Abstract
The heptad repeats (HR1 and HR2) of the spike protein of SARS-CoV are highly conserved regions forming a critical 6-helix bundle during the fusion step of virus entry and are attractive targets of entry inhibitors. In this study, we report that a minimal HR2 peptide, P6 of 23-mer, can block the fusion of SARS-CoV with an IC50 of 1.04 ± 0.22 μM. This finding supports the structural prediction of the deep groove of HR1 trimer as a target for fusion inhibitors, and suggests P6 as a potential lead peptide for future drug development. Moreover, combination of an HR-1 peptide, N46, and its mutated version, N46eg, shows synergistic inhibition with an IC50 of 1.39 ± 0.05 μM and combination index of 0.75 ± 0.15, suggesting a common strategy to achieve promising inhibition by HR1 peptide for other class I envelope viruses.
Keywords: SARS-CoV, Spike, Heptad repeats 1 and 2, Peptide, Inhibitor
Severe acute respiratory syndrome (SARS), a life-threatening disease that has spread to more than 30 countries in 2003, is caused by the SARS coronavirus (SARS-CoV) (Drosten et al., 2003, Ksiazek et al., 2003, Peiris et al., 2004). Despite the end to this epidemic, concerns regarding the possibility of resurgence remain. Since no specific and effective anti-SARS-CoV drug is available, identification of potential targets and development of antiviral agents are critical for the effective treatment and control of the disease (Cinatl et al., 2005, Yeung et al., 2006).
Among different steps of the life cycle of SARS-CoV, viral entry is an attractive target (Moore and Doms, 2003). The entry of enveloped RNA viruses involves the fusion between plasma membrane and viral membrane, which contains the surface envelope glycoprotein and transmembrane fusion protein. Based on the structural and functional similarities, fusion proteins can be grouped into two classes, class I and class II (Eckert and Kim, 2001, Kielian and Rey, 2006, Weissenhorn et al., 1999). Class I enveloped viruses, such as influenza virus, human immunodeficiency virus type 1 (HIV-1), Ebola virus, measles virus and murine hepatitis virus (MHV), utilize a similar and well-studied mechanism of fusion, in which the interaction between two highly conserved heptad repeats (HR1 and HR2 at the N and C-termini, respectively) of the fusion protein results in the formation of a 6-helix bundle structure and fusion of viral and plasma membranes (Eckert and Kim, 2001, Weissenhorn et al., 1999). Presumably through binding to the exposed HR1 in the pre-hairpin intermediate, several HR2 peptides have been reported as effective entry inhibitors, including DP107 and T20 of HIV-1, GP160 of Ebola virus and HR2 of MHV, and developed into drugs, such as enfuvirtide (T-20) (Bosch et al., 2003, Jiang et al., 1993, Kilby et al., 1998, Watanabe et al., 2000, Wild et al., 1994). Recently, combination of HR1-derived peptides of HIV-1 fusion protein, gp41, was reported to show synergistic inhibition and potential therapeutic implications (Gustchina et al., 2006).
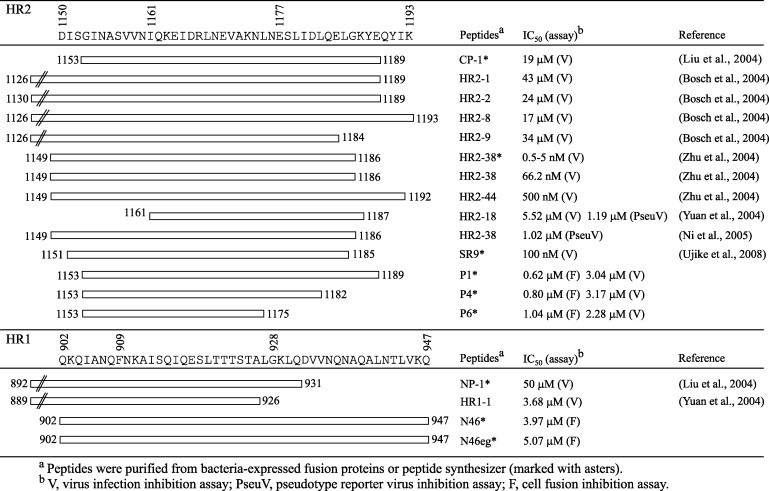
The spike (S) protein of SARS-CoV contains two subunits: S1 and S2, which is a class I fusion protein (Fig. 1A) (Cinatl et al., 2005, Yeung et al., 2006). After binding of S1 subunit to its receptor, angiotensin converting enzyme 2 (ACE2), SARS-CoV enters cells through either the endosomal pathway followed by fusion with endosomal membrane, or direct fusion with cell membrane in the presence of protease (Li et al., 2003, Simmons et al., 2005, Ujike et al., 2008). Several HR1 and HR2 peptides of S2 subunit, such as CP-1, HR2–8, HR2–18, HR2–38 and HR1–1, have been reported to have inhibitory effects with IC50 less than 19 μM (Bosch et al., 2004, Liu et al., 2004, Ni et al., 2005, Ujike et al., 2008, Yuan et al., 2004, Zhu et al., 2004) (Table 1 ). However, most of them were larger than 37-mer in length and the plausible synergistic inhibition on the fusion of SARS-CoV has not been investigated.
