Abstract
Enveloped viruses and cellular transport vesicles share obvious morphological and functional properties. Both are composed of a closed membrane, which is lined with coat proteins and encases cargo. Transmembrane proteins inserted into the membrane define the target membrane area with which the vesicle or virus is destined to fuse. Here we discuss recent insight into the functioning of enveloped viruses in the framework of the “functional module” concept. Vesicular transport is an exemplary case of a functional module, as defined as a part of the proteome that assembles to perform a specific autonomous function in a living cell. Cellular vesicles serve to transport cargo between membranous organelles inside the cell, while enveloped viruses can be seen as carriers of the viral genome delivering their cargo from an infected to an uninfected cell. The turnover of both vesicles and viruses involves an analogous series of submodular events. This comprises assembly of elements, budding from the donor compartment, uncoating and/or maturation, docking to and finally fusion with the target membrane to release the cargo. This modular perception enables us to define submodular building blocks so that mechanisms and elements can be directly compared. It will be analyzed where viruses have developed their own specific strategy, where they share functional schemes with vesicles, and also where they even have “hijacked” complete submodular schemes from the cell. Such a perspective may also include new and more specific approaches to pharmacological interference with virus function, which could avoid some of the most severe side effects.
Keywords: Functional module, Vesicular transport, Virus assembly, Budding, Membrane fusion, Influenza virus
1. Introduction to functional modules
In this contribution we want to compare viruses and cellular vesicles on the level of “functional modules”. This term arose from the notion that major reaction pathways in the living cell are carried out by specific subsets of the proteome. Time-ordered interaction and complex formation within these ensembles of macromolecules facilitate an autonomous function. Functional modules include the important group of macromolecular machines organised as a compact structure, such as the ribosome or the proteasome. However, there are also ensembles that are more dynamic by changing their composition and/or organisation during function. To include the latter group as well, the term “functional module” is used. Common to all modules are a characteristic time domain at which the respective functional cycle proceeds and a certain sequestration from the rest of the cell by spatial limitations, by chemical specificity, and/or by the time domain at which function proceeds.
In terms of systems analysis [1], functional modules mean ensembles of molecular elements that are integral parts of a network but can be separated from its other parts. The separation is not only a method to facilitate the quantitative analysis of interaction data. It is rather supposed to reflect biological reality in the sense of the general definition of module as “a self-contained unit of a system that performs a specific task in support of its major function”. Specific network motifs such as “hubs” or “cliques” can now be assigned to stable or dynamic protein complexes [2]. An alternative to such a “holistic” systems approach is a “bottom up” approach that starts from known properties of the single macromolecules and their interactions and attempts to reconstruct the behaviour of macromolecular assemblies from the behaviour of their elements. This approach is based on the methods and concepts of molecular biology, biochemistry and biophysics. It is naturally limited to small ensembles but has the potential to elucidate how the system's performance arises from the combination of its microscopic molecular properties. This becomes most important when the biological function of the module depends crucially on a specific molecular detail and/or a specific time window. Examples include the γ-secretase module, in which specific key amino acids determine the kinetics and extent of monomer assembly and eventually the toxicity of the whole polymer [3]. Another example are signal transduction modules, where the functional cycle comprises sequential interactions, which are – often with GTPases as timers – only activated in certain time windows. Also mechano-chemical GTPases of the dynamin superfamily employ nucleotide binding and hydrolysis to set a sophisticated time window in which their self-assembly, their membrane remodelling activity and their disassembly are coordinated (Oliver Daumke, personal communication).
Generally, functional modules may show either a stable organisation or dynamic assembly and disassembly during function. The first case includes the macromolecular machines with compact structure, such as the ribosome or the proteasome, while the latter case includes signal transducers and intracellular transport vesicles. Oriented on the machines, we assume that modules go through functional cycles. On receiving an input, only one (or a few) elements are initially activated. Subsequently the elements involved increase in their number and may form transient structures of higher hierarchy, each performing a well-defined subtask. This is the submodular level, which performs regulatory functions such as checking the input, signal amplification or negative feedback, which cannot be performed by single elements. On the modular level the submodular contributions are integrated and the output is generated. Towards the end of the functional cycle more and more elements are deactivated until eventually all elements of the module are back in place again [4].
Vesicular transport of proteins and other molecules in eukaryotic cells fits well the functional module scheme. The different steps are accomplished by submodular entities, which work together in a coordinated process. These include the sorting of cargo, the budding and scission of the vesicle from the donor membrane, the uncoating, and finally the tethering, docking and membrane fusion at the acceptor compartment [4], [5]. Each transport step, e.g. between the endoplasmic reticulum and the Golgi or between the Golgi and the plasma membrane, depends on its own specific protein repertoire [6], [7]. This does not exclude that, as will be discussed below, certain elements such as the Bet3 and p115 proteins can be associated with more than one submodular (or even modular) function [8]. One concept to explain the occurrence of multifunctional proteins might be called “protein parsimony”. In this view, there are simply not enough protein species (only 25,000 genes in humans) to serve all the different functions in a complicated eukaryotic cell under a variety of physiological states. Protein parsimony is especially important for replication of viruses, which is a race against time until the immune system of the host has acquired the ability to stop further spread and transmission of the virus. Thus, there is constant selection pressure for viruses to speed up their replication. One factor to achieve this goal are multifunctional proteins (and thus fewer genes), which allow faster amplification of the viral genome.
The functional cycle of transport vesicles starts with the uptake of cargo that carries an appropriate transport signal and ends when the same cargo is delivered to its acceptor compartment. A functional cycle of the transport module is closed when the cargo has been delivered and the molecules executing the function have been recycled to their starting positions. Analogously, we define the inclusion of the viral genome into virus particles preassembled at membranes of infected cells as the input into the viral module and the release of the genome into the cytoplasm of an uninfected cell as the modular output. In spite of their quite different donor and acceptor compartments, the individual steps of vesicle transport and virus replication can be divided into clearly defined and functionally conserved submodules and follow basically the same series of events: 1. assembly of elements, 2. vesicle budding from the donor membrane, 3. uncoating or maturation, 4. tethering and docking of the vesicle or virus to the acceptor compartment and finally 5. membrane fusion and cargo release. The general buildup of vesicles and viruses is schematically depicted in Fig. 1 and described further below, the processes of the respective functional module are shown in Fig. 2 and covered in-depth through the remainder of the article.
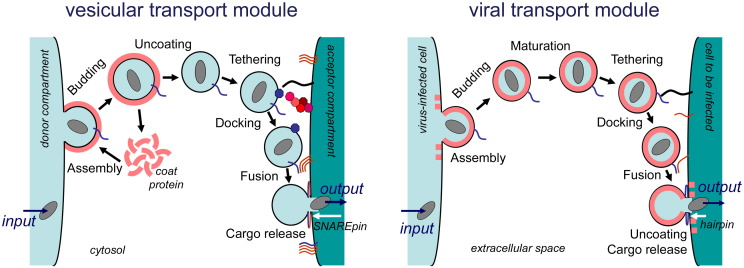
Fig. 1.
Cellular coated vesicles and enveloped viruses: basic composition. Both vesicles and viruses contain a membrane bilayer (thin black circle) derived from the donor membrane, which is lined by a coat (red circle) assembled from soluble, monomeric subunits. Inserted into the bilayer are transmembrane proteins (blue) required for targeting of the vesicle or virus. The interior contains cargo (grey ellipse), either protein or the viral genome. Insets: EM-pictures of COP I vesicles (left, by courtesy of Christoph Rutz and Britta Brügger, Biochemiezentrum, Heidelberg) and of influenza viruses (right, recreated 1918 influenza virus particles, taken from the Centers for Disease Control and Prevention's Public Health Image Library, identification number #8160). Note that the scheme is a drastic simplification to compare analogue structures in viruses and vesicles. Especially the term “coat” often comprises a multitude of different proteins, which might also have additional functions.
Fig. 2.
Functional input/output cycles. The figure illustrates the notion of enveloped viruses as “vesicular carriers of the viral genome”. The input into both vesicular and viral transport modules is the uptake of cargo, either protein or the viral genome, from a donor compartment, the output is the release of cargo into another membrane-encased acceptor compartment. The individual steps of vesicle transport and virus replication are shown to follow a similar sequence of submodular events, comprising assembly of elements, budding from the donor membrane, uncoating of vesicles or maturation of viruses, tethering and docking of the vesicle or virus to the acceptor compartment, membrane fusion, and cargo release and uncoating. Note that some viruses bud into the lumen of membranous organelles of the exocytic pathway (ER/Golgi) and are subsequently secreted by the cell. Likewise, some viruses enter the cell via the endocytic pathway and fuse with the membrane of early and late endosome or with vesicles that transport cargo between them. Maturation of viruses often occurs also inside the cell, e.g. in the exocytic pathway or after endocytic uptake. Furthermore, for herpes viruses the term “maturation” refers to the acquisition of tegument and envelope by the nucleocapsid, which originates from budding of capsid through the inner nuclear membrane and their subsequent fusion with the outer nuclear membrane. See text for details.
Using the concept of functional modules, we will depict the notion that enveloped viruses can be regarded as “vesicular carriers of the viral genome” which transport their cargo, the viral genome and (in the case of negative-stranded RNA and retroviruses) accessory proteins required for its replication, by budding from the donor membrane in the infected cell and fusing with an acceptor membrane in the target cell to be infected. In the following, we will focus on small, enveloped viruses, mainly on influenza virus, but also mention other well-characterized pathogens such as the retrovirus human immunodeficiency virus (HIV; see glossary for a list of abbreviations) or model viruses such as the alphavirus Semliki Forest Virus (SFV) and the vesicular stomatitis virus (VSV). It is our hope that the comparison with vesicular transport will provide insight into molecular mechanisms at the border between cell biology and virology.
2. Similarities and differences between vesicles and enveloped viruses
Morphology is the most obvious level on which enveloped viruses and cellular transport vesicles resemble each other (Fig. 1). Both contain a membrane bilayer with a lipid composition identical (or very similar) to that of the membrane from which the vesicle or virus is derived. Transmembrane proteins are inserted into the bilayer and mediate attachment of the vesicle or virus to and fusion with the acceptor membrane. The bilayer is lined, either on the inside or on the outside, with coat proteins. Cellular vesicles involved in intra-Golgi transport contain a COP I coat, those that promote transport from the ER to the Golgi a COP II coat and for transport from the trans-Golgi-network to the endosome as well as for trafficking within the endocytic pathway a clathrin-coat is required. In the case of viruses the term “coat” comprises a variety of proteins, which might also have other functions. In the simple alphaviruses, such as SFV, the coat is an icosahedral capsid, which has also the function to encase the genome. In HIV, both functions are encoded by the gag-gene, whose product is proteolytically cleaved during virus budding to yield the coat and the capsid. Para- and orthomyxoviruses as well as VSV contain separate genes for the matrix protein, that builds the coat, and for the capsid protein. The large herpes viruses contain an amorphous structure beneath the envelope, called the tegument, which contains a multitude of proteins with a wide variety of functions. These coat proteins often drive the formation of the vesicle or virus by assembly and oligomerization at the budding site, and/or they define the shape of the particle. The interior of the particle contains the cargo, either cellular proteins or the viral genome encased in an icosahedral or helical capsid. Furthermore, cellular transport vesicles and enveloped viruses have almost the same size (50–100 nm and 40–300 nm, respectively), indicating that the curvature of their membranes is similar. A summary of the functional elements of the vesicle module and two representative viruses, influenza virus and HIV, is given in Table 1 .
Table 1.
