Abstract
Severe acute respiratory syndrome (SARS) is an emerging infectious disease caused by a novel coronavirus (SARS-CoV). SARS-CoV spike (S) protein, a type I membrane-bound protein, is essential for the viral attachment to the host cell receptor angiotensin-converting enzyme 2 (ACE2). By screening 312 controlled Chinese medicinal herbs supervised by Committee on Chinese Medicine and Pharmacy at Taiwan, we identified that three widely used Chinese medicinal herbs of the family Polygonaceae inhibited the interaction of SARS-CoV S protein and ACE2. The IC50 values for Radix et Rhizoma Rhei (the root tubers of Rheum officinale Baill.), Radix Polygoni multiflori (the root tubers of Polygonum multiflorum Thunb.), and Caulis Polygoni multiflori (the vines of P. multiflorum Thunb.) ranged from 1 to 10 μg/ml. Emodin, an anthraquinone compound derived from genus Rheum and Polygonum, significantly blocked the S protein and ACE2 interaction in a dose-dependent manner. It also inhibited the infectivity of S protein-pseudotyped retrovirus to Vero E6 cells. These findings suggested that emodin may be considered as a potential lead therapeutic agent in the treatment of SARS.
Abbreviations: SARS, severe acute respiratory syndrome; SARS-CoV, SARS coronavirus; S, spike; ACE2, angiotensin-converting enzyme 2; HIV, human immunodeficiency virus; ELISA, enzyme-linked immunosorbent assay; E. coli, Escherichia coli; SDS-PAGE, sodium dodecyl sulfate-polyacrylamide gel electrophoresis; PBS, phosphate-buffered saline; BSA, bovine serum albumin; IFA, immunofluorescence assay; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; HSV, herpes simplex virus
Keywords: SARS coronavirus, Spike protein, Angiotensin-converting enzyme 2, Emodin
1. Introduction
Severe acute respiratory syndrome (SARS) is a new human disease that results in progressive respiratory failure and death in close to 10% of infected individuals (Ksiazek et al., 2003, Peiris et al., 2003). The etiological agent, SARS coronavirus (SARS-CoV) (Drosten et al., 2003, Fouchier et al., 2003) contains a single-stranded plus-sense RNA genome about 30 kb in length that has a 5′-cap structure and a 3′-polyadenylation tract (Marra et al., 2003, Rota et al., 2003). The genomic organization is typical of coronaviruses, having 14 potential major open reading frames that encode replicase, spike (S), envelope, membrane, and nucleocapsid proteins in the same order as those of other coronaviruses (Tan et al., 2005).
SARS-CoV S protein is a large type I membrane glycoprotein projection from viral envelope (Bosch et al., 2003). SARS-CoV S protein is responsible for binding to cellular receptors and for mediating the fusion of viral and host membranes (Simmons et al., 2004, Tripet et al., 2004). It also contains important virus-neutralizing epitopes that elicit neutralizing antibody in the host species (Hofmann et al., 2004a, Sui et al., 2004). Furthermore, mutations in this gene dramatically affect the virulence, pathogenesis, and host cell tropism (Petit et al., 2005, Yi et al., 2005). Angiotensin-converting enzyme 2 (ACE2) has been identified as a functional receptor for SARS-CoV (Li et al., 2003). Soluble S fragment or ACE2 is able to block S protein-mediated infection (Hofmann et al., 2004b, Moore et al., 2004). Monoclonal antibodies against S protein efficiently neutralize SARS-CoV in vitro and in vivo (Greenough et al., 2005, Sui et al., 2004). Moreover, vaccines that express the S protein induce T cell and neutralizing antibody responses, and protect animals from SARS-CoV infection (Chen et al., 2005, Yang et al., 2004). These findings clearly suggest that blocking the binding of the S protein with its cellular receptor can prevent virus entry.
Recent reports indicate that human immunodeficiency virus protease inhibitors (lopinavir and ritonavir), an anti-psychotic drug (promazine), and an anti-parasitic drug (niclosamide) are effective inhibitors of SARS-CoV infection by inhibiting the SARS-CoV main proteinase (Chan et al., 2003, Wu et al., 2004a, Zhang and Yap, 2004). Natural products, such as glycyrrhizin, baicalin, reserpine, luteolin, ginsenoside-Rb1 and aescin, also abolished SARS-CoV production in vitro; however, the antiviral mechanisms of these compounds remained unclear (Chen et al., 2004, Cinatl et al., 2003a, Wu et al., 2004b, Yi et al., 2004). One of the logical targets of the viral life cycle to inhibit SARS-CoV replication is the step following entry of the infectious virion into the host cell. To discover inhibitors at this stage, we established a biotinylated enzyme-linked immunosorbent assay (ELISA) to evaluate the inhibitory effects of Chinese medicinal herbs on the S protein and ACE2 interaction. Herein we reported the finding that emodin, the likely active component from Rheum officinale and Polygonum multiflorum, blocked both the binding of S protein to ACE2 and the infectivity of S protein-pseudotyped retrovirus to Vero E6 cells.
2. Materials and methods
2.1. Chemicals
Emodin, rhein, and promazine hydrochloride were purchased from Sigma (St. Louis, MO, USA). Chrysin, anthraquinone, and 1,4-bis-(1-anthraquinonylamino)-anthraquinone were purchased from Aldrich (Germany). Emodin (MW = 270.24), rhein (MW = 284.22), chrysin (MW = 254.24), and 1,4-bis-(1-anthraquinonylamino)-anthraquinone (MW = 665.67) were dissolved in dimethyl sulfoxide at 37, 35, 33, and 50 mM, respectively. Anthraquinone (MW = 208.22) was dissolved in ethanol at 5 mM. Promazine hydrochloride (MW = 320.88) was dissolved in water at 1 M. All chemicals were protected from light and stored at −30 °C in small aliquots.
2.2. Purification and biotinylation of recombinant SARS-CoV S protein
The SARS-CoV S gene was cloned into pET-28b(+) (Novagen, Madison, WI, USA) to an N-terminal fusion with six histidine residues. Recombinant S protein was expressed in Escherichia coli (E. coli) BL21(DE3)pLysS strain. The expression and purification of recombinant SARS-CoV S protein were performed as described previously (Ho et al., 2004). Protein was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and quantified with a Bradford method (Bio-Rad, Hercules, CA, USA). Recombinant S protein was biotinylated as described previously (Ho et al., 2006). The unincorporated biotin was removed by centricon-10 filtration (Millipore, Bedford, MA, USA), and the biotinylated S protein was stored at 4 °C until further analysis.
