Abstract
The microarray is a platform with wide-ranging potential in biodefence. Owing to the high level of throughput attainable through miniaturization, microarrays have accelerated the ability to respond in an epidemic or crisis. Extending beyond diagnostics, recent studies have applied microarrays as a research tool towards understanding the etiology and pathogenicity of dangerous pathogens, as well as in vaccine development. The original emphasis was on DNA microarrays, but the range now includes protein, antibody and carbohydrate microarrays, and research groups have exploited this diversity to further extend microarray applications in the area of biodefence. Here, we discuss the impact and contributions of the growing range of microarrays and emphasize the concepts that might shape the future of biodefence research.
Introduction
Natural outbreaks and the wanton use of pathogenic organisms for acts of terror have had tremendous impact on human populations, especially when considering the events over the past decade. Modern travel and trade have further dissipated traditional geographical boundaries that once curbed the spread of disease. Infectious agents capable of spreading from human to human, such as the severe acute respiratory syndrome (SARS) coronavirus, have shown us just how quickly (in a matter of weeks) a threat anywhere could become a threat everywhere [1]. The lessons learnt from SARS, avian flu and the anthrax letter attacks have emphasized the need for improved preparedness for, response to and treatment of both known and emergent biological threats 2, 3. This has also prompted heightened funding initiatives worldwide to build up national and international biodefence capabilities [3].
Among the systems and protocols to be put in place, platforms that facilitate the rapid and accurate identification of agents are particularly vital, both in confirming whether an attack has occurred and in instituting prompt measures to secure public health. The intrinsic ability of microarrays to perform multiplexed, low-volume and sensitive biological assays in a highly scalable manner is a significant advantage in biological threat analysis [4]. Developed in the early to mid 1990s, DNA microarrays stirred a technological revolution that continues to propel genomics research today [5]. Applications included the identification and comparison of mRNA or DNA from across tissues or organisms by hybridization against thousands of oligomeric DNA probes immobilized on planar surfaces, such as glass slides (Figure 1 ). This provided researchers with an unprecedented ability to quantify genome-wide differences in gene expression or sequence changes using minimal amounts of sample. In the context of biodefence, these studies have dramatically extended our capability beyond merely detecting known pathogens; now we are able to rapidly profile and characterize pathogens, as well as identify novel strains or ‘genetic islands’ that manifest changes in virulence (or the evolution of resistance). Such comparative phylogenomic profiling using microarrays facilitates the understanding of pathogen diversification by providing a bird's-eye view of the gene content present or absent within a given microbial genome [6].
Figure 1.
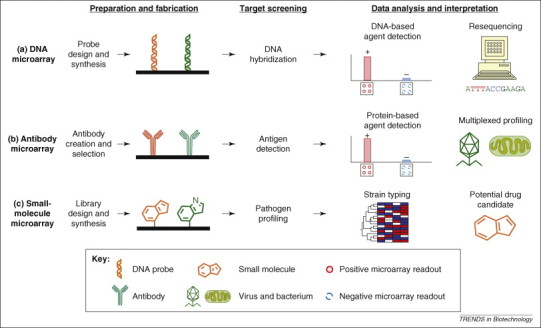
Overview of microarray applications in pathogen detection and biodefence. The main stages of a microarray experiment consist of preparation, fabrication, screening and analysis. Microarrays are distinguished by the type of molecules immobilized in the array; this might include DNA-probe libraries (a), antibody panels (b) or small-molecule libraries (c). (a) As discussed in this article, DNA microarrays can be applied to test for DNA from pathogenic organisms or for the resequencing of pathogen genomes. (b) Antibody microarrays can be used to detect pathogen proteins or antigens that might be present in environmental samples as an indication of contamination or for diagnostic purposes to determine pathogen infection in human tissues. (c) Small-molecule microarrays offer novel approaches for differentiating between pathogens, for example by clustering the binding signatures obtained for each pathogen. They can also be used to identify therapeutics that could potentially disrupt the infection cycle.
Microarray technology has also been brought outside the laboratory to the point-of-care, a development that has widening implications for threat detection. DNA-based detection systems generally rely on the ability of PCR to amplify and fluorescently tag the tiny amounts of target DNA present in the specimen. Advances in miniaturizing this initial PCR step, for instance the development of micro-PCR (μPCR), followed by hybridization and identification against probes on DNA microarrays have spun-off viable chip-based platforms that are able to successfully perform biological threat detection and analysis. Such portable systems, as will be described, are already appearing on the market and competing to attract lucrative biodefence funding. Alternative detection platforms have exploited quantitative PCR in ‘fieldable’ real-time detectors, where positive PCR amplification would indicate the presence of the corresponding target DNA [4]. These have included devices such as GeneXpert (Cepheid), Rapid (Idaho Technologies) or BioSeeq (Smiths Detection), but these systems provide relatively limited throughput [7]. Microarray systems are, by contrast, more definitive and highly scalable because hundreds to tens of thousands of possible DNA elements can be interrogated in a single experiment. Their performance nevertheless hinges on both the adoption of robust panels of probes that can accurately identify DNA from organisms of interest and the successful extraction and amplification of pathogen DNA from the relevant clinical sample or isolates 8, 9.
More recent developments in microarray technologies have significantly broadened the horizon for their use in the biodefence arena well beyond the confines of DNA-based profiling or detection. Newer microarray formats developed at the turn of the 21st century provide a host of other biomolecules that can be presented on chip, including proteins (whole proteomes, enzymes and antibodies) 10, 11, small molecules (drug-like molecules, peptides and carbohydrates) 12, 13 and even whole cells and tissues for simultaneous, multiplexed experimentation [14]. Each format offers unique ways in which microarrays can be harnessed towards improving our understanding of pathogen biology (Figure 1). The microarray paradigm has over the years contributed significantly towards these goals by not only providing a robust tool for molecular diagnostics but also by advancing biodefence capabilities in protection and therapy. This review will describe microarray-based applications for biodefence, as well as exciting new concepts and approaches that will drive future advancement and growth.
