Highlights
-
•
Peptide Nucleic Acids − DNA/RNA analogues.
-
•
Different Modifications on PNA backbone and their effects.
-
•
Neutral backbone − remarkable hybridization properties.
-
•
PNA based biosensors and their diverse biomedical applications.
-
•
Potential antigene and antisense agents.
Keywords: PNA, Antigene, Antisense, Hybridization, PCR, Biosensor
Abstract
Peptide Nucleic Acids (PNAs) are the DNA/RNA analogues in which sugar-phosphate backbone is replaced by N-2-aminoethylglycine repeating units. PNA contains neutral backbone hence due to the absence of electrostatic repulsion, its hybridization shows remarkable stability towards complementary oligonucleotides. PNAs are highly resistant to cleavage by chemicals and enzymes due to the substrate specific nature of enzymes and therefore not degraded inside the cells. PNAs are emerging as new tools in the market due to their applications in antisense and antigene therapies by inhibiting translation and transcription respectively. Hence, several methods based on PNAs have been developed for designing various anticancer and antigene drugs, detection of mutations or modulation of PCR reactions. The duplex homopurine sequence of DNA may also be recognized by PNA, forming firm PNA/DNA/PNA triplex through strand invasion with a looped-out DNA strand. PNAs have also been found to replace DNA probes in varied investigative purposes. There are several disadvantages regarding cellular uptake of PNA, so modifications in PNA backbone or covalent coupling with cell penetrating peptides is necessary to improve its delivery inside the cells. In this review, hybridization properties along with potential applications of PNA in the field of diagnostics and pharmaceuticals are elaborated.
1. Introduction
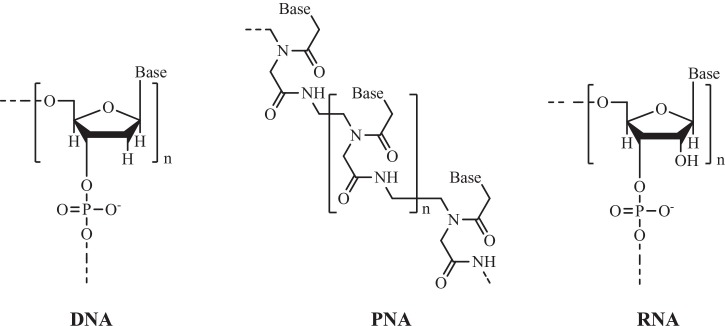
Nucleic acids (DNA/RNA) are the biopolymers which provide fundamental base to the impressive array of complex cellular functions of all known forms of life. These essential building blocks have powerful molecular recognition system, endowing with reliable information transfer by means of various processes such as replication, transcription, and translation. Hence, they play a diverse role in various fields such as research, therapeutics, diagnostics, and nanotechnology. Peptide Nucleic Acids (PNAs), introduced by Nielsen and co-workers in 1991 are DNA/RNA analogues (Nielsen et al., 1991), whereby N-2-aminoethylglycine repeating units replace the sugar-phosphate backbone and the polyamide chain is linked to nucleobases covalently through a carboxymethyl spacer (Fig. 1 ). The attached nucleobases are natural and so through Watson-Crick base pairing can bind to complementary DNA/RNA (Egholm et al., 1993). Since the PNA backbone does not contain any charged phosphate group, the absence of electrostatic repulsion gives rise to a stronger binding between PNA/DNA strands than that of DNA/DNA duplex. Consequently, the thermal stability of PNA/DNA duplexes as compared to natural DNA/DNA double helix of the same length is higher. Moreover, unlike DNA/DNA, PNA/DNA duplex is much less affected by medium with high ionic strength medium. The two strands in PNA/DNA hybrid can be fashioned in either parallel or antiparallel orientation but the later mode grabs higher stability due to the achiral backbone of PNA. The stability of the PNA/DNA duplexes is also found to be highly sensitive to the existence of a single mismatched base pair. Early experiments with homopyrimidine strands (only one repeated pyrimidine base) showed that the T m (“melting” temperature) of a double helix 6-base thymine PNA/adenine DNA was 31 °C as compared to an equivalent 6-base DNA/DNA duplex that denatured at a temperature less than 10 °C. Therefore, PNA probes are very sequence-selective and advanced to DNA probes in single-base mismatch recognition (Anon, 2017). The exceptional sequence selectivity and mismatch intolerance ability of PNA was also proved by comparing the crystal structures of RNA-antisense PNA heteroduplex and PNA-PNA homoduplex with mismatch (Kiliszek et al., 2015). As enzymes are substrate specific so the recognition of PNA neutral backbone is not easy by either nucleases or proteases, making them potentially resistant to enzymatic degradation and their stability over wide pH range. In the lieu of finding relationship of PNA to the origin of life, it was found that PNA-like materials are present in cyanobacteria (Banack et al., 2012). Moreover, the potential of PNA to invade dsDNA has led to the theory that it may have been a prebiotic material (Nielsen, 1993).
Fig. 1.
Structural difference between DNA, PNA and RNA.
Though there are some of the disadvantages with unmodified PNA regarding cellular uptake, covalent coupling of PNA is required with cell penetrating peptides which improves cytosolic delivery. Artificial PNA oligomers which generally require 20–25 bases oligonucleotide probes have been used frequently in diagnosis, molecular biology procedures, antigene and antisense therapies (Marchelli et al., 2011). PNA is neither a nucleic acid nor a protein and it can withstand both nucleases and proteases; so has become a potential biomolecular tool with a superior lifetime for in vivo and in vitro applications such as, molecular diagnostics and antisense therapeutics (Janson and During, 2006). Although many researchers have reviewed the PNA chemistry, (Siddiquee et al., 2015, Veldhoen et al., 2008) their modifications (Rozners, 2012, Sugiyama and Kittaka, 2013, Moccia et al., 2016), properties (Järver et al., 2012) and perspectives in molecular diagnostics (Shi et al., 2015), but in this review we are focusing on various properties of PNA along with special emphasis on its biomedical applications.
1.1. Chemical modifications in PNA backbone
The backbone of the typical PNA monomer has been subjected to a variety of rational modifications with an approach of understanding the structure-activity relations in this class of DNA mimics as well as obtaining PNA oligomers with specifically improved properties for various applications in medicine, diagnostics and molecular biology. The limitations of PNA for such applications include poor membrane permeability, low aqueous solubility, and ambiguity in DNA binding orientation. The strategy behind these modifications includes:
-
1.
Introduction of chirality into the achiral PNA backbone.
-
2.
Rigidify PNA backbone via conformational constraint to pre-organize the PNA structure and entropically drive the duplex formation.
-
3.
Introduction of cationic functional groups to improve water solubility.
-
4.
Modulate nucleobase pairing either by modification of the linker or the nucleobase itself for effective binding at physiological conditions.
Chemical modifications in PNA have been done by introducing different substituents, conformationally constrained cyclic backbones, or modified nucleobases. For instance, the presence of amide group provides rigidity to the structure which was replaced with an amine group, resulting into the reduction of binding affinity which further confirmed the significance of ‘constrained flexibility’ of PNA backbone. In order to increase rigidity, a cyclic group such as cyclohexyl (Sharma et al., 2010) was incorporated but it resulted in decreased binding affinity. On the contrary, introduction of cyclopentyl (Pokorski et al., 2004), (2S,5R)-aminoethyl pipecolyl (Shirude et al., 2004), prolyl (Corradini et al., 2007) derivatives were found to improve binding affinity. Furthermore, charged PNAs have been developed to enhance the solubility and cellular delivery by introducing groups like phosphates (Shiraishi et al., 2008) and guanidium (Katritzky and Narindoshvili, 2008) in the backbone.
Moreover, recognition of single mismatch was greatly improved as compared to that observed for other chiral or achiral PNAs. Thus PNAs having chiral entity possess many properties essential for the detection of point mutations and selective binding in diagnostics and therapeutics. By linking different parts of PNA backbone, the effect of chirality and conformational restriction on the binding affinity of PNA can be determined (Table 1 ).
Table 1.
Different modifications in PNA backbone along with the impact of modifications in properties.
| S. N. | Monomer Structure | Structure | Features | Ref. |
|---|---|---|---|---|
| 1. | Phosphono PNA | 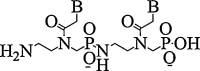 |
Antisense activity | (Shiraishi et al., 2008) |
| 2. | α-Guanyl-ated PNA | 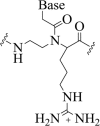 |
Remarkable cellular uptake properties while maintaining Watson-Crick recognition with complementary DNA strand. | (Katritzky and Narindoshvili, 2008, Gupta et al., 2012) |
| 3. | α-Amino methylene PNA | 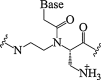 |
Enhanced cellular uptake | (Mitra and Ganesh, 2011, Mitra and Ganesh, 2012) |
| 4. | α-lysine PNA | 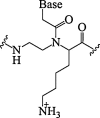 |
PNA-DNA duplex stability. | (Katritzky and Narindoshvili, 2008) |
| 5. | β-methyl PNA (R and S forms) | 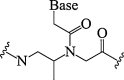 |
PNA bearing S-form chiral units was well suited to form a right-handed hybrid duplex with DNA | (Sugiyama et al., 2011) |
| 6. | Cyclo-pentyl PNA | 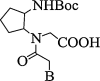 |
Improves the stability of PNA-DNA triplexes and PNA-RNA duplexes for a poly-T PNA. They have a better selectivity for mismatch DNA sequence. | (Pokorski et al., 2004) |
| 7. | Cyclo-hexyl PNA | 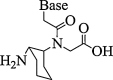 |
(S,S) Isomer can be accommodated more easily in duplex than (R,R) isomer. Inclusion of (R,R) isomer resulted in the drastic decrease in stability of PNA-DNA/RNA complexes. | (Sharma et al., 2010) |
| 8. | Amino-ethyl pipecolyl PNA | 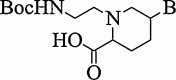 |
Stabilize the resulting complex with complementary DNA. | (Shirude et al., 2004) |
| 9. | Amino-prolyl PNA | 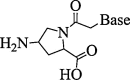 |
Stabilization of derived PNA-DNA hybrids. | (Corradini et al., 2007) |
| 10. | Amino-ethylprolyl PNA | 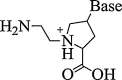 |
They possess remarkable biophysical properties in terms of triplex stability. The mixed pyrimidine hairpin sequences with N-7 guanine and cytosine aep PNA units exhibits directional discrimination in binding to parallel/antiparallel DNA sequences. | (Corradini et al., 2007) |
| 11. | Pyrrolidine PNA | 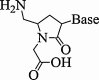 |
(2R,4S) Pyrrolidine PNA decamer formed very stable complexes with both DNA and RNA targets while incorporation of (2S,4S) thymine monomer resulted in a decreased binding efficiency. The fully modified decamer containing (3S,5R) isomer of adenine-9-yl pyrrolidine, was binding rU10 with a little lesser efficiency. | (Corradini et al., 2007) |
| 12. | γ-Amino methylene PNA | 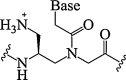 |
Increased PNA-DNA binding. | (Mitra and Ganesh, 2011, Mitra and Ganesh, 2012) |
| 13. | Diethylene glycol PNA | 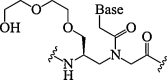 |
Enhanced water solubility & improved hybridization properties. | (Sahu et al., 2011) |
| 14. | Thiol modified PNA | 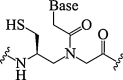 |
No appreciable influence on PNA/RNA duplex. | (de Koning et al., 2006, Gupta, 2017) |
| 15. | γ-Amino propylene PNA | 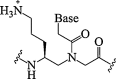 |
Stablized PNA/DNA duplex than corresponding PNA/RNA duplex (ΔTm). Higher thermal stability for PNA/RNA duplex (Tm). | (Kumar and Jain, 2015) |
1.2. Modified nucleobases
Along with backbone modifications, PNA can also be synthesized using non-natural nucleobases which could aid in achieving higher duplex/triplex stabilities and could generate new recognition motifs with potential applications in research and diagnostics. For example, 2,6-diaminopurine offers increased affinity and selectivity for thymine and pseudoisocytosine mimics the C+ recognition pattern for triplex formation (Haaima et al., 1997). 2-Aminopurine can comprise hydrogen bond in the reverse Watson-Crick mode with thymine and uracil and being inherently fluorescent it can further be used to study the kinetics of the hybridization process with complementary nucleic acids (Gangamani and Kumar, 1997). Introduction of fluorescent thiazole orange as nucleobase into PNA afforded a PNA probe that detected hybridization through enhanced fluorescence (Köhler and Seitz, 2003, Jarikote et al., 2005). Hypoxanthine (Sanders et al., 2013), N4-benzoylcytosine (Olsen et al., 2011) and 6-thioguanine (Henrik et al., 1999) represent some more examples of modified nucleobases. Thiouracil along with 2,6-diaminopurine has also been utilized as a base pair in recognizing PNA/DNA which finally led a phenomenon termed as ‘double duplex invasion’. Moreover, introduction of a tricyclic G-clamp (Rajeev et al., 2002, Ortega et al., 2007) modified nucleobase, via both base pairing and base stacking enhanced the duplex stability while increasing the solubility because of the presence of positively charged side chains. Recently, Rozners and coworkers (Li et al., 2010, Rozners, 2012) reported that P and E nucleobases modified PNA monomers were able to isolate pyrimidine interruptions in polypurine tracts of double helical RNA. Another hydrolytically labile nucleobase 5-(acridin-9-ylamino) uracil was prepared by Matarazzoo et al. (Matarazzo et al., 2013) which on hydrolysis produced highly fluorescent acridone and 5-aminouracil. Other modified nucleobases reported are thio-pseudo isocytosine (Devi et al., 2014) for targeting RNA duplex region; 2-amino pyridine (Annoni et al., 2016) as a tool for detecting RNA editing and mono-m-(guanidinoethoxy)phenyl] pyrrolocytosine (Wojciechowski and Hudson, 2009) with enhanced and selective affinity for RNA (Table 2 ).