2.3. Preparation of herb extracts
Chinese medicinal herbs were gifts from Chuang Song Zong Pharmaceutical Co., Ltd (Kaohsiung, Taiwan). Plant sample was ground with the homogenizer to a fine powder. The aqueous extract was prepared by mixing 100 g of each herb powder with 500 ml of deionized water and shaking at 4 °C overnight. The extract was centrifuged at 10,000 × g for 5 min, and the supernatant was evaporated under vacuum to dryness and resuspended in deionized water to a final concentration of 1 mg/ml. The extracts were stored at −20 °C in small aliquots.
2.4. Biotinylated ELISA
The biotinylated ELISA was performed as described previously (Ho et al., 2006). Briefly, microtiter plates (MaxiSorp Nunc-Immum™ plates, Nunc, Denmark) were coated at 4 °C overnight with 50 μl of 0.2 ng/μl ACE2 (R&D Systems, Minneapolis, MN, USA), rinsed with 200 μl washing buffer (0.5% Tween 20 in phosphate-buffered saline (PBS) (137 mM NaCl, 1.4 mM KH2PO4, 4.3 mM Na2HPO4, 2.7 mM KCl, pH 7.2)), and blocked with 200 μl blocking buffer (5% bovine serum albumin (BSA) in washing buffer) by incubating at 37 °C for 30 min. The absorbed protein in each well was challenged with 50 μl of 1 ng/μl biotinylated S protein and incubated at 37 °C for 1 h. After three washes with washing buffer, 50 μl diluted peroxidase-conjugated avidin was added to each well and incubated at 37 °C for 1 h. Following three washes, 50 μl chromogenic substrate, 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) (Sigma, St. Louis, MO, USA), was added to each well and incubated at 37 °C for 15 min. The absorbance was read at 405 nm in an ELISA plate reader (Anthos Labtec Instruments, Austria).
For the competition assay, biotinylated S protein was mixed with various amounts of extracts and incubated at 37 °C with shaking. After a 2-h incubation, the mixture was added to wells, which were coated with ACE2, and incubated at 37 °C for 1 h. Following three washes, peroxidase-conjugated avidin and chromatic substrate were sequentially added. The absorbance was read at 405 nm in an ELISA plate reader. The percent inhibition was calculated by [1 − (OD value of mixture containing extract and S protein/OD value of mixture containing S protein only)] × 100. The IC50 value was determined as the quantity of compound required to inhibit the interaction between S protein and ACE2 at 50%.
2.5. Immunofluorescence assay (IFA)
Vero E6 cells (104 cells) were seeded in 24-well plates containing glass coverslips and incubated at 37 °C for 1 day. The coverslips were then rinsed with PBS, fixed with 3.7% PBS-buffered formaldehyde at room temperature for 30 min, and blocked with 1% BSA at 37 °C for 1 h. After four washes with PBS, biotin-labeled S protein was added to each coverslip and incubated at 4 °C overnight. Following four washes with PBS, diluted fluorescence-conjugated streptavidin (Chemicon, Temecula, CA, USA) was added and incubated at 37 °C for 90 min in the dark. The coverslips were then washed four times with PBS, placed onto glass slides, mounted with fluoromount G (Electron Microscopy Sciences, Hatfield, PA, USA), and observed under a confocal microscope (Leica, Germany).
2.6. Infection with S protein-pseudotyped retrovirus
Recombinant retroviruses expressing a luciferase reporter gene and pseudotyped with S proteins were produced as described previously (Sui et al., 2005). Briefly, 293T cells were cotransfected with a plasmid pcDNA-spike encoding S protein, a plasmid pCMVΔR8.2 encoding HIV-1 Gag-Pol, and a plasmid pHIV-Luc encoding the firefly luciferase reporter gene under control of the HIV-1 long terminal repeat. Forty-eight hours later, viral supernatants were harvested, mixed with various amounts of compound, and incubated at 37 °C with shaking. After a 2-h incubation, the mixture was added to ACE2-expression Vero E6 cells in a 96-well plate. Forty-eight hours postinfection, cells were harvested and the luciferase activity was assayed as previously described (Hsiang et al., 2005). Relative infectivity is calculated as dividing the relative luciferase unit of compound/pseudovirus-infected cells by the relative luciferase unit of pseudovirus-infected cells.
2.7. 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay
Cell viability was monitored by MTT colorimetric assay. Briefly, cells were cultivated in 96-well culture plates. After a 24-h incubation at 37 °C, various amounts of compounds were added to confluent cell monolayers and incubated for another 24 h. One-tenth volume of 5 mg/ml MTT was then added to the culture medium. After a 4-h incubation at 37 °C, equal cell culture volume of 0.04 N HCl in isopropanol was added to dissolve the MTT formazan, and the absorbance value was measured at 570 nm using a microplate reader. Cell viability (%) was calculated by (OD of treated cells/OD of untreated cells) × 100.
2.8. Statistical analysis
Data were presented as mean ± standard error. Student's t test was used for comparisons between two experiments. A value of p < 0.05 was considered statistically significant.
3. Results
3.1. Recombinant S protein binds to ACE2 in a dose-dependent manner
We established a biotinylated ELISA to evaluate the binding efficacy of S protein to ACE2. The 138-kDa recombinant S protein was expressed in soluble form in E. coli and purified to homogeneity (Fig. 1A). The recombinant S protein was biotinylated and its ability to interact with ACE2 coated on ELISA plates was evaluated. As shown in Fig. 1B, the OD value increased as the amount of S protein increased, indicating that recombinant S protein bound to ACE2 in a dose-dependent manner.
Fig. 1.
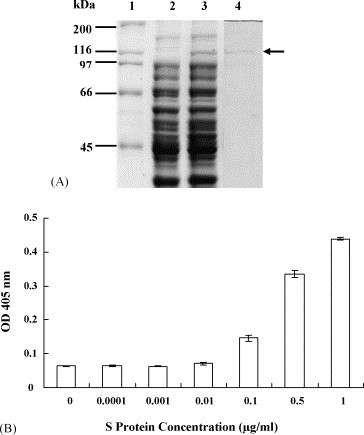
Analysis of SARS-CoV S protein and ACE2 interaction. (A) SDS-PAGE analysis of recombinant SARS-CoV S protein. The preparations of uninduced E. coli (lane 2), induced E. coli (lane 3), and purified recombinant S protein (lane 4) were analyzed by 10% SDS-PAGE and stained by Coomassie brilliant blue. The molecular masses of protein standard (lane 1) are shown at the left. The location of the 138-kDa recombinant S protein is indicated by the arrow. (B) The binding ability of SARS-CoV S protein to ACE2 by biotinylated ELISA. The wells were coated with 1 ng of ACE2 and challenged with various amounts of biotin-labeled S protein. Following three washes, peroxidase-conjugated avidin and chromatic substrate were sequentially added. The absorbance was read at 405 nm in an ELISA plate reader. Values are mean ± standard error of three independent assays.