DNA microarrays in pathogen detection
Probe selection and design is usually an important first step in microarray-based pathogen detection, and many issues associated with probe design for DNA microarrays can impact the overall fidelity of the assay, in particular with regard to levels of specificity and sensitivity attained [15]. These issues and considerations include cross-hybridizations, orthogonal probe binding to target DNA from the specific organism(s) of interest and vice versa, uniformity of annealing temperatures (or GC content) and probe length. The occurrence of false positives and false negatives is highly problematic because they might cause an unwarranted response (and potentially panic) or a missed response (and a lost opportunity for intervention), which are unacceptable when dealing with deadly biological threats. For this reason, biodefence detection platforms strive to reach near perfect accuracies, which are comparable to, if not better than, the usual gold-standard assays – typically culture-based methods. With microarrays, redundancy can, however, be easily engineered into the system to improve accuracy and analytical power simply by over-representation.
The US Centers for Disease Control (CDC) have identified a list of 36 agents that, if spread uncontrollably, have the potential to cause a major public health crisis with heavy morbidity and mortality rates (Box 1 ). These pathogens are further subclassified into Categories A, B and C according to the degree of risk imposed (Table 1 ) [16]. In 2002, Wilson et al. fabricated a customized Affymetrix microarray containing 53 660 probes to detect DNA amplified from 18 different pathogenic microorganisms simultaneously, including pathogens from the US CDC's list of bioterrorism agents, such as Bacillus anthracis (which causes anthrax), Clostridium botulinum (which generates the botulinum toxin), Yersinia pestis (which causes bubonic plague) and the Ebola virus [17]. Specific multiplexed primer sets were designed to amplify unique diagnostic regions specific to each organism for hybridization against tiling arrays comprising 20-mer probes. The highly redundant system facilitated the accurate identification of each organism tested, with over 91% of the probes working as predicted. Impressively, as little as 10 fg, equivalent to the ‘mass’ of just two genomes (or DNA from two cells), of B. anthracis could be detected after multiplexed PCR on these microarrays.
Box 1. Bioterror agent classification.
Biohazardous agents that pose a significant threat require special attention, especially in the implementation of regulatory measures and controls to limit their access and distribution and to prevent them from falling into the wrong hands. In addition, such measures also aim to raise awareness within national healthcare systems and amongst healthcare providers, especially in cases where these bioterror threats are only rarely seen. The US Centers of Disease Control (CDC) have compiled a list of 36 biohazardous agents, which are divided into the categories A, B and C according to the level of threat they impose, and their characteristics are described here. Examples of pathogens from these three categories are provided in Table 1.
Category A (highest priority agents)
The CDC defines Category A agents as those that are of the highest priority for biodefence research. These organisms can be easily disseminated from person to person. They result in high mortality rates and have the potential to cause a major impact on public health. The accidental or deliberate release of these agents could cause public panic and social disruption. These agents thus require particular attention in ensuring public health preparedness because they pose a significant risk to national security.
Category B (second highest priority agents)
The CDC defines Category B agents as those that are moderately easy to disseminate. These pathogens result in moderate morbidity and low mortality. These agents would hence require specific improvements of diagnostic capabilities as well as enhanced measures for disease surveillance.
Category C (third highest priority agents)
The CDC defines Category C agents as those that have the potential for mass dissemination in the future because of their availability, ease of production and dissemination. This category also includes emergent threats that have the potential of causing high morbidity and mortality rates, as well as a major impact on public health.
Further details on the classification and prioritization of biological threat agents are available on the CDC website: http://www.bt.cdc.gov/agent/agentlist-category.asp.
Table 1.
Representative studies that used microarrays for detection and profiling of pathogens from the US CDC Categories A, B and C
| Biological agents | Associated diseases | DNA microarrays |
Non-DNA-based microarrays |
||
|---|---|---|---|---|---|
| Pathogen detection | Resequencing or strain typing | Pathogen detection | Seroprofiling or vaccine or therapeutic discovery | ||
| Category A | |||||
| Bacillus anthracisa | Anthrax | 17, 26 | 20, 28 | 43, 53 | [64] |
| Clostridium botulinuma toxin | Botulism | [17] | -b | - | - |
| Yersinia pestisa | Plague | [17] | 35, 36 | [45] | 62, 63 |
| Variola major | Smallpox | - | - | - | 58, 60, 61 |
| Francisella tularensisa | Tularemia | [26] | 20, 38, 39 | 45, 46, 51, 52 | 57, 58 |
| Ebola virus | Ebola | [17] | [20] | - | - |
| Category B | |||||
| Burkholderia pseudomalleia | Meliodosis | [23] | [37] | [54] | - |
| Burkholderia malleia | Glanders | [23] | [37] | [45] | - |
| Staphylococcusa enterotoxin B | Variety of symptomsc | [24] | - | - | - |
| Category C | |||||
| Avian influenza (H5N1) | Influenza | 24, 26, 45 | [20] | - | 65, 66 |
| SARS-CoV | SARS | 26, 30 | 20, 31, 32 | [49] | 55, 56 |
Abbreviations: H5N1, haemagglutinin 5 and neuramidase 1; SARS-CoV, severe acute respiratory syndrome-associated coronavirus.
Organism types for which DNA microarrays are available from the pathogen functional genomics resource centre (http://pfgrc.tigr.org).
‘-’ = no references identified.
Depending on route of exposure.
Even though culture methods should in theory be able to amplify and detect a single organism, the procedures are usually time-consuming and challenging – detection could take from 18 h to several days depending on the microbe type and even longer for fastidious microbes (e.g. Mycobacterium tuberculosis – a very slow growing microbe) that might require specialized, or as yet undiscovered, laboratory growth conditions. Molecular identification using random PCR followed by multiplexed resolution over a microarray offers a particularly attractive alternative for detection and diagnostics, generating results in 2–6 h once assays are optimized (Figure 2a). Wang et al. [18] developed such a microarray approach for the screening of viral pathogens from across broad viral families. A randomized primer was used to amplify any viral RNA that was present in the sample using reverse transcriptase-PCR followed by hybridization on a microarray comprising 1600 70-mer probes, representing nearly 140 virus genomes. Degeneracy of probes on the microarray and cross-hybridization of certain viruses across expected patterns indicated the emergence of novel, uncharacterized strains, which could hence be identified with the aid of microarrays. Similarly, Sengupta and colleagues [19] developed microarrays with 476 probes to distinguish among various influenza viruses. Primers were designed against characteristic sequences specific to influenza that targeted the viral fusion protein haemagglutinin and the glycosidase neuraminidase segments. Using the Affymetrix respiratory pathogen microarray, Lin et al. [20] detected the respiratory viruses influenza A and adenovirus, which were then further differentiated according to strain and species. This setup made use of a so-called ‘resequencing’ microarray that contained one perfectly matched and three mismatched probes per base and thus was able to identify genetic mutations at the sequence level [20]. The array's large screening capacity meant that both the forward and reverse strands of the DNA targets could be sequenced to provide added sequencing accuracy. Pooled primer pairs have also been optimized for use in detecting both DNA and RNA targets for pathogens that cause upper respiratory tract infections, including viruses (influenza, corona viruses and others) and bacteria (Bordetella pertusis, Streptococcus pyogenes and others) [21].
