Abstract
RNA interference (RNAi) is a natural mechanism for suppressing or silencing expression of aberrant or foreign genes. It is a powerful antiviral strategy that has been widely employed to protect hosts from viral infection. Hepatitis E (HE) is an acute fulminant hepatitis in adults that has particularly high mortality in pregnant women. At this point in time, there is no vaccine or antiviral treatment that is effective against the infectious agent, HEV. The nonstructural polyprotein region possesses an RNA-dependent RNA polymerase (RdRp) that is responsible for the replication of the viral RNA genome. RdRp is therefore regarded as one of the most attractive candidates for RNA interference (RNAi). In the present study, the high efficiency and specificity of siRNA were evaluated by Real-Time quantitative PCR and Western blot assays. Protective effects against HEV infection were achieved in A549 cells and in piglets. In piglets treated with a shRNA-RdRp-1 expression plasmid prior to HEV inoculation, HEV antigens were significantly reduced in the liver, spleen, and kidneys, and the activities of alanine aminotransferase (ALT), aspartate aminotransferase (AST) and total bilirubin (TBIL) were clearly decreased. These results suggested that RNAi is a potentially effective antiviral strategy against HEV replication and infection.
Keywords: Hepatitis E virus, RNA-dependant RNA polymerase, RNA interference, A549 cells, Piglets
1. Introduction
Hepatitis E virus (HEV) is the causative agent of acute fulminant hepatitis E (HE), an enteric zoonotic disease that is transmitted across species in humans, pigs, cats, chicken, deer and rodents (Arankalle et al., 2001, Feagins et al., 2008, Gardner and Luciw, 2008, Huang et al., 2004, Okamoto et al., 2004, Tei et al., 2003, Vitral et al., 2005). Although HEV is transmitted predominantly by a fecal–oral route (Kasorndorkbua et al., 2004), its prevalence is mainly promoted through contaminated water (Krawczynski et al., 2001, Hsieh et al., 1999). HEV outbreaks have occurred several times and have resulted in huge economic losses in developing countries in Asia and Africa, and in Mexico.
HEV is a non-enveloped, single-stranded, positive-strand RNA virus, with a genome length of ∼7.3 kb. The genome contains three major open reading frames (ORFs). ORF1 encodes for nonstructural polyprotein including methyltransferase, protease, RNA helicase, and RNA-dependent RNA polymerase (RdRp)/replicase (Ansari et al., 2000, Jameel, 1999, Panda et al., 2000) RdRp catalyzes viral replication (Agrawal et al., 2001) and plays a key role in the life cycle of RNA viruses. The activity of HEV RdRp has been experimentally demonstrated in previous studies (Agrawal et al., 2001, Ansari et al., 2000, Panda et al., 2000).
Initiation of replication of HEV has been confirmed to require the interaction of 3′ end of HEV genome with RdRp, as well as with the host cell, for synthesis of the minus-strand replicative intermediate (Agrawal et al., 2001). This intermediate acts as a subgenomic promoter for transcription of the structural region mRNA (Jameel, 1999, Nanda et al., 1994). In addition, the nucleotide sequence in RdRp (4254–4560nt) is the most conserved genomic region in 37 HEV strains in four genotypes (Zhai et al., 2006). Therefore, the RdRp gene is expected to be a prime potential target for the development of an antiviral strategy.