Table 2.
Nucleobase modifications and their effects on PNA backbone.
| S. No. | Modified Nucleobase | Structure | Properties | References |
|---|---|---|---|---|
| 1. | 2,6-Diamino purine | 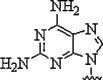 |
Increased affinity and selectivity for thymine | (Haaima et al., 1997) |
| 2. | Pseudoiso-cytosine | 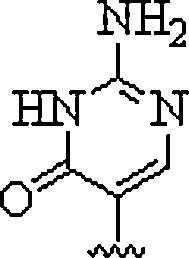 |
Mimics the C+ recognition pattern for triplex formation irrespective of surrounding pH | (Haaima et al., 1997) |
| 3. | 2-Amino purine | 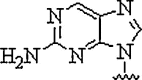 |
Can hydrogen bond with uracil and thymine in the reverse Watson-Crick mode and being inherently fluorescent, can be used to study the kinetics of the hybridization process with complementary nucleic acids | (Gangamani and Kumar, 1997) |
| 4. | Thiazole | 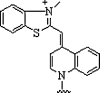 |
Forms PNA probe that fluoresced upon hybridization | (Köhler and Seitz, 2003, Jarikote et al., 2005) |
| 5. | Hypoxanthine | 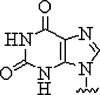 |
Form Watson-Crick base pairs with adenine, cytosine, thymine, and uracil and achieve multimutant specificity | (Sanders et al., 2013) |
| 6. | Thiouracil | 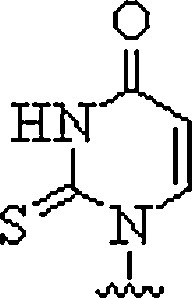 |
Can invade dsDNA in antigene applications | (Haaima et al., 1997) |
| 7. | N4-benzoyl-cytosine | 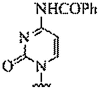 |
candidate for a G-C pseudo-complementary base pair | (Olsen et al., 2011) |
| 8. | 6-Thioguanine | 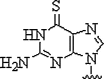 |
Decrease in Tm of 8.5 °C due to PNA:DNA heteroduplex | (Henrik et al., 1999) |
| 9. | G-clamp | 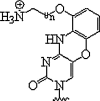 |
Enhanced duplex stability | (Rajeev et al., 2002, Ortega et al., 2007) |
| 10. | P-base |  |
In polypurine tracts of double helical RNA, able to isolate pyrimidine interruptions | (Li et al., 2010, Rozners, 2012) |
| 11. | E-base | 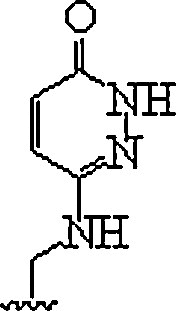 |
In polypurine tracts of double helical RNA, able to isolate pyrimidine interruptions | (Li et al., 2010, Rozners, 2012) |
| 12. | 5(acridin-9-ylamino)uracil | 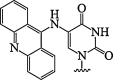 |
Hydrolytically labile modification | (Matarazzo et al., 2013) |
| 13. | Thio-pseudo isocytosine | 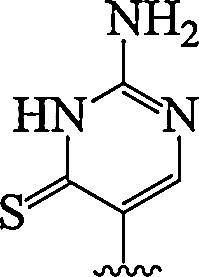 |
Enhanced RNA duplexes recognition | (Devi et al., 2014) |
| 13. | 2-amino pyridine | 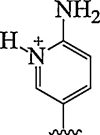 |
Selective triplex formation with RNA duplexes (Adenosine to inosine editing) | (Annoni et al., 2016) |
| 14. | Mono-m-(guanidinoethoxy)phenyl]pyrrolocytosine | 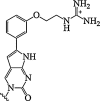 |
Enhanced binding affinity for RNA and an exceptionally high fluorescence quantum yield. | (Wojciechowski and Hudson, 2009) |
2. Applications
The artificially synthesized biopolymer PNA is a powerful biomolecular tool in the molecular genetic diagnostics with a wide range of vital applications. Due to the ability to interact with the selected target with large sequence specificity in a gene sequence, PNAs are of great interest in wide range of applications in medicinal and biotechnological areas. They are highly promising in the expansion of gene therapeutic agents, diagnostic and molecular tools for genetic analysis. It was also indicated by in vitro studies that PNAs could inhibit both transcription and translation of genes to which they have been targeted which guarantee their use as antigene and antisense agents.
2.1. Antigene agents
Antigene therapy using oligonucleotides have targeted various viral agents at the transcription step. In the race of finding potential antiretroviral agents, PNA oligomer (15 bases in length), conjugated to a nuclear localization signal (NLS) was found to significantly suppress HIV-1 replication in chronically infected lymphocytes and macrophages. A decrease of HIV RNA expression in PNA treated infected cells also demonstrated the inhibition of HIV-1 replication during transcription (Pesce et al., 2005).
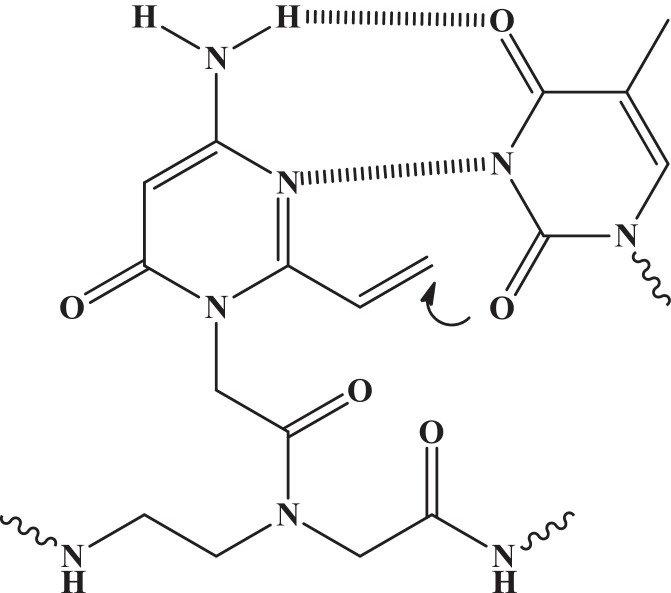
Interstand crosslinking in nucleic acids is one strategy for preparing a stable duplex between PNA and DNA/RNA to be targeted through covalent bond formation either by photolysis or in oxidative conditions. For instance, PNA with 4-amino-6-oxo-2-vinylpyrimidine at terminal position was found to possess excellent and selective crosslinking reactivity with thymine-DNA (Fig. 2 ) and a stable PNA-DNA complex formation was observed (Akisawa et al., 2015). This would make PNA to emerge as a new probe for regulation of the gene expression through sequence specific PNA-DNA interactions.
Fig. 2.
Interaction of 4-amino-6-oxo-2-vinylpyrimidine in PNA with thymine-DNA.
In order to explore the clinical applications in fibro-proliferative disorders, PNA as an antigene reagent was found to decrease mRNA level of the gene, thereby reducing type I collagen production by fibroblast cells (Imamura et al., 2015). In children, rhabdomyosarcomas are chief reason of death by cancer. This is related to the amplification of MYCN which could be decreased in cell lines by RNA interference and antigene PNA (AgPNA) attached to a NLS peptide. Specific nuclear delivery of the PNA was shown by fluorescence microscopy in six human neuroblastoma cell lines: SJ-N-KP and NB-100 (MYCN-nonamplified /less-expressed) ; GI-LI-N and IMR-32 (MYCN-amplified/over-expressed) and GI-CA-N and GI-ME-N (MYCN-unamplified /unexpressed). The results confidently led to the discovery of a PNA-based antigene used as cancer-explicit agent for neuroblastoma with MYCN expression (Tonelli et al., 2012).
The over expression of RAD51, a central protein in the pathway of homologous recombination may result in resistance to chemotherapy. According to a recently published article, PNA conjugated to NLS (PKKKRKV) was found to suppress RAD51 expression in multiple myeloma and act as potential candidate in multiple myeloma chemotherapy (Alagpulinsa et al., 2015). Furthermore, AgPNAs could also be introduced for the purpose to silence expression with mRNA targeted PNAs or siRNAs (Janowski et al., 2006).