3.2. Chinese medicinal herbs block the binding of S protein to ACE2
To evaluate the inhibitory effects of Chinese medicinal herbs on the S protein and ACE2 interaction, we mixed 1 μg of aqueous herbal extracts with 0.05 μg biotin-labeled S protein, incubated the mixture at 37 °C for 2 h, and added to ACE2-coated wells. From a screen of 312 controlled Chinese medicinal herbs supervised by the Committee on Chinese Medicine and Pharmacy at Taiwan, we found that various herbal extracts inhibited the interaction (data not shown). We further divided these herbs into 32 families and compared their level of inhibition with the taxonomic characterization. As shown in Fig. 2 , 25 out of 32 families significantly abolished the interaction between S protein and ACE2. Six families, including Nelumbonaceae, Labiatae, Magnoliaceae, Oleaceae, Lauraceae and Polygonaceae, blocked 60–90% of the binding of S protein to ACE2 at 1 μg. Among these families, Polygonaceae exhibited the highest inhibition on the S protein and ACE2 interaction, with the inhibitory percentage of 86.33 ± 4.55%.
Fig. 2.
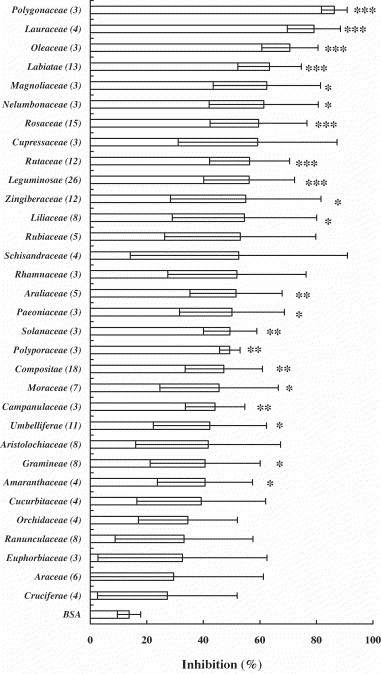
Inhibitory effects of Chinese medicinal herbs on the interaction between SARS-CoV S protein and ACE2. Biotin-labeled S protein (0.05 μg) was mixed with 1 μg of aqueous extracts of herbs, and the mixtures were added to wells, which were coated with 1 ng of ACE2. The biotinylated ELISA was performed as described in Section 2. Results are expressed as inhibition (%) described in Section 2. Numbers in the brackets are the sum of herb species in the family. Values are mean ± standard error of three independent assays. *p < 0.05, **p < 0.01, ***p < 0.001, compared with BSA.
Three kinds of Chinese medicinal herbs belonging to Polygonaceae were further analyzed. Radix et Rhizoma Rhei (Da-Huang) is the root tuber of plant R. officinale Baill. Radix Polygoni multiflori (Ho-Shou-Wu) and Caulis Polygoni multiflori (Yeh-Chiao-Teng) are the root tuber and vine of plant P. multiflorum Thunb., respectively. Preincubation of these herbs with biotin-labeled S protein inhibited the binding of S protein to ACE2 in a dose-dependent manner (Fig. 3 ). The IC50 values for Radix et Rhizoma Rhei, Radix Polygoni multiflori, and Caulis Polygoni multiflori ranged from 1 to 10 μg/ml.
Fig. 3.
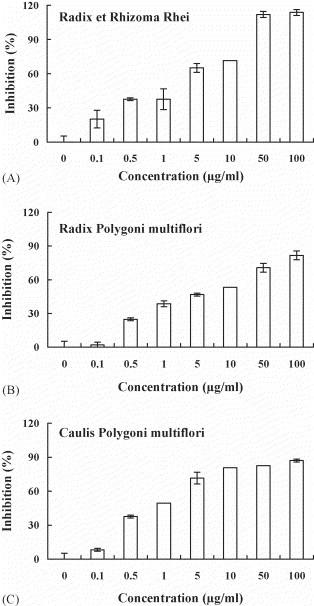
Inhibitory effects of Radix et Rhizoma Rhei, Radix Polygoni multiflori, and Caulis Polygoni multiflori on the SARS-CoV S protein and ACE2 interaction. Biotin-labeled S protein (0.05 μg) was mixed with various amounts of aqueous extracts from Radix et Rhizoma Rhei (A), Radix Polygoni multiflori (B) or Caulis Polygoni multiflori (C), and the mixtures were added to wells, which were coated with 1 ng of ACE2. The biotinylated ELISA was performed as described in Section 2. Results are expressed as inhibition (%) described in Section 2. Values are mean ± standard error of six independent assays.
3.3. Emodin inhibits the interaction between S protein and ACE2
As emodin (1,3,8-trihydroxy-6-methylanthraquinone), rhein (1,8-dihydroxy-3-carboxyl-9,10-anthraquinone), and chrysin (5,7-dihydroxyflavone) are produced in high levels in genus Rheum and Polygonum (Koyama et al., 2003, Nonaka et al., 1977), we investigated whether these compounds were responsible for blocking the binding of S protein to ACE2. Emodin and rhein belong to anthraquinone compounds, while chrysin belongs to a flavonoid compound (Fig. 4 ). As shown in Fig. 5A, emodin blocked the binding of S protein to ACE2 in a dose-dependent manner. The IC50 value of emodin is 200 μM. Preincubation of rhein with biotinylated S protein slightly inhibited the S protein and ACE2 interaction (Fig. 5B). Chrysin exhibited a weak inhibition on the S protein and ACE2 interaction at 400 μM; however, it significantly stimulated the binding of S protein to ACE2 at 50 μM (Fig. 5C). These results suggested that emodin was the likely active constituent of Rheum and Polygonum responsible for blocking the binding of S protein to ACE2.
Fig. 4.
