with the known S gene sequences of PDCoVs in the GenBank database. The PDCoV positive test samples were then further used to detect PEDV and TGEV. To detect PEDV and TGEV from the samples’ RNA, two pair of primers targeting the M gene of PEDV (5′-TGGTGGTCTTTCAATCCTG-3′ and 5′-CTGAGTAGTCGCCGTGTTT-3′) and the N gene of TGEV (5′-CGCTATCGCATGGTGAAG-3′ and 5′-GGATTGTTGCCTGCCTCT-3′) were designed based on the sequence of the JXNC1302 strain (GenBank accession no. KJ526109) and the H155 strain (GenBank accession no. GQ374566) respectively. The expected size of the amplicons are 298 bp for the M gene of PEDV and 324 bp for the N gene of TGEV. The PCR amplifications and products analysis were the same as that of PDCoV.
2.3. Complete genome sequence analysis
Four representative PDCoV Henan strains were selected from four different pig farms that the pigs had severe diarrhea at different times to sequence the complete genome by the Sanger method. These representative PDCoV strains were named as CH-01, HNZK-02, HNZK-04 and HNZK-06. To determine the full-length genomic sequence of the isolated PDCoV strains, 13 pairs of primers were designed based on the HKU15-155 strain (GenBank accession no. JQ065043) (Table 2 ). Viral RNA was extracted according to the aforementioned method. Thirteen overlapping fragments covering the entire viral genome were amplified on the conditions of 95 °C for 5 min, followed by 35 cycles of 95 °C for 1 min, 55 °C for 45 s, 72 °C for 2 min and finally 72 °C for 10 min. The PCR products were purified and cloned into pMD18-T vectors (TaKaRa, Dalian, China) following the manufacturer’s protocol. The positive clone of each amplicon were sequenced twice (Sangon Biotech, Shanghai, China).
Table 2.
Primers used for amplification of the complete genome of PDCoV.
| Primers | Sequence (5’→3’) | Nucleotide position* | Fragment size (bp) |
|---|---|---|---|
| PDC-1-F PDC-3300-R |
ACATGGGGACTAAAGATAAAAATTATAGC GCTCATCGCCTACATCAGTG |
1-29 3281-3300 |
3300 |
| PDC-3091-F PDC-4860-R |
CGGATTTAAAACCACAGACT ACGACTTTACGAGGATGAAT |
3091-3110 4841-4860 |
1770 |
| PDC-4741-F PDC-6420-R |
CTCCTGTACAGGCCTTACAA TCACACGTATAGCCTGCTGA |
4741-4760 6401-6420 |
1680 |
| PDC-6291-F PDC-8041-R |
CTCAATGCAGAAGACCAGTC CAGCTTGGTCTTAAGACTCT |
6291-6310 8041-8060 |
1751 |
| PDC-7920-F PDC-9660-R |
GGTACTGCTTCTGATAAGGAT TAGGTACAGTTGTGAACCGA |
7920-7940 9641-9660 |
1741 |
| PDC-9463-F PDC-10579-R |
TACTCTTCACAGCCTTCAC TTCTTCACAATAGTGTAGCG |
9463-9482 105601-10579 |
1117 |
| PDC-10560-F PDC-12847-R |
CGCTACACTATTGTGAAGAA TTCGTAGGGCTCAAGATA |
10560-10579 12829-12847 |
2288 |
| PDC-12301-F PDC-14952-R |
TCCAGATGACCGTTGCGTAT CCAACAGAGTCGGGTAAA |
12301-12320 14935-14952 |
2652 |
| PDC-14703-F PDC-17185-R |
CACCCATAACGAAGAACC CCGATGAGTGTCGTAGCG |
14703-14720 17168-17185 |
2483 |
| PDC-17165-F PDC-19203-R |
TGCCGCTACGACACTCAT TCCGCTAAAGGAGAATAGGTTG |
17165-17182 19182-19203 |
2039 |
| PDC-18485-F PDC-21079-R |
TGCTACCCAATCTTACAGT GCAAATACTCCGTCTGAAC |
18485-18503 21061-21079 |
2594 |
| PDC-20761-F PDC-23825-R |
GTCTTACCGTGTGAAACCCC AACCACGAGACTGTAAGCAA |
20761-20780 23806-23825 |
3064 |
| PDC-23804-F Oligo(dT) |
TTTTGCTTACAGTCTCGTGGTT TTTTTTTTTTTTTTTTTT |
23804-23825 25432 |
1629 |
Nucleotide position of each primer was numbered based on the HKU15-155 strain (GenBank accession no. JQ065043).
2.4. Sequence alignment and phylogenetic analyses
Complete sequences of identified PDCoV strains in this study were obtained by assembling overlapping contigs followed by trimming off primer sequence, and then used in sequence alignment and phylogenetic analyses with other PDCoV reference strains (the representative strains of USA, China, South Korea, Thailand, Lao and Vietnam). The information of the four isolate strains and other reference strains of PDCoV are shown in Table 3 . The nucleotide and deduced amino acid sequences were aligned and analyzed using the Clustal W program of DNAStar 7.0 green (DNAstar, Madison, WI). Phylogenetic trees were constructed using the neighbor-joining method in MEGA 6.0 software (http://www.megasoftware.net/) with bootstrap analysis of 1000 replicates.