Fig. 1.
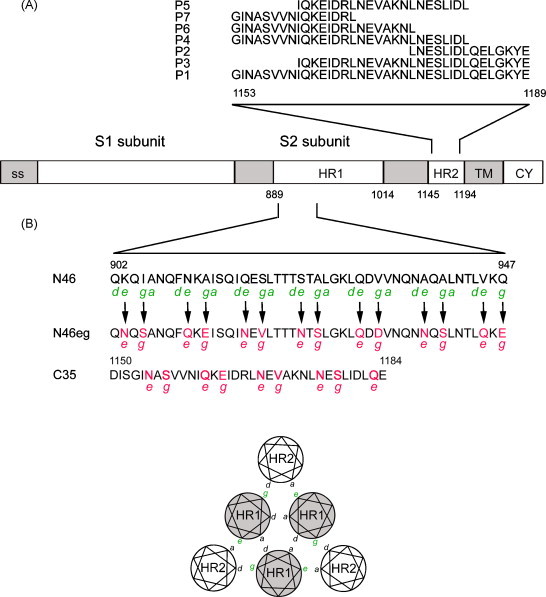
Schematic drawing of S protein of SARS-CoV and the designed peptides. (A) The S2 subunit contains two heptad repeats (HR1, HR2). The amino acid sequences of peptides (P1–P7) in HR2 are shown. ss, signal sequence. TM, transmembrane domain. CY, cytoplasmic tail. The numbers of amino acid residues are indicated. (B) The amino acid sequences of the HR1 peptide (N46) and its derivative (N46eg), designed based on residues at the e and g positions of the interacting HR2 peptide (C35), are shown (Xu et al., 2004). The helical wheel representation of three HR1 and three HR2 helices in the 6-helix bundle is shown.
Table 1.
Summary of inhibitory peptides derived from HR1 and HR2 of the S protein of SARS-CoV.
To explore the possibility of minimizing the length of HR2 and HR1 inhibitory peptides, we first examine one of the smallest core structures of S protein resolved by X-ray crystallography, which consists of three HR1 (N46) and three HR2 (C35) helices (Xu et al., 2004). In the HR2 region, we design 6 shortened peptides based on CP-1, the first reported HR2 inhibitory peptide of 37-mer, which overlaps largely with C35 (Liu et al., 2004). The sequences of CP-1, designated as P1, and its truncated versions at the N-terminus (P3, P2), C-terminus (P4, P6, P7) and both ends (P5) are shown in Fig. 1A.