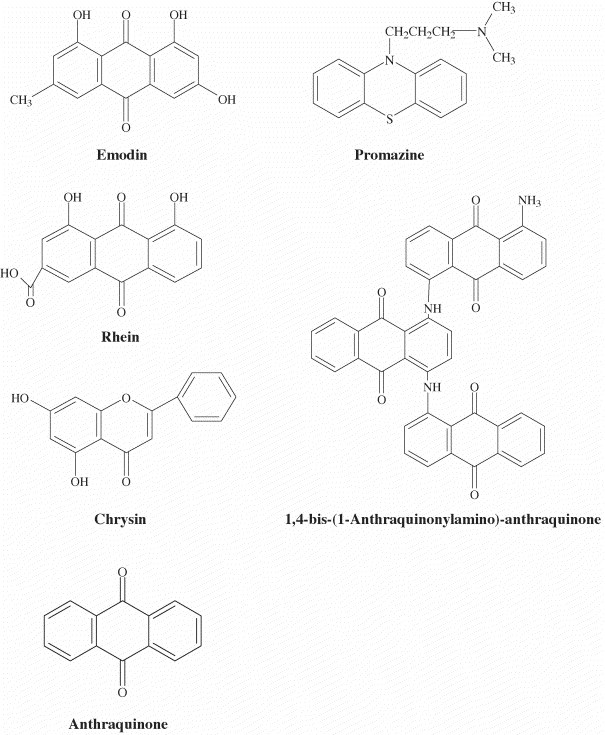
Chemical structures of compounds used in this study.
Fig. 5.
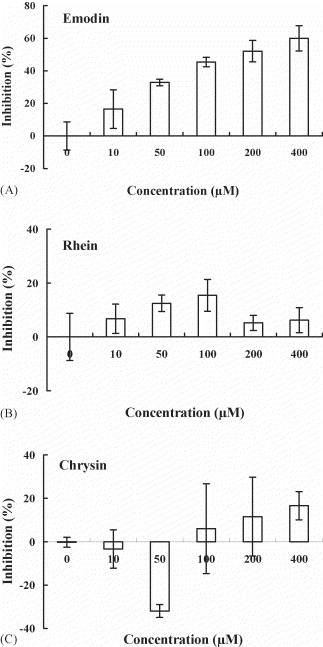
Inhibitory effects of emodin, rhein, and chrysin on the interaction between SARS-CoV S protein and ACE2. Biotin-labeled S protein (0.05 μg) was mixed with various amounts of emodin (A), rhein (B) or chrysin (C), and the mixtures were added to wells, which were coated with 1 ng of ACE2. The biotinylated ELISA was performed as described in Section 2. Results are expressed as inhibition (%) described in Section 2. Values are mean ± standard error of six independent assays.
Emodin is an anthraquinone compound consists of three cyclic rings. The anti-psychotic drug promazine, which has been shown to exhibit the significant effect in inhibiting the replication of SARS-CoV (Zhang and Yap, 2004), shared a similar structure with emodin (Fig. 4). Therefore, we compared the inhibitory effects of emodin, promazine, and two other anthraquinone compounds on the S protein and ACE2 interaction. These compounds all showed inhibition on the binding of S protein to ACE2 (Fig. 6 ); however, the inhibitory activities of anthraquinone and 1,4-bis-(1-anthraquinonylamino)-anthraquinone were not significant. As compared to emodin, promazine exhibited the highest inhibition; however, the differences between emodin and promazine were not significant. These findings suggested that emodin and promazine might be capable of inhibiting SARS-CoV infectivity by blocking the S protein and ACE2 interaction.
Fig. 6.
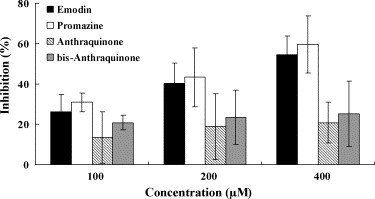
Inhibitory effects of emodin and emodin-like compounds on the SARS-CoV S protein and ACE2 interaction. Biotin-labeled S protein (0.05 μg) was mixed with various amounts of emodin, promazine, anthraquinone or 1,4-bis-(1-anthraquinonylamino)-anthraquinone, and the mixtures were added to wells, which were coated with 1 ng of ACE2. The biotinylated ELISA was performed as described in Section 2. Results are expressed as inhibition (%) described in Section 2. Values are mean ± standard error of six independent assays.
3.4. Emodin abolishes the infectivity of protein-pseudotyped retrovirus to Vero E6 cells
The inhibitory potential of emodin on the SARS-CoV S protein and Vero E6 cell interaction was further evaluated by IFA. Emodin and promazine displayed no cytotoxicity in the concentration used in IFA. No cellular morphological change has been observed in emodin or promazine-treated cells. Vero E6 cells treated with BSA showed negative result, while cells treated with biotin-labeled S protein purified from E. coli or baculovirus showed the strong fluorescence (Fig. 7 ). Treatment of Vero E6 cells with either emodin/biotinylated S protein or promazine/biotinylated S protein diminished the cell-associated fluorescence. These results indicated that emodin and promazine were able to block the S protein and Vero E6 cell interaction.
Fig. 7.
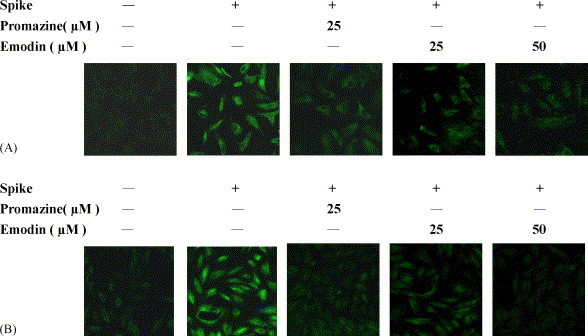
Inhibitory effects of emodin and promazine on the interaction between SARS-CoV S protein and Vero E6 cells. Vero E6 cells were cultured on glass coverslips and treated with biotin-labeled S protein in the presence of various amounts of compounds. Recombinant S protein was purified from E. coli (A) or baculovirus (B). After a 16-h incubation at 4 °C, cells were stained with fluorescence-conjugated streptavidin and evaluated under a confocal microscope. Magnification, 400×.
S protein-pseudotyped retrovirus infectivity was also used to evaluate the inhibitory potential of emodin. Vero E6 cells transfected with the plasmid encoding ACE2 were infected with S protein-pseudotyped retrovirus in the presence or absence of compounds. Emodin inhibited the S protein-pseudotyped retrovirus infectivity in a dose-dependent manner (Fig. 8 ). The percent inhibition of emodin at 50 μM was 94.12 ± 5.90%. Because Vero cells treated with 50 μM emodin remained 82.4 ± 3.8% viability, the anti-SARS-CoV activity of emodin was not due to toxicity. Promazine also inhibited the S protein-pseudotyped retrovirus infectivity in a dose-dependent manner. Promazine exhibited the cytotoxicity effect at higher concentration (>25 μM). Thus, the possibility that the antiviral activity of promazine was due to toxicity could not be excluded. However, the relative infectivity was 0.23 ± 0.2 when pseudovirus was treated with 5 μM promazine. At the same concentration, cell viability remained 95.6 ± 7.7% by MTT assay. Thus, the anti-SARS-CoV activity of promazine at 5 μM was not due to toxicity. Our findings indicated that emodin was a novel anti-SARS-CoV compound that was able to block the SARS-CoV S protein binding to Vero E6 cells.