Table 3.
The isolate strains and other reference strains of PDCoV used for sequence alignment and phylogenetic analysis.
| Strain | Accession no. | Country | Isolated time |
|---|---|---|---|
| CH-01 | KX443143 | China/Hebi | 2016 |
| HNZK-02 | MH708123 | China/Zhoukou | 2018 |
| HNZK-04 | MH708124 | China/Zhoukou | 2018 |
| HNZK-06 | MH708125 | China/Zhoukou | 2018 |
| CHN-AH-2004 | KP757890 | China/Anhui | 2004 |
| CHJXNI2 | KR131621 | China/Jiangxi | 2015 |
| CHN-HB-2014 | KP757891 | China/Hebei | 2014 |
| CHN-JS-2014 | KP757892 | China/Jiangsu | 2014 |
| CHN-HN-2014 | KT336560 | China/Henan | 2014 |
| CH/JXJGS01/2016 | KY293677 | China/Jiangxi | 2016 |
| CH/SXD1/2015 | KT021234 | China | 2015 |
| HKU15-155 | JQ065043 | China/Hong Kong | 2010 |
| HKU15-44 | JQ065042 | China/Hong Kong | 2009 |
| Ohio137 | KJ601780 | USA | 2014 |
| Illinois134 | KJ601778 | USA | 2014 |
| Illinois121 | KJ481931 | USA | 2014 |
| Indiana453 | KR265851 | USA | 2014 |
| Minnesota159 | KR265859 | USA | 2014 |
| Minnesota292 | KR265864 | USA | 2014 |
| Minnesota | KR265853 | USA | 2013 |
| NorthCarolina452 | KR265858 | USA | 2014 |
| Arkansas61 | KR150443 | USA | 2015 |
| Michigan/8977 | KM012168 | USA | 2014 |
| Nebraska209 | KR265860 | USA | 2014 |
| OhioCVM1 | KJ769231 | USA | 2014 |
| NE3579 | KJ584359 | USA | 2014 |
| USA/IA/2014 | KJ567050 | USA | 2014 |
| PA3148 | KJ584358 | USA | 2014 |
| MI6148 | KJ620016 | USA | 2014 |
| OH1987 | KJ462462 | USA | 2014 |
| SD3424 | KJ584356 | USA | 2014 |
| Swine/Thailand/S5025 | KU051656 | Thailand | 2015 |
| PDCoV/2016/Lao | KX118627 | Lao | 2016 |
| Swine/Vietnam/Binh21/2015 | KX834352 | Vietnam | 2015 |
| KNU16-07 | KY364365.1 | South Korea | 2014 |
| KNU14-04 | KM820765 | South Korea | 2014 |
2.5. Nucleotide sequence accession numbers
The complete genome sequences of PDCoV CH-01, HNZK-02, HNZK-04 and HNZK-06 strains were deposited in GenBank under accession numbers KX443143, MH708123, MH708124 and MH708125, respectively.
3. Results
3.1. Prevalence of PDCoV in clinical samples from diarrhoeic pigs
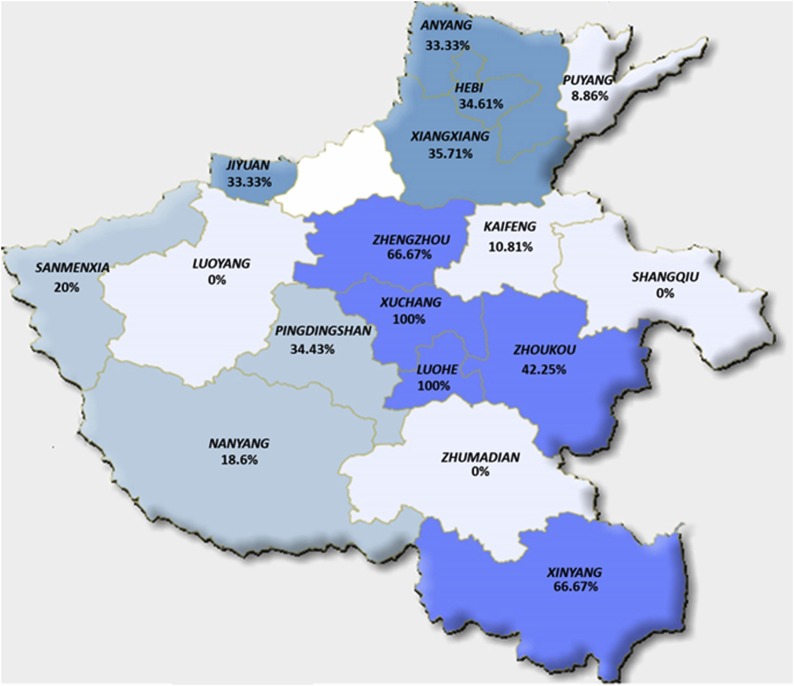
In total, 430 porcine clinical samples with diarrhea were collected from 17 regions in Henan province from April 2015 to March 2018 for PDCoV detection. The results showed that 101 PDCoV positive samples were identified from the 430 suspected samples and the total positive proportion was 23.49% (101/430) as shown in Table 4 . In addition, PDCoV could be detected in sows’ sera samples with the positive proportion of 25.59%. The positive proportion of PDCoV in different regions since April 2015 was shown in Fig. 1 . PDCoV was detected out in 14 regions out of the 17 regions, including the regions of Xuchang (100%), Luohe (100%), Zhengzhou (66.67%) and Zhoukou (42.25%), implying this novel coronavirus existed widely in pig farms in Henan province (Fig. 1).