Comparison of the structural elements of cellular transport vesicles with two enveloped viruses, influenza and human immunodeficiency virus.
| Cellular vesicles | Influenza virus | Human immunodeficiency virus (HIV) | |
|---|---|---|---|
| Transmembrane proteins | v-SNARE (different variants for each transport step) | Hemagglutinin (HA, attachment and fusion), neuraminidase (NA, detachment after budding) | gp160 with subunits gp120 (attachment) and gp41 (fusion) |
| Task: attachment to and fusion with acceptor membrane | |||
| Coat proteins | Clathrin or COP I or COP II | Matrix protein (M1) | gag (four subunits) |
| Task: polymerization, budding, morphology | |||
| Cargo | Proteins: membrane or lumenal | Genome plus associated proteins: arranged in eight different ribonucleoparticles (vRNP) | Genome plus associated proteins: arranged in one segment |
| Task: signalling to initiate budding | |||
| Membrane bilayer | Derived from different donor membranes, various composition | Derived from subdomains of the plasma membrane, enriched in raft-lipids | Derived from subdomains of the plasma membrane, enriched in raft-lipids |
| Task: recruitment of coat, envelope formation |
Since the high protein concentration in the cell's cytosol severely restricts diffusion of large size particles, such as vesicles or viruses (or submodular structures, such as capsids), an active mechanism is required for their transport over long distances. For that purpose both vesicles and viruses rely on interactions with the cytoskeleton. In particular, directed polymerization of actin or the actin–myosin system facilitates transport underneath the plasma membrane and probably inside the nucleus. Transport from the cell's periphery to the nucleus (or more precisely to the perinuclear microtubuli organising center) and vice versa is achieved by the motor proteins dynein and kinesin, respectively, which move particles along microtubules. Viruses do not only misuse the cytoskeleton of the cell for their own purpose, but also hijack its vesicular transport system. During virus entry the endocytic pathway is used for virus transport towards the nucleus, whereas the exocytic pathway is utilized to deliver spike proteins to the plasma membrane or to secrete virus particles that bud into the lumen of exocytic organelles, such as the ER or the Golgi [9], [10].
There are, of course, also obvious differences between cellular modules and virus particles. First of all, vesicles and viruses perform their tasks in different environments, i.e. transport of cargo within a cell or between cells, respectively. Therefore, vesicles bud into the cell's cytosol, whereas enveloped viruses bud out of the cytosol into the extracellular space or into a topologically equivalent compartment, such as the lumen of the endoplasmic reticulum (ER) or of the Golgi apparatus.
The different topology of budding has certain consequences: since budding of both vesicles and viruses requires assembly of coat proteins synthesized in the cytosol, the resulting cellular transport vesicles have their coat on the outside whereas virus particles possess a coat lining the inside of its membrane. As a further consequence cellular transport vesicles must uncoat prior to fusion to expose the elements required for the recognition of the acceptor compartment. In contrast, uncoating of virus particles can occur only during or after fusion of viral and cellular membranes (Fig. 2).
Another difference between vesicles and viruses arises from the environment in which they perform their function. Budded virus particles are metabolically inactive, crystalline entities in search of suitable target cells. During transport within or between organisms they encounter changing, sometimes harmful environmental conditions, and the main function of the viral module is to protect the virus genome against those influences. In contrast, cellular transport vesicles operate within the same environment, the cell's cytosol, which also provides energy and metabolites. As a consequence, vesicles are functionally active after budding, e.g. they hydrolyze GTP and uncoat, which is an obligatory series of events mandatory for their task of protein transport.
Furthermore, after cargo has been delivered to the acceptor membrane, the elements of vesicular carriers are recycled to their start position, which allows further cycles of cargo transport with the same elements. In contrast, elements of the viral module perform their function usually only once, recycling (in the sense of re-use of an element after performing its function) does not occur. Even more, elements of the viral module are amplified after delivery of the cargo using the machinery of the freshly infected host cells. Although negative feedback loops occur at the submodular level, they are absent for the whole replication cycle of viruses. This ensures that a maximal number of virus particles are produced, at the expense of cellular integrity. Thus, the original notion of a module as a unit that performs a task in support of the major function of the system, i.e. in that case an infected tissue, organ or organism, does not apply for viruses. On the contrary, viruses subvert cellular (sub)modular processes for their purpose and their buildup as a functional module.
Since the elements of a cellular module are recycled to perform their function multiple times, the system must be recharged with energy at one step of its cycle. ATPases or especially small GTPases fulfil this function by binding and subsequently hydrolyzing the respective nucleotidetriphosphate. Although small G-proteins play pivotal roles for almost every cellular module, structural proteins with nucleotidetriphosphate hydrolyzing activity (with the exception of RNA- or DNA-polymerases) are apparently absent from virus particles. As a consequence, input of energy into the functional cycle of a virus occurs only during synthesis of proteins, which must therefore be equipped with sufficient conformational energy to perform their various functions. Since most viral proteins or submodules (again with the exception of polymerases) perform their function only once, input of additional energy and thus the integration of GTPases into a viral module are not required.
The absence of GTPases in viral modules might be also due to a decisive difference in the tasks cellular and viral modules perform. In vesicular transport, the hydrolyzing activity of small GTPases is used to uncoat the vesicle in the cytosol. In contrast, uncoating of viruses occurs after their entry into a freshly infected target cell. Assuming a similar function, i.e. uncoating, for a hypothetical viral GTPase, high-energy GTP would have to be integrated into virus particles. However, the labile phosphodiester bonds of GTP would be prone to hydrolyzation in the harsh environmental conditions that the virus particles can encounter during their passage from one organism to another. Hence, virus uncoating – which ought to occur only after entering the target cell – would be disturbed. To prevent this, viruses have adopted other strategies than “own” GTPases to uncoat their genome in freshly infected cells as will be described below.
3. Comparison of the functional cycle of transport vesicles and enveloped viruses
3.1. Assembly
3.1.1. Principal requirements for budding of vesicles and enveloped virus particles
In current textbooks, description of the replication cycle of a virus usually begins with the binding of a virus particle to a receptor present on a suitable target cell and ends with the release of virus particles assembled from newly synthesized components. Here we start our description of the replicative cycle of a virus with the assembly step to facilitate its comparison with the functional cycle of a cellular transport vesicle.
To create a functional vesicle or virus several elements of the module must work together. Since transport is a vectorial process, the membrane from which the vesicle or virus is supposed to bud must be defined and cargo and components of the prospective vesicle must be specifically transported to the budding site. The assembly of the components often occurs via semistable submodular intermediates each with a specific function. Certain elements must check the input into the module, i.e. select the appropriate cargo and enrich it inside the vesicle or virus. Besides the cargo other elements must be incorporated into the particle and exposed on its surface to mediate recognition of and fusion with the correct target membrane. The budding process itself needs an element which physically drives the curvature of membranes to generate a bud. This is often achieved by assembly and polymerization of cytosolic coat proteins on the membrane. Other elements serve to select the site on the membrane where the coat proteins should polymerize. After formation of a bud the coat proteins (or other elements) must catalyze the fission of the vesicle/virus from the donor membrane. Finally, the activity of all elements must be orchestrated such that only functional vesicles, i.e. those that contain all required elements and the cargo, are released from the membrane [6], [11]. In the case of viruses, selection of their elements is less precise since the majority of released particles of many virus families are non-infectious, i.e. they lack at least one of the necessary components. This functional imperfection of virus budding is overcome by the sheer abundance of particles released by an infected cell (several thousands). For that purpose viruses can exploit the resources of the infected cell, whereas a similar wasteful strategy for vesicular transport would probably be detrimental for the cell's physiology. The elements which perform the described functions will now be discussed and compared for cellular transport vesicles and enveloped viruses.
3.1.2. Assembly of virus particles from submodular structures
Most enveloped virus particles are assembled from at least two submodular structures, the genome complexed to proteins (usually called capsid) and the viral envelope containing spike glycoproteins (often embedded in specific lipids), and additionally in most cases a peripheral membrane protein (often designated “matrix protein”) lining the inner leaflet of the membrane bilayer and representing the coat. In the process of assembly of the virus particle, the molecular interactions involved increasingly gain complexity and ultimately lead to the formation of the viral module from submodular structures as described below and depicted in Fig. 3 for influenza virus as an example.
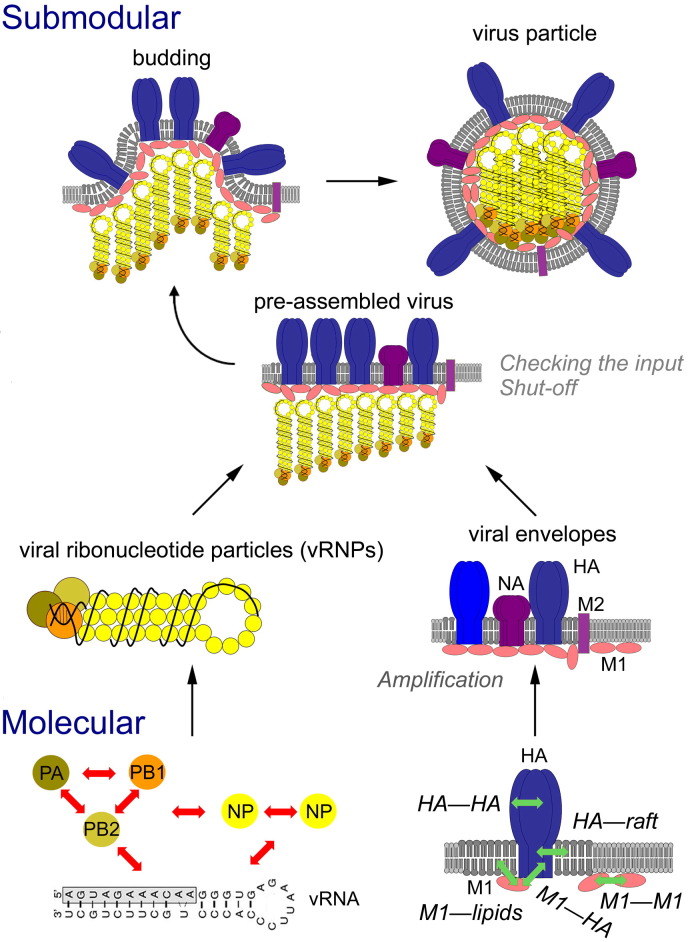
Fig. 3.
Example of an input submodule: assembly of influenza virus. Molecular level: molecular interactions (left; red arrows) between the viral RNA, the three polymerase proteins (PA, PB1, PB2) and the structural protein NP build the viral ribonucleoprotein-particle (vRNP). The glycoprotein HA (right) forms trimers, which associate with rafts in the plasma membrane. The coat protein M1 associates with membranes, where it binds weakly to the cytoplasmic tail of HA; these molecular interactions are illustrated by green arrows. Submodular level: oligomerization of M1 strengthens the weak interactions with HA and draws M1 to the viral budzone, preassembled viral envelopes. This submodular entity contains also the second viral glycoprotein neuraminidase (NA) and a few copies of the viral proton channel M2. Finally, the vRNPs bind to M1, a complete virus particle buds from the membrane and is released. The hallmarks of functional modules (grey lettering) are described in the text, 3.1, 3.2.
The segmented genome of influenza virus, which is arranged in ribonucleoparticles (vRNPs), is assembled in the nucleus, while the surface glycoproteins hemagglutinin (HA) and neuraminidase (NA) are processed in the secretory pathway. vRNPs represent one submodular structure with a clearly defined inventory of elements and a characteristic shape: for vRNPs to be formed, the viral RNA segments first form panhandle-like structures by base pairing, which then interact with the nucleoprotein NP and the three subunits of the viral polymerase, PA, PB1 and PB2 [12], [13] (Fig. 3, bottom left). The matrix protein M1 participates in transport of vRNPs out of the nucleus, but also localizes peripherally to cellular membranes. Meanwhile, the glycoproteins HA and NA first form homooligomers in the endoplasmic reticulum (ER), which are transported to the plasma membrane and enriched in specific membrane subdomains termed membrane rafts (Fig. 3, right, see below). The cytoplasmic tails of HA and NA are believed to bind to M1 via weak interactions, which are amplified by oligomerization of M1 and which draw M1 to membrane rafts. As a consequence of these molecular interactions, cellular proteins are displaced causing the formation of the second submodular structure, namely, viral envelopes preassembled at the budding site. Finally, vRNPs bind to M1 causing assembly of the complete virus module and its budding from the membrane [14], [15].