2.2. Antisense agents
Antisense oligonucleotides are nucleic acids that bind to the complementary mRNA sequence of the target gene and thereby reduce or block the production of the associated protein. There has been a recent revival of concern in the application of antisense oligonucleotides to treat or prevent numerous genetic disorders or infections. It is done by using different ways to prevent disease onset or arrest disease progression and the first clinical trials for amyotrophic lateral sclerosis and spinal muscular atrophy showing promising results (Rigo et al., 2012). Acute lung injury (ALI) is a complex disorder of acute tenderness that causes degradation of the lung endothelial and epithelial barriers causing hypoxemia, dyspnoea and pulmonary oedema. Radiolabelled antisense PNA-YR9 oligodeoxynucleotide hybrid bound with cationic knedel-like nanoparticle can detect iNOS mRNA in vitro and in vivo which demonstrated their use in diagnosis of ALI during early stages (Shen et al., 2013).
2.2.1. Antibacterial agents and antibiotics
One of the serious problem which is emerging at an alarming rate is the increasing multidrug resistant bacteria. These bacterial pathogens exhibit resistance towards conventional antibiotics due to excessive use of these antibiotics and resistance genes transfer within the bacteria. We desperately require new antibacterial drugs for the treatment of patients infected with drug resistant bacteria. Antisense PNAs as antibacterial agents were found to treat infections caused by multidrug resistant bacteria. Pseudomonas aeruginosa is one of the most common and clinically important pathogens due to its resistance to an extensive variety of antibiotics. PNA antisense conjugated to the carrier peptide (RXR)4 can be a therapeutic platform capable of targeting a variety of genes in P. aeruginosa (Maekawa et al., 2015). Acinetobacter baumannii causes common and severe community and hospital acquired infections. Wang et al. (Wang et al., 2014) established antisense PNAs to carry out gene specific inhibition effects in multiresistant drug against A. baumannii. The described PNAs conjugated to the (KFF)3K peptide targeted the growth of essential gene gyrA in A. baumannii and showed strong inhibition effects with lowest inhibitory and bactericidal concentration of 5 and 10 μM, respectively.
Recent studies also revealed the potential of duplex DNA-invading γPNAs for the identification of both bacterial and fungal pathogens directly from blood without any need for culturing which is time consuming. This approach would help in improving patient’s health by fast and appropriate antimicrobial intervention (Nölling et al., 2016). Gorska et al. (Górska et al., 2016) scanned various target sites in 16S RNA that could be bound with PNA oligomers and one of the tested site verified the bacterial growth inhibition by a PNA oligomer. PNA as antisense molecules when conjugated to a cell penetrating peptide (CPP, i.e. octa-d-lysine) is used as a potential tool for Plasmodium falciparum gene silencing and in the development of potential and highly specific antimalarial agents (Kolevzon et al., 2014). Antisense peptide nucleic acids (APNAs) could also be effectively used to restore vulnerability to β-lactams in methicillin-resistant Staphylococcus aureus and Staphylococcus pseudintermedius (Goh et al., 2015). It was also shown that APNA conjugated with antimicrobial peptides could effectively target acpP gene (responsible for growth of fatty acid chain in the biosynthesis of fatty acid) in E. coli with MIC values of 2–4 μM (Hansen et al., 2016). The ability of APNAs conjugated to the (KFF)3K CPP in Salmonella enterica serovar Typhimurium was explored to target possible essential genes (ligA, rpoA, rpoD, engA, tsf, and kdtA). It was concluded that the anti-rpoA and −rpoD PNAs showed highest strength and can be used as antisense agent for down regulating biofilm formation in P. aeruginosa PAO1 as well as inhibit the inflammatory response. It provides great opportunity for the development of new tool for microbial genetic treatment (Soofi and Seleem, 2012, Hu et al., 2011, Montagner et al., 2017).
Sadeghizadeh et al. (Sadeghizadeh et al., 2009) studied the potential of peptide-PNA as antibacterial agent and also considered the uptake efficiency of dendritic cells (DCs) for PNA treated bacteria. It was found that peptide-PNA may emerge as an excellent candidate in immunological and DNA vaccine. APNAs also targeted essential genes infected with Klebsiella pneumoniae (10^4 CFU) without any significant toxicity to the human cells (Kurupati et al., 2007).
Campylobacter jejuni is the chief pathogen that enters the human body through food causing millions of cases of infection. A resistance-nodulation-cell division (RND)-type multidrug, CmeABC efflux pump resulted into the resistance to potential antibiotics in Campylobacter. For its inhibition, developed PNAs were conjugated to the translational initiation regions of CmeABC and amongst them, CmeA gene was found to be the best target by antisense PNA (Table 3 ) (Oh et al., 2014).
Table 3.
CmeA-PNAs used.
| PNA | Sequence | Target sequences of PNAs against cmeA |
|---|---|---|
| CmeA-PNA1 | KFFKFFKFFK-TGCCTTGAAAAA | aaaaagttccgt |
| CmeA-PNA2 | KFFKFFKFFK-TTTTGCCTTGAA | aagttccgtttt |
| CmeA-PNA3 | KFFKFFKFFK-TGGTTTTGCCTT | ttccgttttggt |
| CmeA-PNA4 | KFFKFFKFFK-TCATGGTTTTGC | cgttttggtact |
| CmeA-PNA5 | KFFKFFKFFK-ATTTCATGGTTT | tttggtacttta |
| CmeA-PNA6 | KFFKFFKFFK-AATAATTTCATG | gtactttaataa |
Note:- The ribosome binding sites, most subjected to antisense inhibition by PNA are underlined, and the start codon is in bold.
2.2.2. Anticancer agents
Micro-RNA consists of 20–24 nucleotides, are regulatory RNA and increase or decrease in their expression have been found to be associated with various diseases. MicroRNA-155 (miR-155) regulates various pathways associated with immune-regulation and multiplication of the cell but it’s over expression causes various tumors. A conjugate of encapsulated APNAs inhibited the over expression of miR-155 which led to the development of potential therapeutic option for cancer (Babar et al., 2012).
B-cell leukemia-2 (bcl-2) is a proto-oncogene and its protein product is a chief apoptosis inhibitor in non-Hodgkin's lymphoma. The overexpression of gene bcl-2 was found to exhibit strong resistance to chemotherapy & radiations. A radiolabeled 177Lu DOTA-anti-bcl-2-Tyr3-octreotate PNA-peptide conjugate was designed which resulted in 51% decline in expression of bcl-2 protein in lymphocytic lymphoma cell line Mec-1 cells through antisense activity along with radiologic effects (Balkin et al., 2014). Another designed APNA as per sequence of MUC1 gene was transfected by liposome into human gastric cancer cells (MKN-45) and the down regulation of MUC1 gene expression successfully inhibited the invasion of tumor cell (Tao et al., 2012). Survivin mRNA APNA (Zhao et al., 2011a, Zhao et al., 2011b) and c-myc mRNA APNA (Zhang et al., 2008, Zhao et al., 2008) labeled with 99Tcm could be effectively used as prospective tumor imaging agents. They might also be used to differentiate tumor from inflammation and also in early diagnosis of colorectal cancer. In another citation, it was also concluded that 125I labeled APNA could inhibit the overexpression of CerbB-2 proto-oncogene in human ovarian epithelial cancer cell lines SKOV3. This inhibition of the gene was done on the mRNA translation and protein expression level (Huang et al., 2010). Moreover, APNA was also found to block mitochondrial DNA transcript promoter of lung cancer SPC-A1 and changes its physiological characteristics (Wang et al., 2009a). In addition to this, PNAs as antisense agents can also provide a new approach to be used to suppress nonsense terminations which are responsible for a large number of hereditary genetic disorders and cancer (Kulyte et al., 2005).
2.2.3. Antiviral agents
PRF-1 signal is responsible for synthesizing RNA replicase polyproteins in virus for genome replication, especially for severe acute respiratory syndrome coronavirus replication. Lee et al. (Ahn et al., 2011) evaluated the potential antiviral effect of APNAs with RNA sequence on the PRF-1 signal as target.
Hepatitis B virus (HBV) is emerging as a major risk to the human health as it can cause chronic hepatitis, carcinoma and cirrhosis. Recently, an APNA was synthesized which targeted the HBV direct repeat (DR) sequences. Tat-PNA-DR efficiently inhibited the replication process of HBV in HepG2.2.15 cells in vitro and in vivo. Interestingly, Tat-PNA-DR was also found to exhibit less cellular toxicity and hemolysis (Zeng et al., 2016).
2.3. PNA Probes in Polymerase Chain Reaction (PCR)
In medical and biological research laboratories, polymerase chain reaction (PCR) is a frequent technique to amplify a DNA target sequence, generating thousands of copies for a variety of biomedical applications. PNA-mediated PCR clamping was also used for the amplification and consequently identification of minor allele in blood chimerism whereas direct sequencing of ordinary PCR product could not identify it (Sano et al., 2014).
Jeong et al. (Jeong et al., 2015) had utilized PNA probe to detect carbapenemase genes by preparing PNA probe-based multiplex real-time PCR kits. They focused their study on most prevailing carbapenemase genes by collecting data from 318 g-negative clinical isolates in 36 diverse hospital laboratories situated in Korea. A high sensitivity and specificity (above 99.0%) with good detection limit, stable reproducibility, no cross reactivity and a long shelf-life (more than eight months) suggested the potential application of PNA based multiplex real-time PCR kits and aid the control of carbapenemase genes. Furthermore, PNA mediated QPCR technique was developed by Machnik and group (Machnik et al., 2014) for the ultrasensitive detection of multiple Sclerosis-associated retrovirus (MSRV) which belongs to the human endogenous retrovirus-W (HERV-W) family. They utilized the complementary PNA probe which did not obstruct the amplification of MSRV. The balancing PNA probe used in this assay selectively blocked the increase of ERVWE1 (another member of HERV-W family), resulting in the specific detection of MSRV. Now-a-days, PNA probe based PCR techniques are quite demanding in the scientific community for the ultrasensitive detection of different kinds of genetic drivers (Lee et al., 2014).
PNA clamping was used in detecting mutation in gene GNAS1 that causes McCune-Albright syndrome, a disorder which affects bones, skin and endocrine tissues of patients (Mukai et al., 2009). PNA clamping was also found to show higher and accurate detection rate for epidermal growth factor receptor (EGFR) gene mutations as compared to direct sequencing in patients with non-small cell lung cancer (Yoon et al., 2015). In addition, PNA-LNA PCR clamp assay was used to detect the status of gene mutation of EGFR recognized in non-small cell lung cancer in pleural fluid samples (Soh et al., 2006, Chen et al., 2014).
2.4. Use of PNA probes in Fluorescence In Situ Hybridization (PNA-FISH)
Techniques like PNA-FISH are used for the specific and selective identification of S. aureus (Hermsen et al., 2008). PNA-FISH methodology was also used for the diagnosis of bacterial vaginosis which is a common vaginal infection. The method was found to be time saving, highly specific and provided a valuable and trustful diagnostic tool (Machado et al., 2015). PNA-FISH was found to be useful for the detection of pathogenic oomycete Aphanomyces invadans which is responsible for ulcerative mycosis, a skin disease caused by a fungus-like agent of wild and cultured fish (Vandersea et al., 2006). In another study, PNA probe-FISH assay was used for detecting mycobacteria with great specificity and sensitivity which helps in the fast and effective treatment of tuberculosis (Lee et al., 2015).