Table 4.
The detection result of PDCoV epidemic in different years and seasons in Henan province, China.
| Seasons | Sample number | Positive number of PDCoV |
|---|---|---|
| 2015 | 59 | 1 |
| Spring | 27 | 0 |
| Summer | 13 | 0 |
| Autumn | 14 | 0 |
| Winter | 16 | 3 |
| 2016 | 92 | 22 |
| Spring | 34 | 11 |
| Summer | 18 | 4 |
| Autumn | 15 | 0 |
| Winter | 29 | 14 |
| 2017 | 224 | 33 |
| Spring | 132 | 19 |
| Summer | 32 | 0 |
| Autumn | 29 | 0 |
| Winter | 55 | 37 |
| 2018 | 55 | 45 |
| Spring | 16 | 13 |
| Total | 430 | 101 |
Fig. 1.
Map of PDCoV distribution in pigs in 17 regions of Henan province. The positive rates of PDCoV infection were labeled in the regions. The different color scales represent different positive rates and the regions with dark colors represent high positive rates for PDCoV infection.
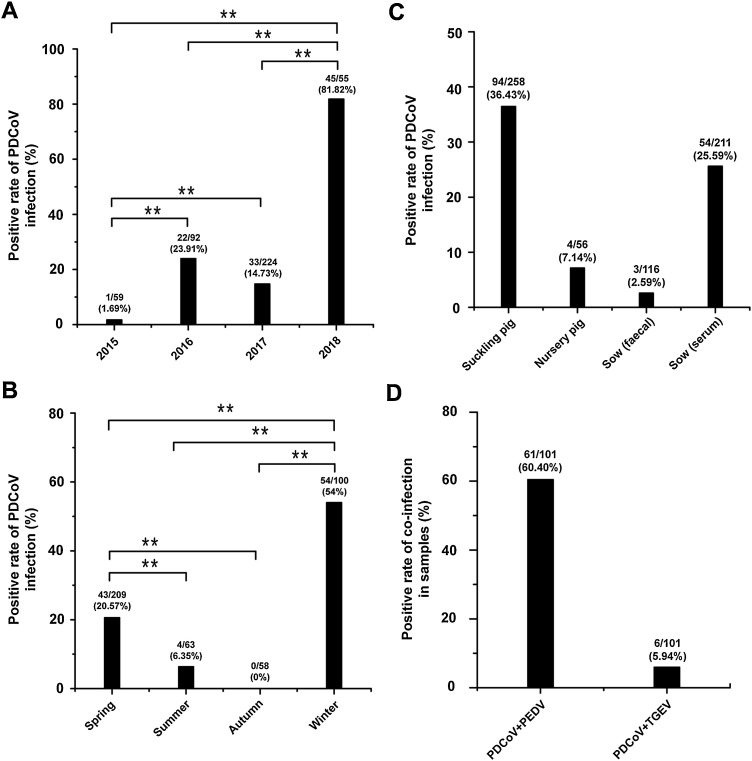
The positive proportions of PDCoV in different years and seasons were analyzed and shown in Fig. 2 A–B. The results indicated that the prevalence of PDCoV in the diarrheal pigs was increasing from 2015 (1.69%) to 2018 (81.82%). The PDCoV positive proportion in spring (20.57%) and winter (54%) was obviously higher than that in summer (6.35%) and autumn (0%) (Fig. 2B). Moreover, the positive proportion of PDCoV infection in suckling piglets was 36.43%, which was the highest at different stages of pigsin Henan province. PDCoV infection was relatively low in nursery pigs with the positive proportion of 7.14%. PDCoV positive proportion was the lowest in sows faeces (2.59%) in our current study (Fig. 2C). Our detection also found that co-infections of PDCoV with PEDV were common in Henan diarrhoeic pigs. 61 out of 101 PDCoV positive samples were proved to exist PEDV, with the positive proportion of 60.40%. In addition, TGEV was identified in 6 out of 101 (5.94%) PDCoV positive samples (Fig. 2D). These data also suggested that the co-infections of PDCoV with PEDV and TGEV often occurred and had caused great damage to local pig farms.
Fig. 2.