Our cell fusion inhibition assay is modified from a previously described β-galactosidase reporter gene-based SARS-CoV cell fusion assay (Xiao et al., 2003, Huentelman et al., 2004). VRC(S)8304, a plasmid containing the S gene of SARS-CoV with humanized codon usage, is designated as S(h) in this study (Huang et al., 2004). To construct pCDNA3HA-ACE2, total RNA extracted from VeroE6 cells by the Qiagen RNeasy mini kit (Qiagen, Valencia, CA) was subjected to RT using random primers, followed by PCR using the primer pair (NotI-ACE2, 5′-AAGGAAAAAAGCGGCCGCCGATGTCAAGCTCTTCC-3′ and XhoI-ACE2, 5′-CCCGCTC GAGCTAAAAGGAGGTCTGAAC-3′), digestion of the product with NotI and XhoI, and cloning into respective sites of pCDNA3-HA (Invitrogen, Carlsbad, CA). The construct was confirmed by sequencing both junctions of the insert with T7 and SP6 primers. HeLa cells (1 × 106) transfected with 2 μg of S(h) (cells #1) by use of lipofectamine 2000 (Invitrogen, Carsblad, CA) were infected with recombinant vaccinia virus expressing β-galactosidase (vCB21R) at a multiplicity of infection (MOI) of 1 at 6 h post-transfection. HeLa cells (1 × 106) transfected with 2 μg of pCDNA3HA-ACE2 (cells #2) were infected with recombinant vaccinia virus expressing T7 polymerase (vTF7.3) at a MOI of 1. After adsorption at 37 °C for 2 h and replacement with fresh medium, both cells were incubated at 28 °C overnight, washed with 1× PBS once, trypsinized, washed with 1× PBS twice, and resuspended in DMEM containing 10% FCS. Both cells #1 and #2 (1 × 105 cells, each well) were added into 96-well in duplicates, pre-incubated with or without different concentrations of peptides (50 μl per well) at 37 °C for 20 min, and co-cultured at 37 °C for 3 h. Cell lysates were measured for β-galactosidase activity using the Galcto-Start kit (Applied Biosystems, Bedford, MA). The percentage of fusion is the ratio of β-galactosidase activity in the presence of peptide to that in the absence of peptide.
As shown in Fig. 2A and B, co-culture of the HeLa cells expressing S protein and β-galactosidase (cells #1) and those expressing ACE2 and T7 polymerase (cells #2) results in syncytia formation and a rise in β-galactosidase activity (63,987 ± 1198 RLu/s). In contrast, different controls of cells #1 and cells #2 with or without transfection and/or infection had very low signals, demonstrating the specificity of the cell fusion assay. Initial testing of the 7 peptides at high concentrations (25 and 9 μM) reveals that P1 and 2 truncated peptides (P4, P6) have inhibitory effects (Fig. 2C). The inhibitory activities of P1, P4 and P6 were further tested at lower concentrations, and their similar dose–response curves are shown in Fig. 2D. Compared with the IC50 of P1, 0.62 ± 0.20 μM, the IC50 of P4 is 0.80 ± 0.21 μM and that of P6 is 1.04 ± 0.22 μM, suggesting that truncation of 14 residues from the C-terminus of P1 still has good inhibitory activity (Fig. 1A). In contrast, P7 and P5, which have further truncation of P6 at the C-terminus and of P4 at the N-terminus, respectively, have mild or no inhibitory effect at 25 and 9 μM (Fig. 2C). There findings suggest that P6, a 23-mer peptide, is the minimal inhibitory peptide in this region.
Fig. 2.
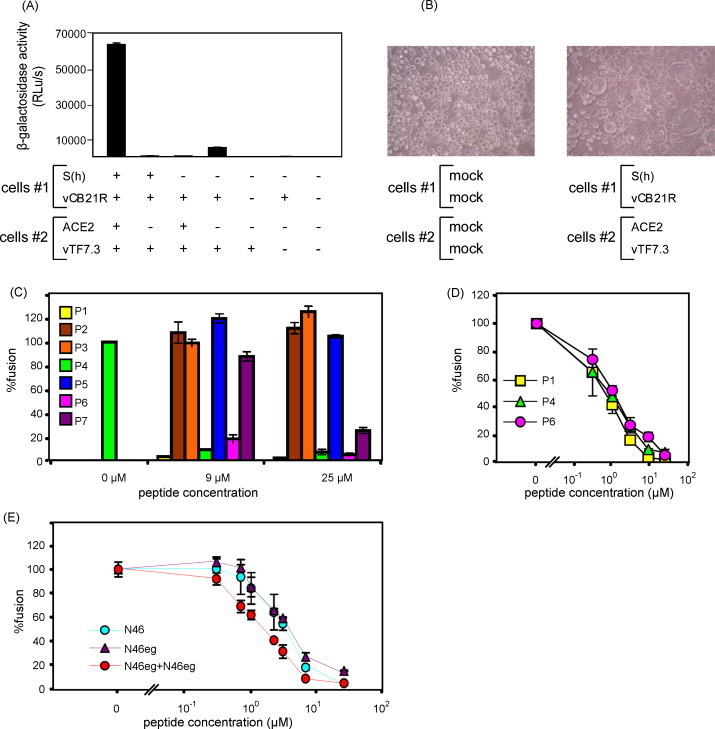
Inhibition of the S protein-mediated cell fusion by HR2 and HR1 peptides. (A) The cell fusion assay and (B) syncytia formation. β-Galactosidase activities were measured for co-cultured HeLa cells #1, cells #2 and various controls with (+) or without (−) transfection and/or infection. Syncytia formation was observed under light microscope. (C) Inhibitory effects of HR2 peptides (P1–P7) at high concentrations (25 and 9 μM). (D, E) Inhibitory effects of HR2 peptides (P1, P4 and P6), HR1 peptides (N46, N46eg) and their combination (N46 plus N46eg, molar ratio 1:1) at various concentrations. Data are means ± standard errors of duplicate wells. One representative experiment of more than three is shown.