Figure 2.
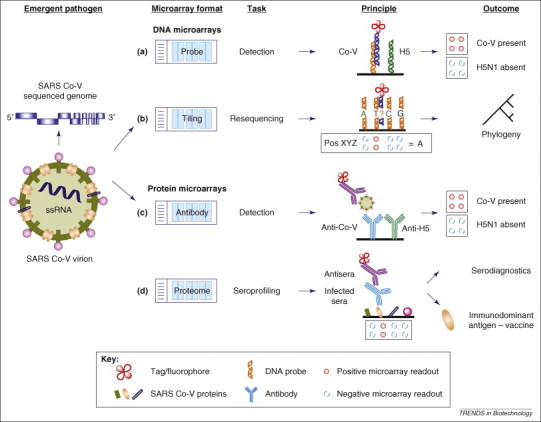
The use of microarrays for studying emergent pathogens, exemplified by severe acute respiratory syndrome (SARS)-associated coronavirus and avian influenza (H5N1). Various approaches have been developed for detection and diagnostics with DNA and protein microarrays. (a) Specific DNA probes, for instance Co-V (orange) for SARS and H5 (green) for H5N1, can be used to detect the presence of pathogen DNA or RNA using PCR or reverse transcriptase (RT)-PCR, thus enabling multiple pathogens to be detected simultaneously [26]. (b) Tiling arrays can be used to resequence pathogens so that any mutations in evolving pathogens can be rapidly detected and tracked. As illustrated, four probes, one for each nucleotide base, are used to determine the genotype at a given loci. A set of probes designed for an arbitrary position XYZ is shown to reveal the unknown base to be adenine (A). Multiple probe sets are ‘tiled’ across the whole pathogen genome and, upon hybridization and analysis, can confer the complete sequence information of the pathogen with great accuracy 17, 23, 25. (c) Antibody arrays make use of the sandwich immunoassay to screen for the presence of a pathogen. As depicted, the pathogen sample is first applied to the microarray, followed by the reporter antibody. As a result, the pathogen is sandwiched in between two antibodies – an immobilized antibody (blue) and a tagged antibody (purple) capable of reporting the presence of a pathogen on the microarray 43, 45. (d) Proteome microarrays, which contain immobilized proteins from the target pathogen, can be screened against sera obtained from infected individuals. If the individual has been exposed to the pathogen, the sera will contain antibodies (blue) against specific antigens of the pathogens that will react with the immobilized protein and that can be detected using tagged secondary antibodies (purple) [49]. This procedure also facilitates the identification of specific immunodominant antigens present in the pathogen proteome. These proteins are considered to be largely responsible for triggering the host immune response and thus have a high potential for the development of vaccines against this particular pathogen.
The small subunit ribosomal RNA (ssu rRNA) gene is used widely as a microbial marker for taxonomy and species classification [22]. DeSantis and colleagues [23] generated a high-density tiling microarray with 62 358 oligonucleotide probes of ssu rRNA with sufficient coverage to detect 18 different orders of microbes from environmental samples, as well as novel variants that exhibited mutations in their ssu rRNA. The array fluorescence intensities correlated well with the spiked pathogen concentrations, providing a means of quantifying the pathogen levels in the original sample. A similar method was used to detect Staphylococci. Out of 201 isolates taken from 33 Staphylococci species, 185 were correctly assigned by using only 16S rRNA sequences on microarrays, conferring a sensitivity of 92% [24]. The ability to refine and expand on the existing probe set, for example by the incorporation of additional informative genetic loci, is a key advantage provided by the DNA microarray platform to increase redundancy and hence improve assay performance and fidelity.
Miller and colleagues [25] applied microarrays in clinical diagnostics and were able to identify pathogens in a panel of 36 patient specimens with a 94% accuracy score, calculated from 76% sensitivity and 100% specificity. The microarray was designed to detect up to 35 RNA viruses, including the SARS coronavirus and dengue viruses, using 40-mer probes. A total of 53 555 such probes in replicates of seven were printed on Nimblegen tiling microarrays. The authors studied ways to minimize PCR bias to ensure uniform amplification and developed a unique statistical approach to infer pathogen identity from the resulting microarray fingerprints. Apart from the high-density microarrays described here, more focused biodefence platforms have been developed for selected panels of environmental [26] or blood-borne pathogens [27], using in-house microarrays that comprise only several hundred relevant probes. Such panels are designed to selectively identify targets of interest with enough redundancy to reduce the occurrence of false positives, which is infrequently controlled in other diagnostic platforms that often use either one or a handful of primer sets specific to each pathogen of interest (as is the case in the GeneXpert, Rapid and Cepheid real time PCR platforms).
DNA microarrays in pathogen profiling
There are several instructive examples where DNA microarrays have advanced our understanding of disease etiology and epidemiology through their use in studies into molecular evolution, host–pathogen interactions and modes of virulence. In the following section, we will review salient studies that have provided such fundamental insight, focusing on agents from the CDC Categories A to C.