Detection of PDCoV infection in pigs in Henan, China. (A) The total positive rates of PDCoV infection in diarrhea pigs from 2015 to 2018 in Henan. Two asterisks indicate significant differences between groups (p < 0.01 as determined by chi-square test). (B) The positive rates of PDCoV infection in diarrhea pigs at each season from 2015 to 2018 in Henan. Two asterisks indicate significant differences between groups (p < 0.01 as determined by chi-square test). (C) The positive rate of PDCoV infection in pig herds at different stages of growth. The corresponding positive rates of PDCoV infection were labeled in the histogram. (D) The positive rate of PDCoV co-infection with PEDV and TGEV in 101 PDCoV positive pig herds. The corresponding positive rates of co-infection were labeled in the histogram.
3.2. Complete genomic characterization of PDCoV strains in Henan, China
The complete genome sequence of four representative PDCoV strains (CH-01, HNZK-02, HNZK-04, HNZK-06) were determined using the designed primers. The results showed that the entire genome of PDCoV CH-01 strain comprised 25,404 nt in length with the identical order of a 539-nt 5′ untranslated region (UTR), 18,788-nt open reading frame 1a/1b (ORF1a/1b), 3,480-nt S gene, 252-nt E gene, 654-nt M gene, 285-nt NS6 gene, 1,029-nt N gene, 603-nt NS7 gene and 390-nt 3′ UTR. The entire genome of the other three representative PDCoV strains (HNZK-02, HNZK-04, HNZK-06) comprised 25,419 nt in length and containing a 18,803-nt ORF1a/1b, which had a 6-nt and 9-nt insert at position 1739 and 2804 nt when compared with CH-01 strain (the nt positions were numbered according to the entire gene of PDCoV CH-01 sequence). The four sequenced PDCoV S genes had a 3 nucleotides deletion at position 52 amino acid (aa) when compared with the US strains and early Chinese strains (CHN-AH-2004, HKU15-44), and these nucleotides deletion were also existed in some Chinese PDCoV strains (CHJXNI2, CHN-HB-2014, CHN-HN-2014, CHN-JS-2014, HKU15-155). Compared with the other PDCoV strains, PDCoV CH-01 strain had 5 aa mutations in S protein, these changed positions were as following: His changed to Gln at position 99, His changed to Gln at position 148, Asp changed to Ala at position 260, lle changed to Leu at position 636, and Phe changed to Leu at position 929 (the aa positions were numbered according to the S gene of PDCoV CH-01 sequence).
The PDCoV complete genome sequences examined in this study shared 98.4%–100% nucleotide identities with each other, and they shared 98.1%–99.4% nucleotide identities and 98.1%˜99.4% aa homology with other PDCoV strains available in GenBank. The S and N gene of PDCoV CH-01 strain had the lowest nucleotide similarity with the other PDCoV strain, which shared 83.9%˜95.6% and 98.4%˜99.2% nucleotide identity with others, respectively.
3.3. Phylogenetic analysis of PDCoV Henan strains
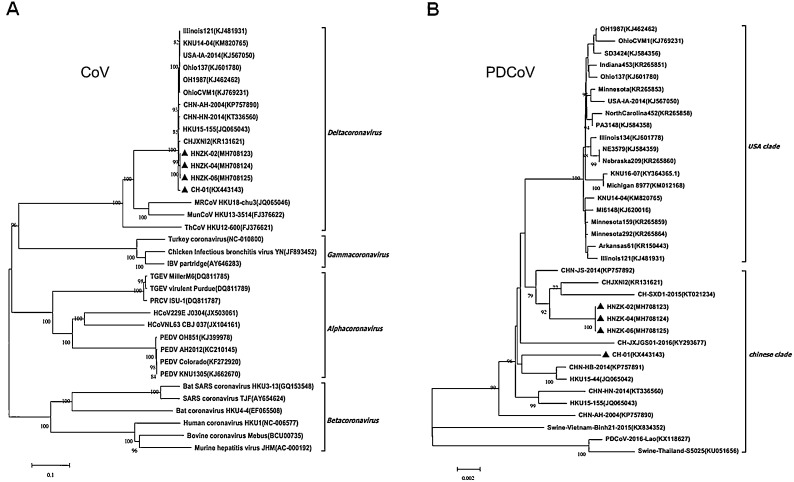
To study the genetic relationships of identified PDCoV strains in this study with other reference PDCoV strains, phylogenetic analyses were first carried out based on the complete genome of the four sequenced PDCoV strains and other coronaviruses obtained from GenBank. The results showed that the sequenced Henan PDCoV strains were clustered into the deltacoronavirus group, forming a branch with other ten swine-origin PDCoV strains, which was distinct from the bird-origin deltacoronaviruses (Fig. 3 A). Further phylogenetic analysis based on the complete genomes of PDCoV indicated that PDCoV CH-01 strain isolated in Henan was grouped within the Chinese clade and closely related to CHN-HB-2014 strain (KP757891) and HKU15-44 strain (JQ065042). (Fig. 3B). However, the PDCoV HNZK-02, HNZK-04 and HNZK-06 strains were located in the same separate subcluster and more related to PDCoV CHJXNI2 strain (KR131621) and CH-SXD1-2015 strain (KT021234), indicating that the origin of these isolated strains may be different (Fig. 3B).
Fig. 3.