Paramyxo- and rhabdoviruses follow the same scheme as outlined for influenza viruses with formation of helical vRNPs in the cytosol or nucleus, and assembly of the coat and packaging of the genome at the plasma membrane [16], [17]. In contrast, due to the large amount of capsid-like particles present in the cytoplasm of infected cells, one model for budding of SFV suggests that assembly of its capsid and integration of the viral genome occur inside the cell. However, since virus mutants defective in intracellular capsid formation can still form functional particles, it is possible that the capsid-like particles in the cytoplasm are dead end products and that, even for wild-type SFV, assembly of infectious viruses occurs at the plasma membrane [18], [19]. Some submodular structures are not only transient intracellular building blocks of virus particles, but stable functional units on their own. An example are cells infected with hepatitis B virus (HBV), which secrete subviral lipoprotein particles, viral envelopes devoid of the capsid, at a 10,000fold higher concentration than virus particles, probably as decoys for the immune system [20].
3.1.3. Definition of the budding site and transport of components to the membrane
The coat lining the membrane of a vesicle or of a virus is assembled from a cytosolic pool of monomers. In principle, assembly of the coat could occur on any cellular membrane, yet vesicles or viruses bud only from specific membranes. Thus, the coat proteins must recognize other elements (proteins or lipids) already present in the membrane from which the vesicle or virus should bud.
The viral budding site is often defined by specific localization of viral glycoproteins at certain membranes or membrane microdomains. In polarized epithelial cells viral glycoproteins are transported either to the apical (HA of influenza virus) or basolateral membrane (G of VSV) and as a consequence influenza virus buds only from apical membranes, whereas VSV buds from the basolateral membrane [15]. (For other viruses with a polarized budding phenotype, the coat protein may be the determining factor.) Viruses which bud into intracellular compartments (coronaviruses, flaviviruses, HBV) contain glycoproteins with intrinsic signals for their retention in the ER or Golgi causing their accumulation in the membrane of the respective organelle [21]. Many viral glycoproteins (HA and NA of influenza virus, gp160 of HIV) which are transported to the plasma membrane associate with membrane rafts [22], microdomains enriched in cholesterol and glycolipids that are thought to serve as an platform for assembly of virus particles by enriching viral proteins (and excluding others), thereby facilitating specifically the protein–protein interactions required for budding. As a consequence the envelope of these virus particles closely resembles the lipid composition of membrane rafts [23].
With the exception of coronaviruses (see below), the coat proteins do not contain membrane-spanning regions and must thus associate with membranes by other means. The Gag protein of HIV contains a membrane binding signal composed of a myristoylated glycine and an adjacent stretch of basic amino acids at its N-terminus. After cotranslational myristoylation, the fatty acid is initially hidden inside a hydrophobic cleft of the monomeric protein. Gag then binds via electrostatic interactions to phosphatidylinositol-4,5-bisphosphate (PtdIns(4,5)P2), a specific component of the plasma membrane. Binding to the membrane facilitates multimerization of Gag and exposes myristate, which now inserts into the lipid bilayer (myristyl switch). The 2′, unsaturated acyl chain of PtdIns(4,5)P2 binds to the now empty hydrophobic groove of Gag. The mutual exchange of a fatty acid between PtdIns(4,5)P2 and myristoylated Gag causes a stable association between both molecules [24]. An interesting speculation suggests that Gag bound to PtdIns(4,5)P2 is then targeted to membrane rafts, where it can associate with gp160 [25], [26], [27].
Binding to specific lipids has not been demonstrated so far for the coat proteins of other viruses. The matrix proteins of ortho- and paramyxoviruses (including influenza virus M1) have intrinsic membrane binding properties, probably mediated by rather unspecific electrostatic and hydrophobic interactions [28]. They accumulate at various membranes, such as ER, Golgi and the plasma membrane and they might then be drawn to the budding site by interactions with the cytoplasmic tails of viral spike proteins. The capsid protein of hepatitis C has no intrinsic membrane targeting domain, but is synthesized together with the glycoproteins as a polyprotein precursor at the ER. Upon proteolytical cleavage, both the glycoproteins and the capsid protein remain associated with the ER membrane ensuring colocalization of proteins prior to budding [21], [29].
In the case of cellular transport vesicles, small G-proteins of the ras-superfamily, such as Arf or the closely related Sar1, define the budding site. They are recruited from the cytosol by binding to specific transmembrane proteins localized in the donor compartment and are activated by guanine-nucleotide exchange factors (GEFs). The GEF causes exchange of GDP with GTP in the G-protein, leading to exposure of its hydrophobic N-terminus, which is either myristoylated or intrinsically hydrophobic, and insertion into the membrane. This anchors the G-protein to specific membranes and serves as a platform for assembly of the coat proteins [30]. The assembly site of clathrin vesicles is additionally regulated by PtdIns(4,5)P2 and other phosphatidylinositol lipids are likely to orchestrate assembly of further cellular transport vesicles [31].
In summary, the budding site of viruses or vesicles is defined by elements having intrinsic signals for accumulation in certain membranes or membrane domains, either transmembrane proteins with intrinsic targeting signals and/or lipids present only in certain membranes. The budding sites selected in this manner are recognized by cytosolic coat proteins, which, however, might also have intrinsic signals for binding to specific membranes. While the regulation by PtdIns(4,5)P2 appears to be a common feature, the actual membrane binding elements are different: vesicles use the myristoyl anchors of GTPases which also provide the energy for the time-ordered and repetitive functional cycles, while viruses have other proteins with diverse membrane binding structures.
3.1.4. Input check: selection of cargo
Budding of viruses or vesicles containing a random selection of all components present in the donor compartment would result in a very inefficient mechanism of transport. Thus, the input into the transport module, either protein or the viral genome, must be checked to ensure that the desired elements are enriched in the vesicle or virus. Furthermore, both viruses and vesicles must incorporate other elements into their membrane that facilitate targeting to and fusion with the target membrane. As already mentioned above, the input check of viruses tends to be less precise compared to vesicles since many released virus particles are non-infectious. Likewise, the formation of virus-like particles, containing a minimal number of proteins, but lacking the genome, is possible (see below). However, budding of vesicles is also not completely faultless, but missorted cargo often contains a retrieval signal, such as KDEL or KKXX, returning proteins that have left the compartment in which they reside [32].
Checking the input is especially demanding in the case of viruses with a positive-stranded RNA genome, which must select their genome out of the many mRNA molecules of the cell. For example, retroviral genomic RNA, which bears all hallmarks of cellular mRNA, constitutes much less than 1% of an infected cell's cytoplasmic RNA population. A component of the viral coat must therefore recognize unique features in the viral genome. In retroviruses these packaging signals are several hundred nucleotides long, sometimes discontinuous stretches of RNA, which are absent in spliced or subgenomic viral RNA molecules. The NC domain of the Gag-precursor binds to the packaging signal via a zinc-binding motif and adjacent basic amino acids. NC is inherently flexible, and it is therefore possible that different NC molecules bind to different regions of the packaging signal, which is too long to be bound by a single NC molecule. As retroviruses contain a diploid genome, specific base pairing between loop sequences of individual RNA molecules ensures packaging of dimers into virus particles, which is important for reverse transcription in a freshly infected cell [33].
Another, even more demanding packaging problem appears with viruses containing a segmented genome. For instance, newly formed influenza virus particles must contain at least one copy of each of its eight vRNPs to be infectious. A random packaging mechanism, in which any eight RNA segments were incorporated into virus particles, would yield a maximum of one infectious particle for every 400 (8!/88) assembled. As several thousands of virus particles are released by one infected cell, a random mechanism of packaging in principle would not preclude production of viruses capable of infecting other cells [33]. Nevertheless, recent cryo-electron microscopy (EM) tomography has shown that each virus particle contains exactly eight vRNPs, arranged as a specific pattern, seven in a circle surrounding one in the center. Each vRNP visible in the tomogram had a different length showing that eight different genes are present in a virus particle [34]. These argues in favour of a highly selective mechanism of genome packaging, but the individual packaging signals present in each vRNP still need to be identified.
Besides the genome, budding virus particles must also incorporate spike proteins to be infectious, but have to exclude most cellular membrane proteins. The viral proteins contribute only 1% (or less) of the surface proteins of the infected cell and yet the budded virus particles contain just a few, if any, cellular proteins. Initial budding models have proposed that very specific interactions between the cytoplasmic tails of viral transmembrane proteins and the viral coat ensure the integrity of virus particles [35], but this has been proven so far only for budding of SFV as described below. Despite many efforts, binding sites for cytoplasmic tails of viral spike proteins have not been identified in coat proteins of other viruses, suggesting that they are of low affinity [36]. It is also noteworthy in this regard that during a mixed infection of a host cell with various unrelated viruses, so-called pseudotypes can appear. These are virus particles that contain the genome and the coat protein of one virus and the envelope proteins of another. The largest numbers of examples of pseudotyping were reported for vesicular stomatitis virus (VSV) and other viruses containing a matrix protein [37]. Thus, viral coat proteins have the remarkable ability to discriminate between cellular and viral transmembrane proteins, although the latter do not possess sequence homology in the region which may possibly interact with the coat. A possible solution to this paradoxon would be a spatial separation of viral from cellular proteins within the membrane. Rafts could perform this function since many viral glycoproteins are present in detergent-resistant-membranes, which are considered the biochemical correlate of membrane rafts [38]. However, this correlation does not seem to hold true in all cases [39], [40], [41] and other microdomains in the membrane, besides rafts, may be responsible. Upon expression at the plasma membrane, HA is neither randomly distributed, nor accumulates only in domains with the size of a raft. Instead HA molecules form irregular clusters on length scales from 40 nm up to many micrometers. Thus, HA (and possibly other viral glycopoteins) somehow accumulate in microdomains of the membrane or even induce their formation, which leads to their separation from most cellular proteins [22], [42], [43], [44]. Alternatively, binding of the matrix protein to the inner leaflet of the bilayer might modify the biophysical properties of the membrane such that microdomains are formed that are recognized mainly by viral glycoproteins.
In the case of cellular transport vesicles, the cargo proteins are usually capable of binding directly to the element that drives the budding reaction, i.e. the coat proteins. Transmembrane proteins supposed to be retrieved from the Golgi to the ER contain a cytoplasmic KKXX sequence that binds to a yet unknown component of the COP I coat. The majority of cargo proteins that travel along the exocytic pathway are concentrated in COP II vesicles prior to export from the ER. Transmembrane proteins possessing di-acidic, di-basic or short hydrophobic motifs in their tail bind to the sec24 subunit of the COP II coat. The diversity of signals explains the ability of COP II to package a wide variety of proteins. Endocytosis of surface receptors by clathrin-coated vesicles requires adaptor proteins connecting the receptor to the coat. The adaptors recognize sorting signals in the cytoplasmic tail of receptors, such as critical tyrosines, di-leucines or conjugated ubiquitin. If soluble components need to be transported, transmembrane cargo receptors, such as the KDEL-receptor for retrieval of lumenal proteins from the Golgi to the ER, are required, which must also be incorporated into the vesicle. Likewise, certain vesicular (v-) SNARE-proteins must be included to allow fusion of the vesicle only with the anticipated acceptor membrane. SNAREs involved in ER-to-Golgi transport bind to distinct sites of the Sec24 subunit of the COP II coat ensuring that vesicle budding and subsequent fusion are mechanistically integrated [6], [30].