Recently, Rosso and group (Rosso et al., 2015) developed an assay for the specified detection at a single cell level of BCR-ABLT315I mutation that causes chronic myelogenous leukemia. They prepared the assay using PNA technology coupled to immune-fluorescence microscopy, thereby improving the diagnostic resolution and clonal prevalence study.
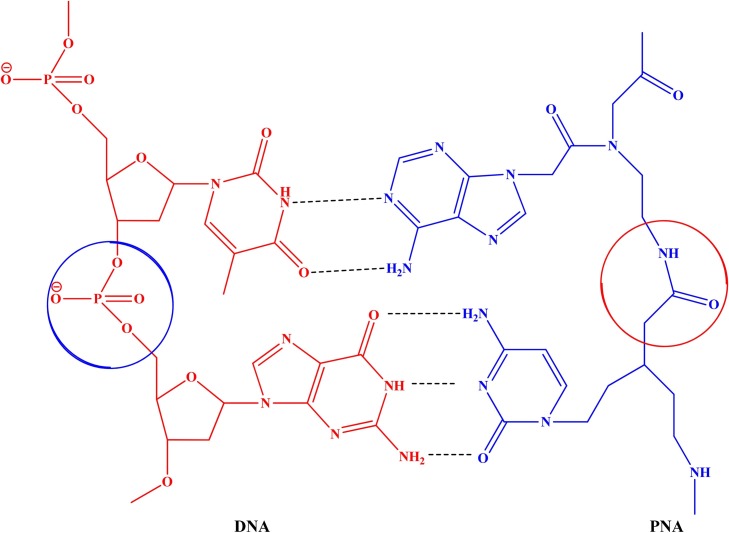
2.5. PNA based Biosensors
During past few decades, biosensor technology has gained considerable attention in playing a significant analytical role in different industrial and academic applications especially in certain areas like medical, environmental, food, quarantine control, safety, security, as well as defense. Singh et al. (Singh et al., 2010) reviewed the use of PNA based biosensors that provide rapid, selective, sensitive, cost effective, simple, and precise detection of DNA hybridization. These biosensors are quite functional in perceiving targeted genes which are responsible for particular disease. Fig. 3 shows PNA-DNA hybridization used for detection of DNA sequence. The biosensor functionalized with PNA probes based on a double tilted fiber Bragg grating (DTFBG) was used for the detection of direct label-free DNA with good sensitivity for a solution having concentration as low as 10 nM. Moreover, the sensor could discriminate single-nucleotide polymorphism (SNP) of the DNA strand if mismatched DNA strands were used. This strategy of electrochemical biosensor based on PNA probe can also be applied to other target analytes like proteins or contaminants (Candiani et al., 2012). It was also useful in the detection of target DNA sequence and single nucleotide mutation in p53 tumor suppressor gene (Raoof et al., 2011).
Fig. 3.
DNA-PNA hybridization which helps in detection of DNA sequence via signal amplification process.
Hejazi et al. (Hejazi et al., 2010) reported an electrochemical DNA biosensor consisting of PNA probe and 6-mercapto-1-hexanol. The hepatitis C virus genotype 3a core/E1 region based 14-mer PNA probe was covalently attached on the electrode. A double stranded PNA-DNA was produced by the selective hybridization of this probe with a complementary sequence in solution which was further used to detect the target DNA sequence. High reproducibility, specificity along with good detection limit 5.7 × 10−11 M described the hybridization of PNA probe as a good detection technique.
Furthermore, ion-sensitive field-effect transistor (IS-FET) based biosensors show potential applications in various human disease diagnostics. Due to highly specific and selective binding nature of PNA at low ionic strength, PNA modified IS-FET biosensor has been developed. It is found to be useful in the detection of hybridization of negatively charged DNA with PNA (Uno et al., 2007). In this biosensor, PNA was immobilized on silicon nitride gate insulator via reaction between thiol group of cysteine in PNA and succinimide group of N-(6-maleimidocaproyloxy)succinimide. It showed a good correlation between Kd and Tm of PNA-DNA duplexes. Upon PNA probe immobilization, the biosensor was used to monitor the PNA-DNA hybridization event efficiently in situ and in real time. Furthermore, 5-trifluoromethyl-derived biosensor was prepared by incorporating trifluoromethyl and 3,3-bis(trifluoromethyl)-4,4,4-trifluorobut-1-ynyl groups on PNA strand. The applicability of biosensor was studied by 19F NMR spectroscopy due to the appearance of distinctive 19F resonance shifts in NMR upon PNA targeting to complementary DNA/RNA. This biosensor monitored the inter-conversions from parallel DNA/PNA complex to an anti-parallel RNA/PNA complex and from a PNA/PNA complex to two different DNA/PNA complexes (i.e. double-duplex invasion) (Kiviniemi et al., 2013).
In lieu of finding integrated, portable and economical devices for DNA recognition, a highly sensitive electrochemical DNA sensor that does not require probe immobilization has been developed (Xuan et al., 2012). This method took advantage of the strand-displacement activity of a DNA polymerase and the difference in diffusivity between free ferrocene-labeled (Fc-PNA) PNA and DNA hybridized Fc-PNA towards a negatively charged ITO electrode surface. This resulted in a 2–3 orders of magnitude improvement in detection sensitivity as compared with earlier reported immobilization free DNA biosensors. The superior sensitivity in combination with the operation convenience made this method a promising alternative for development of integrated, miniaturize, portable and low cost DNA detection devices.
2.6. PNA/DNA hybridization with Nanoparticles
Colorimetric assay is another important method of DNA detection which is based on the aggregation of unmodified metallic nanoparticles (Kanjanawarut and Su, 2009). In this hybridization based method, charged neutral PNA were used as a “coagulant” of citrate anion-coated particles and the assay was validated using gold (Au) and silver (Ag) nanoparticles (NPs). It was found that for discriminating single-base-mismatch, the Ag NPs possess greater sensitivity than Au NPs, further improving the accuracy of result with the use of the multiple aggregation signatures from the two types of nanoparticles. SPR imaging enhanced by ultrasensitive nanoparticles also uses PNA probes for detecting single-nucleotide mismatches in DNA sequences with greater selectivity (D’Agata et al., 2008).
In another research article, an electrochemical biosensor was reported for the fast analysis of nucleic acids as well as nuclease activity on both electroactive bionanoparticles and nucleic acids (Kerman et al., 2008). In this process, the detection of SNPs was done by using ferrocene-conjugated chitosan nanoparticles (Chi-Fc) as an electroactive marker of hybridization. The hybridized PNA-DNA gold electrode (AuE) surface enabled the electrostatic attachment of Chi-Fc to the negatively charged phosphate backbone of DNA resulting into a sharp electrochemical oxidation signal from ferrocene at ∼0.30 V. The nonspecifically adsorbed SNP DNA was effectively removed by exposing the surface to a Nuclease S1, a specific nuclease for ssDNA. Then the enzymatic digestion with successive exposure of the surface to Chi-Fc signified the presence of SNPs with a good detection limit of 1 fM (S/N = 3).
Recently, a group has utilized PNA/DNA hybridization directly in solution using unmodified Au NPs for dengue virus detection (Rahman et al., 2014). In this novel optical detection method, neutral charged PNA was coated on AuNPs surface causing immediate aggregation in solution. When the PNA probe got hybridized to the negatively charged phosphate backbone of complementary target DNA, it resulted in to the formation of negatively charged complexes which got adsorbed on the surface of AuNPs. The process could be seen by naked eyes due to the immediate change in color as well as through UV–vis adsorption spectra. The group also showed good selectivity and specificity of the above label-free colorimetric method, indicating it as a successful method in solution too without any immobilization strategy.
PNA-modified magnetic nanoparticles were prepared from 4-pyridyldithiol derivative of PNA attached to 3-mercapropropyloxysilane coated magnetic nanoparticles. These were found to hybridize with the ssDNA target efficiently and did not show any affinity for non-complementary DNA. This approach may provide a means of direct and label-free gene analysis (Wang et al., 2006). Immobilization of PNA on super paramagnetic iron oxide nanoparticles showed its potential for sequence selective DNA recognition. These magnetic nanosensors could be easily delivered to a desired target by simply applying controlled external electromagnetic field, providing a novel method of selective DNA recognition. These generated PNA-nanoconjugates were found to possess great potential for varied biomedical applications (Prencipe et al., 2009).
2.7. PNA/PNA duplexes
PNA having chiral entity possess many properties essential for the detection of point mutations and selective binding in diagnostics and therapeutics. Totsingan and group (Totsingan et al., 2010) synthesized and studied the chiral optical properties of different oligomers of PNA/PNA duplexes, where different amino acids were attached at the terminal of a single strand of each duplex. In this statistical theoretical model, it was observed that the PNA/PNA double helix had access to a dynamic ensemble of conformational states in solution which varied as a function of oligomer length. This was in accordance with the joint properties of the challenging conformations of the double helix which further showed the importance of chiral optical properties of different oligomers. PNA duplex (PNA zipper) can also bring two polypeptides or protein domains in close proximity. This ability of dimerization was tested with PNA zipper-GCN4 that was known to bind both the TRE and CRE DNA sites, but it was found not to bind TRE and CRE mutants as the native GCN4 containing even a single base mutation. A good correlation was found between the capability of the PNA zipper-GCN4 to fold into α helixes and to bind DNA (Pensato et al., 2010).
2.8. PNA encoded libraries
Researchers have demonstrated the application of combinatorial self assembly of PNA-encoded carbohydrate fragments in accessing the complex glycan arrays (Huang et al., 2011). These PNA-tagged glycan arrays were prepared from native oligosaccharides in a two to three step process. In this protocol of glycan immobilization, the homogeneous distribution of ligands was obtained within a microarray spot. This further offered the use of microarrays in higher degree than the available standard contact printing method. In another article, the good compatibility along with standard solid phase synthesis capability of PNA encoded chemical libraries was utilized to synthesize libraries of peptides, heterocycles and glycoconjugates and further diverse screening formats were used to yield specific ligands (Zambaldo et al., 2015). Furthermore, PNA encoded synthesis of a 10,000-member hetero-glycoconjugate library was also reported and an extremely miniaturized synthetic format was used to analyze microarray of diverse lectins (Novoa et al., 2014). Recently, Decurtins and group (Decurtins et al., 2016) have utilized DNA encoded chemical libraries in automated screening for small organic ligands.
2.9. PNA based photosensitive compound
Hybridization of PNA with DNA gives rise to the formation of PNA based photosensitive compounds which have noteworthy potential in research and diagnostics, medicines, biotechnology and nanotechnology (Stafforst and Hilvert, 2010). These hybridized PNA molecules act as photochromic compounds by their modification with a single azobenzene, the most studied class of photochromic molecules. As per the study, these photochromic compounds could act as light-activated switches with high spatial and temporal resolution for controlling structure, suggestive of a new and generic way of placing nucleic acids based bio- and nanotechnologies under dynamic control by light (Bergen et al., 2016). In another article, novel complex was found to be formed between cyanine dye, ‘3,3′-diethylthiacarbocyanine iodide’, target genomic DNA and a complementary PNA probe. This complex worked as a photosensitizer that enhances photobleaching of dye molecules which rapidly detected the nucleic acid (Wang et al., 2009b).