One important application of microarrays is in the resequencing of pathogen genomes, which has been successfully applied to interrogate inter-species and/or inter-strain differences (Figure 2b). This provides a vital alternative to traditional shotgun sequencing approaches because it is quicker and more cost-effective; however, the disadvantage is that the sequence information for the organism of interest must available beforehand. This is not a severe limitation given that many pathogenic organisms have or are being sequenced, and with the advances in de novo sequencing capabilities, new pathogens such as SARS can be readily sequenced in a matter of weeks from the time of discovery [28]. Subsequent resequencing using microarrays would only take several days. Early experiments on resequencing arrays showed high rates of false discovery of up to 12–45% [29], but experimental improvements and the development of robust algorithms, such as the ABACUS (adaptive background genotype calling scheme) software package [30], have now greatly improved sequencing quality.
Adopting this approach for variation analysis, Wong and colleagues [31] tracked strains of the SARS coronavirus that were rapidly resequenced using high-density microarrays. A total of 383 102 probes (27-mers) were designed to cover the complete 29.7 kb genome on Nimblegen microarrays [31]. Virus samples from cell cultures and patient tissues could be amplified by reverse-transcriptase PCR using optimized primers and resequenced with accuracy greater than 99.99%. The platform was used to confirm that a lone case of a SARS virus infection of a graduate student had most likely come from a laboratory source because a unique 47-bp deletion was detected that was not present in other SARS isolates. Wang et al. [32] also developed a SARS microarray that helped to characterize a novel coronavirus variant. The microarray platform is thus ideal in virus tracking should outbreaks occur and, furthermore, is readily applicable to other pathogens with manageably sized genomes, including smallpox [33]. In an impressive demonstration that extended beyond the smaller viral genomes, Zwick et al. [29] developed resequencing microarrays for the anthrax bacterium; a total of 115 resequencing arrays, each capable of calling 29 212 bases, gave 92.6% coverage of the 3.3 Mb B. anthracis genome.
Evolutionary perspectives lend unique insight into the genetic changes that result in differences in pathogenicity. Plague-causing Y. pestis is estimated to have diverged from the non-lethal Yersinia pseudotuberculosis an estimated 1500–20 000 years ago [34]. Bearing identical 16S rRNA sequences, more comprehensive genetic analysis was required to understand the differences in disease morbidity manifested by these two bacterial species, as well as in the different modes of transmission to humans: Y. pestis by flea bites and Y. pseudotuberculosis by food or water. A microarray specific for Y. pestis revealed three unique genomic islands and the inactivation of several genes, including those encoding the cell-adhesion proteins adhesin (yadA) and invasion (inv), as well as the O-antigen biosynthetic operon. These unique characteristics potentially led to Y. pestis’ enhanced virulence and its adaptation from an enteric organism (Y. pseudotuberculosis) to a mammalian blood-borne pathogen [35]. Equivalent findings were reported by Hinchliffe et al. [36] using a similar Y. pestis gene-specific microarray. These also showed that, of the three major Y. pestis subspecies, the Antiqua and Mediaevalis biovars showed greater divergence from the Orientalis strains [36].
Similarly, variation analysis has been performed on Burkholderia pseudomallei microarrays to differentiate amongst virulent and avirulent Burkholderia species, specifically bioterror agents such as B. pseudomallei (which causes meliodosis), B. mallei (which causes equine glanders) and the avirulent (but closely related) B. thailandensis [37]. In an interesting approach to reveal molecular mechanisms for pathogenesis, Weiss and colleagues [38] developed a microarray-based negative-screening approach for Francisella and obtained surviving bacterial samples from infected mice to reveal genes responsible for bacterial survival and growth in vivo. In a related study, genome-wide microarrays also revealed loci that are only present in the highly virulent Franscicella tularensis subspecies tularensis strain [39]. These loci could be used to identify potential pathogenicity islands as well as to provide unique genetic markers that could be used for strain typing and identification. Microarrays have also been applied to distinguish among variants of influenza A that are resistant to the antiviral drug amantidine by detecting mutations in the viral ion channel M2 protein [40]. Such molecular forensics experiments using microarrays have extended our capabilities to profile new or emergent threats. The differences identified might provide an understanding of the causes of increased virulence, as well as reveal vulnerabilities that can be exploited for building therapeutic defences.
Operational use of DNA microarrays
Beyond laboratory-based applications, DNA microarrays have been tested successfully in epidemic outbreak surveillance (EOS). Project Silent Guardian was a 10-week trial launched by the Naval Research Laboratory during the 2005 US Presidential Inauguration [41]. The challenge was to take laboratory-based microarray technology to a production-scale system capable of operationally screening up to 300 samples per day. A custom DNA microarray was designed with the capability of detecting 20 natural (including avian flu) and biothreat agents with strain-level resolution. In total, 10 000 samples were collected and screened by civilian and military laboratory personnel within the stipulated period. The trial included blinded samples that were spiked with pathogens, which were all successfully detected. This exercise showcased the feasibility of implementing microarrays as a screening tool for EOS and demonstrated their use as a rapid and robust means of sample processing and pathogen identification. Recommendations from the study included the development of a more user-friendly system with greater automation of the steps involved. These features are being incorporated by several recently launched commercial platforms. Microarray-based platforms for biothreat screening from Akonni Biosystems (http://www.akonni.com) and Veredus Laboratories (http://www.vereduslabs.com) are now on the market for multiplexed threat detection. New advances include (i) greater integration of the sample preparation (which is considered to be one of the major bottlenecks), (ii) the use of μPCR and (iii) the development of small handheld microfluidic chips and portable readers to take microarray technology to the point-of-care. Veredus has also recently launched an influenza test chip that enables the amplification and discrimination of influenza A and B subtypes, including avian flu (H5N1), within just two hours. This great improvement in detection time has been attributed to the μPCR step, which, through rapid thermocycling, significantly reduces the duration of PCR to under 30 min. Similar devices might be developed in the future for protein and other types of non-DNA microarrays for applications in serodiagnostics.
The National Institute of Allergy and Infectious Diseases (NIAID) and the J. Craig Venter Institute fund a programme that enables researchers to obtain, through an approval process and material transfer agreement (MTA), pathogen DNA microarrays for select Category A–C threats at no cost to promote and accelerate research on these pathogens. The Pathogen Functional Genomics Resource Centre (PFGRC) currently fabricates and distributes a range of these 70-mer microarrays covering 39 agents, including B. anthracis, Coronaviruses, F. tularensis, Y. pestis and B. pseudomallei (http://pfgrc.tigr.org) (Table 1). Such collaborative and valuable initiatives augur favourably for the future of the biodefence community worldwide and will hopefully lead to greater sharing of resources in the pursuit of knowledge on global and regional pathogens.