In conclusion, the input into a virus particle or vesicle is checked by specific interactions of coat components already present at the site of assembly (often a specific membrane subdomain) and components of the input. However, the exact nature of these interactions is, especially for budding of virus particles, in most cases quite unclear. Whatever the exact mechanism might be, the coat proteins must somehow recognize that the binding sites are filled. The assembly process is then switched off and a signal must be delivered that a given preformed vesicle or virus particle is now ready for budding.
3.2. Budding
To release a vesicle or virus, some elements of the module must bend the membrane, executing either a pushing or a pulling force on the bilayer, which leads to the formation of a bud. The bud must then close (scission of the vesicle) and be released from the donor membrane, reactions which may (or may not) require other elements of the module.
3.2.1. Determinants of virus budding
Budding of a virus may either depend on inner coat proteins or viral envelope glycoproteins, or both components must work together [21]. This can be experimentally determined by expression of structural proteins of the respective virus in eukaryotic cells and identification of the minimal set of proteins required for release of virus-like particles (VLPs), enveloped vesicles with the same density and appearance as complete virus particles. For most retroviruses, including HIV, paramyxo-, rhabdo- and filoviruses, the expression of coat proteins alone leads to the formation of VLPs, showing that assembly of the coat protein at the membrane is the driving force for virus budding. However, coexpression of viral glycoproteins often enhances release of virus particles [16], [17], [45], [46].
For influenza virus it was found recently that expression of the coat protein M1 – which had previously been assumed to be the driving force for influenza virus budding – is per se not sufficient to produce VLPs nor is it required for VLP formation. Instead, the major determinant for VLP formation are the cytoplasmic tails of the glycoproteins NA and especially HA [47].
In other viruses (HBV, corona- and flavivirus), viral glycoproteins alone catalyze budding of VLPs from a cell indicating that they can execute a pulling force on the membrane. The M protein of coronaviruses, which provides the driving force for budding, contains only a short extraviral domain but three transmembrane segments. Yet it self-assembles to higher-order complexes indicating that lateral interactions occur within the membrane. The glycoproteins of flaviviruses do not project as visible spikes from the envelope, but instead form a rigid shell which lies flat on the envelope such that it can execute force on the bilayer [20], [48], [49].
SFV requires interactions between glycoproteins and coat proteins for budding. By combining atomic structures with cryo-EM it was demonstrated that SFV contains two rigid protein coats or shells, both with icosahedral symmetry. The capsid composed of the C-protein lines the internal side of the membrane, whereas lateral interactions between viral spikes form the outer shell. The spikes are composed of E1/E2 heterodimers, which are grouped into 80 trimers. E1 is located at the periphery of each trimer and mediates interactions between spikes [50]. Each SFV-particle contains a fixed set of viral proteins, i.e. 240 copies of the E1/E2 dimer and 240 copies of the capsid protein. According to the enwrapping model of virus budding, the capsid preassembles in the cytosol and when it approaches the membrane, it interacts via a hydrophobic pocket in the C-protein with a Tyr-X-Leu peptide present in the cytoplasmic tail of the E2 glycoprotein. If all of its 240 binding sites are filled the capsid is completely enwrapped by the E1/E2 containing envelope and as a consequence is released from the membrane. Thus, sequential C–E2 interactions force the membrane around the capsid. The alternative model suggests that preassembled glycoprotein complexes cause binding of the C-protein and facilitate capsid formation. Whatever the exact mechanism might be, a very precise shut-off mechanism for virus assembly must exist to yield virus particles with a defined molecular structure [19]. This unique budding mechanism might explain why SFV does not use membrane rafts for enrichment of viral proteins.
Most other enveloped viruses are not composed of a strictly defined number of proteins. As a consequence of having a variable number of structural components, different virus strains (or even a population of virus particles released from the same cell) exhibit variations in their morphology. This is especially evident for influenza viruses, which can form filamentous and spherical particles. Other viruses, such as filoviruses, are quite homogeneous in their morphology, e.g. forming long filamentous particles each with the same diameter, but the length of virus particles can vary substantially between individual virus particles.
Although there is in most cases some variability in the exact composition of the viral module, the shut-off principles, which determine when exactly a newly formed virus particle is completed, remain to be elucidated. Shut-off is achieved by the interplay between cargo and coat components. In the case of the bullet-shaped VSV the vRNPs associate with the M protein at the plasma membrane of infected cells, which leads to compaction of the vRNP into a skeleton, a tightly coiled complex. Directly after enclosure of the nucleocapsid, the virus buds are separated from the host cell. The size of the virus particle depends on the length of the helical nucleocapsid, attachment of foreign nucleic acid sequences to viral genes yields longer virus particles. The concerted action of the submodular viral components ensures that a functional virus is formed [16].
Morphogenesis of herpes viruses, which are among the most complex virus particles, requires two budding events. Capsids preformed in the nucleus are translocated to the cytoplasm by budding at the inner nuclear membrane followed by fusion of the primary envelope with the outer nuclear membrane. Final envelopment, including the acquisition of dozens of tegument and envelope proteins, occurs by budding into Golgi-derived vesicles. The two budding events do not only differ in their intracellular location, but also in the viral proteins involved. As an example, two membrane proteins essential for budding at the inner nuclear membrane are absent from mature virus particles. Furthermore, capsids direct budding at the inner nuclear membrane, but do not trigger secondary envelopment. Likewise, the glycoproteins which mediate membrane fusion during cell entry of herpes viruses are not required for fusion of the primary envelope with the outer nuclear membrane, indicating that not only both budding processes, but also fusion events are mechanistically different [51].
The exact way of how a virus particle is formed is, in most cases, not well-characterized. On the whole, it can be stated that budding mechanisms vary for different virus families, but that in most cases the proteins which drive the budding reaction are engaged in lateral interactions. They thereby pack into lattices to form a cage-like construction at the outer surface of, within, or underneath the membrane and thus resemble budding of cellular transport vesicles. The budding mechanism of influenza virus may be unique since intense lateral interactions between its spike proteins, the driving force for budding, apparently do not occur. It was speculated that the accumulation of viral glycoproteins in membrane rafts may induce curvature of the membrane sufficient for spontaneous vesicle formation [47].
3.2.2. Virus budding by hijacking the cellular ESCRT module
However, assembly of viral proteins at the budding site is often not sufficient for release of virus particles. Budding may also require cellular functions, which are recruited to the budding site by viral proteins. This mechanism was first found for the Gag protein of the retrovirus HIV-1, where a so-called “late domain” (L-domain), which affects late steps in virus budding, was identified. Viruses with a mutation in this domain assemble and bud, but remain linked to the infected cell by a thin bridge of continuous membrane bilayer, i.e. scission of virus particles does not occur. Viral late domains are also present in proteins of other viruses (but probably not in influenza viruses) and consist of short amino acid sequences, such as P(S/T)AP (Gag of HIV) or PPEY (M of filoviruses), which are partially interchangeable between viral proteins. Each L-domain binds to a different cellular protein of the family of “class E VPS factors” (VPS = vesicular protein sorting), which assemble to so-called ESCRT complexes (endosomal sorting complexes required for transport). In the cellular context ESCRT complexes are involved in sorting of proteins into vesicles that bud into the lumen of endosomes thereby forming multivesicular bodies (MVB). No detailed mechanistic understanding of the budding process is available yet, but ESCRT proteins must mediate similar functions as coat proteins, e.g. sorting of cargo proteins, induction of membrane curvature, and membrane fission. However, in contrast to coated vesicles, which bud into the cytosol, ESCRT-mediated budding occurs, like budding of viruses, out of the cytosol. Thus, many enveloped viruses have hijacked a cellular budding machinery (or parts thereof) for their own purpose making use of a well-established and fine-tuned cellular module. This interplay leads to the formation of new submodules using viral and cellular components that maintain modular regulation properties of the original cellular module, but lead to formation of the viral module [21], [47], [52].
The minimal set of elements required for budding and release of a cellular transport vesicle has been experimentally defined in vitro with purified components for COP I and COP II vesicles. In both cases addition of a small G-protein (Arf1 or Sar1) preloaded with GTP and the subunits of the coat (coatomer plus cytoplasmic tails of cargo receptors or sec24/24 and sec13/31) to liposomes leads to the formation and release of vesicles. Thus, assembly of cytosolic coat protein at membranes and their subsequent polymerization mediated by the small G-protein drives the budding reaction, further elements for scission of the vesicle are apparently not required [53], [54]. In contrast, release of clathrin-coated vesicles requires the large GTPase dynamin, which forms a ring around the neck of the budding vesicle, and GTP-hydrolysis provides the energy for scission [55]. Overall, the budding of coated vesicles bears the closest similarities to those viruses where assembly of cytosolic coat proteins on the surface of a membrane and their subsequent polymerization are the driving force for budding. This includes the retro-, rhabdo-, filo- and paramyxoviruses. The second principle of virus budding (by polymerization of proteins within the membrane) is apparently also used by cells. Endocytosis of caveolae is probably executed by polymerisation of caveolin, a protein that has two cytoplasmic domains and inserts into membranes with a hairpin configuration. Binding of caveolin to cholesterol, a raft-lipid, is also required for endocytosis, emphasising that not only proteins, but also lipids are key players in budding of vesicles and viruses [56].
In summary, virus assembly shares features with the assembly of coated transport vesicles. Both require the bringing together of many components – transmembrane proteins, coat proteins and the cargo – at specific sites of cellular membranes so that an ordered series of specific interactions leads to recognition and assembly. Often the components to be assembled are concentrated and segregated from the other cellular components so that the appropriate interactions become more likely. Most of the required interactions are probably rather weak since it must be easy to disassemble the module after it has fulfilled its function. Recognition of weak signals is enhanced by cooperativity, e.g. oligomerisation of coat proteins, and assembly of the components in a two-dimensional milieu, e.g. at a membrane or even at a specific microdomain of a membrane. This enhances the effect of the individual recognition steps so that a weak preference for interaction between two components becomes amplified by cooperation among many. Consequently, assembly in two dimensions must use a hierarchy of sorting steps and mechanisms for regulation and control that allow selection of the correct components and exclusion of incorrect ones.
3.3. Maturation of viruses and uncoating of vesicles
We have so far discussed assembly and budding of the viral module, a process in which the biological activities of the viral submodules and elements are switched off rather than activated as in the case of cellular functional modules of the cell. For instance, all enzymatic activities of the viral genome of influenza viruses are inhibited by assembly of vRNPs into a compact structure and their subsequent transport out of the nucleus. A key role in this process is played by the viral M1 protein. M1 is transported into the nucleus, where it binds to the vRNPs and mediates (in collaboration with the viral nonstructural protein NS2/NEP) their export through the nuclear pore to the budding site. The transcription activity of the vRNPs is not resumed before M1 is released from vRNPs in the low-pH environment of the target cell's endosome (see below). Thus, binding of M1 to vRNPs and subsequent release can be regarded as a molecular switch regulating the activity of the influenza virus genome [14], [57].
Likewise, upon budding of influenza virus particles, the lipids of the viral envelope become immobile and, as a consequence, the lateral mobility of the viral glycoproteins HA and NA in the membrane is restricted. The formation of ordered lipid domains in the membrane requires enrichment of cholesterol and of lipids containing unsaturated fatty acids in budding virus particles, which is probably mediated by S-acylation of the cytoplasmic tail of HA [58]. The saturated fatty acids may collect other saturated-chain lipids and cholesterol when HA is concentrated in the plasma membrane before budding. Since ordered lipid domains are especially prominent at ambient temperature they are thought to stabilize virus particles against environmental conditions, which is especially critical for viruses spread by airborne transmission [59]. Thus, unlike vesicles, which are processed after release from their donor compartment, the elements of a virus generally remain in an inactive, “frozen” state after virus release to protect the virus genome against harmful environmental conditions. The virus then needs to be “reanimated” upon encounter of an infectable cell.