2.10. PNA microarray
Researchers have also demonstrated a new on chip rapid technology based on PNA microarray as a powerful tool for cancer research, diagnosis and extrapolative assessment. In this technique, the unlabeled RNA was hybridized to the PNA microarray. This was further labeled by enzymic ligation of pCp-Cy3 on the chip, resulting into the analysis of expression profiles with high fidelity. This assay showed high reproducibility and low cross-hybridization for miRNAs belonging to the let-7 and miR-181 families, which differ by a single nucleotide (Kim et al., 2012). While using microarray technology for different biological and medical studies, arrays of PNAs were prepared as well as their microRNAs capture abilities were evaluated. It was found that the sensitivity of 10-mer PNA probes was much higher than that of the corresponding DNA probes in targeting miRNAs (Endoh et al., 2009).
2.11. Loop structures
PNAs when interact with complementary dsDNAs by displacement of strand, the loop induced by the PNA can be recognized by the RNA polymerase. Thus, PNA can act as artificial transcription promoters describing PNA as a specific gene activating reagents and its use in drugs (Møllegaard et al., 1994). Shimada and group (Shimada et al., 2013) prepared a PNA conjugate having bipyridine unit into the loop moiety of PNAs. This conjugate was specifically designed to structure a PNA/DNA bimolecular triplex and a hairpin. It was observed that on addition of Cu2+, the thermal stability of hairpin structure was hardly affected whereas the stability of PNA/DNA bimolecular triplex structure was drastically destabilized. This was due to its complexation with Cu2+ that changed the conformation of bipyridine unit and decreased the Tm value by 17.4 °C resulting into 1000-fold decrease in the binding constant. Hence, the formed loop moiety could further be utilized as an allosteric DNA carrier which released DNA in the presence of certain concentration of metal ion. Kundu and group (Kundu et al., 2012) developed a simple method to study the dynamic properties of modified nucleotides. They determined the binding constant between PNA and a hairpin-structured DNA through an affinity capillary electrophoresis method. In another report, small cyclic PNAs (Upert et al., 2012) and guanidine based PNAs (Gupta et al., 2012) were found to inhibit viral replication while targeting human immunodeficiency virus-1 (HIV-1) transactivation responsive element (TAR)RNA loop. These modified short PNA sequences were found to be highly specific, stable in biological media and possessed enhanced cellular delivery which proved them to be potential anti-HIV agents. These cyclic PNAs were also observed to target HIV-1 TAR RNA loop through strong interactions with their TAR RNA target. This was found to be even more specific than APNA (Upert et al., 2007).
It was also shown by researchers that guanine rich PNAs selectively formed PNA-DNA or PNA-RNA heteroquadruplex structures which may emerge as potential anticancer agents. It was found that γ-PNA binding to complementary DNA and RNA was favored thermodynamically which resulted in a stronger binding than unmodified PNA. It might be due to the fact that the L-modified structure matched the preferred right handed helicity of DNA and RNA duplexes (Lusvarghi et al., 2009).
2.12. Diverse applications
Recently, it was shown by researchers that γ-S-carboxyethyl-T10 PNA formed a stable PNA2-DNA triplex as compared to its α-analogue (Kirillova et al., 2015). PNAs are also found to form stable triplex invasion complexes, and also conventional triplexes with the dsDNA target. This sequence-selective recognition might occur by helix invasion via triplex P-loop formation or by major groove triplex binding (Bentin et al., 2006).
Cao and group (Cao et al., 2015) explored the single-molecule AFM measurements to probe PNA-DNA hybridization where a stable hybrid was formed between PNA sequence of six thymine bases and complementary DNA sequence. It was observed that a very high force (∼148 pN) was required to rupture the hybrid than that of <100 pN to unbind the native short DNA duplex. The energetic and kinetic aspects of PNA–DNA interaction were portrayed by applying single molecular force study and estimating various kinetic parameters such as thermal force scale (fβ∼17.5 pN), distance to the energy barrier for dissociation (Δx ∼ 0.23 nm) and dissociation rate constant at zero force (koff ∼ 2.5 s−1). The results indicated the stronger binding affinity in PNA-DNA than DNA-DNA duplexes as well as sensitivity and specificity of PNA–DNA hybridization to a single base mismatch.
In addition to above, the fluorescent intensity of nucleobase by adding vinyl group at 8 position of guanine was utilized for differentiating the hybridization states of specific double strands with DNA, RNA, and PNA as well as quadruplex forming RNA/PNA oligomers (Muellar et al., 2012). Moreover, it was also shown that dual labeled PNA probes can effectively be used for mutation detection through melting point analysis (Hur et al., 2015). Another concept of PNA based hybrid hydrogels was introduced by Chu and co-workers. Multiple PNAs were grafted on water soluble polymer, N-(2-hydroxypropyl)methacrylamide which underwent further crosslinking upon linker DNA addition to produce two types of hydrogels. These hydrogels could be exploited as novel biomaterials for varied pharmaceutical applications (Chu et al., 2015).
3. Conclusion
PNAs, synthesized in 1991 are the DNA/RNA analogues which have been found to possess significant applications in molecular genetics and biotechnology. Due to its increased stability in biological fluids and high binding affinity, the PNA applications are attaining interest in vast field of research and development. PNA can be utilized as biological probe as well as in discovering electrifying innovative and influential detection methods, viz. sensors, detection of mutations and modulation of PCR reactions. In this review, we have focused on various reported applications of backbone (un)modified PNA. The hybrid structures have varied biological and chemical properties which could make PNA’s great impact especially as antigene and antisense agents and probe-based assays which directed PNA as a major strike for the medical professionals in the coming century.
Acknowledgements
Author A. Gupta gratefully acknowledges the financial assistance from Science and Engineering Research Board (DST), New Delhi. All the authors made a significant contribution in this review.
References
- Ahn D.-G., Lee W., Choi J.-K., Kim S.-J., Plant E.P., Almazan F., Taylor D.R., Enjuanes L., Oh J.-W. Interference of ribosomal frameshifting by antisense peptide nucleic acids suppresses SARS coronavirus replication. Antiviral Res. 2011;91:1–10. doi: 10.1016/j.antiviral.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akisawa T., Ishizawa Y., Nagatsugi F. Synthesis of peptide nucleic acids containing a crosslinking agent and evaluation of their reactivities. Molecules. 2015;20:4708–4719. doi: 10.3390/molecules20034708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alagpulinsa D.A., Yaccoby S., Ayyadevara S., Reis R.J.S. A peptide nucleic acid targeting nuclear RAD51 sensitizes multiple myeloma cells to melphalan treatment. Cancer Biol. Ther. 2015;16:976–986. doi: 10.1080/15384047.2015.1040951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annoni C., Endoh T., Hnedzko D., Rozners E., Sugimoto N. Triplex-forming peptide nucleic acid modified with 2-aminopyridine as a new tool for detection of A-to-I editing. Chem. Commun. 2016;52:7935–7938. doi: 10.1039/c6cc02164f. [DOI] [PubMed] [Google Scholar]
- http://en.wikipedia.org/wiki/Peptidenucleicacid.
- Babar I.A., Cheng C.J., Booth C.J., Liang X., Weidhaas J.B., Saltzman W.M., Slack F.J. Nanoparticle-based therapy in an in vivo microRNA-155 (miR-155)-dependent mouse model of lymphoma. Proc. Natl. Acad. Sci. U. S. A. 2012;109:E1695–E1704. doi: 10.1073/pnas.1201516109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balkin E.R., Liu D., Jia F., Ruthengael V.C., Shaffer S.M., Miller W.H., Lewis M.R. Comparative biodistributions and dosimetry of [177Lu]DOTA-anti-bcl-2-PNA-Tyr3-octreotate and [177Lu]DOTA-Tyr3-octreotate in a mouse model of B-cell lymphoma/leukemia. Nucl. Med. Biol. 2014;41:36–42. doi: 10.1016/j.nucmedbio.2013.10.006. [DOI] [PubMed] [Google Scholar]
- Banack S.A., Metcalf J.S., Jiang L., Craighead D., Ilag L.L., Cox P.A. Cyanobacteria produce N-(2-aminoethyl)glycine, a backbone for peptide nucleic acids which may have been the first genetic molecules for life on earth. PLoS One. 2012;7:e49043. doi: 10.1371/journal.pone.0049043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentin T., Hansen G.I., Nielsen P.E. Structural diversity of target-specific Homopyrimidine peptide nucleic acid?dsDNA complexes. Nucl. Acids Res. 2006;34:5790–5799. doi: 10.1093/nar/gkl736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergen A., Rudiuk S., Morel M., Saux T.L., Ihmels H., Baigl D. Photodependent melting of unmodified DNA using a photosensitive intercalator: a new and generic tool for photoreversible assembly of DNA nanostructures at constant temperature. Nano Lett. 2016;16:773–780. doi: 10.1021/acs.nanolett.5b04762. [DOI] [PubMed] [Google Scholar]
- Candiani A., Sozzi M., Cucinotta A., Selleri S., Veneziano R., Corradini R., Marchelli R., Childs P., Pissadakis S. Optical fiber ring cavity sensor for label-free DNA detection. IEEE J. Sel. Top. Quantum Electron. 2012;18:1176–1183. [Google Scholar]
- Cao M., Deng L., Xu H. Study of PNA–DNA hybridization by AFM-based single- molecule force spectroscopy. Coll. Surf. A. 2015;470:46–51. [Google Scholar]