In the HR1 region, a dose–response curve is also found for N46 with an IC50 of 3.97 ± 1.40 μM. Since the IC50 of N46 is 6 fold higher than that of P1, N46 is not further shortened (Fig. 2E). Instead, we designed another peptide, N46eg, based on the predicted interaction between residues at the e and g positions of HR1 with those at the a and d positions of HR2, respectively, in the 6-helix bundle structure (Fig. 1B). The residues at the e and g positions of N46 are replaced by those at the e and g positions of C35, respectively, to obtain peptide N46eg. Similar to that of N46, the IC50 of N46eg is 5.07 ± 0.17 μM (Fig. 2E). We next examined whether combination of N46 and N46eg has a synergistic effect. Compared with the dose–response curve of N46 or N46eg alone, a shift of the curve to left is found for combination of N46 and N46eg (Fig. 2E). The IC50 of N46 plus N46eg at the molar ratio of 1:1 is 1.39 ± 0.05 μM, which is lower than that of either N46 or N46eg alone. The IC50 is also lower than that of 46 plus N46eg at a molar ratio of 1:5 or 1:10 (data not shown). The dose reduction indices of N46 and N46eg (molar ratio 1:1) are 2.91 and 3.55, respectively (Chou and Talalay, 1984). The combination index is 0.75 ± 0.15, suggesting moderate synergism (Chou, 1991).
In this study, we report that P6 as well as combination of N46 and N46eg can efficiently block the fusion of SARS-CoV. The inhibitory effects of these peptides are unlikely due to their cytotoxicity, since examining these peptides by a previously described MTT assay reveals no discernable cytotoxicity on HeLa or VeroE6 cells at the highest concentrations (25 μM) tested (data not shown) (Lai et al., 2008). While different HR1 and HR2 peptides of several class I envelope viruses have been reported as entry inhibitors (Bosch et al., 2003, Jiang et al., 1993, Kilby et al., 1998, Watanabe et al., 2000, Wild et al., 1994), the 23-mer peptide of P6, to our knowledge, is the smallest inhibitory peptide for class I envelope viruses. To examine the biophysical properties of P6 and its interaction with HR1 peptide, circular-dichroism (CD) spectra were analyzed on a spectropolarimeter (model J-720, Jasco Inc., Japan) in the range from 200 to 260 nm at 25 °C with a bandwidth of 1.0 nm, resolution of 0.1 nm, response time of 4.0 s and scanning speed of 100 nm/min. Each spectrum was an average of 4 scans of peptide at 20 μM under identical conditions and was analyzed by the program J7STDANL (Jasco Inc., Japan). After mixing P6 with N46 in equimolar concentrations, P6/N46 complex shows a CD spectrum of double minima with a clear negative peak at 208 nm and small negative peak at 220 nm (Fig. 3B), which is similar to that of the P1/N46 complex (Fig. 3A), suggesting that both P6 and P1 interact with N46 and form an α-helical conformation. In contrast, mixing N46eg with P1 or P6 in equimolar concentrations reveals CD spectra as predicted by two peptides together without interaction (Fig. 3C).
Fig. 3.