Nevertheless, freshly budded viruses are often exposed to simple maturation steps, carried out by cellular or viral enzymes, which are required to allow their release from the infected cell and to prepare the particle for subsequent steps of docking at and fusion with their target membrane. Virus surface component mediating attachment to target cell must be prevented from binding with the virus receptors on the cell from which the virus is exiting. This can be achieved by virus-induced down-regulation of the virus receptor on the cell surface, by limiting the budding to a distinct part of the cell surface or by an enzymatic reaction. The latter is found in the case of influenza virus, where viral HA interacts with cellular surface glycoproteins containing sialic acid. This step is required for binding of viral particles to a cell to be infected, but disadvantageous when freshly assembled virus particles are to be released from an infected cell. The second viral glycoprotein NA cleaves sialic acid from every cellular glycoprotein and glycolipid it encounters during its transit through the exocytic pathway to the plasma membrane and thus destroys the cellular receptor on the infected cell. Likewise, NA cleaves sialic acid also from viral HA to avoid aggregation of virus particles and loss of infectivity [60].
Maturation of viruses during or after budding often involves proteolytic cleavage of structural proteins, which prepares the virus for subsequent fusion or uncoating steps. The coat protein of HIV is synthesized largely as a Gag-precursor containing the matrix protein (MA), the capsid protein (CA), the nucleocapsid protein (NC) and the p6 protein (containing the recognition motif for elements of the cellular ESCRT-complex, see above) as one large polypeptide chain. Smaller amounts (5%) of a Gag–Pol fusion protein containing also a protease (PR) and the polymerase are also made. After its synthesis the polyprotein binds to the plasma membrane and polymerizes, which initiates budding of virus particles as described above. The low autoproteolytic activity of the Gag-precursor cleaves the polyprotein into the mentioned subunits. This releases free protease, which is more potent and rapidly attacks further Gag-precursors causing the release of more protease. Thus, a positive feedback mechanism exists until every polyprotein is cleaved. Upon cleavage, the spherical Gag shell typical of the immature particle undergoes structural rearrangement to form a morphologically distinct mature particle, which can even be recognized in EM-pictures. Whereas immature particles have a disorganised central core, the matrix protein lines the inner side of the envelope in mature virus particles, and the capsid protein forms a cone-shaped core containing the genome complexed to the nucleocapsid protein and the polymerase. Proteolytic cleavage of the Gag-precursor is not required for budding, but prepares the virus for the subsequent uncoating of its genetic material in a freshly infected cell. This process must be regulated such that the initial proteolytic attack does not occur in the cytoplasm, but only during or shortly after the budding of the virus particle [26], [61].
Another drastic maturation step occurs in flaviviruses after their budding, affecting the external protein shell composed of the E and preM-glycoproteins. Members of this virus family bud from the cytosol into the lumen of the ER and are subsequently secreted by the cell. During passage of virus particles through the trans-Golgi-network (TGN), preM is cleaved at a stretch of basic amino acids by a cellular protease from the furin-family. This changes the arrangement of the protein shell, the 60 trimers composed of pre M–E heterodimers are restructured to form 90 homodimers, which is an essential prerequiste to allow the E-protein to perform its fusion activity in the target cell [49].
Proteolytic cleavage of glycoproteins by trypsin-like proteases at either monobasic or polybasic sites occurs for many enveloped viruses, such as myxo- and alphaviruses, affecting either a protein in a complex with a fusion protein or the fusion protein itself. Proteolytic cleavage is an essential step of virus maturation since it primes the fusion protein for subsequent activation at the cell the virus is about to infect. Proteins with a polybasic stretch of amino acids at the cleavage site are proteolyzed inside every cell by a ubiquitous protease of the furin-family present in the TGN. Fusion proteins with a monobasic cleavage site are not recognized by this protease; therefore, they have to be cleaved by an extracellular protease, which usually takes place in the organism to be infected [62]. The fusion proteins of other viruses (e.g. SARS corona virus, Ebola) are cleaved after uptake into the endosome by proteases of the cathepsin family [63], [64].
In the case of cellular transport vesicles, the function of viral fusion proteins is fulfilled by SNARE-proteins, C-tail anchored transmembrane proteins with their N-terminal domain facing the cytosol (see below). To expose the v-SNAREs, transport vesicles must lose their coat. Small G-proteins present in the vesicle, Arf or Sar, hydrolyze GTP, which causes retraction of their hydrophobic N-termini from the membrane and detachment of the G-proteins from the vesicle membrane. This destablilizes the coat, which is disassembled, and its constituents are released into the cytosol, where they serve as a pool for the formation of new vesicles (recycling). It is still unclear how the GTPase activity is regulated, but it must contain a timing scheme which prevents disassembly of the coat before the vesicle has completely budded from the membrane.
3.4. Tethering and docking to the target membrane
The biologically “frozen” state of the virus after budding is overcome when the virus binds to a suitable receptor on the surface of a target cell. Often only a few hydrogen bonds are involved in binding [65], but the reaction triggers a cascade of events leading to membrane fusion, disassembly of virus particles, cargo release, disassembly of its submodules containing the nucleic acids, and finally to transcription and replication of the viral genome. These events must be temporally coordinated for successful infection of cells: for instance, the infectivity of the virus would be irreversibly destroyed if the fusion activity were triggered prior to virus binding to the target cell. Thus, recognition of the cellular receptor by the viral glycoprotein is a modular switch that activates the further processing events of the module, which are schematically depicted in Fig. 4 .
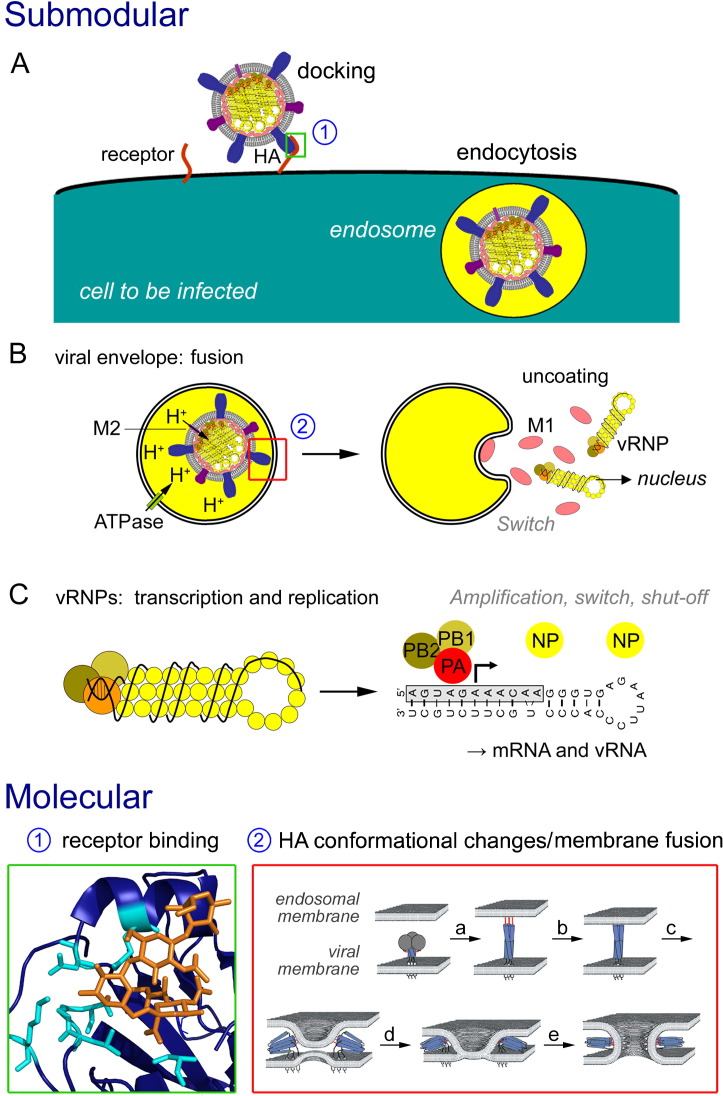
Fig. 4.
Example of an output submodule: cell entry and disassembly of influenza virus. Submodular level: (A) The virus particle docks to a receptor on the target cell, thus causing endocytosis of the receptor with bound particle. Molecular details of receptor binding, framed in the green box, are shown in the lower part of the figure. (B) The low pH in the endosome (arising from ATPases in the endosomal membrane) activates HA, which catalyzes fusion of viral and endosomal membrane (red box, see the blow up at the bottom). The viral membrane contains the proton channel M2 which acidifies the interior of the virus particle, causing displacement of M1 from vRNPs (switch). vRNPs are released through the fusion pore into the cytoplasm of the cell. (C) In the nucleus, the compact structure of the vRNPs is partially disassembled allowing synthesis of mRNA and vRNA. The hallmarks of functional modules (grey lettering) are described in the text, 3.4, 3.5, 3.6. Molecular level: green box: interaction of HA with a sialic acid containing receptor. The peptide-backbone of HA (blue) with the amino acids (cyan) involved in recognition of the substrate sialic acid-galactose-N-acetyl-glucosamine (orange). Figure modified with Pymol from pdb file 2WR7, [84]. Red box: conformational changes in HA leading to fusion of the viral with the endosomal membrane. The fusion peptide (red) becomes exposed on the molecule's surface after acidification (a) and inserts into the cellular membrane (b). A second conformational change then bends HA (c), which leads to hemifusion with exchange of lipids (d) and opening of a fusion pore (e). For clarity, the receptor-binding HA1 subunit (grey) is omitted in b to e. Picture modified from Ref. [85].
The presence of a suitable receptor on the cell surface is recognized by a viral glycoprotein and is one of the factors that determine the type of cell a certain virus can infect. Some receptors are present on many cell types, such as glycoproteins and glycolipids containing terminal N-acetyl neuraminic acid (sialic acid) recognized by influenza virus HA (see Fig. 4, green box, for the molecular details of this interaction). However, avian and human viruses have a different preference for the type of linkage that attaches the sialic acid moiety to the penultimate sugar of the carbohydate chain, galactose. Human viruses preferentially recognize an α2,6 linkage, whereas avian viruses require an α2,3 bond. Since the matching receptors are distributed correspondingly in human and avian epithelial cells, the type of receptor represents a species barrier an avian virus, e.g. the H5N1 virus, has to overcome to become a human pathogen [65], [66]. Other viruses recognize even more specific receptors on the target cell, such as the nicotinic acetylcholine receptor present on motoneurons, which is used by the G-protein of rabies virus, or CD4 present on T-helper cells, which is recognized by gp160 of HIV. Successful infection of T-helper cells by HIV requires a second component, a member of the chemokine receptor family. Thus, HIV uses a “two step mode” of cell binding; other viruses, e.g. herpes viruses, also require two types of receptors to which they bind with low and high affinity, respectively. This two step mode of binding thus resembles reversible tethering and irreversible docking of cellular vesicles. Binding to the first receptor allows viruses to sample whether the second, high-affinity receptor is present — if no receptor is found virus particles are discharged. Release of unproductively bound viruses may also be exerted by a receptor-destroying activity present in one of the viral glycoproteins. An example is the neuraminidase of influenza and paramyxoviruses, which removes virus particles from their receptor if subsequent steps of virus entry do not follow. This is especially important during virus invasion into an organism since virus particles have to make their way through the sialic acid containing mucus present in the airways until they encounter a susceptible cell. Thus, the input signal for the switch that activates the virus might be turned off if subsequent steps of virus entry do not occur [67], [68].
Cellular transport vesicles also use a two step binding mode to attach to their acceptor compartment. Each membranous compartment is equipped with specific tethering complexes, either large multisubunit particles, such as TRAPP or the exocyst, or long putative coiled-coil proteins, such as p115. Tethering complexes interact with appropriate transport vesicles, where they activate a member of the family of rab-proteins, small GTPases of the ras-superfamily. Activated rab then interacts with its downstream effectors, which triggers a cascade of events that prime the subsequently acting SNARE-proteins to fulfil their function, docking and fusion [69], [70].