Fig. 8.
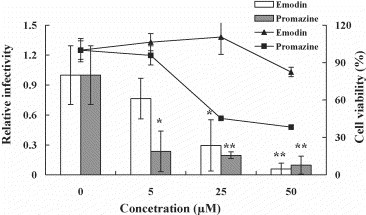
Inhibitory effects of emodin and promazine on the SARS-CoV S protein-pseudotyped retrovirus infectivity. S protein-pseudotyped retroviruses were mixed with various amounts of emodin and promazine, and then incubated at 37 °C with shaking. After a 2-h incubation, the mixtures were inoculated with Vero E6 cells transfected with the plasmid encoding ACE2. The luciferase activity of cell lysate was assayed 2 days postinfection. The cell viability was determined after a 24-h treatment with emodin or promazine. The bars and lines represent the relative infectivity and cell viability (%), respectively, described in Section 2. Values are mean ± standard error of three independent assays. *p < 0.05, **p < 0.01, compared with untreated cells.
4. Discussion
Various clinically approved drugs, including nucleoside analogs, interferons, HIV protease inhibitors, anti-psychotic drug, anti-parasitic drug and antibiotics, have been screened for their anti-SARS-CoV effects (Cinatle et al., 2005, Tan et al., 2004, Wu et al., 2004a). Interferons and ribavirin exhibit anti-SARS-CoV effects at high concentrations; however, the significant cytotoxic effects of these compounds are also observed (Cinatl et al., 2003b). Promazine and niclosamide have been shown to exhibit significant effects in inhibiting the replication of SARS-CoV by abolishing the main proteinase activity (Zhang and Yap, 2004). In addition to clinically approved drugs, some components of traditional Chinese medicine are found to be effective inhibitors of SARS-CoV replication (Wu et al., 2004b). For examples, glycyrrhizin, the bioactive compounds of licorice root, is one of the first compounds found to be active against SARS-CoV in vitro (Cinatl et al., 2003a). Sinigrin, a phenolic compound derived from Isatis indigotica root, exhibits the anti-SARS-CoV potential by inhibiting SARS-CoV main proteinase activity (Lin et al., 2005). Baicaline, a flavonoid derived from Scutellaria baicalensis, inhibits SARS-CoV replication in vitro and in vivo (Chen et al., 2004). These findings might explain some beneficial effects of traditional Chinese medicine observed in SARS patients (Zhang et al., 2004). It also suggested the anti-SARS-CoV potential of natural products from Chinese medicinal herbs.
By screening 312 herbs, we identified three widely used Chinese medicinal herbs of the family Polygonaceae exhibited the inhibitory efficacy on the SARS-CoV S protein and ACE2 interaction. Radix et Rhizoma Rhei is widely used in Southeast Asian folk medicine to alleviate liver and kidney damage (Agarwal et al., 2000, Arosio et al., 2000). Radix Polygoni multiflori and Caulis Polygoni multiflori are the most famous tonic traditional medicines in China and Japan, and clinically used for the treatment of coronary heart disease hyperlipidemia, neurosis, and other diseases commonly associated with aging (Xiao et al., 1993). Radix et Rhizoma Rhei has been shown to inhibit herpes simplex virus (HSV) infection by blocking both the attachment and penetration processes (Hsiang et al., 2001). Radix Polygoni multiflori and Caulis Polygoni multiflori have never been identified to exhibit the antiviral effects. In this study, we demonstrated that these herbs might be capable of inhibiting SARS-CoV infection by blockade of the binding of SARS-CoV S protein to cellular receptors.
The genus Rheum and Polygonum are sources of a wide range of phenolic compounds, flavonoids, anthraquinone, stilbenes, and tannins (Nonaka et al., 1977). Anthraquinones, derived from the methanolic extract of Radix et Rhizoma Rhei, have been shown to disrupt the viral envelope, resulting in the abolishment of viral adsorption (Sydiskis et al., 1991). Several studies indicated that anthraquinones exhibited anti-HIV, anti-human cytomegalovirus, anti-HSV, and anti-Epstein-Barr virus activities (Barnard et al., 1992, Konoshima et al., 1989, Schinazi et al., 1990). Recent study further indicated that anthraquinone show antiviral activity but only emodin is a virucidal agent (Alves et al., 2004). Emodin is the active constituent deriving from genus Rheum and Polygonum. Emodin possesses antibacterial, diuretic, and vasorelaxant effects (Huang et al., 1991, Koyama et al., 1988, Zhou and Chen, 1988). It also exhibits anti-inflammatory, anti-proliferative, and anti-carcinogenic properties (Chen et al., 2002, Kumar et al., 1998). Emodin is suspected to exhibit its antiviral activity by inhibiting casein kinase 2, which is exploited by many viruses for the phosphorylation of proteins that are essential to their life cycle (Battistutta et al., 2000, Guerra and Issinger, 1999). It also has been demonstrated that emodin disrupted the lipid bilayer, resulting in the inactivation of enveloped virus (Sydiskis et al., 1991). In this study, we demonstrated that emodin was able to block the SARS-CoV S protein and ACE2 interaction. Preincubation of emodin with S protein or S protein-pseudotyped retrovirus also abolished the SARS-CoV and Vero E6 cell interaction. These findings suggested that in addition to disrupting the viral envelope, emodin might abolish SARS-CoV infection by competing the binding site of S protein with ACE2.
Emodin is an anthraquinone compound consisting of three cyclic rings. Promazine, which has demonstrated anti-SARS-CoV effect, is also a phenolic compound consisting of three cyclic rings. Based on the structural similarity, we wondered whether other anthraquinones or phenolic compounds also exhibited the anti-SARS effects. By comparing the inhibitory efficacy of promazine, anthraquinone, 1,4-bis-(1-anthraquinonylamino)-anthraquinone with emodin, we found that anthraquinone and 1,4-bis-(1-anthraquinonylamino)-anthraquinone slightly inhibited the S protein and ACE2 interaction. Emodin and promazine blocked the S protein and ACE2 in a dose-dependent manner. These results suggested that the side chains but not the anthraquinone skeleton has a great impact on the S protein and ACE2 binding. These findings also indicated that promazine exhibited anti-SARS effect by inhibiting both the virus entry and protein processing.