- Chen Y.-L., Lu C.-C., Yang S.-C., Su W.-P., Lin Y.-L., Chen W.-L., Huang W., Su W.-C., Chow N.-H., Ho C.-L. Verification of wild-type EGFR status in non–small cell lung carcinomas using a mutant-enriched PCR on selected cases. J. Mol. Diagn. 2014;16:486–494. doi: 10.1016/j.jmoldx.2014.05.007. [DOI] [PubMed] [Google Scholar]
- Chu T.W., Feng J.Y., Yang J.Y., Kopeček J. Hybrid polymeric hydrogels via peptide nucleic acid (PNA)/DNA complexation. J. Controlled Release. 2015;220:608–616. doi: 10.1016/j.jconrel.2015.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corradini R., Sforza S., Tedeschi T., Totsingan F., Marchelli R. Peptide nucleic acids with a structurally biased backbone: effects of conformational constraints and stereochemistry. Curr. Top. Med. Chem. 2007;7:681–694. doi: 10.2174/156802607780487759. [DOI] [PubMed] [Google Scholar]
- D'Agata R., Corradini R., Grasso G., Marchelli R., Spoto G. Ultrasensitive detection of DNA by PNA and nanoparticle-Enhanced surface plasmon resonance imaging. Chem. Bio. Chem. 2008;9:2067–2070. doi: 10.1002/cbic.200800310. [DOI] [PubMed] [Google Scholar]
- de Koning M.C., Petersen L., Weterings J.J., Overhand M., van der Marel G.A., Filippov D.V. Synthesis of thiol modified peptide nucleic acids designed for post-assembly conjugation reactions. Tetrahedron. 2006;62:3248–3258. [Google Scholar]
- Decurtins W., Wichert M., Franzini R.M., Buller F., Stravs M.A., Zhang Y., Neri D., Scheuermann J. Automated screening for small organic ligands using DNA-encoded chemical libraries. Nat. Protoc. 2016;11:764–780. doi: 10.1038/nprot.2016.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devi G., Yuan Z., Lu Y., Zhao Y., Chen G. Incorporation of thio-pseudoisocytosine into triplex-forming peptide nucleic acids for enhanced recognition of RNA duplexes. Nucl. Acids Res. 2014;42:4008–4018. doi: 10.1093/nar/gkt1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egholm M., Buchardt O., Christensen L., Behrens C., Freier S.M., Driver D.A., Berg R.H., Kim S.K., Norden B., Nielsen P.E. PNA hybridizes to complementary oligonucleotides obeying the Watson- Crick hydrogen bonding rules. Nature. 1993;365:566–568. doi: 10.1038/365566a0. [DOI] [PubMed] [Google Scholar]
- Endoh T., Kitamatsu M., Sisido M., Ohtsuki T. PNA arrays for miRNA detection. Chem. Lett. 2009;38:438–439. [Google Scholar]
- Górska A., Zagrajek A.M., Równicki M., Trylska J. Scanning of 16S ribosomal RNA for peptide nucleic acid targets. J. Phys. Chem. B. 2016;120:8369–8378. doi: 10.1021/acs.jpcb.6b02081. [DOI] [PubMed] [Google Scholar]
- Gangamani B.P., Kumar V.A. 2-Aminopurine peptide nucleic acids (2-apPNA): intrinsic fluorescent PNA analogues for probing PNA–DNA interaction dynamics. Chem. Commun. 1997:1913–1914. [Google Scholar]
- Goh S., Loeffler A., Lloyd D.H., Nair S.P., Good L. Oxacillin sensitization of methicillin-resistant staphylococcus aureus and methicillin-resistant staphylococcus pseudintermedius by antisense peptide nucleic acids in vitro. BMC Microbiol. 2015;15:262. doi: 10.1186/s12866-015-0599-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta P., Muse O., Rozners E. Recognition of double-stranded RNA by guanidine-modified peptide nucleic acids. Biochemistry. 2012;51:63–73. doi: 10.1021/bi201570a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A. Synthesis and characterization of novel α-monomers of peptide nucleic acid. J. Appl. Res. Technol. 2017;15:122–131. [Google Scholar]
- Haaima G., Hansen H.F., Christensen L., Dahl O., Nielsen P.E. Increased DNA binding and sequence discrimination of PNA oligomers containing 2,6-diaminopurine. Nucleic Acids Res. 1997;25:4639–4643. doi: 10.1093/nar/25.22.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen A.M., Bonke G.Larsen, Yavari C.J., Nielsen N., Franzyk P.E. Antibacterial peptide nucleic acid-Antimicrobial peptide (PNA-AMP) conjugates: antisense targeting of fatty acid biosynthesis. Bioconjugate Chem. 2016 doi: 10.1021/acs.bioconjchem.6b00013. [DOI] [PubMed] [Google Scholar]
- Hejazi M.S., Pournaghi-Azar M.H., Ahour F. Electrochemical detection of short sequences of hepatitis C 3a virus using a peptide nucleic acid-assembled gold electrode. Anal. Biochem. 2010;399:118–124. doi: 10.1016/j.ab.2009.11.019. [DOI] [PubMed] [Google Scholar]
- Henrik F., Hansen L.C., Dahl O., Nielsen P.E. 6-Thioguanine in peptide nucleic acids: synthesis and hybridization properties. Nucleosides Nucleotides. 1999;18:5–9. [Google Scholar]
- Hermsen E.D., Shull S.S., Klepser D.G., Iwen P.C., Armbrust A., Garrett J., Freifeld A.G., Rupp M.E. Pharmacoeconomic analysis of microbiologic techniques for differentiating staphylococci directly from blood culture bottles. J. Clin. Microbiol. 2008;46:2924–2929. doi: 10.1128/JCM.00623-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Xia Y., Xiong Y., Li X., Su X. Inhibition of biofilm formation by the antisense peptide nucleic acids targeted at the motA gene in Pseudomonas aeruginosa PAO1 strain. World J. Microbiol. Biotechnol. 2011;27:1981–1987. [Google Scholar]
- Huang H-m., Zhang B., Hu J-m. Depressed expression of CerbB-2 mRNA and Her-2 protein in human ovarian cancer Cell line SKOV3 by 125I labeled antisense peptide nucleic acid. Suzhou Daxue Xuebao, Yixueban. 2010;30:1014–1017. [Google Scholar]
- Huang K.-T., Gorska K., Alvarez S., Barluenga S., Winssinger N. Combinatorial self-assembly of glycan fragments into microarrays. Chem. Bio. Chem. 2011;12:56–60. doi: 10.1002/cbic.201000567. [DOI] [PubMed] [Google Scholar]
- Hur D., Kim M.S., Song M., Jung J., Park H. Detection of genetic variation using dual-labeled peptide nucleic acid (PNA) probe-based melting point analysis. Biol. Proced. Online. 2015;17:14. doi: 10.1186/s12575-015-0027-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura Y., Tsuboi S., Sugiyama T., Kittaka A., Shin Y. A peptide nucleic acid to reduce type I collagen production by fibroblast cells. Open J. Med. Chem. 2015;5:1–8. [Google Scholar]
- Järver P., Coursindel T., Andaloussi S.E.L., Godfrey C., Wood M.J.A., Gait M.J. Peptide-mediated cell and in vivo delivery of antisense oligonucleotides and siRNA. Mol. Ther. Nucl. Acids. 2012;1:e27. doi: 10.1038/mtna.2012.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janowski B.A., Hu J., Corey D.R. Silencing gene expression by targeting chromosomal DNA with antigene peptide nucleic acids and duplex RNAs. Nat. Protoc. 2006;1:436–443. doi: 10.1038/nprot.2006.64. [DOI] [PubMed] [Google Scholar]
- Janson C.G., During M.J. Biomolecules. Springer; US: 2006. Peptide nucleic acids, morpholinos and related antisense. [Google Scholar]
- Jarikote D.V., Köhler O., Socher E., Seitz O. Divergent and linear solid-phase synthesis of PNA containing thiazole orange as artificial base. Eur. J. Org. Chem. 2005;2005:3187–3195. [Google Scholar]
- Jeong S., Kim J.O., Jeong S.H., Bae I.K., Song W. Evaluation of peptide nucleic acid-mediated multiplex real-time PCR kits for rapid detection of carbapenemase genes in gram-negative clinical isolates. J. Microbiol. Methods. 2015;113:4–9. doi: 10.1016/j.mimet.2015.03.019. [DOI] [PubMed] [Google Scholar]
- Köhler O., Seitz O. Thiazole orange as fluorescent universal base in peptide nucleic acids. Chem. Commun. 2003:2938–2939. doi: 10.1039/b308299g. [DOI] [PubMed] [Google Scholar]
- Kanjanawarut R., Su X. Coloimetric detection of DNA using unmodified metallic nanoparticles and peptide nucleic acid probes. Anal. Chem. 2009;81:6122–6129. doi: 10.1021/ac900525k. [DOI] [PubMed] [Google Scholar]
- Katritzky A.R., Narindoshvili T. Chiral peptide nucleic acid monomers (PNAM) with modified backbones. Org. Biomol. Chem. 2008;6:3171–3176. doi: 10.1039/b806141f. [DOI] [PubMed] [Google Scholar]
- Kerman K., Saito M., Tamiya E. Electroactive chitosan nanoparticles for the detection of single-nucleotide polymorphisms using peptide nucleic acids. Anal. Bioanal. Chem. 2008;391:2759–2767. doi: 10.1007/s00216-008-2204-8. [DOI] [PubMed] [Google Scholar]
- Kiliszek A., Banaszak K., Dauter Z., Rypniewski W. The first crystal structures of RNA-PNA duplexes and a PNA-PNA duplex containing mismatches-toward antisensetherapy against TREDs. Nucl. Acids Res. 2015;44(4):1937–1943. doi: 10.1093/nar/gkv1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Choi J.-j., Cho M., Park H. A PNA microarray platform for miRNA expression profiling using on-chip labeling technology. BioChip J. 2012;6:25–33. [Google Scholar]
- Kirillova Y., Boyarskaya N., Dezhenkov A., Tankevich M., Prokhorov I., Varizhuk A., Eremin S., Esipov D., Smirnov I., Pozmogova G. Polyanionic carboxyethyl peptide nucleic acids (ce-PNAs): synthesis and DNA binding. PLoS One. 2015;10:e0140468. doi: 10.1371/journal.pone.0140468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiviniemi A., Murtola M., Ingman P., Virta P. Synthesis of fluorine-labeled peptide nucleic acid building blocks as sensors for the 19F NMR spectroscopic detection of different hybridization modes. J. Org. Chem. 2013;78:5153–5159. doi: 10.1021/jo400014y. [DOI] [PubMed] [Google Scholar]
- Kolevzon N., Nasereddin A., Naik S., Yavin E., Dzikowski R., Hakimi M.A. Use of peptide nucleic acids to manipulate gene expression in the malaria parasite plasmodium falciparum. PLoS One. 2014;9:e86802. doi: 10.1371/journal.pone.0086802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulyte A., Dryselius R., Karlsson J., Good L. Gene selective suppression of nonsense termination using antisense agents. Biochim. Biophys. Acta Gene Struct. Expr. 2005;1730:165–172. doi: 10.1016/j.bbaexp.2005.07.004. [DOI] [PubMed] [Google Scholar]
- Kumar P., Jain D.R. Cγ-Aminopropylene peptide nucleic acid (amp-PNA): chiral cationic PNAs with superior PNA:DNA/RNA duplex stability and cellular uptake. Tetrahedron. 2015;71:3378–3384. [Google Scholar]
- Kundu L.M., Tsukada H., Matsuoka Y., Kanayama N., Takarada T., Maeda M. Estimation of binding constants of peptide nucleic acid and secondary-structured DNA by affinity capillary electrophoresis. Anal. Chem. 2012;84:5204–5209. doi: 10.1021/ac301025m. [DOI] [PubMed] [Google Scholar]