Each type of vesicle is equipped with a specific v-SNARE-protein recognizing only cognate t-SNAREs (“t” standing for “target”) present in the acceptor compartment with which the vesicle is destined to fuse as described below. Thus, tethers, rabs and SNAREs collaborate in a sequential series of events to ensure that cellular membranes fuse at the correct time and place. However, it does not universally hold true that each module or submodule has its unique protein inventory. For example, the TRAPP subunit Bet3, which we know to be associated to the submodular function of vesicle tethering, is also found to interact with subunits of the COP II vesicle coat. Since the coat is removed from the vesicle before tethering, this means that Bet3 must be associated to more than one submodular function [8].
3.5. Membrane fusion catalyzed by SNAREs and viral fusion proteins
In order to release the cargo (protein or viral genome), the membrane envelope of the vesicle or virus has to fuse with the target membrane, which results in a continuous connection between the vesicle/virus lumen and the target compartment.
There are two pathways into the cell's cytoplasm used by enveloped viruses, each associated with a different activation mode of the fusion activity. Some viruses, such as retroviruses and paramyxoviruses, fuse directly with the plasma membrane of the cell, and this activity is triggered by binding to a cellular receptor. The viral fusion protein might be identical with (retroviruses) or different from (paramyxoviruses) the receptor-binding protein. In the latter case a signal generated by binding to the cellular receptor must be transmitted to the fusion protein. Other viruses, such as influenza, are taken up by endocytic vesicles, which they use as a ferry for their transport from the periphery of the cell to a perinuclear location. However, since endosomes ultimately fuse with lysosomes, the virus has to leave the compartment early enough to avoid degradation by hydrolytic enzymes. The increasingly acidic pH generated by a membrane proton pump V-ATPase in the endosome triggers a conformational change in HA that executes fusion of viral and endosomal membranes thereby allowing access of vRNPs to the cytoplasm [67], [68], [71], [72]. Enveloped viruses often exhibit flexibility in cell entry, being able to use different pathways. Influenza viruses uses both clathrin-dependent and -independent endocytosis for uptake and HIV, which can fuse with the plasma membrane, can also utilize endocytotic vesicles for cell entry [73], [74].
Elucidation of the X-ray structure of the hemagglutinin at neutral and mildly acidic pH, which are equivalent to the HA structure before and after membrane fusion, has led to a model on how conformational changes of HA execute membrane fusion. An internal hydrophobic segment (fusion peptide) at the N-terminus of the subunit HA2, which is buried inside the trimeric structure at neutral pH, becomes exposed on the molecule's surface after acidification. It is thought to insert into the cellular membrane allowing the molecule to exert force on the bilayer. A second conformational change then bends HA thereby drawing the fusion peptide towards the transmembrane region. The second conformational change requires two so-called “heptad repeats”, amphipathic helices in the vicinity of the fusion peptide and the transmembrane region, which interact to form a stable coiled-coil domain. The two conformational changes resemble the opening and subsequent closing of one blade of a pocket knife. This leads to a close apposition of viral and endosomal membranes, hemifusion with exchange of lipids, opening of a fusion pore and eventually complete merger of both lipid bilayers [65]. (See Fig. 4, red box, for detailed series of events.) Many other viral fusion proteins, e.g. the F-protein of paramyxoviruses and gp160 of HIV, have a similar helical coiled-coil structure as influenza virus HA after fusion suggesting that they cause membrane fusion by a similar mechanism. Other viral fusion proteins do not exhibit a helical coiled-coil structure after fusion, but the basic principle of their fusion mechanism is the same: exposure of a fusion peptide that is hidden in the unactivated state, its insertion into the cellular membrane and a second conformational change, which pulls on and finally fuses the membranes [75]. Membrane fusion is not an achievement of fusion proteins alone, since specific lipids must be present at the fusion site to allow negative and positive curvature of the bilayer required at the different steps of membrane merger [76].
Cellular SNAREs do not contain initially hidden fusion peptides, but catalyze membrane fusion by the formation of a complex composed of the v-SNARE and several t-SNAREs anchored in the target membrane. This SNARE-complex forms a coiled-coil structure similar to that of HA after fusion. The function of the viral fusion peptide, i.e. insertion into the target membrane to execute force, is carried out by the transmembrane region of the t-SNAREs, and the formation of the SNARE-complex tightly connects and finally fuses both membranes (zippering-mechanism). Thus, although viral and cellular fusion proteins have no homology in their primary structure, the mechanism how they execute membrane fusion is surprisingly similar [7], [77]. However, SNARE-proteins are recycled for further functional cycles after the SNARE-complex has been disassembled by the ATPase NSF [7], whereas viral fusion proteins are dispensable after successful delivery of the viral genome into the cytoplasm.
3.6. Uncoating of the virus and cargo release
Viruses must be assembled in such a way that this assembly can be reversed later on in subsequent infections. Thus, a control mechanism, i.e. a switch must exist to shift the equilibrium of metastable interactions to assembly in the cell the virus is about to leave and to disassembly in a newly infected cell. During influenza virus infection this is achieved by chemically different environments upon assembly and disassembly. Assembly of influenza virus occurs at the neutral pH of the cytoplasm, whereas disassembly is accomplished through the low pH which the virus encounters in the endosome. The protons in the endosome trigger an irreversible conformational change in the hemagglutinin, which thereby gets activated to catalyze fusion of viral and endosomal membranes (see above). At the same time the interior of virus particles is acidified through the proton-channel activity of the third viral envelope protein, M2 [78]. The protons induce the dissociation of M1 from vRNPs, which are then released through the fusion pore into the cytosol [57]. Thus, upon infection, the fragmentation of the viral module into submodules and single components is achieved via spatial sequestration and chemical specificity (see Fig. 4).
Due to intrinsic targeting signals, vRNPs are transported to the nucleus, where their compact structure is opened and partially disassembled. From the vRNA the three polymerase subunits synthesize messenger RNA for amplification of viral elements and (via a cRNA intermediate) new genomic vRNA. The switch from mRNA to vRNA synthesis is regulated by the number of NP-molecules, which bind to elongating RNA-strands thereby allowing the polymerase to make a vRNA copy. Thus, in the absence of NP, as it occurs when the viral genome has just entered the cell, mRNAs (and subsequently viral proteins) are made, whereas later in infection, when NP (and other viral proteins) are abundant, a switch to genome replication occurs. The submodular activity of transcription and replication is finally switched off by export of vRNPs from the nucleus, which is regulated by binding to M1 and which requires also the nonstructural protein NS2/NEP [13].
Alphaviruses, such as SFV, use an entirely different strategy involving other cellular modules. The 60S subunit of the ribosome has high-affinity binding sites for the viral capsid protein. The interaction with the ribosome disassembles incoming capsids thereby liberating the viral RNA, which can be instantly used for the synthesis of nonstructural proteins. When synthesis of structural protein starts later in infection, abundantly expressed capsid proteins saturate the binding site on the ribosomal subunits, which thus can no longer interfere with assembly of progeny capsids. Alternatively, the structure of freshly assembled capsids may be different from incoming capsids such that only the latter can associate with ribosomes [19].
Cellular transport vesicles also use a difference in the pH of the donor and acceptor compartment to discharge their cargo, similarly to the processes outlined above for influenza virus. Endocytic vesicles contain a proton pump which acidifies their lumen after internalization. Upon drop in pH the receptors dissociate from their ligands and are recycled back to the cell surface. Likewise, the KDEL-receptor binds its ligand optimally at acidic pH, which suggests that pH differences between the Golgi and ER might control the association and dissociation of the KDEL ligands [30].
4. A modular perspective of pharmacological intervention
Most currently available antiviral drugs target individual viral enzymes, such as the neuraminidase of influenza virus, the protease and transcriptase of HIV and thymidine-kinases of herpes viruses [60], [79]. As cells have enzymes performing similar functions these drugs often interact which their cellular counterpart leading to unwanted side effects. Most importantly, a single point-mutation in its target often prevents binding of these drugs. Since the high mutation rate observed for all viral genomes supplies such mutations with a high frequency, viruses which avoid the inhibitory action of these drugs emerge rapidly.
The modular character of viral assembly makes it a multistep process of increasing hierarchy and complexity. Based on this property, new targets for pharmacological intervention may arise, which do not inhibit enzymatic activities, but block another function of the viral module, for example cell entry. A step in this direction is the drug Fuzeon®, which was licensed in 2003 as a new class of antiretroviral drugs for HIV therapy. Fuzeon, a peptide that corresponds in its sequence to one of the mentioned heptad repeats of the fusion protein gp41, prevents the second conformational change of the glycoprotein, which executes membrane fusion. The efficiency of such drugs can even be increased by linking them to cholesterol, which targets the molecule to raft domains where HIV fusion occurs [80]. However, the application of Fuzeon has led to the appearance of resistant virus strains, since the binding site of the peptide in the other heptad repeat region can change without affecting the fusion activity of the protein [81].
The overall function of a functional module always relies on the fine-tuned interplay between different submodules. The idea would therefore be to interfere with this interplay, for example by inhibiting interactions between the components of the viral coat. Interesting in this respect is a study describing a peptide that binds to the capsid-domain of gag of HIV thereby inhibiting assembly of immature and mature capsids in the test tube [24], [82]. Such drugs might be even better if they block the interaction surface between two different components of a virus particle, for example between different proteins of a viral coat or between spike proteins and coat proteins. The use of such therapeuticals might be less prone to the formation of resistant virus strains, since this would require several mutations on different submodules or molecular components, which must simultaneously occur to confer drug resistance. This has not been sufficiently exploited so far in the search for antiviral drugs and offers a new field of intervention.
Likewise, a better understanding of the interplay between viral and cellular modules might also lead to the development of novel antiviral drugs. Viruses do make use of cellular modules, such as signal transduction cascades (activated by binding of viruses to a receptor on the cell surface), ribosomes for protein synthesis and vesicles for transport of newly synthesized glycoproteins to the viral assembly site. Inhibition of such non-redundant cellular modules would block replication of a virus, but would obviously also disturb vital functions of the cell. An exception might be the various vesicular transport pathways, such as phagocytosis, macropinocytosis, clathrin-, caveolae or raft-mediated endocytosis, which a cell uses to internalize extracellular compounds [67], [68]. Most, if not all, viruses hijack one of these pathways to gain entry into the host cell. The exact composition of the endocytic modules and submodules is currently an emerging area of research. Protein kinases, which selectively control one of several different entry pathways, have been identified as promising drug targets. The strategy is based on the assumption that cells are robust enough to allow temporal inactivation of one of their endocytic pathways, whereas simple viruses are not prepared to use backup strategies if their obligatory entry pathway is blocked. Furthermore, since genetically unrelated viruses might use the same entry pathway, it is even imaginable that broad-spectrum antivirals might be developed [83]. Thus, the search for such drugs might be very promising, but requires a better understanding of the modular features of the viral life cycle and the fine-tuned interplay between viral and cellular function.
5. Conclusion
In spite of distinctive differences, common principles govern vesicular and viral function. The similarity does not arise from the same or similar elements which are used — there is not even a high degree of sequence homology between any of the analogous elements that constitute the vesicular and viral modules. The similarity rather arises from the basic composition and functionality of the underlying structures and their dynamics. For example, it is a decisive property of biological membranes, indispensable to life, that they allow the selective and controlled passage of cargo. To transit the membranes that sequester intracellular compartments from one another, life has invented proteins to induce membrane bending, using the fluidity of the lipid bilayer. The virus mimics this ability by employing virus fusion proteins (which replace the vesicular SNAREs). The “fraud” which viruses commit is not on the level of the same or similar molecular inventory but on the level of submodular function. We have seen that this extends to the essential properties and hallmarks of cellular modules: Functional input–output cycles, spatial sequestration of elements, amplification of signals and negative feedback occur also during the replicative cycle of an enveloped virus. The occurrence of the same schemes in two entirely different domains of life may imply that these submodular functions are indeed a critical level of biological organisation [1], reflecting general biochemical and biophysical design principles.