5. Conclusion
By screening 312 herbs, we identified three widely used Chinese medicinal herbs of the family Polygonaceae which inhibited the SARS-CoV S protein and ACE2 interaction. Emodin, the major components of the genus Rheum and Polygonum, is the likely active constituent responsible for blocking both the binding of SARS-CoV S protein to ACE2 and the infectivity of S protein-pseudotyped retrovirus to Vero E6 cells. These findings suggested that emodin was a novel anti-SARS-CoV compound and might be considered as a potential therapeutic agent in the treatment of SARS.
Acknowledgments
We thank Prof. M. Farzan for providing plasmids for the construction of pseudovirus. We thank Miss Y.C. Wei for her technical assistance. This work was supported by grants from National Science Council (NSC 92-2751-B-039-001-Y and NSC 92-2751-B-039-005-Y) and China Medical University (CMU93-M-05 and CMU94-117), Taiwan, ROC.
References
- Agarwal S.K., Singh S.S., Verma S., Kumar S. Antifungal activity of anthraquinone derivatives from Rheum emodi. J. Ethnopharmacol. 2000;72:43–46. doi: 10.1016/s0378-8741(00)00195-1. [DOI] [PubMed] [Google Scholar]
- Alves D.S., Pe′rez-Fons L., Estepa A., Micol V. Membrane-related effects underlying the biological activity of the anthraquinones emodin and barbaloin. Biochem. Pharmacol. 2004;68:549–561. doi: 10.1016/j.bcp.2004.04.012. [DOI] [PubMed] [Google Scholar]
- Arosio B., Gagliano N., Fusaro L.M., Parmeggiani L., Tagliabue J., Galetti P., DeCastri D., Moscheni C., Annoni G. Aloe-Emodin quinone pretreatment reduces acute liver injury induced by carbon tetrachloride. Pharmacol. Toxicol. 2000;87:229–233. doi: 10.1034/j.1600-0773.2000.d01-79.x. [DOI] [PubMed] [Google Scholar]
- Barnard D.L., Huffman J.H., Morris J.L.B., Wood S.G., Hughes B.G., Sidwell R.W. Evaluation of the antiviral activity of anthraquinones, anthrones and anthraquinone derivatives against human cytomegalovirus. Antiviral Res. 1992;17:63–77. doi: 10.1016/0166-3542(92)90091-i. [DOI] [PubMed] [Google Scholar]
- Battistutta R., Sarno S., DeMoliner E., Papinutto E., Zanotti G., Pinna L.A. The replacement of ATP by the competitive inhibitor emodin induces conformational modifications in the catalytic site of protein kinase CK2. J. Biol. Chem. 2000;275:29618–29622. doi: 10.1074/jbc.M004257200. [DOI] [PubMed] [Google Scholar]
- Bosch B.J., van der Zee R., de Haan C.A., Rottier P.J. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 2003;77:8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K.S., Lai S.T., Chu C.M., Tsui E., Tam C.Y., Wong M.M., Tse M.W., Que T.L., Peiris J.S., Sung J., Wong V.C., Yuen K.Y. Treatment of severe acute respiratory syndrome with lopinavir/ritonavir: a multicentre retrospective matched cohort study. Hong Kong Med. J. 2003;9:399–406. [PubMed] [Google Scholar]
- Chen F., Chan K.H., Jiang Y., Kao R.Y., Lu H.T., Fan K.W., Cheng V.C., Tsui W.H., Hung I.F., Lee T.S., Guan Y., Peiris J.S., Yuen K.Y. In vitro susceptibility of 10 clinical isolates of SARS coronavirus to selected antiviral compounds. J. Clin. Virol. 2004;31:69–75. doi: 10.1016/j.jcv.2004.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.C., Shen S.C., Lee W.R., Hsu F.L., Lin H.Y., Ko C.H., Tseng S.W. Emodin induces apoptosis in human promyeloleukemic HL-60 cells accompanied by activation of caspase 3 cascade but independent of reactive oxygen species production. Biochem. Pharmacol. 2002;64:1713–1724. doi: 10.1016/s0006-2952(02)01386-2. [DOI] [PubMed] [Google Scholar]
- Chen Z., Zhang L., Qin C., Ba L., Yi C.E., Zhang F., Wei Q., He T., Yu W., Yu J., Gao H., Tu X., Gettie A., Farzan M., Yuen K.Y., Ho D.D. Recombinant modified vaccinia virus Ankara expressing the spike glycoprotein of severe acute respiratory syndrome coronavirus induces protective neutralizing antibodies primarily targeting the receptor binding region. J. Virol. 2005;79:2678–2688. doi: 10.1128/JVI.79.5.2678-2688.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinatle J., Jr., Michaelis M., Hoever G., Preiser W., Doerr H.W. Development of antiviral therapy for severe acute respiratory syndrome. Antiviral Res. 2005;66:81–97. doi: 10.1016/j.antiviral.2005.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinatl J., Jr., Morgenstern B., Bauer G., Chandra P., Rabenau H., Doerr H.W. Glycyrrhizin, an active component of liquorice roots, and replication of SARS-associated coronavirus. Lancet. 2003;361:2045–2046. doi: 10.1016/S0140-6736(03)13615-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinatl J., Jr., Morgenstern B., Bauer G., Chandra P., Rabenau H., Doerr H.W. Treatment of SARS with human interferons. Lancet. 2003;362:293–294. doi: 10.1016/S0140-6736(03)13973-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., Berger A., Burguiere A.M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.C., Muller S., Rickerts V., Sturmer M., Vieth S., Klenk H.D., Osterhaus A.D., Schmitz H., Doerr H.W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- Fouchier R.A., Kuiken T., Schutten M., van Amerongen G., van Doornum G.J.J., van den Hoogen B.G., Peiris M., Lim W., Stohr K., Osterhaus A.D.M.E. Koch's postulates fulfilled for SARS virus. Nature. 2003;423:240. doi: 10.1038/423240a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenough T.C., Babcock G.J., Roberts A., Hernandez H.J., Thomas W.D., Coccia J.A., Graziano R.F., Srinivasan M., Lowy I., Finberg R.W., Subbarao K., Vogel L., Somasundaran M., Luzuriaga K., Sullivan J.L., Ambrosino D.M. Development and characterization of a severe acute respiratory syndrome-associated coronavirus-neutralizing human monoclonal antibody that provides effective immunoprophylaxis in mice. J. Infect. Dis. 2005;191:507–514. doi: 10.1086/427242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra B., Issinger O.G. Protein kinase CK2 and its role in cellular proliferation, development and pathology. Electrophoresis. 