Emerging roles of protein, peptide and carbohydrate microarrays
Newer microarray formats have already significantly extended the scope of microarray applications. The advantages of parallelization, miniaturization and automation hold true for whichever biomolecule is presented on high-density microarrays. It was initially difficult to believe that proteins would retain their functionality once anchored on a solid support. However, Schreiber and colleagues [42] erased this concern by developing the first small-molecule microarray in 1999 and the first protein microarray in 2000, simultaneously demonstrating that proteins could retain their activity despite being covalently attached to glass slides. Soon afterwards, other array types were developed, such as cell arrays, carbohydrate arrays and proteome arrays. Some of these newer applications have brought vital capabilities to pathogen detection and biodefence, as will be described below. As before, examples are provided for key biothreat agents and are categorized according to their consequent applications – either in detection or profiling.
Pathogen detection using non-DNA-based microarrays
Antibodies have become an established denominator for disease detection, through the time-honoured use of enzyme-linked immunosorbent assays (ELISAs), agglutination and lateral-flow assays. As a next-generation tool, antibody microarrays are increasing in popularity, and not without reason, for they offer unparalleled throughput, minimal reagent consumption and sensitive detection of multiple targets simultaneously [10]. The accuracy conferred is nevertheless closely linked with the quality of antibodies employed, and there are issues relating to cross-reactivity and antibody availability, both of which are particularly crucial for finer and more accurate strain typing. Notwithstanding these matters, a plethora of antibody microarrays have been developed that have growing potential for clinical, biothreat and point-of-care applications (Figure 2c). Rucker et al. [43] applied antibody microarrays for the detection of toxins with low-nanomolar sensitivities, such as anthrax lethal factor (LF, a metalloendopeptidase), protective antigen (PA) and tetanus toxins (endopeptidases), through the co-application of a competitive fluorescently-labelled toxin reporter. In this setting, the samples can be tested without the need for labelling by monitoring the depletion and competitive displacement of reporter signals generated by the labelled reporter. Thirty-five antibodies were also arrayed to subtype the 20 most common Salmonella serovars [44], and a similar study was undertaken for Escherichia coli [45]. In an impressive demonstration of the detection throughput attainable, Huelseweh and colleagues [46] built an antibody microarray to detect several Category A and B agents simultaneously, including F. tularensis, Y. pestis, Brucella melitensis (which causes brucellosis) and B. mallei, using sandwich immunoassays (Figure 2c).
Conversely, antigen microarrays have also been used to identify seropositive individuals by using the presence of defensive antibodies in the serum as a means of detecting exposure to a pathogen. Pathogen proteins are typically orthologously expressed using high-throughput cloning and expression and then affinity purified to provide protein sets. In special cases, it might be necessary to consider the post-translational modifications or the way the pathogen presents itself to the host to identify and use the correct immunogenic protein variants in serodiagnostics. It is also desirable to purify the protein and check for efficient and generally uniform immobilization on the microarrays as a quality control, especially when comparisons are made across slides. In one such example, Sundaresh et al. [47] narrowed down an initial panel of 1741 F. tularensis antigens to just the top 244 in terms of their ability to predictively discriminate seropositive sera and then used these antigens to establish a diagnostic microarray comprising whole proteins. Mezzasoma et al. [48] also developed protein microarrays for simultaneous diagnostics using parasitic and viral antigens. Zhu et al. [49] monitored the antibody profiles of SARS patients using protein microarrays containing 82 purified coronavirus proteins. Antibodies present in human serum samples were detected on the protein microarrays with fluorescently labelled goat anti-human immunoglobulins. It was found that immunoreactivity against the coronavirus nucleocapsid proteins remained high for 120–320 days post infection. This provided a means to check for exposure long after infection might have occurred. Over 400 human sera samples were screened with an accuracy of 91% in differentiating infected and normal specimens [49]. Nevertheless, caution should be applied in protein-based diagnostics because incubation periods vary greatly amongst diseases. During the incubation period, the patient might not express antibodies at detectable levels but might still be capable of spreading the infection. In the case of SARS, the incubation period (the time between exposure and onset of symptoms) has a median of 4–5 days (up to a maximum of 10 days). Antibodies might be detectable only 6–7 days after first symptoms are presented, but PCR testing might be able to pick up the virus more quickly, just 1–2 days after symptoms become apparent. It might become possible in the future to integrate DNA- and protein-based microarray methods to extend the range of rapid clinical diagnostics from detection of current pathogen infections to testing for exposure long after the actual infection has occurred.
As an alternative, short peptides have also been applied for serodiagnostics, and this was facilitated by the robust and routine procedures established to chemically synthesize peptide sequences up to 50 amino acids in length. Peptide microarrays have been applied for detecting immunoreactive sera against SARS, amongst other pathogens, enabling the detection of low-picomolar concentrations of antibodies [50]. Lipopolysaccharide, carbohydrate-based and whole-cell microarrays have also been used for antibody-based detection of pathogens such as F. tularensis 51, 52, B. anthracis [53] and B. pseudomallei [54]. The surface antigens on these microbes are frequently responsible for immunoreactivity, so these methods rely on detecting the presence of such immunogenic antigens. Further improvements in fabrication techniques and greater knowledge of surface chemistries might lead to the development of hybrid microarray platforms in which multiple types of antigens, such as peptides, proteins and carbohydrates, are presented simultaneously to detect host antibodies with improved efficiency and sensitivity.
Pathogen profiling using non-DNA-based microarrays
Protein microarrays with a high pathogen proteome content offer a valuable platform for high-throughput serology. Proteins that are immunoreactive to patient sera represent antigens that can be applied as markers in serodiagnostics or as therapeutic proteins for vaccine development. In one such example, the immunogenic epitopes of the SARS coronavirus were traced to the C-terminal fragments of the nucleocapsid protein by screening against 52 human sera samples from infected individuals (Figure 2d) 55, 56.