Acknowledgements
The work carried out in the authors' laboratories was supported by the Deutsche Forschungsgemeinschaft (SFB 740, “From molecules to modules: organisation and dynamics of functional units in cells” and SPP 1175, “Dynamics of cellular membranes and their exploitation by viruses”) and by the EU (Marie Curie network “Virus entry”). We thank Andreas Herrmann and Udo Heinemann for helpful discussions.
Glossary
- Arf
ADP-ribosylation factor, member of a family of small G-proteins (GTPases) involved in assembly and budding of COP I and clathrin vesicles.
- Clathrin
Coat protein mediating the formation of endocytotic vesicles at the plasma membrane and at the trans-Golgi network.
- COP I
Coat protein complex I, mediating the formation of vesicles for retrograde transport from the cis-Golgi to the ER and probably through the Golgi.
- COP II
Coat protein complex II, protein complex mediating the formation of vesicles for anterograde vesicle transport from the ER to the Golgi.
- ESCRT
Endosomal sorting complexes required for transport, protein complexes involved in the budding of vesicles into endosomes (formation of multivesicular bodies) and the budding of some viruses, e.g. HIV.
- Gag
Group-specific antigen, precursor protein of (among others) HIV matrix protein (MA, N-terminally myristoylated), capsid and nucleocapsid proteins (CA and NC, respectively), cleaved into its subunits by the viral protease.
- gp160
160 kDa glycoprotein, precursor of HIV surface glycoproteins gp120 (mediating binding of the virus to its cell surface receptor CD4) and gp41 (fusion with the cell membrane of the target cell).
- HA
Hemagglutinin, surface glycoprotein of influenza virus mediating attachment to the target cell (binding to sialic acid) and fusion with the target membrane.
- HIV
Human immunodeficiency virus, a retrovirus (positive-stranded RNA genome transcribed into DNA upon infection). Surface glycoprotein: gp160; coat protein: Gag.
- M1
Matrix protein of influenza virus, coat protein lining the inner surface of the virion and organiser of virus assembly.
- NA
Neuraminidase, surface glycoprotein of influenza virus, cleaves sialic acid from glycoproteins and thus mediates virus release.
- PtdIns(4,5)P2
Phosphatidylinositol-4,5-bisphosphate, a lipid of the plasma membrane serving as a substrate for the formation of second messengers (signal transduction) and docking site for proteins.
- Sar1
Small G-protein (GTPase) involved in the assembly and budding of COP II vesicles.
- SFV
Semliki Forest virus, an alphavirus with single-stranded positive-sense RNA genome in an icosahedral capsid and 80 trimers of E1/E2 embedded in the virus envelope.
- SNARE
Soluble N-ethylmaleimide sensitive factor attachment protein receptor, coiled-coil SNARE-complex between a v-SNARE on the vesicle and t-SNAREs on the target membrane mediates vesicle attachment and membrane fusion.
- VLP
Virus-like particle, cargoless non-infectious vesicle with similar size and density as a virus, produced by cells upon expression of the viral components necessary for budding.
- vRNP
Viral ribonucleoprotein-particle, complex of viral RNA, nucleoprotein (NP) and polymerases.
- VSV
Vesicular stomatitis virus, a rhabdovirus with single-stranded negative-sense RNA genome, surface glycoprotein G, coat/matrix protein M, and helical nucleocapsid.
References
- 1.Hartwell L.H., Hopfield J.J., Leibler S., Murray A.W. From molecular to modular cell biology. Nature. 1999;402:C47–C52. doi: 10.1038/35011540. [DOI] [PubMed] [Google Scholar]
- 2.Alexander R.P., Kim P.M., Emonet T., Gerstein M.B. Understanding modularity in molecular networks requires dynamics. Sci. Signal. 2009;2:e44. doi: 10.1126/scisignal.281pe44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harmeier A., Wozny C., Rost B.R., Munter L.M., Hua H., Georgiev O., Beyermann M., Hildebrand P.W., Weise C., Schaffner W., Schmitz D., Multhaup G. Role of amyloid-beta glycine 33 in oligomerization, toxicity, and neuronal plasticity. J. Neurosci. 2009;29:7582–7590. doi: 10.1523/JNEUROSCI.1336-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hofmann K.P., Spahn C.M., Heinrich R., Heinemann U. Building functional modules from molecular interactions. Trends Biochem. Sci. 2006;31:497–508. doi: 10.1016/j.tibs.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 5.Kummel D., Heinemann U. Diversity in structure and function of tethering complexes: evidence for different mechanisms in vesicular transport regulation. Curr. Protein Pept. Sci. 2008;9:197–209. doi: 10.2174/138920308783955252. [DOI] [PubMed] [Google Scholar]
- 6.Bonifacino J.S., Glick B.S. The mechanisms of vesicle budding and fusion. Cell. 2004;116:153–166. doi: 10.1016/s0092-8674(03)01079-1. [DOI] [PubMed] [Google Scholar]
- 7.Jahn R., Scheller R.H. SNAREs — engines for membrane fusion. Nat. Rev., Mol. Cell Biol. 2006;7:631–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- 8.Cai H., Yu S., Menon S., Cai Y., Lazarova D., Fu C., Reinisch K., Hay J.C., Ferro-Novick S. TRAPPI tethers COPII vesicles by binding the coat subunit Sec23. Nature. 2007;445:941–944. doi: 10.1038/nature05527. [DOI] [PubMed] [Google Scholar]
- 9.Hehnly H., Stamnes M. Regulating cytoskeleton-based vesicle motility. FEBS Lett. 2007;581:2112–2118. doi: 10.1016/j.febslet.2007.01.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Radtke K., Dohner K., Sodeik B. Viral interactions with the cytoskeleton: a hitchhiker's guide to the cell. Cell Microbiol. 2006;8:387–400. doi: 10.1111/j.1462-5822.2005.00679.x. [DOI] [PubMed] [Google Scholar]
- 11.Rothman J.E., Wieland F.T. Protein sorting by transport vesicles. Science. 1996;272:227–234. doi: 10.1126/science.272.5259.227. [DOI] [PubMed] [Google Scholar]
- 12.Martin-Benito J., Area E., Ortega J., Llorca O., Valpuesta J.M., Carrascosa J.L., Ortin J. Three-dimensional reconstruction of a recombinant influenza virus ribonucleoprotein particle. EMBO Rep. 2001;2:313–317. doi: 10.1093/embo-reports/kve063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Portela A., Digard P. The influenza virus nucleoprotein: a multifunctional RNA-binding protein pivotal to virus replication. J. Gen. Virol. 2002;83:723–734. doi: 10.1099/0022-1317-83-4-723. [DOI] [PubMed] [Google Scholar]
- 14.Nayak D.P., Hui E.K., Barman S. Assembly and budding of influenza virus. Virus Res. 2004;106:147–165. doi: 10.1016/j.virusres.2004.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmitt A.P., Lamb R.A. Escaping from the cell: assembly and budding of negative-strand RNA viruses. Curr. Top. Microbiol. Immunol. 2004;283:145–196. doi: 10.1007/978-3-662-06099-5_5. [DOI] [PubMed] [Google Scholar]
- 16.Jayakar H.R., Jeetendra E., Whitt M.A. Rhabdovirus assembly and budding. Virus Res. 2004;106:117–132. doi: 10.1016/j.virusres.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 17.Takimoto T., Portner A. Molecular mechanism of paramyxovirus budding. Virus Res. 2004;106:133–145. doi: 10.1016/j.virusres.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 18.Forsell K., Xing L., Kozlovska T., Cheng R.H., Garoff H. Membrane proteins organize a symmetrical virus. EMBO J. 2000;19:5081–5091. doi: 10.1093/emboj/19.19.5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garoff H., Sjoberg M., Cheng R.H. Budding of alphaviruses. Virus Res. 2004;106:103–116. doi: 10.1016/j.virusres.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 20.Bruss V. Envelopment of the hepatitis B virus nucleocapsid. Virus Res. 2004;106:199–209. doi: 10.1016/j.virusres.2004.08.016. [DOI] [PubMed] [Google Scholar]
- 21.Welsch S., Muller B., Krausslich H.G. More than one door — budding of enveloped viruses through cellular membranes. FEBS Lett. 2007;581:2089–2097. doi: 10.1016/j.febslet.2007.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simons K., Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 23.Brugger B., Glass B., Haberkant P., Leibrecht I., Wieland F.T., Krausslich H.G. The HIV lipidome: a raft with an unusual composition. Proc. Natl. Acad. Sci. U. S. A. 2006;103:2641–2646. doi: 10.1073/pnas.0511136103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saad J.S., Miller J., Tai J., Kim A., Ghanam R.H., Summers M.F. Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly. Proc. Natl. Acad. Sci. U. S. A. 2006;103:11364–11369. doi: 10.1073/pnas.0602818103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Freed E.O. HIV-1 Gag: flipped out for PI(4, 5)P(2) Proc. Natl. Acad. Sci. U. S. A. 2006;103:11101–11102. doi: 10.1073/pnas.0604715103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ganser-Pornillos B.K., Yeager M., Sundquist W.I. The structural biology of HIV assembly. Curr. Opin. Struct. Biol. 2008;18:203–217. doi: 10.1016/j.sbi.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Resh M.D. Intracellular trafficking of HIV-1 Gag: how Gag interacts with cell membranes and makes viral particles. AIDS Rev. 2005;7:84–91. [PubMed] [Google Scholar]
- 28.Thaa B., Herrmann A., Veit M. The polybasic region is not essential for membrane binding of the matrix protein M1 of influenza virus. Virology. 2009;383:150–155. doi: 10.1016/j.virol.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 29.Roingeard P., Hourioux C., Blanchard E., Brand D., Ait-Goughoulte M. Hepatitis C virus ultrastructure and morphogenesis. Biol. Cell. 2004;96:103–108. doi: 10.1016/j.biolcel.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 30.Lee M.C., Miller E.A., Goldberg J., Orci L., Schekman R. Bi-directional protein transport between the ER and Golgi. Annu. Rev. Cell Dev. Biol. 2004;20:87–123. doi: 10.1146/annurev.cellbio.20.010403.105307. [DOI] [PubMed] [Google Scholar]
- 31.Haucke V., Di Paolo G. Lipids and lipid modifications in the regulation of membrane traffic. Curr. Opin. Cell Biol. 2007;19:426–435. doi: 10.1016/j.ceb.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pelham H.R. Sorting and retrieval between the endoplasmic reticulum and Golgi apparatus. Curr. Opin. Cell Biol. 1995;7:530–535. doi: 10.1016/0955-0674(95)80010-7. [DOI] [PubMed] [Google Scholar]
- 33.Flint S.J., Enquist L.W., Skalka A.M., editors. Principles of Virology. second edition ed. Blackwell; 2003. [Google Scholar]
- 34.Fujii Y., Goto H., Watanabe T., Yoshida T., Kawaoka Y. Selective incorporation of influenza virus RNA segments into virions. Proc. Natl. Acad. Sci. U. S. A. 2003;100:2002–2007. doi: 10.1073/pnas.0437772100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simons K., Garoff H. The budding mechanisms of enveloped animal viruses. J. Gen. Virol. 1980;50:1–21. doi: 10.1099/0022-1317-50-1-1. [DOI] [PubMed] [Google Scholar]