Phylogenetic analysis based on the complete genome sequences of PDCoV strains. (A) The nucleotide sequences of the full-length genomes from four coronavirus genera (Alphacoronavirus, Betacoronavirus, Gammacoronavirus and Deltacoronavirus). (B). Phylogenetic analyses of complete nucleotide sequences of 4 PDCoVs from this study and some published PDCoV sequences in Genbank. The phylogenetic tree was constructed from the aligned nucleotide sequences using the neighbour-joining method with MEGA 6.06 software (http://www.megasoftware.net). Bootstrap values were calculated with 1000 replicates. Reference sequences obtained from GenBank are indicated by strain names and GenBank accession numbers. The PDCoV isolates identified in this study are indicated with black triangles. Scale bars indicate nucleotide substitutions per site.
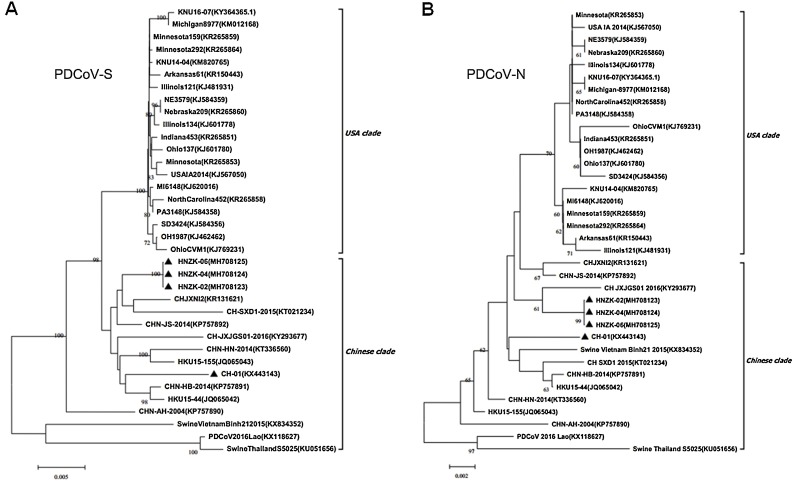
As shown in Fig. 4 , phylogenetic trees were also constructed using the full-length PDCoV S and N genes of the sequenced Henan strains and other reference PDCoV strains. Phylogenetic analyses of the PDCoV N genes revealed that they belonged to the same groups as in the tree based on the PDCoV complete genomes. Phylogenetic analysis based on the entire S genes of PDCoV showed that CH-01 strain was closely related with CHN-HB-2014 and HKU15-44 strains (Fig. 4A). As the phylogenetic tree based on the PDCoV complete genome, the other 3 sequenced PDCoV Henan strains (HNZK-02, HNZK-04 and HNZK-06) were still closely related each other and located in the same separate subcluster, and they also showed a close relationship with CHJXNI2 and CH-SXD1-2015 strains. (Fig. 4B). Altogether, the phylogenetic data indicated that these sequenced strains of PDCoV in Henan were consistent with epidemic strains in China and might evolve from different origins.
Fig. 4.
Phylogenetic analysis using the S genes (A) and the N genes (B) from different PDCoV strains. Reference sequences obtained from GenBank are indicated by strain names and GenBank accession numbers. The S and N genes from PDCoV isolates identified in this study are indicated with black triangles. The phylogenetic tree was constructed as Fig. 3.
4. Discussion
Piglet diarrhea is an important disease that threatens the pig industry development, and the coronaviruses are currently the main pathogens that caused piglet diarrhea. PDCoV is a novel swine intestine coronavirus and has caused serious economic losses for pig industry (Hu et al., 2015). So far, PDCoV has been reported in several countries and the pathogenicity for this novel virus has been studied using the neonatal piglets (Wang et al., 2014a; Wang et al., 2015; Dong et al., 2016; Hu et al., 2016). However, little regarding the molecular epidemiology of PDCoV has been reported in Henan province, a major pig-raising region of China. In the present study, we investigated the PDCoV prevalence in the diarrheal pigs in different regions and analyzed the full-length genome sequence properties of PDCoV strains in Henan province, which might provide more insight about the pigs diarrhea outbreaks in China.
In our study, we found that the PDCoV prevalence in Henan diarrheal pigs was high, which is similar to the other independent reports from the China (Song et al., 2015; Mai et al., 2018). RT-PCR detection of 430 porcine diarrhea samples collected from April 2015 to March 2018 revealed that the total positive rate of PDCoV infection in Henan pigs was 23.49%. Previous reports have shown that the prevalence of PDCoV in diarrheal pigs in Guangdong and Jiangxi were 21.8% and 33.71%, respectively (Song et al., 2015; Mai et al., 2018). In addition, the molecular-based detection also showed a relatively high prevalence of PDCoV infection in South Korea pigs and the total positive rate was 19.03% (Jang et al., 2017). The prevalence of PDCoV in Tibetan diarrheal pigs surrounding the Qinghai-Tibet Plateau of China was significantly lower than that in other provinces and the positive rate was only 3.70% (Wang et al., 2018). For the limitation of the collected sample sizes, especially in the regions of Xu Chang (3), Luo He (3), An Yang (6), Ji Yuan (6) and Xin Yang (6), the PDCoV existence in the pig herds need to be verified by evaluating more samples. So we will continue to surveil the PDCoV prevalence in these regions. In addition, we only have collected the samples from pigs with diarrhea but not collected the samples from pigs without such symptoms to detect PDCoV infection. We will also collect the samples from pigs without diarrhea to fully evaluate the prevalence of PDCoV in Henan pig herds.