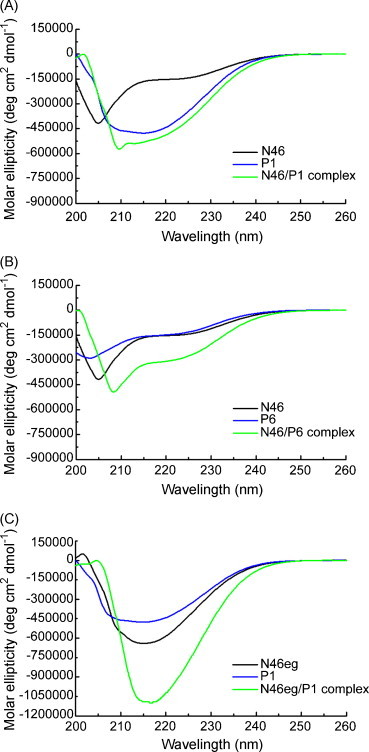
Secondary structure of HR1, HR2 and HR1/HR2 complex by CD analysis. CD spectra of (A) P1, N46 and P1/N46 complex (B) P6, N46 and P6/N46 complex and (C) P1, N46eg and P1/N46eg complex in equimolar concentrations were analyzed at 25 °C by a spectropolarimeter.
Our simple and convenient cell fusion inhibition assay can be used to screen and identify entry inhibitors of SARS-CoV. The requirement of trypsin treatment in this assay suggests that it mimics the entry from cell surface by direct fusion between viral and cell membranes (Simmons et al., 2005, Ujike et al., 2008). Using a similar assay, Huentelman et al. have recently reported a novel ACE2 inhibitor, N-(2-aminoethyl)-1 aziridine-ethanamine, as fusion inhibitor for SARS-CoV (Huentelman et al., 2004). To examine the inhibitory effect of the peptides identified by the cell fusion inhibition assay on SARS-CoV, we carried out a previously described SARS-CoV infectivity assay for three HR1 peptides at the BSL-3 facility (Hsueh et al., 2003, Hsueh et al., 2004). Briefly, VeroE6 cells (1 × 104 cells, each well) prepared in 96-well were infected with 75 TCID50 of SARS-CoV TW01 strain, which were pre-incubated with or without different concentration of peptides in quadricates at 37 °C for 1 h. After incubation at 37 °C for 72 h, cytopathic effects were observed under light microscope, and the IC50 was determined (Welkos and O’Brien, 1994). As shown in Table 1, the IC50 of P1, P4 and P6 are 3.04 ± 0.06 μM, 3.17 ± 0.24 μM and 2.28 ± 0.81 μM, respectively, verifying their inhibitory effect on SARS-CoV.
X-ray crystallographic studies of fusion core structures of S protein have shown that the N-terminus (residue 1150–1160) and C-terminus (residue 1178 and beyond) of HR2 formed two extended conformations and that a 5-turn α-helix at the center (residues 1160–1177) packed against a relatively deep groove (residues 909–928) of HR1 trimer, which was highly conserved and predicted as a potential target for fusion inhibitor (Duquerroy et al., 2005, Supekar et al., 2004, Xu et al., 2004). Consistent with this, strong inhibitory activity retains in HR2 peptides with C-terminal truncation up to residue 1175 (P4, P6) but not in those beyond residue 1175 (P7) (Fig. 1, Fig. 2). Moreover, the inhibitory effect is completely lost in the HR2 peptides with N-terminal truncation (P2, P3) but not in those with C-terminal truncation (P4, P6), suggesting that the extended region of HR2 at the N-terminus is more important than that at the C-terminus.
Table 1 summarizes different HR1 and HR2 inhibitory peptides for SARS-CoV in this and previous studies (Bosch et al., 2004, Liu et al., 2004, Ni et al., 2005, Ujike et al., 2008, Yuan et al., 2004, Zhu et al., 2004). For HR2 peptides, the IC50 ranged from 0.5 nM to 43 μM depending not only on the location and length but also on the type of the peptides (synthetic or bacteria-expressed) and assay used. Using the same assay, the inhibitory activity of HR2-18 was totally lost by adding 3 residues at its N-terminus (HR2-17) (Yuan et al., 2004). The IC50 of the synthetic HR2-38 (0.5–5 nM) was lower that that of the bacteria-expressed HR2-38 (66.2 nM) (Zhu et al., 2004). Using different assays, the IC50 of HR2-18 determined by the infectivity assay (5.52 μM) was higher than that by the pseudotype reporter virus assay (1.19 μM) (Yuan et al., 2004). Moreover, the IC50 of P1 (0.62 μM and 3.04 μM) in our study was slightly higher than the IC50 of HR2-38 (0.5 nM to 1.02 μM), a peptide of similar length and location, but was lower than that of CP-1 (19 μM) in another study (Liu et al., 2004, Ni et al., 2005, Zhu et al., 2004). Further studies using the same assay for different peptides are warranted to reconcile these discrepancies. Nontheless, all the inhibitory HR2 peptides reported thus far cover a common region defined by residues 1161 and 1175, corresponding to the central helix of HR2 (Supekar et al., 2004, Xu et al., 2004) (Table 1). Similarly, all the inhibitory HR1 peptides cover a region (residues 902–926) corresponding to the deep groove of HR1. These findings support the prediction that the deep groove in HR1 and the interacting central helix of HR2 are important targets for fusion inhibitors (Duquerroy et al., 2005, Supekar et al., 2004, Xu et al., 2004). In this regard, the minimal HR2 inhibitory peptide, P6, which covers the central helix and part of the N-terminal extended region, could be a potential lead peptide for future drug development.