Vaccines were previously considered as the best strategy for prevention of HEV infection, because all four HEV genotypes (1–4) have a single serotype (Mushahwar, 2008, Panda et al., 2007). However, earlier evidence has documented that, despite the presence of antibodies to HEV capsid protein, HEV is still capable of causing infection (Panda et al., 1995, Yarbough, 1999). Moreover, although vaccine-induced antibody might attenuate HEV infection, it may not prevent virus excretion in the feces (Panda et al., 1995). The reasons are that the protective neutralizing antibodies either do not develop or are only short-lived (Jameel, 1999, Yarbough, 1999). Recently, a purified polypeptide vaccine has been produced in insect cells infected with a recombinant baculovirus containing a truncated HEV genomic sequence encoding the capsid antigen, and has completed a phase 3 trial (Abreu, 2007, Krawczynski, 2007, Mushahwar, 2008, Shrestha et al., 2007). However, the safety and efficacy of this vaccine is doubtful (Tacke and Trautwein, 2007). First of all, the subjects were exclusively male, and it is pregnant women infected with HEV who have the highest mortality; secondly, the inconsistent data in humans and in rhesus monkeys (Krawczynski, 2007, Tacke and Trautwein, 2007) and finally, pigs can still suffer from HEV within 4–7 days post-infection, can transmit the virus as early as the first week post-infection and can continue to do so for at least several weeks (Halbur et al., 2001). However, the current vaccines cannot provide complete protection prior to 6 months (Shrestha et al., 2007). Thus, it is urgent to develop a more effective and safer therapeutic strategy for HEV control.
RNAi is a self-defense mechanism present in eukaryotic cells, which specifically prevents infections induced by viruses (Plasterk, 2002). It can inhibit the expression of crucial viral proteins by directly targeting viral mRNA for degradation (Gitlin et al., 2002, Shabalina and Koonin, 2008). It has been widely used to suppress viral replication by post-transcriptional regulation of gene expression, and has been successfully applied in infection inhibition studies of some viruses, such as human immunodeficiency virus (HIV) (Kumar et al., 2008, Ludwig, 2008, Saayman et al., 2008, ter Brake et al., 2009), hepatitis C virus (HCV) (DeMarini et al., 2003, Lupberger et al., 2008) as well as severe acute respiratory syndrome-associated coronavirus (SARS-Cov) (Lu et al., 2004, Ni et al., 2005). In the present study, RNAi as a potential antiviral therapeutic strategy targeting the RdRp gene was used to inhibit HEV replication in vitro in A549 cells and in vivo in piglets.
2. Materials and methods
2.1. Target sequences and plasmid construction
The positive HEV characterized as genotype 4 (GenBank accession no. AY594199), isolated from swine feces from Shanghai suburbs, China, was used as template in a reverse transcription nested polymerase chain reaction PCR (RT-nPCR) to amplify RdRp gene (717 base pairs). The external forward primer and reverse primer were 5′-AGCTCGGCCCTATTAGTGTCA-3′ and 5′-TTCCGATCAGGTTATGTAC-3′ respectively. The internal forward primer and reverse primer were 5′-GAGCTTTTTGAGCTTGTG-3′ and 5′-CCCAATAGGCCTAAAATCTAC-3′, respectively. RT-nPCR analysis was conducted using an AMV Reverse Transcriptase XL for RT-PCR (Takara, Japan) according to the manufacturer's directions. The reverse transcription reaction protocol was performed at 42 °C for 30 min, 86 °C for 15 s. The 2 μl cDNA that resulted was amplified by nested PCR at 94 °C for 2 min, followed by 94 °C for 30 s, 42 °C for 30 s and 72 °C for 1 min, and repeated for 29 cycles. The PCR products were detected on an agarose gel containing 0.5 μg/ml ethidium bromide. The amplified DNA fragment was inserted into the multicloning site of a eukaryotic expression vector pcDNA3.1 containing the reporter gene of enhanced green fluorescence protein (eGFP), using standard cloning procedures with the restriction sites of BamHI (30 °C digestion) and BstXI (45 °C digestion). The eGFP gene was located downstream of the target genes.
2.2. Design and synthesis of siRNA
Four small interfering RNA (siRNA) targeting the RdRp gene was designed according to Qiagen's guidelines (http://www.qiagen.com) and the sequences are shown in Table 1 . The siRNA duplexes have been designed using the Hiperformance design Algorithm licensed from Novartis AG, integrated with a stringent in-house homology analysis tool. The highest-ranking siRNA duplexes generated by the algorithm were chosen as representing the best combination of activity and specificity. Scrambled siRNA, constructed from a random sequence heterology with the HEV sequence, served as a negative control for identification of the specificity of HEV siRNA. GAPDH siRNA served as a control in the experiments to confirm that the transfection procedure and cell cultures supported gene silencing. The siRNAs were diluted with RNase-free buffer to obtain a 20 μM solution. The solution was denatured by heating at 90 °C for 1 min, and then incubated at 37 °C for 60 min, and either used immediately or stored at −20 °C in an RNase-free environment.