To address the bottleneck of protein expression, Davies and colleagues 57, 58 applied PCR recombinant cloning to accelerate the process of cloning and expression. This enabled more than 190 proteins from vaccinia virus and over 1700 proteins from F. tularensis to be expressed and profiled using microarrays 57, 58. These arrays have also helped to identify antibodies against the vaccinia virus H3L envelope protein, which confers protection in vivo (in a mouse model) [59]. The vaccinia protein microarrays have also been used to derive antibody profiles in humans inoculated with the licenced Dryvax® smallpox vaccine [60], and these profiles were found to be very similar to those induced by the attenuated modified vaccinia virus Ankara strain, an alternative vaccine candidate [61]. Various protein microarrays have similarly been generated for identification of the immunodominant antigens of Y. pestis 62, 63.
Other microarray platforms have demonstrated valuable potential in biodefence. For instance, small-molecule microarrays have been applied to functionally screen anthrax LF through activity-based binding signatures against a library of 1400 immobilized peptide-hydroxamate inhibitors. Putative drug candidates were discovered that bound selectively to the anthrax LF (with a low dissociation constant, K D = 0.81 μM) and inhibited its activity in vitro [64]. Carbohydrate microarrays have also been developed to elucidate the receptor preferences of influenza viruses. Stevens and colleagues 65, 66 applied glycan arrays to establish the α2-3 and α2-6 binding selectivity of the H5 and H1 haemagglutinins, which determine influenza virulence. The selectivity was altered by specific mutations in the protein, providing a useful way of functionally monitoring viral adaptation. Non-DNA microarray formats have thus contributed unique ways in which we can explore pathogen biology and develop countermeasures in the event of an outbreak.
Concluding remarks
It is clear that microarrays have significantly strengthened our biodefence capabilities. Although the upstream cost of library creation and microarray fabrication, as well as the initial infrastructure investment of several hundreds of thousands of dollars (for microarray spotters and scanners), might represent significant hurdles for small laboratories with tight budgets, the provision of printed microarray slides through commercial or research avenues heralds a favourable trend towards the reduction of exclusivity. Greater access to the technology, such as the increased availability of microarrays together with established array designs, is helping researchers worldwide to move more quickly towards the downstream application phase, for example to perform regional studies of endemic variants.
The applications of microarrays described here are broad-ranging and span from DNA- or protein-based detection of pathogens to epidemic outbreak surveillance and vaccine development, thus showcasing the full spectrum of microarray potential (Table 1). The progress made in the past several years has brought microarrays to the forefront of rapid diagnostics and medical research. Further integration of upstream sample preparation with downstream data processing is expected to transform microarrays into compact, field expedient solutions for analysis and monitoring in the near future. The large assortments of biomolecules now utilized in microarrays provide novel opportunities in molecular forensics and comparative profiling, especially for the newer and less well-understood pathogens. As we equip ourselves with better capabilities and countermeasures against potential biological threats, we hope to become more agile and responsive whenever such threats emerge in the future. Microarrays have improved our confidence in this respect and will continue to play a decisive part in biodefence research and technology.
Acknowledgements
The authors acknowledge funding support from DSO National Laboratories.
Glossary
- Biodefence
defensive measures against biological threats, including natural/emerging pathogens and bioterror agents, that have significant potential to endanger public health
- Detection
identifying the presence of target pathogen(s) from clinical or environmental samples.
- Diagnostics
tests used to detect a medical condition, for example to test for the causative pathogen responsible for an infection.
- Sandwich immunoassay
a biochemical assay for detecting the presence and/or abundance of a target substance using the antibody–antigen reaction. Two antibodies are used; the first is immobilized and the other, free antibody carries a reporter group, thereby providing a positive readout when the target is recognized and ‘sandwiched’ between the two antibodies.
- Sensitivity
probability of a positive result when the pathogen is indeed present; high sensitivity is related to a low Type 2 error.
- Serovars
a group of microorganisms distinguished by the presence of specific surface antigens.
- Specificity
probability of a negative result when the pathogen is not present; high specificity is related to a low Type 1 error.
- Tiling arrays
tiling arrays are a type of DNA microarray in which short probe segments that have been designed to cover the entire genome are used. The extent of probe overlap will translate to the mapping resolution and might range from highly overlapped probes across each individual nucleotide base (for resequencing applications) to non-overlapping probes (for genome-wide expression analysis).
- Vaccinia virus
a poxvirus that is closely related to the virus that causes cowpox. It was the first human vaccine and has been extensively used for vaccination against smallpox.
References
- 1.Chinese SARS Molecular Epidemiology Consortium Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science. 2004;303:1666–1669. doi: 10.1126/science.1092002. [DOI] [PubMed] [Google Scholar]
- 2.Demirev P.A. Chemical and biological weapons: current concepts for future defenses. Johns Hopkins APL Tech. Dig. 2005;26:321–333. [Google Scholar]
- 3.Trull M.C. Turning biodefense dollars into products. Nat. Biotechnol. 2007;25:179–184. doi: 10.1038/nbt0207-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lim D.V. Current and developing technologies for monitoring agents of bioterrorism and biowarfare. Clin. Microbiol. Rev. 2005;18:583–607. doi: 10.1128/CMR.18.4.583-607.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Venkatasubbarao S. Microarrays – status and prospects. Trends Biotechnol. 2004;22:630–637. doi: 10.1016/j.tibtech.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 6.Dorrell N. Comparative phylogenomics of pathogenic bacteria by microarray analysis. Curr. Opin. Microbiol. 2005;8:620–626. doi: 10.1016/j.mib.2005.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ivnitski D. Nucleic acid approaches for detection and identification of biological warfare and infectious disease agents. Biotechniques. 2003;35:862–869. doi: 10.2144/03354ss03. [DOI] [PubMed] [Google Scholar]