1999;20:391–408. doi: 10.1002/(SICI)1522-2683(19990201)20:2<391::AID-ELPS391>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Ho T.Y., Wu S.L., Chen J.C., Wei Y.C., Cheng S.E., Chang Y.H., Liu H.J., Hsiang C.Y. Design and biological activities of novel inhibitory peptides for SARS-CoV spike protein and angiotensin-converting enzyme 2 interaction. Antiviral Res. 2006;69:70–76. doi: 10.1016/j.antiviral.2005.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho T.Y., Wu S.L., Cheng S.E., Wei Y.C., Huang S.P., Hsiang C.Y. Antigenicity and receptor-binding ability of recombinant SARS coronavirus spike protein. Biochem. Biophys. Res. Commun. 2004;313:938–947. doi: 10.1016/j.bbrc.2003.11.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann H., Hattermann K., Marzi A., Gramberg T., Geiner M., Krumbiegel M., Kuate S., Uberla K., Niedrig M., Pohlmann S.H. S protein of severe acute respiratory syndrome-associated coronavirus mediates entry into hepatoma cell lines and is targeted by neutralizing antibodies in infected patients. J. Virol. 2004;78:6134–6142. doi: 10.1128/JVI.78.12.6134-6142.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann H., Geier M., Marzi A., Krumbiegel M., Peipp M., Fey G.H., Gramberg T., Pohlmann S. Susceptibility to SARS coronavirus S protein-driven infection correlates with expression of angiotensin converting enzyme 2 and infection can be blocked by soluble receptor. Biochem. Biophys. Res. Commun. 2004;319:1216–1221. doi: 10.1016/j.bbrc.2004.05.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiang C.Y., Hsieh C.L., Wu S.L., Lai I.L., Ho T.Y. Inhibitory effect of anti-pyretic and anti-inflammatory herbs on herpes simplex virus replication. Am. J. Chin. Med. 2001;29:459–467. doi: 10.1142/S0192415X01000472. [DOI] [PubMed] [Google Scholar]
- Hsiang C.Y., Wu S.L., Ho T.Y. Morin inhibited 12-O-tetradecanoylphorbol-13-acetate-induced hepatocellular transformation via activator protein 1 signaling pathway and cell cycle progression. Biochem. Pharmacol. 2005;69:1603–1611. doi: 10.1016/j.bcp.2005.03.008. [DOI] [PubMed] [Google Scholar]
- Huang H.C., Chu S.H., Chao P.D. Vasorelaxants from Chinese herbs, emodin and scoparone, possess immunosuppressive properties. Eur. J. Pharmacol. 1991;198:211–213. doi: 10.1016/0014-2999(91)90624-y. [DOI] [PubMed] [Google Scholar]
- Konoshima T., Kozuka M., Koyama J., Okatani T., Tagahara K. Studies on inhibitors of skin tumor promotion. VI. Inhibitory effects of quinones on Epstein-Barr virus activation. J. Nat. Prod. 1989;52:987–995. doi: 10.1021/np50065a012. [DOI] [PubMed] [Google Scholar]
- Koyama J., Morita I., Kawanishi K., Tagahara K., Kobayashi N. Capillary electrophoresis for simultaneous determination of emodin, chrysophanol, and their 8-beta-d-glucosides. Chem. Pharm. Bull. (Tokyo) 2003;51:418–420. doi: 10.1248/cpb.51.418. [DOI] [PubMed] [Google Scholar]
- Koyama M., Kelly T.R., Watanabe K.A. Novel type of potential anticancer agents derived from chrysophanol and emodin. Some structure–activity relationship studies. J. Med. Chem. 1988;31:283–284. doi: 10.1021/jm00397a002. [DOI] [PubMed] [Google Scholar]
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A., Humphrey C.D., Shieh W., Guarner J., Paddock C.D., Rota P., Fields B., DeRisi J., Yang J., Cox N., Hughes J.M., LeDuc J.W., Bellini W.J., Anderson L.J. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- Kumar A., Dhawan S., Aggarwal B.B. Emodin (3-methyl-1,6,8-trihydroxyanthraquinone) inhibits TNF-induced NF-κB activation, IκB degradation, and expression of cell surface adhesion proteins in human vascular endothelial cells. Oncogene. 1998;17:913–918. doi: 10.1038/sj.onc.1201998. [DOI] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.W., Tsai F.J., Tsai C.H., Lai C.C., Wan L., Ho T.Y., Hsieh C.C., Chao P.D. Anti-SARS coronavirus 3C-like protease effects of Isatis indigotica root and plant-derived phenolic compounds. Antiviral Res. 2005;68:36–42. doi: 10.1016/j.antiviral.2005.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S., Khattra J., Asano J.K., Barber S.A., Chan S.Y., Cloutier A., Coughlin S.M., Freeman D., Girn N., Griffith O.L., Leach S.R., Mayo M., McDonald H., Montgomery S.B., Pandoh P.K., Petrescu A.S., Robertson A.G., Schein J.E., Siddiqui A., Smailus D.E., Stott J.M., Yang G.S., Plummer F., Andonov A., Artsob H., Bastien N., Bernard K., Booth T.F., Bowness D., Czub M., Drebot M., Fernando L., Flick R., Garbutt M., Gray M., Grolla A., Jones S., Feldmann H., Meyers A., Kabani A., Li Y., Normand S., Stroher U., Tipples G.A., Tyler S., Vogrig R., Ward D., Watson B., Brunham R.C., Krajden M., Petric M., Skowronski D.M., Upton C., Roper R.L. The Genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- Moore M.J., Dorfman T., Li W., Wong S.K., Li Y., Kuhn J.H., Coderre J., Vasilieva N., Han Z., Greenough T.C., Farzan M., Choe H. Retroviruses pseudotyped with the severe acute respiratory syndrome coronavirus spike protein efficiently infect cells expressing angiotensin-converting enzyme 2. J. Virol. 2004;78:10628–10635. doi: 10.1128/JVI.78.19.10628-10635.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka G., Minami M., Nishioka I. Studies on rhubarb (Rhei rhizoma). III. Stilbene glycosides. Chem. Pharm. Bull. (Tokyo) 1977;25:2300–2305. doi: 10.1248/cpb.25.2300. [DOI] [PubMed] [Google Scholar]