Comparing the inhibitory effects between HR1 and HR2 peptides of HIV-1, MHV, Ebola virus and SARS-CoV in most studies, HR1 peptides were weaker than HR2 peptides (Wild et al., 1994, Watanabe et al., 2000, Bosch et al., 2003, Bosch et al., 2004, Liu et al., 2004, Zhu et al., 2004). Using a strategy employed in HIV-1, we design a novel HR1-derived peptide, N46eg, which has a mean IC50 of 5.07 μM. Interestingly, combination of N46 and N46eg shows synergistic inhibition with a mean IC50 of 1.39 μM. In the case of HIV-1, the mechanism of synergistic inhibition by the HR1 homologue, NCCG-gp41, and the mutated HR1 peptide, N36Mut(e,g), was attributed to different targets of the two inhibitors; the former interacts with HR2 and the latter interacts with HR1 in the inner core through its intact residues at the a and d positions but not with HR2 because of its mutated residues at the e and g positions (Bewley et al., 2002, Gustchina et al., 2006). It is conceivable that similar scenario may occur in SARS-CoV and account for the synergistic inhibition by N46 and N46eg, though the exact mechanism remains to be investigated. Nonetheless, our findings of synergistic inhibition between an HR1 peptide and a mutated HR1 peptide, the second such observation, suggest that this can be a common strategy of achieving good inhibition for other class I envelope viruses, especially in the situation of fighting against an emerging and life-threatening infectious disease.
Acknowledgments
We thank Drs. Gary J. Nabel and Edward A. Berger at the National Institutes of Health, Bethesda, Maryland for kindly providing VRC(S)8304 and vTF7-3/vCB21R, respectively. This work was supported by the National Science Council Taiwan (NSC96-2321-B-002-027).
References
- Bewley C.A., Louis J.M., Ghirlando R., Clore G.M. Design of a novel peptide inhibitor of HIV fusion that disrupts the internal trimeric coiled coil of gp41. J. Biol. Chem. 2002;277:14238–14245. doi: 10.1074/jbc.M201453200. [DOI] [PubMed] [Google Scholar]
- Bosch B.J., Martina B.E., Van Der Z.R., Lepault J., Haijema B.J., Versluis C., Heck A.J., de Groot R., Osterhaus A.D., Rottier P.J. Severe acute respiratory syndrome coronavirus (SARS-CoV) infection inhibition using spike protein heptad repeat-derived peptides. Proc. Natl. Acad. Sci. U.S.A. 2004;101:8455–8460. doi: 10.1073/pnas.0400576101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch B.J., van der Z.R., de Haan C.A., Rottier P.J. The coronavirus spike protein is a class I virus fusion prote: structural and functional characterization of the fusion core complex. J. Virol. 2003;77:8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou T.C., Talalay P. Quantitative analysis of dose–effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv. Enzyme Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- Chou T.C. The median-effect principle and the combination index for quantitation of synergism and antagonism. In: Chou T.C., Rideout D.C., editors. Synergism and Antagonism in Chemistry. Academic Press; San Diego: 1991. pp. 61–102. [Google Scholar]
- Cinatl J., Jr., Michaelis M., Hoever G., Preiser W., Doerr H.W. Development of antiviral therapy for severe acute respiratory syndrome. Antiviral Res. 2005;66:81–97. doi: 10.1016/j.antiviral.2005.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C., Gunther S., Preiser W., Van der W.S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., Berger A., Burguiere A.M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.C., Muller S., Rickerts V., Sturmer M., Vieth S., Klenk H.D., Osterhaus A.D., Schmitz H., Doerr H.W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- Duquerroy S., Vigouroux A., Rottier P.J., Rey F.A., Bosch B.J. Central ions and lateral asparagine/glutamine zippers stabilize the post-fusion hairpin conformation of the SARS coronavirus spike glycoprotein. Virology. 2005;335:276–285. doi: 10.1016/j.virol.2005.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert D.M., Kim P.S. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 2001;70:777–810. doi: 10.1146/annurev.biochem.70.1.777. [DOI] [PubMed] [Google Scholar]