- Kurupati P., Tan K.S.W., Kumarasinghe G. Poh, C. L. inhibition of gene expression and growth by antisense peptide nucleic acids in a multiresistant–lactamase-producing klebsiella pneumoniae strain. Antimicrob. Agents Chemother. 2007;51:805–811. doi: 10.1128/AAC.00709-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B., Lee B., Han G., Kwon M.J., Han J., Choi Y.-L. KRAS mutation detection in non-small cell lung cancer using a peptide nucleic acid-mediated polymerase chain reaction clamping method and comparative validation with next-generation sequencing. Korean J. Pathol. 2014;48:100–107. doi: 10.4132/KoreanJPathol.2014.48.2.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.H., Kim S.Y., Kim H.H., Lee E.Y., Chang C.L. Evaluation of peptide nucleic acid probe-based fluorescence in situ hybridization for the detection of mycobacterium tuberculosis complex and nontuberculous mycobacteria in clinical respiratory specimens. Ann. Clin. Microbiol. 2015;18:37–43. [Google Scholar]
- Li M., Zengeya T., Rozners E. Short peptide nucleic acids bind strongly to homopurine tract of double helical RNA at pH 5.5. J. Am. Chem. Soc. 2010;132:8676–8681. doi: 10.1021/ja101384k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusvarghi S., Murphy C.T., Roy S., Tanious F.A., Sacui I., Wilson W.D., Ly D.H., Armitage B.A. Loop and backbone modifications of peptide nucleic acid improve G-quadruplex binding selectivity. J. Am. Chem. Soc. 2009;131:18415–18424. doi: 10.1021/ja907250j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møllegaard N.E., Buchardt O., Egholm M., Nielsen P.E. Peptide nucleic acid.DNA strand displacement loops as artificial transcription promoters. Proc. Natl. Acad. Sci. U. S. A. 1994;91:3892–3895. doi: 10.1073/pnas.91.9.3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado A., Castro J., Cereija T., Almeida C., Cerca N. Diagnosis of bacterial vaginosis by a new multiplex peptide nucleic acid fluorescence in situ hybridization method. Peer J. 2015;3:e780. doi: 10.7717/peerj.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machnik G., Łabuzek K., Skudrzyk E., Rekowski P., Ruczyński J., Wojciechowska M., Mucha P., Giri S., Okopień B. A peptide nucleic acid (PNA)-mediated polymerase chain reaction clamping allows the selective inhibition of the ERVWE1 gene amplification. Mol. Cell. Probes. 2014;28:237–241. doi: 10.1016/j.mcp.2014.04.003. [DOI] [PubMed] [Google Scholar]
- Maekawa K., Azuma M., Okuno Y., Tsukamoto T., Nishiguchi K., Setsukinai K., Maki H., Numata Y., Takemoto H., Rokushima M. Antisense peptide nucleic acid-peptide conjugates for functional analyses of genes in pseudomonas aeruginosa. Bioorg. Med. Chem. 2015:30097–30103. doi: 10.1016/j.bmc.2015.10.020. (pii: S0968-0896) [DOI] [PubMed] [Google Scholar]
- Marchelli R., Corradini R., Manicardi A. Gene modulation by peptide nucleic acids (PNAs) targeting microRNAs (miRs) In: You Y., editor. Targets in Gene Therapy. InTech; 2011. pp. 29–46. [Google Scholar]
- Matarazzo Augusto, Moustafa Mohamed E., Hudson Robert H.E. 5-(Acridin-9-ylamino)uracil − a hydrolytically labile nucleobase modification in peptide nucleic acid. Can. J. Chem. 2013;91:1202–1206. [Google Scholar]
- Mitra R., Ganesh K.N. PNAs grafted with (α/γ, R/S)-aminomethylene pendants: regio and stereo specific effects on DNA binding and improved cell uptake. Chem. Commun. 2011;47:1198–1200. doi: 10.1039/c0cc03988h. [DOI] [PubMed] [Google Scholar]
- Mitra R., Ganesh K.N. Aminomethylene peptide nucleic acid (am-PNA): synthesis, regio-/stereospecific DNA binding, and differential cell uptake of (α/γ, R/S)am-PNA analogues. J. Org. Chem. 2012;77:5696–5704. doi: 10.1021/jo300860f. [DOI] [PubMed] [Google Scholar]
- Moccia M., Adamo M.F.A., Saviano M. Insights on chiral, backbone modified peptide nucleic acids: properties and biological activity. Artif. DNA: PNA & XNA. 2016;5(3):e1107176. doi: 10.1080/1949095X.2015.1107176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagner G., Bezzerri V., Cabrini G., Fabbri E., Borgatti M., Lampronti I., Finotti A., Nielsen P.E., Gambari R. An antisense peptide nucleic acid against Pseudomonas aeruginosa inhibiting bacterial-induced inflammatory responses in the cystic fibrosis IB3-1 cellular model system. Int. J. Biol. Macromol. 2017;S0141-8130-16:30671–30677. doi: 10.1016/j.ijbiomac.2017.02.011. [DOI] [PubMed] [Google Scholar]
- Muellar S., Strohmeier J., Diederichsen U. 8-Vinylguanine nucleo amino acid: a fluorescent PNA building block. Org. Lett. 2012;14(6):1382–1385. doi: 10.1021/ol3000603. [DOI] [PubMed] [Google Scholar]
- Mukai N., Matsuo K., Suzuki S., Maimaiti M. Gene diagnosis using peripheral blood sample in patients with McCune-Albright syndrome. Kenkyu Nenpo – Seicho Kagaku Kyokai. 2009;32:233–235. [Google Scholar]
- Nölling J., Rapireddy S., Amburg J.I., Crawford E.M., Prakash R.A., Rabson A.R., Tang Y.-W., Singer A. Duplex DNA-invading γ-modified peptide nucleic acids enable rapid identification of bloodstream infections in whole blood. Mbio. 2016;7:e00345–16. doi: 10.1128/mBio.00345-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen P.E., Egholm M., Berg R.H., Buchardt O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science. 1991;254:1497–1500. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- Nielsen P.E. Peptide nucleic acid (PNA): a model structure for the primordial genetic material? Orig. Life Evol. Biosph. 1993;23:323–327. doi: 10.1007/BF01582083. [DOI] [PubMed] [Google Scholar]
- Novoa A., Machida T., Barluenga S., Imberty A., Winssinger N. PNA-encoded synthesis (PES) of a 10 000-member hetero-glycoconjugate library and microarray analysis of diverse lectins. Chem. Bio. Chem. 2014;15:2058–2065. doi: 10.1002/cbic.201402280. [DOI] [PubMed] [Google Scholar]
- Oh E., Zhang Q., Jeon B. Target optimization for peptide nucleic acid (PNA)-mediated antisense inhibition of the CmeABC multidrug efflux pump in Campylobacter jejuni. J. Antimicrob. Chemother. 2014;69:375–380. doi: 10.1093/jac/dkt381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen A.G., Dahl O., Petersen A.B., Nielsen J., Nielsen P.E. A novel pseudo-complementary PNA G-C base pair. Artif. DNA PNA XNA. 2011;2:33–37. doi: 10.4161/adna.2.1.15554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega J.-A., Blas J.R., Orozco M., Grandas A., Pedroso E., Robles J. Binding affinities of oligonucleotides and PNAs containing phenoxazine and G-Clamp cytosine analogues are unusually sequence-dependent. Org. Lett. 2007;9:4503–4506. doi: 10.1021/ol701826x. [DOI] [PubMed] [Google Scholar]
- Pensato S., Renda M., Leccia F., Saviano M., D’Andrea L.D., Pedone C., Pedone P.V., Romanelli A. PNA zipper as a dimerization tool: development of a bZip mimic. Biopolymers. 2010;93:434–441. doi: 10.1002/bip.21357. [DOI] [PubMed] [Google Scholar]
- Pesce C.D., Bolacchi F., Bongiovanni B., Cisotta F., Capozzi M., Diviacco S., Quadrifoglio F., Mango R., Novelli G., Mossa G., Esposito C., Ombres D., Rocchi G., Bergamini A. Anti-gene peptide nucleic acid targeted to proviral HIV-1 DNA inhibits in vitro HIV-1 replication. Antiviral Res. 2005;66:13–22. doi: 10.1016/j.antiviral.2004.12.001. [DOI] [PubMed] [Google Scholar]
- Pokorski J.K., Witschi M.A., Purnell B.L., Appella D.H. (S,S)-trans-Cyclopentane-constrained peptide nucleic acids: a general backbone modification that improves binding affinity and sequence specificity. J. Am.Chem. Soc. 2004;126:15067–15073. doi: 10.1021/ja046280q. [DOI] [PubMed] [Google Scholar]
- Prencipe G., Maiorana S., Verderio P., Colombo M., Fermo P., Caneva E., Prosperi D., Licandro E. Magnetic peptide nucleic acids for DNA targeting. Chem. Commun. 2009;40:6017–6019. doi: 10.1039/b911449a. [DOI] [PubMed] [Google Scholar]
- Rahman S.A., Saadun R., Azmi N.E., Ariffin N., Abdullah J., Yusof N.A., Sidek H., Hajian R. Label-free dengue detection utilizing PNA/DNA hybridization based on the aggregation process of unmodified gold nanoparticles. J. Nanomater. 2014;2014:5–839286. [Google Scholar]
- Rajeev K.G., Maier M.A., Lesnik E.A., Manoharan M. High-affinity peptide nucleic acid oligomers containing tricyclic cytosine analogues! Org. Lett. 2002;4:4395–4398. doi: 10.1021/ol027026a. [DOI] [PubMed] [Google Scholar]
- Raoof J.B., Ojani R., Golabi S.M., Hamidi-Asl E., Hejazi M.S. Preparation of an electrochemical PNA biosensor for detection of target DNA sequence and single nucleotide mutation on p53 tumor suppressor gene corresponding oligonucleotide. Sens. Actuators B. 2011;157:195–201. [Google Scholar]
- Rigo F., Hua Y., Krainer A.R., Bennett C.F. Antisense based therapy for the treatment of spinal muscular atrophy. J. Cell Biol. 2012;199:21–25. doi: 10.1083/jcb.201207087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso V., Bracco E., Pedrola R., Carturan S., Signorino E., Petiti J., Calabrese C., Nicoli P., Gobbi M.D., Gaidano V., Gallo D., Ulisciani S., Fava C., Rege-Cambrin G., Frassoni F., Saglio G., Cilloni D. Detection of BCR-ABL T315I mutation by peptide nucleic acid directed PCR clamping and by peptide nucleic acid FISH. Biomark. Res. 2015;3 doi: 10.1186/s40364-015-0039-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozners E. Recent advances in chemical modification of peptide nucleic acids. J. Nucl. Acids. 2012;2012:518162–518168. doi: 10.1155/2012/518162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadeghizadeh M., Nikravesh A., Behmanesh M. Good, L. cellular morphology and immunologic properties of escherichia coli treated with antimicrobial antisense peptide nucleic acid. Iran. J. Pathol. 2009;4:13–18. [Google Scholar]