- 8.Stenger D.A. Potential applications of DNA microarrays in biodefense-related diagnostics. Curr. Opin. Biotechnol. 2002;13:208–212. doi: 10.1016/s0958-1669(02)00321-x. [DOI] [PubMed] [Google Scholar]
- 9.Bodrossy L., Sessitsch A. Oligonucleotide microarrays in microbial diagnostics. Curr. Opin. Microbiol. 2004;7:245–254. doi: 10.1016/j.mib.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 10.Angenendt P. Progress in protein and antibody microarray technology. Drug Discov. Today. 2005;10:503–511. doi: 10.1016/S1359-6446(05)03392-1. [DOI] [PubMed] [Google Scholar]
- 11.Bertone P., Snyder M. Advances in functional protein microarray technology. FEBS J. 2005;272:5400–5411. doi: 10.1111/j.1742-4658.2005.04970.x. [DOI] [PubMed] [Google Scholar]
- 12.Uttamchandani M. Small molecule microarrays: recent advances and applications. Curr. Opin. Chem. Biol. 2005;9:4–13. doi: 10.1016/j.cbpa.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 13.Uttamchandani M., Yao S.Q. Peptide microarrays: Next generation biochips for detection, diagnostics and high-throughput screening. Curr. Pharm. Des. 2008;14:2428–2438. doi: 10.2174/138161208785777450. [DOI] [PubMed] [Google Scholar]
- 14.Maynard J.A. Microarrays in infection and immunity. Curr. Opin. Chem. Biol. 2007;11:306–315. doi: 10.1016/j.cbpa.2007.01.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loy A., Bodrossy L. Highly parallel microbial diagnostics using oligonucleotide microarrays. Clin. Chim. Acta. 2006;363:106–119. doi: 10.1016/j.cccn.2005.05.041. [DOI] [PubMed] [Google Scholar]
- 16.Yadav P., Blaine L. Microbiological threats to homeland security. IEEE Eng. Med. Biol. Mag. 2004;23:136–141. doi: 10.1109/memb.2004.1297185. [DOI] [PubMed] [Google Scholar]
- 17.Wilson W.J. Sequence-specific identification of 18 pathogenic microorganisms using microarray technology. Mol. Cell. Probes. 2002;16:119–127. doi: 10.1006/mcpr.2001.0397. [DOI] [PubMed] [Google Scholar]
- 18.Wang D. Microarray-based detection and genotyping of viral pathogens. Proc. Natl. Acad. Sci. U. S. A. 2002;99:15687–15692. doi: 10.1073/pnas.242579699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sengupta S. Molecular detection and identification of influenza viruses by oligonucleotide microarray hybridization. J. Clin. Microbiol. 2003;41:4542–4550. doi: 10.1128/JCM.41.10.4542-4550.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin B. Broad-spectrum respiratory tract pathogen identification using resequencing DNA microarrays. Genome Res. 2006;16:527–535. doi: 10.1101/gr.4337206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lodes M.J. Identification of upper respiratory tract pathogens using electrochemical detection on an oligonucleotide microarray. PLoS One. 2007;2:e924. doi: 10.1371/journal.pone.0000924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hashsham S.A. Potential of DNA microarrays for developing parallel detection tools (PDTs) for microorganisms relevant to biodefense and related research needs. Biosens. Bioelectron. 2004;20:668–683. doi: 10.1016/j.bios.2004.06.032. [DOI] [PubMed] [Google Scholar]
- 23.Desantis T.Z. Rapid quantification and taxonomic classification of environmental DNA from both prokaryotic and eukaryotic origins using a microarray. FEMS Microbiol. Lett. 2005;245:271–278. doi: 10.1016/j.femsle.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 24.Couzinet S. High-density DNA probe arrays for identification of staphylococci to the species level. J. Microbiol. Methods. 2005;61:201–208. doi: 10.1016/j.mimet.2004.11.019. [DOI] [PubMed] [Google Scholar]
- 25.Wong C.W. Optimization and clinical validation of a pathogen detection microarray. Genome Biol. 2007;8:R93. doi: 10.1186/gb-2007-8-5-r93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sergeev N. Multipathogen oligonucleotide microarray for environmental and biodefense applications. Biosens. Bioelectron. 2004;20:684–698. doi: 10.1016/j.bios.2004.04.030. [DOI] [PubMed] [Google Scholar]
- 27.Tomioka K. A multiplex polymerase chain reaction microarray assay to detect bioterror pathogens in blood. J. Mol. Diagn. 2005;7:486–494. doi: 10.1016/S1525-1578(10)60579-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marra M.A. The genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 29.Zwick M.E. Microarray-based resequencing of multiple Bacillus anthracus isolates. Genome Biol. 2004;6:R10. doi: 10.1186/gb-2004-6-1-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cutler D.J. High-throughput variation detection and genotyping using microarrays. Genome Res. 2001;11:1913–1925. doi: 10.1101/gr.197201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wong C.W. Tracking the evolution of the SARS coronavirus using high-throughput, high-density resequencing arrays. Genome Res. 2004;14:398–405. doi: 10.1101/gr.2141004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang D. Viral discovery and sequence recovery using DNA microarrays. PLoS Biol. 2003;1:e2. doi: 10.1371/journal.pbio.0000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sulaiman I.M. GeneChip resequencing of the smallpox virus genome can identify novel strains: a biodefense application. J. Clin. Microbiol. 2007;45:358–363. doi: 10.1128/JCM.01848-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Achtman M. Microevolution and history of the plague bacillus, Yersinia pestis. Proc. Natl. Acad. Sci. U. S. A. 2004;101:17837–17842. doi: 10.1073/pnas.0408026101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou D. DNA microarray analysis of genome dynamics in Yersinia pestis: insights into bacterial genome microevolution and niche adaptation. J. Bacteriol. 2004;186:5138–5146. doi: 10.1128/JB.186.15.5138-5146.