Table 1.
Nucleotide sequences of siRNAs targeting HEV RdRp.
| siRNA | Nucleotide sequence |
|---|---|
| si-RdRp-1 | |
| Sense | 5′-UUUCUCAAUAGCACGGAACdTdT-3′ |
| Antisense | 5′-GUUCCGUGCUAUUGAGAAAdAdA-3′ |
| si-RdRp-2 | |
| Sense | 5′-UUAUCCGAGACACAUCACGdTdT-3′ |
| Antisense | 5′-CGUGAUGUGUCUCGGAUAAdGdA-3′ |
| si-RdRp-3 | |
| Sense | 5′-UGUCUCACCAGUUGUAAACdTdT-3′ |
| Antisense | 5′-GUUUACAACUGGUGAGACAdTdG-3′ |
| si-RdRp-4 | |
| Sense | 5′-UGAUUAUACACUCUAACCCdTdT-3′ |
| Antisense | 5′-GGGUUAGAGUGUAUAAUCAdAdG-3′ |
| Scrambled | |
| Sense | 5′-UUCUCCGAACGUGUCACGUdTdT-3′ |
| Antisense | 5′-ACGUGACACGUUCGGAGAAdTdT-3′ |
The corresponding small hairpin RNA (shRNA) expression vectors containing a human U6 polymerase III (Pol III) promoter and neomycin resistance gene (NEOr) (McIntyre and Fanning, 2006, Saayman et al., 2008) were constructed and extracted by the Endo-free plasmid Maxi kit (Omega, America).
2.3. Virus challenge in A549 cell
2.3.1. Cell culture and plasmid transfection
A549 cell line was used in this study. Cells were trypsinized and used at 0.5–2.0 × 105 per well, diluted in 500 μl antibiotic-free growth medium in 24-well plates. Cells were 80–90% confluent at the time of transfection. One microgram of pcDNA3.1-RdRp-eGFP recombinant plasmid and each of 20 nM siRNAs (0.5 μl) were transfected using 1.5 μl Lipofectamine 2000 (Invitrogen, America) according to the manufacturer's instructions. Four to six hours post-transfection, the cell culture was replaced with fresh growth medium. The optimal combination of four parameters for each cell line, including the highest transfection efficiency, the lowest non-specific effects, the conditions for the most efficient delivery of siRNA, and the concentration of Lipofectamine 2000, was first determined in preliminary experimentation. Cells were incubated at 37 °C for 48 h for mRNA expression level observation, and for 72 h for protein expression level determination. The transfection and suppression efficiencies were assessed by measuring the eGFP fluorescence intensity of transfected cells with an inverted fluorescence microscope (Nikon TE2000, Japan). The pcDNA3.1-eGFP empty vector plasmid was transfected with each siRNA to test non-specific effects and scrambled siRNA was used to establish the specificity. GAPDH siRNA was used as a criterion of the efficiency of inhibition and as a control to determine any cytotoxicity induced by transfection.
2.3.2. Virus challenge in A549 cells
HEV has been successfully cultured in A549 and PLC/PRF/5 line cells, as previously described (Huang et al., 1999, Lorenzo et al., 2008, Tanaka et al., 2007) and only A549 cells show a typical cytopathic effect (CPE) induced by the virus. HEV positive feces suspension was centrifuged at 12,000 × g at 4 °C for 10 min, and filtered through 0.22 μm microfilters, followed by treatment with penicillin and streptomycin for 1 h. The virus was 10-fold serially diluted, and each dilution was added to A549 cells in triplicate. After 3 days infection, the viral 50% cell culture infectious dose (CCDI50) was calculated using the Reed and Muench Method. For HEV infection challenge, A549 cells were transfected with each siRNA, as described above, 24 h before inoculation. The cell was challenged with HEV at 200 CCID50 for a 1 h inoculation, and then the virus was replaced with fresh growth medium. The infection was observed every 12 h. Cell transfection efficiency was evaluated by transfection of Fluor-labeled GAPDH siRNA. The cells were harvested 48 h post-infection for Real-Time qPCR analysis and 72 h post-infection for Western blot assay.