RT-PCRs detection indicated that the prevalence of PDCoV in the diarrheal pigs was increasing year by year from 2015 to 2018, leading to the increase of prevalence and severity of diarrhea. Our result also suggested that the positive rate of PDCoV infection in spring and winter was striking higher than the positive rate in summer and autumn and the prevalence of PDCoV in the diarrheal pigs reached a high level in the winter, indicating that this novel virus infection mainly occurred in the cold season. Furthermore, the detection showed that the positive rate of PDCoV infection in suckling piglets was highest and the rate was 36.43%. Interestingly, our results also showed that PDCoV can be discovered in sows and the positive rates were 2.59% in faecal samples and 25.59% in serum samples, indicating that the PDCoV can infect sows and infection rate was increasing. Previous reports showed that PEDV can infect the pigs of all growth stages, including sows and caused severe vomiting and watery diarrhea (Yu et al., 2018). However, PDCoV mainly infected the piglets but rarely infected the sows in previous reports (Liu et al., 2018; Wang et al., 2018). Previous report suggested that the PDCoVs were only detected in Tibetan pigs under 1 month of age but PEDV could detected in pigs of different ages (Wang et al., 2018). A recent report found that the PDCoV can infect the sows and the infection rate was up to 22.4% in sows (Mai et al., 2018). In this study, we also proved this result, indicated that we will need to continue monitoring the infection of PDCoV in sows.
The co-infections of PDCoV with PEDV and TGEV in natural infections were common in previous reports (Song et al., 2015; Jang et al., 2017; Mai et al., 2018). Our detection also found that co-infections of PDCoV with PEDV were common in Henan diarrheal pigs and 61 of 101 (60.40%) PDCoV positive samples were proved to be positive for PEDV. The co-infections of diarrhea coronavirus have caused severe mortality in piglets (Song et al., 2015) and further study will needed to elucidate the prevalence of these viruses in Henan province.
Homology analysis showed that the complete genome sequence of PDCoV CH-01 strain shared 98.1%˜99.4% nucleotide identity with all reference strains described in previous reports. Furthermore, the analysis results showed that the S and N gene of PDCoV CH-01 shared 83.9%˜95.6% and 98.4%˜99.2% nucleotide identity with the reference strains, respectively, indicating that the S gene had a high genetic diversity and strong evolutionary of PDCoV in pigs. At present, coronaviruses are divided into four genera based on the conserved domains, including Alpha-, Beta-, Gamma- and Deltacoronavirus(Chan et al., 2013; Ma et al., 2015). Phylogenetic analyses based on the complete genome showed that the Henan PDCoV strains were clustered into the porcine deltacoronavirus group and located in the Chinese clade. Further phylogenetic analysis based on the full genomes of PDCoV indicated that the CH-01 was closely related to CHN-HB-2014 strain and HKU15-44 strain. However, the PDCoV HNZK-02, HNZK-04 and HNZK-06 strains were more related to CHJXNI2 strain and CH-SXD1-2015 strain. The results indicated that these sequenced strains might evolve from the different Chinese strains. The reference strains CHN-HB-2014, CHJXNI2 and CH-SXD1-2015 were detected from the piglets in recently years, indicated that these sequenced PDCoV Henan strains might evolve from the latest Chinese epidemic strains. In addition, the phylogenetic tree based on S and N genes suggested that Henan strains were still clustered into Chinese clade.
In conclusion, diarrheal samples collected from the pigs in Henan were detected using RT-PCR assay and evaluated for the prevalence of PDCoV in the diarrheal pigs. Our results indicated that the monoinfection of PDCoV and co-infection with PEDV in Henan pigs were common and the novel virus existed in pigs of different ages, including piglets and sows. Genetic and phylogenetic analyses suggested that the sequenced strains from Henan pigs were closely related to Chinese strains and might evolve from the latest epidemic strains in China. These results would provide insight into the prevalence and evolution of PDCoV in China and are beneficial to develop new vaccines.
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgments
This study was supported by funds from the National Key Research and Development Program of China (2016YFD0500102 and 2018YFD0500100), National Natural Science Foundation of China (No. 31772773 and U1704231).