- 36.Jin H., Leser G.P., Zhang J., Lamb R.A. Influenza virus hemagglutinin and neuraminidase cytoplasmic tails control particle shape. EMBO J. 1997;16:1236–1247. doi: 10.1093/emboj/16.6.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zavada J. The pseudotypic paradox. J. Gen. Virol. 1982;63(Pt 1):15–24. doi: 10.1099/0022-1317-63-1-15. [DOI] [PubMed] [Google Scholar]
- 38.Briggs J.A., Wilk T., Fuller S.D. Do lipid rafts mediate virus assembly and pseudotyping? J. Gen. Virol. 2003;84:757–768. doi: 10.1099/vir.0.18779-0. [DOI] [PubMed] [Google Scholar]
- 39.Edidin M. The state of lipid rafts: from model membranes to cells. Annu. Rev. Biophys. Biomol. Struct. 2003;32:257–283. doi: 10.1146/annurev.biophys.32.110601.142439. [DOI] [PubMed] [Google Scholar]
- 40.Heerklotz H., Szadkowska H., Anderson T., Seelig J. The sensitivity of lipid domains to small perturbations demonstrated by the effect of Triton. J. Mol. Biol. 2003;329:793–799. doi: 10.1016/s0022-2836(03)00504-7. [DOI] [PubMed] [Google Scholar]
- 41.Munro S. Lipid rafts: elusive or illusive? Cell. 2003;115:377–388. doi: 10.1016/s0092-8674(03)00882-1. [DOI] [PubMed] [Google Scholar]
- 42.Engel S., Scolari S., Thaa B., Krebs N., Korte T., Herrmann A., Veit M. FLIM-FRET and FRAP reveal association of influenza virus hemagglutinin with membrane rafts. Biochem. J. 2009;425:567–573. doi: 10.1042/BJ20091388. [DOI] [PubMed] [Google Scholar]
- 43.Hess S.T., Gould T.J., Gudheti M.V., Maas S.A., Mills K.D., Zimmerberg J. Dynamic clustered distribution of hemagglutinin resolved at 40 nm in living cell membranes discriminates between raft theories. Proc. Natl. Acad. Sci. U. S. A. 2007;104:17370–17375. doi: 10.1073/pnas.0708066104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scolari S., Engel S., Krebs N., Plazzo A.P., De Almeida R.F., Prieto M., Veit M., Herrmann A. Lateral distribution of the transmembrane domain of influenza virus hemagglutinin revealed by time-resolved fluorescence imaging. J. Biol. Chem. 2009;284:15708–15716. doi: 10.1074/jbc.M900437200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Demirov D.G., Freed E.O. Retrovirus budding. Virus Res. 2004;106:87–102. doi: 10.1016/j.virusres.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 46.Dolnik O., Kolesnikova L., Becker S. Filoviruses: interactions with the host cell. Cell. Mol. Life Sci. 2008;65:756–776. doi: 10.1007/s00018-007-7406-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen B.J., Lamb R.A. Mechanisms for enveloped virus budding: can some viruses do without an ESCRT? Virology. 2008;372:221–232. doi: 10.1016/j.virol.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu D.X., Yuan Q., Liao Y. Coronavirus envelope protein: a small membrane protein with multiple functions. Cell. Mol. Life Sci. 2007;64:2043–2048. doi: 10.1007/s00018-007-7103-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mukhopadhyay S., Kuhn R.J., Rossmann M.G. A structural perspective of the flavivirus life cycle. Nat. Rev. Microbiol. 2005;3:13–22. doi: 10.1038/nrmicro1067. [DOI] [PubMed] [Google Scholar]
- 50.Lescar J., Roussel A., Wien M.W., Navaza J., Fuller S.D., Wengler G., Rey F.A. The Fusion glycoprotein shell of Semliki Forest virus: an icosahedral assembly primed for fusogenic activation at endosomal pH. Cell. 2001;105:137–148. doi: 10.1016/s0092-8674(01)00303-8. [DOI] [PubMed] [Google Scholar]
- 51.Mettenleiter T.C., Klupp B.G., Granzow H. Herpesvirus assembly: a tale of two membranes. Curr. Opin. Microbiol. 2006;9:423–429. doi: 10.1016/j.mib.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 52.Hurley J.H., Emr S.D. The ESCRT complexes: structure and mechanism of a membrane-trafficking network. Annu. Rev. Biophys. Biomol. Struct. 2006;35:277–298. doi: 10.1146/annurev.biophys.35.040405.102126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bremser M., Nickel W., Schweikert M., Ravazzola M., Amherdt M., Hughes C.A., Sollner T.H., Rothman J.E., Wieland F.T. Coupling of coat assembly and vesicle budding to packaging of putative cargo receptors. Cell. 1999;96:495–506. doi: 10.1016/s0092-8674(00)80654-6. [DOI] [PubMed] [Google Scholar]
- 54.Matsuoka K., Orci L., Amherdt M., Bednarek S.Y., Hamamoto S., Schekman R., Yeung T. COPII-coated vesicle formation reconstituted with purified coat proteins and chemically defined liposomes. Cell. 1998;93:263–275. doi: 10.1016/s0092-8674(00)81577-9. [DOI] [PubMed] [Google Scholar]
- 55.Ungewickell E.J., Hinrichsen L. Endocytosis: clathrin-mediated membrane budding. Curr. Opin. Cell Biol. 2007;19:417–425. doi: 10.1016/j.ceb.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 56.Bauer M., Pelkmans L. A new paradigm for membrane-organizing and -shaping scaffolds. FEBS Lett. 2006;580:5559–5564. doi: 10.1016/j.febslet.2006.08.077. [DOI] [PubMed] [Google Scholar]
- 57.Martin K., Helenius A. Nuclear transport of influenza virus ribonucleoproteins: the viral matrix protein (M1) promotes export and inhibits import. Cell. 1991;67:117–130. doi: 10.1016/0092-8674(91)90576-k. [DOI] [PubMed] [Google Scholar]
- 58.Kordyukova L.V., Serebryakova M.V., Baratova L.A., Veit M. S acylation of the hemagglutinin of influenza viruses: mass spectrometry reveals site-specific attachment of stearic acid to a transmembrane cysteine. J. Virol. 2008;82:9288–9292. doi: 10.1128/JVI.00704-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Polozov I.V., Bezrukov L., Gawrisch K., Zimmerberg J. Progressive ordering with decreasing temperature of the phospholipids of influenza virus. Nat. Chem. Biol. 2008;4:248–255. doi: 10.1038/nchembio.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.De Clercq E. Antiviral agents active against influenza A viruses. Nat. Rev. Drug Discov. 2006;5:1015–1025. doi: 10.1038/nrd2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bukrinskaya A. HIV-1 matrix protein: a mysterious regulator of the viral life cycle. Virus Res. 2007;124:1–11. doi: 10.1016/j.virusres.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 62.Garten W., Klenk H.D. Understanding influenza virus pathogenicity. Trends Microbiol. 1999;7:99–100. doi: 10.1016/s0966-842x(99)01460-2. [DOI] [PubMed] [Google Scholar]
- 63.Bosch B.J., Bartelink W., Rottier P.J. Cathepsin L functionally cleaves the severe acute respiratory syndrome coronavirus class I fusion protein upstream of rather than adjacent to the fusion peptide. J. Virol. 2008;82:8887–8890. doi: 10.1128/JVI.00415-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chandran K., Sullivan N.J., Felbor U., Whelan S.P., Cunningham J.M. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science. 2005;308:1643–1645. doi: 10.1126/science.1110656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Skehel J.J., Wiley D.C. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 66.Horimoto T., Kawaoka Y. Influenza: lessons from past pandemics, warnings from current incidents. Nat. Rev. Microbiol. 2005;3:591–600. doi: 10.1038/nrmicro1208. [DOI] [PubMed] [Google Scholar]
- 67.Marsh M., Helenius A. Virus entry: open sesame. Cell. 2006;124:729–740. doi: 10.1016/j.cell.2006.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Smith A.E., Helenius A. How viruses enter animal cells. Science. 2004;304:237–242. doi: 10.1126/science.1094823. [DOI] [PubMed] [Google Scholar]
- 69.Cai H., Reinisch K., Ferro-Novick S. Coats, tethers, Rabs, and SNAREs work together to mediate the intracellular destination of a transport vesicle. Dev. Cell. 2007;12:671–682. doi: 10.1016/j.devcel.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 70.Markgraf D.F., Peplowska K., Ungermann C. Rab cascades and tethering factors in the endomembrane system. FEBS Lett. 2007;581:2125–2130. doi: 10.1016/j.febslet.2007.01.090. [DOI] [PubMed] [Google Scholar]
- 71.Malsam J., Kreye S., Sollner T.H. Membrane fusion: SNAREs and regulation. Cell. Mol. Life Sci. 2008;65:2814–2832. doi: 10.1007/s00018-008-8352-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sollner T.H. Intracellular and viral membrane fusion: a uniting mechanism. Curr. Opin. Cell Biol. 2004;16:429–435. doi: 10.1016/j.ceb.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 73.Lakadamyali M., Rust M.J., Zhuang X. Endocytosis of influenza viruses. Microbes Infect. 2004;6:929–936. doi: 10.1016/j.micinf.2004.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Miyauchi K., Kim Y., Latinovic O., Morozov V., Melikyan G.B. HIV enters cells via endocytosis and dynamin-dependent fusion with endosomes. Cell. 2009;137:433–444. doi: 10.1016/j.cell.2009.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kielian M., Rey F.A. Virus membrane-fusion proteins: more than one way to make a hairpin. Nat. Rev. Microbiol. 2006;4:67–76. doi: 10.1038/nrmicro1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chernomordik L.V., Kozlov M.M. Mechanics of membrane fusion. Nat. Struct. Mol. Biol. 2008;15:675–683. doi: 10.1038/nsmb.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wickner W., Schekman R. Membrane fusion. Nat. Struct. Mol. Biol. 2008;15:658–664. doi: 10.1038/nsmb.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pinto L.H., Lamb R.A. The M2 proton channels of influenza A and B viruses. J. Biol. Chem. 2006;281:8997–9000. doi: 10.1074/jbc.R500020200. [DOI] [PubMed] [Google Scholar]
- 79.De Clercq E. Antivirals and antiviral strategies. Nat. Rev. Microbiol. 2004;2:704–720. doi: 10.1038/nrmicro975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rajendran L., Knolker H.J., Simons K. Subcellular targeting strategies for drug design and delivery. Nat. Rev. Drug Discov. 2010;9:29–42. doi: 10.1038/nrd2897. [DOI] [PubMed] [Google Scholar]
- 81.Naider F., Anglister J. Peptides in the treatment of AIDS. Curr. Opin. Struct. Biol. 2009;19:473–482. doi: 10.1016/j.sbi.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sticht J., Humbert M., Findlow S., Bodem J., Muller B., Dietrich U., Werner J., Krausslich H.G. A peptide inhibitor of HIV-1 assembly in vitro. Nat. Struct. Mol. Biol. 2005;12:671–677. doi: 10.1038/nsmb964. [DOI] [PubMed] [Google Scholar]
- 83.Damm E.M., Pelkmans L. Systems biology of virus entry in mammalian cells. Cell Microbiol. 2006;8:1219–1227. doi: 10.1111/j.1462-5822.2006.00745.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu J., Stevens D.J., Haire L.F., Walker P.A., Coombs P.J., Russell R.J., Gamblin S.J., Skehel J.J. Structures of receptor complexes formed by hemagglutinins from the Asian Influenza pandemic of 1957. Proc. Natl. Acad. Sci. U. S. A. 2009;106:17175–17180. doi: 10.1073/pnas.0906849106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Earp L.J., Delos S.E., Park H.E., White J.M. The many mechanisms of viral membrane fusion proteins. Curr. Top. Microbiol. Immunol. 2005;285:25–66. doi: 10.1007/3-540-26764-6_2. [DOI] [PMC free article] [PubMed] [Google Scholar]