2.3.3. Real-Time qPCR analysis
Forty-eight hours post-inoculation, the culture medium was removed, the cells were lysed and the total RNA was isolated by Trizol (Invitrogen, America) according to the manufacturer's directions. A reverse transcription analysis was carried out using an AMV Reverse Transcriptase XL for Real-Time PCR (Takara, Japan) according to the manufacturer's directions. The protocol of reverse transcription reaction was performed as described above, except the forward primer was 5′-GGTGGTTTCTGGGGTGAC-3′, and reverse primer was 5′-AGGGGTTGGTTGGATGAA-3′. The synthesized first strand cDNA (2 μl) was added as a template for Real-Time qPCR using Premix Ex Taq™ (perfect Real-Time, Takara, Japan) according to the manufacturer's directions. The probe was 5′-TGATTCTCAGCCCTTCGC-3′ according to Jothikumar et al. (2006). The mixtures were reacted at 95 °C for 30 s, followed by 95 °C for 5 s and 60 °C for 31 s repeated for 39 cycles. The product was expected to be 79 bp. The housekeeping gene GAPDH served as a loading control. The Real-Time qPCR analysis was performed in the ABI PRISM 7000 Real-Time PCR System (America). All procedures were performed in triplicate and data are expressed as means (±S.D.). The percentage of HEV mRNA expression was analyzed by compared with normal control.
2.3.4. Western blot
A549 cells were harvested 72 h post-inoculation and lysed with lysis buffer (50 mM Tris–Cl, pH 8.0, 150 mM NaCl, 1% Triton-X 100, 1 μg/ml aprotinin, and 100 μg/ml phenylmethylsulfonyl fluoride). An equivalent amount of total protein was separated by 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) and electrophoretically transferred onto a nitrocellulose membrane. Non-specific-binding sites were blocked with 5% skim milk, and the membrane was incubated with primary antibodies at 4 °C overnight. The primary antibody was HEV ORF2 (ABR, America, 1:500 dilutions). A goat-rabbit IgG conjugated with HRP was used as secondary antibody (Promega, America, 1:500 dilutions). The GAPDH protein served as a loading control. The bands were exposed to X-ray films with SuperSignal West Pico Trial Kit (Pierce, America).
2.4. Virus challenge in piglets
Twenty-one 1–2 weeks piglets (500–800 g) were purchased from Shanghai Jiao Tong University and maintained in a pathogen-free animal facility. The study protocol was approved by Animal Care and Use Committee (ACUC) of Shanghai Jiao Tong University. We followed guidelines of the Shanghai Jiao Tong University during this study. All piglets were tested for anti-HEV IgM and IgG by ELISA (Wantai, China). Piglets confirmed seronegative were included in the study. Twenty-one piglets were randomly distributed into seven groups, three piglets per group. Group 1 was the normal control, one piglet in this group was injected intravenously with pcDNA3.1-eGFP plasmid to serve as the reporter to identify shRNA plasmids expressed in vivo; Group 2 was injected intravenously with sterile PBS; Groups 3–7 was injected intravenously with 200 μg each of five plasmids, shRNA-RdRp-1, shRNA-RdRp-2, shRNA-RdRp-3, shRNA-RdRp-4 or shRNA-scrambled, respectively (Table 2 ). All plasmids were diluted with sterile PBS. Piglets in Groups 2–7 were intravenously inoculated with 1 ml HEV (1 ml per pig) 24 h later. The feces were collected daily for HEV RNA detection.
Table 2.