- Sahu B., Sacui I., Rapireddy S., Zanotti K.J., Bahal R., Armitage B.A., Ly D.H. Synthesis and characterization of conformationally preorganized, (R)-diethylene glycol-containing γ-peptide nucleic acids with superior hybridization properties and water solubility. J. Org. Chem. 2011;76:5614–5627. doi: 10.1021/jo200482d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders J.M., Wampole M.E., Chen C.P., Sethi D., Singh A., Dupradeau F.Y., Wang F., Gray B.D., Thakur M.L., Wickstrom E. Effects of hypoxanthine substitution in peptide nucleic acids targeting KRAS2 oncogenic mRNA molecules: theory and experiment. J. Phys. Chem. B. 2013;117:11584–11595. doi: 10.1021/jp4064966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano R., Takahashi Y., Nakajima T., Yoshii M., Kubo R., Takahashi K., Kominato Y., Takeshita H., Yasuda T., Tsuneyama H., Uchikawa M., Isa K., Ogasawara K. ABO chimerism with a minor allele detected by the peptide nucleic acid-mediated polymerase chain reaction clamping method. Blood Transfus. 2014;12:431–434. doi: 10.2450/2014.0162-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S., Sonavane U.B., Joshi R.R. Molecular dynamics simulations of cyclohexyl modified peptide nucleic acids (PNA) J. Biomol Struct Dyn. 2010;27:663–676. doi: 10.1080/07391102.2010.10508580. [DOI] [PubMed] [Google Scholar]
- Shen Y., Shrestha R., Ibricevic A., Gunsten S.P., Welch M.J., Wooley K.L., Brody S.L., Taylor J.-S.A., Liu Y. Antisense peptide nucleic acid-functionalized cationic nanocomplex for in vivo mRNA detection. Interface Focus. 2013;3:20120059. doi: 10.1098/rsfs.2012.0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Yang F., Li W., Zhao W., Nie K., Dong B., Liu Z. A review: fabrications: detections and applications of peptide nucleic acids (PNAs) microarray. Biosens. Bioelectron. 2015;6:481–489. doi: 10.1016/j.bios.2014.12.010. [DOI] [PubMed] [Google Scholar]
- Shimada H., Sakurai T., Kitamura Y., Matsuura H., Ihara T. Metallo-regulation of the bimolecular triplex formation of a peptide nucleic acid. Dalton Trans. 2013;42:16006–16013. doi: 10.1039/c3dt51386f. [DOI] [PubMed] [Google Scholar]
- Shiraishi T., Hamzavi R., Nielsen P.E. Subnanomolar antisense activity of phosphonate-peptide nucleic acid (PNA) conjugates delivered by cationic lipids to HeLa cells. Nucl. Acids Res. 2008;36:4424–4432. doi: 10.1093/nar/gkn401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirude P., Kumar V.A., Ganesh K.N. Chimeric peptide nucleic acids incorporating 2S, 5R)-aminoethyl pipecolyl units: synthesis and DNA binding studies. Tettrahedron Lett. 2004;45:3085–3088. [Google Scholar]
- Siddiquee S., Rovina K., Azriah A. A review of peptide nucleic acid. Adv. Tech. Biol. Med. 2015;3 (1000131-10) [Google Scholar]
- Singh R.P., Oh B.-K., Choi J.-W. Applications of peptide nucleic acid towards development of nanobiosensor arrays. Bioelectrochem. 2010;79:153–161. doi: 10.1016/j.bioelechem.2010.02.004. [DOI] [PubMed] [Google Scholar]
- Soh J., Toyooka S., Aoe K., Asano H., Ichihara S., Katayama H., Hiraki A., Kiura K., Aoe M., Sano Y., Sugi K., Shimizu N., Date H. Usefulness of EGFR mutation screening in pleural fluid to predict the clinical outcome of gefitinib treated patients with lung cancer. Int. J. Cancer. 2006;119:2353–2358. doi: 10.1002/ijc.22190. [DOI] [PubMed] [Google Scholar]
- Soofi M.A., Seleem M.N. Targeting essential genes in salmonella enterica serovar typhimurium with antisense peptide nucleic acid. Antimicrob. Agents Chemother. 2012;56:6407–6409. doi: 10.1128/AAC.01437-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stafforst T., Hilvert D. Modulating PNA/DNA hybridization by light. Angew. Chem. 2010;49:9998–10001. doi: 10.1002/anie.201004548. [DOI] [PubMed] [Google Scholar]
- Sugiyama T., Kittaka A. Chiral peptide nucleic acids with a substituent in the N-(2-aminoethy)glycine backbone. Molecules. 2013;18:287–310. doi: 10.3390/molecules18010287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T., Imamura Y., Demizu Y., Kurihara M., Takano M., Kittaka A. β-PNA: peptide nucleic acid (PNA) with a chiral center at the b-position of the PNA backbone. Biorg. Med. Chem. Lett. 2011;21:7317–7320. doi: 10.1016/j.bmcl.2011.10.017. [DOI] [PubMed] [Google Scholar]
- Tao C.-h., Chen T.-f., Yadav P.K., Wu R.-j., Qiu H.-j., Wu W., Liu Z.-j. The role of mucin gene 1 mediated anti-MKN-45 cell invasion. Zhonghua Xiaohua Zazhi. 2012;32:175–179. [Google Scholar]
- Tonelli R., McIntyre A., Camerin C., Walters Z.S., Leo K.D., Selfe J., Purgato S., Missiaglia E., Tortori A., Renshaw Astolfi A., Taylor K.R., Serravalle S., Bishop R., Nanni C., Valentijn L.J., Faccini A., Leuschner I., Formica S., Reis-Filho J.S., Ambrosini V., Thway K., Franzoni M., Summersgill B., Marchelli R., Hrelia P., Cantelli-Forti G., Fanti S., Corradini R., Pession A., Shipley J. Antitumor Activity of Sustained N-Myc reduction in rhabdomyosarcomas and transcriptional block by antigene therapy. J. Clin. Cancer Res. 2012;18:796–807. doi: 10.1158/1078-0432.CCR-11-1981. [DOI] [PubMed] [Google Scholar]
- Totsingan F., Jain V., Bracken W.C., Faccini A., Tedeschi T., Marchelli R., Corradini R., Kallenbach N.R., Green M.M. Conformational heterogeneity in PNA:PNA duplexes. Macromolecules. 2010;43:2692–2703. [Google Scholar]
- Uno T., Tabata H., Kawai T. Peptide-nucleic acid-modified ion-sensitive field-effect transistor-based biosensor for direct detection of DNA hybridization. Anal. Chem. 2007;79:52–59. doi: 10.1021/ac060273y. [DOI] [PubMed] [Google Scholar]
- Upert G., Mehiri M., Giorgio A.D., Condom R., Patino N. Solid-phase synthesis and thermal denaturation study of cyclic PNAs targeting the HIV-1 TAR RNA loop. Bioorg. Med. Chem. Lett. 2007;17:6026–6030. doi: 10.1016/j.bmcl.2007.07.030. [DOI] [PubMed] [Google Scholar]
- Upert G., Giorgio A.D., Upadhyay A., Manvar D., Pandey N., Pandey V.N., Patino N. Inhibition of HIV replication by cyclic and hairpin PNAs targeting the HIV-1 TAR RNA loop. J. Nucl. Acids. 2012:591025–591029. doi: 10.1155/2012/591025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandersea M.W., Litaker R., Wayne Yonnish B., Sosa E., Landsberg J.H., Pullinger C., Moon-Butzin P., Green J., Morris J.A., Kator H. Molecular assays for detecting aphanomyces invadans in ulcerative mycotic fish lesions. Appl. Environ. Microbiol. 2006;72:1551–1557. doi: 10.1128/AEM.72.2.1551-1557.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldhoen S., Laufer S.D., Restle T. Review Recent developments in peptide-based nucleic acid delivery. Int. J. Mol. Sci. 2008;9:1276–1320. doi: 10.3390/ijms9071276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Shen H., Feng J., Yang H. PNA-Modified magnetic nanoparticles and their hybridization with single-stranded DNA target: surface enhanced raman scatterings study. Microchim. Acta. 2006;153:15–20. [Google Scholar]
- Wang G.H., Huang G.J., Yu S.C., Zhang Z.Y., Qian G.S. The effect of blocking the expression of mtDNA on lung cancer SPC-A1 cell by antisense peptide nucleic acid. Chongqing Yike Daxue Xuebao. 2009;34:1457–1461. [Google Scholar]
- Wang M., Holmes-Davis R., Rafinski Z., Jedrzejewska B., Choi K.Y., Zwick M., Bupp C., Izmailov A., Paczkowski J., Warner B., Koshinsky H. Accelerated photobleaching of a cyanine dye in the presence of a ternary target DNA, PNA probe, dye catalytic complex: a molecular diagnostic. Anal. Chem. 2009;81:2043–2052. doi: 10.1021/ac702519k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., He Y., Xia Y., Wang L., Liang S. Inhibition of gene expression and growth of multidrug-resistant acinetobacter baumannii by antisense peptide nucleic acids. Mol. Biol. Rep. 2014;41:7535–7541. doi: 10.1007/s11033-014-3643-2. [DOI] [PubMed] [Google Scholar]
- Wojciechowski F., Hudson R.H.E. Peptide nucleic acid containing a meta-substituted phenylpyrrolo cytosine exhibits a fluorescence response and increased binding affinity toward RNA. Org. Lett. 2009;11:4878–4881. doi: 10.1021/ol9019474. [DOI] [PubMed] [Google Scholar]
- Xuan F., Luo X., Hsing I.-M. Sensitive immobilization-free electrochemical DNA sensor based on isothermal circular strand displacement polymerization reaction. Biosens. Bioelectron. 2012;35:230–234. doi: 10.1016/j.bios.2012.02.054. [DOI] [PubMed] [Google Scholar]
- Yoon S.-H., Choi Y.-D., Oh I.J., Kim K.S., Choi H., Chang J., Shin H.J., Park C.-K., Kim Y.C. Peptide nucleic acid clamping versus direct sequencing for the detection of EGFR gene mutation in patients with non-small cell lung cancer. Cancer Res. Treat. 2015;47:661–669. doi: 10.4143/crt.2014.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambaldo C., Barluenga S., Winssinger N. PNA-encoded chemical libraries. Curr. Opin. Chem. Biol. 2015;26:8–15. doi: 10.1016/j.cbpa.2015.01.005. [DOI] [PubMed] [Google Scholar]
- Zeng Z., Han S., Hong W., Lang Y., Li F., Liu Y., Li Z., Wu Y., Li W., Zhang X., Cao Z. A tat-conjugated peptide nucleic acid tat-PNA-DR inhibits hepatitis B virus replication in vitro and In vivo by targeting LTR direct repeats of HBV RNA. Mol. Ther. Nucl. Acids. 2016;5:e295. doi: 10.1038/mtna.2016.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Zhao X., Wang J., Zhang J., Wang Y., Sun L., Dai C., Li D., Jiang Z. Experimental study on imaging of 99Tcm labeled c-myc mRNA antisense PNA in colorectal cancer tumor-bearing nude mice. Zhonghua Heyixue Zazhi. 2008;28:181–183. [Google Scholar]
- Zhao X., Zhang Z., Wang J., Zhang J., Wang Y. Preparation of 99Tcm labeled c-myc mRNA antisense peptide nucleic acid and its biodistribution in tumor-bearing nude mice. Zhonghua Heyixue Zazhi. 2008;28:304–307. [Google Scholar]
- Zhao X., Liu Y., Dai M., Ren X., Wang J., Zhang J., Wang Y., Zhang Z., Zhao X., Dai C. Comparative study on imaging of 99Tcm-survivin mRNA antisense peptide nucleic acid in tumor and inflammation animal models. Zhonghua Heyixue Zazhi. 2011;31:364–367. [Google Scholar]
- Zhao X., Dai M., Liu Y., Wang J., Zhang J., Wang Y., Zhang Z., Dai C., Li D. Preparation of 99Tcm labeled survivin mRNA antisense PNA and gene imaging in nude mice bearing lung carcinoma A549 xenografts. Zhonghua Heyixue Zazhi. 2011;31:339–343. [Google Scholar]