- Peiris J.S.M., Lai S.T., Poon L.L.M., Guan Y., Yam L.Y.C., Lim W., Nicholls J., Yee W.K.S., Yan W.W., Cheung M.T., Cheng V.C.C., Chan K.H., Tsang D.N.S., Yung R.W.H., Ng T.K., Yuen K.Y., Members of the SARS Study Group Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit C.M., Melancon J.M., Chouljenko V.N., Colgrove R., Farzan M., Knipe D.M., Kousoulas K.G. Genetic analysis of the SARS-coronavirus spike glycoprotein functional domains involved in cell-surface expression and cell-to-cell fusion. Virology. 2005;341:215–230. doi: 10.1016/j.virol.2005.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.H., Tong S., Tamin A., Lowe L., Frace M., DeRisi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., Gunther S., Osterhaus A.D., Drosten C., Pallansch M.A., Anderson L.J., Bellini W.J. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- Schinazi R.F., Chu C.K., Babu J.R., Oswald B.J., Saalmann V., Cannon D.L., Eriksson B.F.H., Nasr M. Anthraquinones as a new class of antiviral agents against human immunodeficiency virus. Antiviral Res. 1990;13:265–272. doi: 10.1016/0166-3542(90)90071-e. [DOI] [PubMed] [Google Scholar]
- Simmons G., Reeves J.D., Rennekamp A.J., Amberg S.M., Piefer A.J., Bates P. Characterization of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spike glycoprotein-mediated viral entry. Proc. Natl. Acad. Sci. U.S.A. 2004;101:4240–4245. doi: 10.1073/pnas.0306446101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui J., Li W., Murakami A., Tamin A., Matthews L.J., Wong S.K., Moore M.J., Tallarico A.S., Olurinde M., Choe H., Anderson L.J., Bellini W.J., Farzan M., Marasco W.A. Potent neutralization of severe acute respiratory syndrome (SARS) coronavirus by a human mAb to S1 protein that blocks receptor association. Proc. Natl. Acad. Sci. U.S.A. 2004;101:2536–2541. doi: 10.1073/pnas.0307140101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui J., Li W., Roberts A., Matthews L.J., Murakami A., Vogel L., Wong S.K., Subbarao K., Farzan M., Marasco W.A. Evaluation of human monoclonal antibody 80R for immunoprophylaxis of severe acute respiratory syndrome by an animal study, epitope mapping, and analysis of spike variants. J. Virol. 2005;79:5900–5906. doi: 10.1128/JVI.79.10.5900-5906.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sydiskis R.J., Owen D.G., Lohr J.L., Rosler K.H.A., Blomster R.N. Inactivation of enveloped viruses by anthraquinones extracted from plants. Antimicrob. Agents Chemother. 1991;35:2463–2466. doi: 10.1128/aac.35.12.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan E.L., Ooi E.E., Lin C.Y., Tan H.C., Ling A.E., Lim B., Stanton L.W. Inhibition of SARS coronavirus infection in vitro with clinically approved antiviral drugs. Emerg. Infect. Dis. 2004;10:581–586. doi: 10.3201/eid1004.030458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y.J., Lim S.G., Hong W. Characterization of viral proteins encoded by the SARS-coronavirus genome. Antiviral Res. 2005;65:69–78. doi: 10.1016/j.antiviral.2004.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripet B., Howard M.W., Jobling M., Holmes R.K., Holmes K.V., Hodges R.S. Structural characterization of the SARS-coronavirus spike S fusion protein core. J. Biol. Chem. 2004;279:20836–20849. doi: 10.1074/jbc.M400759200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.J., Jan J.T., Chen C.M., Hsieh H.P., Hwang D.R., Liu H.W., Liu C.Y., Huang H.W., Chen S.C., Hong C.F., Lin R.K., Chao Y.S., Hsu J.T. Inhibition of severe acute respiratory syndrome coronavirus replication by niclosamide. Antimicrob. Agents Chemother. 2004;48:2693–2696. doi: 10.1128/AAC.48.7.2693-2696.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.Y., Jan J.T., Ma S.H., Kuo C.J., Juan H.F., Cheng Y.S., Hsu H.H., Huang H.C., Wu D., Brik A., Liang F.S., Liu R.S., Fang J.M., Chen S.T., Liang P.H., Wong C.H. Small molecules targeting severe acute respiratory syndrome human coronavirus. Proc. Natl. Acad. Sci. U.S.A. 2004;101:10012–10017. doi: 10.1073/pnas.0403596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao P.G., Xing S.T., Wang L.W. Immunological aspects of Chinese medicinal plants as antiageing drugs. J. Ethnopharmacol. 1993;38:167–175. doi: 10.1016/0378-8741(93)90012-t. [DOI] [PubMed] [Google Scholar]
- Yang Z.Y., Kong W.P., Huang Y., Roberts A., Murphy B.R., Subbarao K., Nabel G.J. A DNA vaccine induces SARS coronavirus neutralization and protective immunity in mice. Nature. 2004;428:561–564. doi: 10.1038/nature02463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi C.E., Ba L., Zhang L., Ho D.D., Chen Z. Single amino acid substitutions in the severe acute respiratory syndrome coronavirus spike glycoprotein determine viral entry and immunogenicity of a major neutralizing domain. J. Virol. 2005;79:11638–11646. doi: 10.1128/JVI.79.18.11638-11646.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi L., Li Z., Yuan K., Qu X., Chen J., Wang G., Zhang H., Luo H., Zhu L., Jiang P., Chen L., Shen Y., Luo M., Zuo G., Hu J., Duan D., Nie Y., Shi X., Wang W., Han Y., Li T., Liu Y., Ding M., Deng H., Xu X. Small molecules blocking the entry of severe acute respiratory syndrome coronavirus into host cells. J. Virol. 2004;78:11334–11339. doi: 10.1128/JVI.78.20.11334-11339.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M.M., Liu X.M., He L. Effect of integrated traditional Chinese and Western medicine on SARS: a review of clinical evidence. World J. Gastroenterol. 2004;10:3500–3505. doi: 10.3748/wjg.v10.i23.3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.W., Yap Y.L. Old drugs as lead compounds for a new disease? Binding analysis of SARS coronavirus main proteinase with HIV, psychotic and parasite drugs. Bioorg. Med. Chem. 2004;12:2517–2521. doi: 10.1016/j.bmc.2004.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X.M., Chen Q.H. Biochemical study of Chinese rhubarb. XXII. Inhibitory effect of anthraquinone derivatives on Na+-K+-ATPase of the rabbit renal medulla and their diuretic action. Acta Pharmacol. Sin. 1988;23:17–20. [PubMed] [Google Scholar]