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hinchliffe S.J. Application of DNA microarrays to study the evolutionary genomics of Yersinia pestis and Yersinia pseudotuberculosis. Genome Res. 2003;13:2018–2029. doi: 10.1101/gr.1507303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ong C. Patterns of large-scale genomic variation in virulent and avirulent Burkholderia species. Genome Res. 2004;14:2295–2307. doi: 10.1101/gr.1608904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weiss D.S. In vivo negative selection screen identifies genes required for Francisella virulence. Proc. Natl. Acad. Sci. U. S. A. 2007;104:6037–6042. doi: 10.1073/pnas.0609675104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Broekhuijsen M. Genome-wide DNA microarray analysis of Francisella tularensis strains demonstrates extensive genetic conservation within the species but identifies regions that are unique to the highly virulent F. tularensis subsp. tularensis. J. Clin. Microbiol. 2003;41:2924–2931. doi: 10.1128/JCM.41.7.2924-2931.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Townsend M.B. Detection of adamantane-resistant influenza on a microarray. J. Clin. Virol. 2008;42:117–123. doi: 10.1016/j.jcv.2007.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stenger, D. (2006). NRL uses microarray technology to detect natural and bio-threat pathogens during Silent Guardian project. 106th American Society for Microbiology General Meeting, 2006 May 21–25, Orlando, Session 246/Y, Paper Y-018 (http://www.asm.org/Media/index.asp?bid=42925)
- 42.MacBeath G., Schreiber S.L. Printing proteins as microarrays for high-throughput function determination. Science. 2000;289:1760–1763. doi: 10.1126/science.289.5485.1760. [DOI] [PubMed] [Google Scholar]
- 43.Rucker V.C. Antibody microarrays for native toxin detection. Anal. Biochem. 2005;339:262–270. doi: 10.1016/j.ab.2005.01.030. [DOI] [PubMed] [Google Scholar]
- 44.Cai H.Y. Development of a novel protein microarray method for serotyping Salmonella enterica strains. J. Clin. Microbiol. 2005;43:3427–3430. doi: 10.1128/JCM.43.7.3427-3430.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anjum M.F. Use of miniaturized protein arrays for Escherichia coli O serotyping. Clin. Vaccine Immunol. 2006;13:561–567. doi: 10.1128/CVI.13.5.561-567.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huelseweh B. A simple and rapid protein array based method for the simultaneous detection of biowarfare agents. Proteomics. 2006;6:2972–2981. doi: 10.1002/pmic.200500721. [DOI] [PubMed] [Google Scholar]
- 47.Sundaresh S. From protein microarrays to diagnostic antigen discovery: a study of the pathogen Francisella tularensis. Bioinformatics. 2007;23:i508–i518. doi: 10.1093/bioinformatics/btm207. [DOI] [PubMed] [Google Scholar]
- 48.Mezzasoma L. Antigen microarrays for serodiagnosis of infectious diseases. Clin. Chem. 2002;48:121–130. [PubMed] [Google Scholar]
- 49.Zhu H. Severe acute respiratory syndrome diagnostics using a coronavirus protein microarray. Proc. Natl. Acad. Sci. U. S. A. 2006;103:4011–4016. doi: 10.1073/pnas.0510921103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Andresen H. Functional peptide microarrays for specific and sensitive antibody diagnostics. Proteomics. 2006;6:1376–1384. doi: 10.1002/pmic.200500343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thirumalapura N.R. Lipopolysaccharide microarrays for the detection of antibodies. J. Immunol. Methods. 2005;298:73–81. doi: 10.1016/j.jim.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 52.Thirumalapura N.R. Bacterial cell microarrays for the detection and characterization of antibodies against surface antigens. J. Immunol. Methods. 2006;309:48–54. doi: 10.1016/j.jim.2005.11.016. [DOI] [PubMed] [Google Scholar]
- 53.Wang D. Photogenerated glycan arrays identify immunogenic sugar moieties of Bacillus anthracis exosporium. Proteomics. 2007;7:180–184. doi: 10.1002/pmic.200600478. [DOI] [PubMed] [Google Scholar]
- 54.Parthasarathy N. Polysaccharide microarray technology for the detection of Burkholderia pseudomallei and Burkholderia mallei antibodies. Diagn. Microbiol. Infect. Dis. 2006;56:329–332. doi: 10.1016/j.diagmicrobio.2006.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen Z. Antigenicity analysis of different regions of the severe acute respiratory syndrome coronavirus nucleocapsid protein. Clin. Chem. 2004;50:988–995. doi: 10.1373/clinchem.2004.031096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lu D.D. Screening of specific antigens for SARS clinical diagnosis using a protein microarray. Analyst. 2005;130:474–482. doi: 10.1039/b415888a. [DOI] [PubMed] [Google Scholar]
- 57.Eyles J.E. Immunodominant Francisella tularensis antigens identified using proteome microarray. Proteomics. 2007;7:2172–2183. doi: 10.1002/pmic.200600985. [DOI] [PubMed] [Google Scholar]
- 58.Davies D.H. Profiling the humoral immune response to infection by using proteome microarrays: high-throughput vaccine and diagnostic antigen discovery. Proc. Natl. Acad. Sci. U. S. A. 2005;102:547–552. doi: 10.1073/pnas.0408782102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Davies D.H. Vaccinia virus H3L envelope protein is a major target of neutralizing antibodies in humans and elicits protection against lethal challenge in mice. J. Virol. 2005;79:11724–11733. doi: 10.1128/JVI.79.18.11724-11733.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Davies D.H. Proteome-wide analysis of the serological response to vaccinia and smallpox. Proteomics. 2007;7:1678–1686. doi: 10.1002/pmic.200600926. [DOI] [PubMed] [Google Scholar]
- 61.Davies D.H. Antibody profiling by proteome microarray reveals the immunogenicity of the attenuated smallpox vaccine modified vaccinia virus ankara is comparable to that of Dryvax. J. Virol. 2008;82:652–663. doi: 10.1128/JVI.01706-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li B. Protein microarray for profiling antibody responses to Yersinia pestis live vaccine. Infect. Immun. 2005;73:3734–3739. doi: 10.1128/IAI.73.6.3734-3739.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen Z. Quorum sensing affects virulence-associated proteins F1, LcrV, KatY and pH6 etc. of Yersinia pestis as revealed by protein microarray-based antibody profiling. Microbes Infect. 2006;8:2501–2508. doi: 10.1016/j.micinf.2006.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Uttamchandani M. Quantitative inhibitor fingerprinting of metalloproteases using small molecule microarrays. J. Am. Chem. Soc. 2007;129:13110–13117. doi: 10.1021/ja073914v. [DOI] [PubMed] [Google Scholar]
- 65.Stevens J. Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science. 2006;312:404–410. doi: 10.1126/science.1124513. [DOI] [PubMed] [Google Scholar]
- 66.Stevens J. Glycan microarray technologies: tools to survey host specificity of influenza viruses. Nat. Rev. Microbiol. 2006;4:857–864. doi: 10.1038/nrmicro1530. [DOI] [PMC free article] [PubMed] [Google Scholar]