References
- Ajayi T., Dara R., Misener M., Pasma T., Moser L., Poljak Z. 2018. Herd-level Prevalence and Incidence of Porcine Epidemic Diarrhoea Virus (PEDV) and Porcine Deltacoronavirus (PDCoV) in Swine Herds in Ontario, Canada. Transboundary and Emerging Diseases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F., To K.K., Tse H., Jin D.Y., Yuen K.Y. Interspecies transmission and emergence of novel viruses: lessons from bats and birds. Trends Microbiol. 2013;21:544–555. doi: 10.1016/j.tim.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong B.Q., Liu W., Fan X.H., Vijaykrishna D., Tang X.C., Gao F., Li L.F., Li G.J., Zhang J.X., Yang L.Q., Poon L.L., Zhang S.Y., Peiris J.S., Smith G.J., Chen H., Guan Y. Detection of a novel and highly divergent coronavirus from asian leopard cats and Chinese ferret badgers in Southern China. J. Virol. 2007;81:6920–6926. doi: 10.1128/JVI.00299-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong N., Fang L., Zeng S., Sun Q., Chen H., Xiao S. Porcine Deltacoronavirus in Mainland China. Emerg. Infect. Dis. 2015;21:2254–2255. doi: 10.3201/eid2112.150283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong N., Fang L., Yang H., Liu H., Du T., Fang P., Wang D., Chen H., Xiao S. Isolation, genomic characterization, and pathogenicity of a Chinese porcine deltacoronavirus strain CHN-HN-2014. Vet. Microbiol. 2016;196:98–106. doi: 10.1016/j.vetmic.2016.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang P., Fang L., Liu X., Hong Y., Wang Y., Dong N., Ma P., Bi J., Wang D., Xiao S. Identification and subcellular localization of porcine deltacoronavirus accessory protein NS6. Virology. 2016;499:170–177. doi: 10.1016/j.virol.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong L., Li J., Zhou Q., Xu Z., Chen L., Zhang Y., Xue C., Wen Z., Cao Y. A new Bat-HKU2-like coronavirus in swine, China, 2017. Emerg. Infect. Dis. 2017;23 doi: 10.3201/eid2309.170915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham R.L., Baric R.S. Recombination, reservoirs, and the modular spike: mechanisms of coronavirus cross-species transmission. J. Virol. 2010;84:3134–3146. doi: 10.1128/JVI.01394-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H., Jung K., Vlasova A.N., Chepngeno J., Lu Z., Wang Q., Saif L.J. Isolation and characterization of porcine deltacoronavirus from pigs with diarrhea in the United States. J. Clin. Microbiol. 2015;53:1537–1548. doi: 10.1128/JCM.00031-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H., Jung K., Vlasova A.N., Saif L.J. Experimental infection of gnotobiotic pigs with the cell-culture-adapted porcine deltacoronavirus strain OH-FD22. Arch. Virol. 2016;161:3421–3434. doi: 10.1007/s00705-016-3056-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H., Jung K., Wang Q., Saif L.J., Vlasova A.N. Development of a one-step RT-PCR assay for detection of pancoronaviruses (α-, β-, γ-, and δ-coronaviruses) using newly designed degenerate primers for porcine and avian `fecal samples. J. Virol. Methods. 2018;256:116–122. doi: 10.1016/j.jviromet.2018.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janetanakit T., Lumyai M., Bunpapong N., Boonyapisitsopa S., Chaiyawong S., Nonthabenjawan N., Kesdaengsakonwut S., Amonsin A. Porcine deltacoronavirus, Thailand, 2015. Emerg. Infect. Dis. 2016;22:757–759. doi: 10.3201/eid2204.151852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang G., Lee K.K., Kim S.H., Lee C. Prevalence, complete genome sequencing and phylogenetic analysis of porcine deltacoronavirus in South Korea, 2014-2016. Transbound. Emerg. Dis. 2017;64:1364–1370. doi: 10.1111/tbed.12690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K., Hu H., Saif L.J. Porcine deltacoronavirus induces apoptosis in swine testicular and LLC porcine kidney cell lines in vitro but not in infected intestinal enterocytes in vivo. Vet. Microbiol. 2016;182:57–63. doi: 10.1016/j.vetmic.2015.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K., Hu H., Saif L.J. Porcine deltacoronavirus infection: Etiology, cell culture for virus isolation and propagation, molecular epidemiology and pathogenesis. Virus Res. 2016;226:50–59. doi: 10.1016/j.virusres.2016.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Lee C. Complete genome characterization of korean porcine deltacoronavirus strain KOR/KNU14-04/2014. Genome Announc. 2014;2 doi: 10.1128/genomeA.01191-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Lee C. Functional characterization and proteomic analysis of the nucleocapsid protein of porcine deltacoronavirus. Virus Res. 2015;208:136–145. doi: 10.1016/j.virusres.2015.