- Gustchina E., Louis J.M., Bewley C.A., Clore G.M. Synergistic inhibition of HIV-1 envelope-mediated membrane fusion by inhibitors targeting the N and C-terminal heptad repeats of gp41. J. Mol. Biol. 2006;364:283–289. doi: 10.1016/j.jmb.2006.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Yang Z.Y., Kong W.P., Nabel G.J. Generation of synthetic severe acute respiratory syndrome coronavirus pseudoparticles: implications for assembly and vaccine production. J. Virol. 2004;78:12557–12565. doi: 10.1128/JVI.78.22.12557-12565.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huentelman M.J., Zubcevic J., Hernandez Prada J.A., Xiao X., Dimitrov D.S., Raizada M.K., Ostrov D.A. Structure-based discovery of a novel angiotensin-converting enzyme 2 inhibitor. Hypertension. 2004;44:903–906. doi: 10.1161/01.HYP.0000146120.29648.36. [DOI] [PubMed] [Google Scholar]
- Hsueh P.R., Huang L.M., Chen P.J., Kao C.L., Yang P.C. Chronological evolution of IgM, IgA, IgG and neutralization antibodies after infection with SARS-associated coronavirus. Clin. Microbiol. Infect. 2004;10:1062–1066. doi: 10.1111/j.1469-0691.2004.01009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsueh P.R., Hsiao C.H., Yeh S.H., Wang W.K., Chen P.J., Wang J.T., Chang S.C., Kao C.L., Yang P.C. Microbiologic characteristics, serologic responses, and clinical manifestations in severe acute respiratory syndrome. Taiwan. Emerg. Infect. Dis. 2003;9:1163–1167. doi: 10.3201/eid0909.030367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S., Lin K., Strick N., Neurath A.R. HIV-1 inhibition by a peptide. Nature. 1993;365:113. doi: 10.1038/365113a0. [DOI] [PubMed] [Google Scholar]
- Kielian M., Rey F.A. Virus membrane-fusion proteins: more than one way to make a hairpin. Nat. Rev. Microbiol. 2006;4:67–76. doi: 10.1038/nrmicro1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilby J.M., Hopkins S., Venetta T.M., DiMassimo B., Cloud G.A., Lee J.Y., Alldredge L., Hunter E., Lambert D., Bolognesi D., Matthews T., Johnson M.R., Nowak M.A., Shaw G.M., Saag M.S. Potent suppression of HIV-1 replication in humans by T-20, a peptide inhibitor of gp41-mediated virus entry. Nat. Med. 1998;4:1302–1307. doi: 10.1038/3293. [DOI] [PubMed] [Google Scholar]
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A.E., Humphrey C.D., Shieh W.J., Guarner J., Paddock C.D., Rota P., Fields B., DeRisi J., Yang J.Y., Cox N., Hughes J.M., LeDuc J.W., Bellini W.J., Anderson L.J. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- Lai C.Y., Hu H.P., King C.C., Wang W.K. Incorporation of dengue virus replicon into virus-like particles by a cell line stably expressing precursor membrane and envelope proteins of dengue virus type 2. J. Biomed. Sci. 2008;15:15–27. doi: 10.1007/s11373-007-9204-0. [DOI] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enztme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Xiao G., Chen Y., He Y., Niu J., Escalante C.R., Xiong H., Farmar J., Debnath A.K., Tien P., Jiang S. Interaction between heptad repeat 1 and 2 regions in spike protein of SARS-associated coronavirus: implications for virus fusogenic mechanism and identification of fusion inhibitors. Lancet. 2004;363:938–947. doi: 10.1016/S0140-6736(04)15788-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore J.P., Doms R.W. The entry of entry inhibitors: a fusion of science and medicine. Proc. Natl. Acad. Sci. U.S.A. 2003;100:10598–10602. doi: 10.1073/pnas.1932511100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni L., Zhu J., Zhang J., Yan M., Gao G.F., Tien P. Design of recombinant protein-based SARS-CoV entry inhibitors targeting the heptad-repeat regions of the spike protein S2 domain. Biochem. Biophys. Res. Commun. 