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B.J., Zuo Y.Z., Gu W.Y., Luo S.X., Shi Q.K., Hou L.S., Zhong F., Fan J.H. Isolation and phylogenetic analysis of porcine deltacoronavirus from pigs with diarrhoea in Hebei province, China. Transbound. Emerg. Dis. 2018;65:874–882. doi: 10.1111/tbed.12821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Zhang Y., Liang X., Lou F., Oglesbee M., Krakowka S., Li J. Origin, evolution, and virulence of porcine deltacoronaviruses in the United States. mBio. 2015;6 doi: 10.1128/mBio.00064-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai K., Feng J., Chen G., Li D., Zhou L., Bai Y., Wu Q., Ma J. The detection and phylogenetic analysis of porcine deltacoronavirus from Guangdong Province in Southern China. Transbound. Emerg. Dis. 2018;65:166–173. doi: 10.1111/tbed.12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marthaler D., Raymond L., Jiang Y., Collins J., Rossow K., Rovira A. Rapid detection, complete genome sequencing, and phylogenetic analysis of porcine deltacoronavirus. Emerging Infect. Dis. 2014;20:1347–1350. doi: 10.3201/eid2008.140526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y., Tian X., Qin P., Wang B., Zhao P., Yang Y.L., Wang L., Wang D., Song Y., Zhang X., Huang Y.W. Discovery of a novel swine enteric alphacoronavirus (SeACoV) in southern China. Vet. Microbiol. 2017;211:15–21. doi: 10.1016/j.vetmic.2017.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeng-Chuto K., Lorsirigool A., Temeeyasen G., Vui D.T., Stott C.J., Madapong A., Tripipat T., Wegner M., Intrakamhaeng M., Chongcharoen W., Tantituvanont A., Kaewprommal P., Piriyapongsa J., Nilubol D. Different lineage of porcine deltacoronavirus in Thailand, Vietnam and Lao PDR in 2015. Transbound. Emerg. Dis. 2017;64:3–10. doi: 10.1111/tbed.12585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J., Zheng Y., Yang Y., Liu C., Geng Q., Tai W., Du L., Zhou Y., Zhang W., Li F. Cryo-EM structure of porcine delta coronavirus spike protein in the pre-fusion state. J. Virol. 2017 doi: 10.1128/JVI.01556-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha A., Gauger P., Zhang J., Yoon K.J., Harmon K. PCR-based retrospective evaluation of diagnostic samples for emergence of porcine deltacoronavirus in US swine. Vet. Microbiol. 2015;179:296–298. doi: 10.1016/j.vetmic.2015.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song D., Zhou X., Peng Q., Chen Y., Zhang F., Huang T., Zhang T., Li A., Huang D., Wu Q., He H., Tang Y. Newly emerged porcine deltacoronavirus associated with diarrhoea in swine in China: identification, prevalence and full-length genome sequence analysis. Transbound. Emerg. Dis. 2015;62:575–580. doi: 10.1111/tbed.12399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Byrum B., Zhang Y. Detection and genetic characterization of deltacoronavirus in pigs, Ohio, USA, 2014. Emerging Infect. Dis. 2014;20:1227–1230. doi: 10.3201/eid2007.140296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Byrum B., Zhang Y. Porcine coronavirus HKU15 detected in 9 US states, 2014. Emerging Infect. Dis. 2014;20:1594–1595. doi: 10.3201/eid2009.140756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.W., Yue H., Fang W., Huang Y.W. Complete genome sequence of porcine deltacoronavirus strain CH/Sichuan/S27/2012 from Mainland China. Genome Announc. 2015;3 doi: 10.1128/genomeA.00945-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Wang Y., Baloch A.R., Pan Y., Tian L., Xu F., Shivaramu S., Chen S., Zeng Q. Detection and genetic characterization of porcine deltacoronavirus in Tibetan pigs surrounding the Qinghai-Tibet Plateau of China. Transbound. Emerg. Dis. 2018;65:363–369. doi: 10.1111/tbed.12819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Huang Y., Lau S.K., Yuen K.Y. Coronavirus genomics and bioinformatics analysis. Viruses. 2010;2:1804–1820. doi: 10.3390/v2081803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Lam C.S., Lau C.C., Tsang A.K., Lau J.H., Bai R., Teng J.L., Tsang C.C., Wang M., Zheng B.J., Chan K.H., Yuen K.Y. Discovery of seven novel mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Chai X., Cheng Y., Xing G., Liao A., Du L., Wang Y., Lei J., Gu J., Zhou J. Molecular characteristics of the spike gene of porcine epidemic diarrhoea virus strains in Eastern China in 2016. Virus Res. 2018;247:47–54. doi: 10.1016/j.virusres.2018.01.013. [DOI] [PubMed] [Google Scholar]
- Zhai S.L., Wei W.K., Li X.P., Wen X.H., Zhou X., Zhang H., Lv D.H., Li F., Wang D. Occurrence and sequence analysis of porcine deltacoronaviruses in southern China. Virol. J. 2016;13:136. doi: 10.1186/s12985-016-0591-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Fan H., Lan T., Yang X.L., Shi W.F., Zhang W., Zhu Y., Zhang Y.W., Xie Q.M., Mani S., Zheng X.S., Li B., Li J.M., Guo H., Pei G.Q., An X.P., Chen J.W., Zhou L., Mai K.J., Wu Z.X., Li D., Anderson D.E., Zhang L.B., Li S.Y., Mi Z.Q., He T.T., Cong F., Guo P.J., Huang R., Luo Y., Liu X.L., Chen J., Huang Y., Sun Q., Zhang X.L., Wang Y.Y., Xing S.Z., Chen Y.S., Sun Y., Li J., Daszak P., Wang L.F., Shi Z.L., Tong Y.G., Ma J.Y. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature. 2018;556:255–258. doi: 10.1038/s41586-018-0010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]