2005;330:39–45. doi: 10.1016/j.bbrc.2005.02.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris J.S., Guan Y., Yuen K.Y. Severe acute respiratory syndrome. Nat. Med. 2004;10:S88–S97. doi: 10.1038/nm1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons G., Gosalla D.N., Renneckamp A.J., Reeves J.D., Diamond S.L., Bates P. Inhibitors of cathepsin L prevent SARS-CoV entry. Proc. Natl. Acad. Sci. U.S.A. 2005;102:11876–11881. doi: 10.1073/pnas.0505577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supekar V.M., Bruckmann C., Ingallinella P., Bianchi E., Pessi A., Carfi A. Structure of a proteolytically resistant core from the severe acute respiratory syndrome coronavirus S2 fusion protein. Proc. Natl. Acad. Sci. U.S.A. 2004;101:17958–17963. doi: 10.1073/pnas.0406128102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ujike M., Nishikawa H., Otaka A., Yamamoto N., Yamamoto N., Matsuoka M., Kodama E., Fujii N., Taguchi F. HR peptides block protease-mediated direct entry from cell surface of SARS-CoV but not entry via endosomal pathway. J. Virol. 2008;82:588–592. doi: 10.1128/JVI.01697-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S., Takada A., Watanabe T., Ito H., Kida H., Kawaoka Y. Functional importance of the coiled-coil of the Ebola virus glycoprotein. J. Virol. 2000;74:10194–10201. doi: 10.1128/jvi.74.21.10194-10201.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welkos S., O’Brien A. Determination of median lethal and infectious doses in animal model systems. Methods Enzymol. 1994;235:29–39. doi: 10.1016/0076-6879(94)35128-7. [DOI] [PubMed] [Google Scholar]
- Weissenhorn W., Dessen A., Calder L.J., Harrison S.C., Skehel J.J., Wiley D.C. Structural basis for membrane fusion by enveloped viruses. Mol. Membr. Biol. 1999;16:3–9. doi: 10.1080/096876899294706. [DOI] [PubMed] [Google Scholar]
- Wild C.T., Shugars D.C., Greenwell T.K., McDanal C.B., Matthews T.J. Peptides corresponding to a predictive α-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc. Natl. Acad. Sci. 1994;91:9770–9774. doi: 10.1073/pnas.91.21.9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X., Chakraborti S., Dimitrov A.S., Gramatikoff K., Dimitrov D.S. The SARS-CoV S glycoprotein: expression and functional characterization. Biochem. Biophys. Res. Commun. 2003;312:1159–1164. doi: 10.1016/j.bbrc.2003.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Lou Z., Liu Y., Pang H., Tien P., Gao G.F., Rao Z. Crystal structure of severe acute respiratory syndrome coronavirus spike protein fusion core. J. Biol. Chem. 2004;279:49414–49419. doi: 10.1074/jbc.M408782200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung K.S., Yamanaka G.A., Meanwell N.A. Severe acute respiratory syndrome coronavirus entry into host cells: opportunities for therapeutic intervention. Med. Res. Rev. 2006;26:414–433. doi: 10.1002/med.20055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan K., Yi L., Chen J., Qu X., Qing T., Rao X., Jiang P., Hu J., Xiong Z., Nie Y., Shi X., Wang W., Ling C., Yin X., Fan K., Lai L., Ding M., Deng H. Suppression of SARS-CoV entry by peptides corresponding to heptad regions on spike glycoprotein. Biochem. Biophys. Res. Commun. 2004;319:746–752. doi: 10.1016/j.bbrc.2004.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Xiao G., Xu Y., Yuan F., Zheng C., Liu Y., Yan H., Cole D.K., Bell J.I., Rao Z., Tien P., Gao G.F. Following the rule: formation of the 6-helix bundle of the fusion core from severe acute respiratory syndrome coronavirus spike protein and identification of potent peptide inhibitors. Biochem. Biophys. Res. Commun. 2004;319:283–288. doi: 10.1016/j.bbrc.2004.04.141. [DOI] [PMC free article] [PubMed] [Google Scholar]