Biochemical, virological, histological and serological changes in piglets.
| Group 1 | Group 2 | Group 3 | Group 4 | Group 5 | Group 6 | Group 7 | |
|---|---|---|---|---|---|---|---|
| Injection | None | PBS | sh-1 | sh-2 | sh-3 | sh-4 | sh-scr |
| Inoculation | None | HEV | HEV | HEV | HEV | HEV | HEV |
| Clinical | None | Severe | None | Mild | Mild | Severe | Severe |
| Antibodies | |||||||
| IgM/IgG | N/N | P(3/3)/N | N/N | N/N | N/N | P(1/3)/N | P(3/3)/N |
| ALT | — | ↑ | — | ↑ | ↑ | ↑ | ↑ |
| AST | — | ↑ | — | ↑ | — | ↑ | ↑ |
| TBIL | — | ↑ | — | — | — | — | ↑ |
| Antigens | |||||||
| 1–4 Day | N | N | N | N | N | N | N |
| 5 Day | N | P (2/3) | N | N | N | P (1/3) | P (1/3) |
| 6 Day | N | P (3/3) | N | P (1/3) | P (1/3) | P (1/3) | P (3/3) |
| 7 Day | N | P (3/3) | N | P (1/3) | P (1/3) | P(1/3) | P (3/3) |
| Liver | N | P (3/3) | Less | Less | Less | P (1/3) | P (3/3) |
| Spleen | N | P (3/3) | Less | Less | Less | P (1/3) | P (3/3) |
| Kidney | N | P (3/3) | N | Less | N | P (1/3) | P (3/3) |
| Histological | None | Severe | Mild | Mild | Mild | Severe | severe |
Injection, sh-1: shRNA-RdRp-1 injection; sh-2: shRNA-RdRp-2 injection; sh-3: shRNA-RdRp-3 injection; sh-4: shRNA-RdRp-4 injection; sh-scr: shRNA-scrambled injection. Clinical, none: showing no clinical disease symptoms; severe: showing acute hepatitis with icteric viral hepatitis or diarrhea; mild: inapparent infection. IgG/IgM: HEV antibodies (IgG and IgM) determined by ELISA 7 days post-inoculation, N: negative; P: positive; P (1/3): one of three piglets was positive. ALT, AST, TBIL, —: no significant changes; ↑: significantly elevated compared with normal control. Antigens, N: HEV RNA negative detected by RT-PCR; P: HEV RNA positive detected by RT-PCR. Liver, spleen, kidney, N: HEV antigen negative; P: HEV antigen positive; Less: HEV antigens were less intense than in positive infection. Histological, N: no histological changes; severe: severe histological injuries; mild: mild injuries.
Piglets were humanely euthanized, following the guidelines of the Care and Use of Laboratory Animals, at 7 days post-inoculation when all the piglets in Group 2 were confirmed to be passing HEV RNA in feces. Blood was collected for RT-nPCR detection, ELISA tests, and enzyme activity assays by automated biochemistry analyzer. Liver, spleen, and kidney were collected for RT-nPCR detection (stored at −80 °C until used), for indirect immunofluorescence observation (frozen and cut into 6 μm sections), and for histopathological examination (fixed immediately in 10% neutral buffered formalin).
2.4.1. Anti-HEV antibody determination
HEV IgM and IgG were separately determined using commercial ELISA kits (Wantai, China), according to the manufacturer's directions. The kit contains both positive and negative controls. The cutoff value for IgM assay was determined based on 0.25 (0.22 for IgG) plus the mean OD450 values of sera from normal control piglets (±standard deviation).
2.4.2. HEV RNA detection
The total RNA of the feces was extracted by Trizol (Invitrogen, America), according to the manufacturer's instructions. The reverse transcription PCR was performed as described above.
2.4.3. HEV antigens observation
Frozen tissues were cut into 6 μm sections for indirect immunofluorescence observation. HEV-specific ORF2 primary antibody (ABR, America, 1:500 dilution), whose recombined protein is completely conserved in four genotypes (1–4), designed to detect capsid proteins of HEV genotypes (1–4), was added to sections and incubated at 37 °C for 30 min. After washing with phosphate buffered solution (PBS), FITC-labelled goat anti-rabbit secondary antibody (Dingguo, China, 1:500 dilution) was added and incubated at 37 °C for 30 min. After washing with PBS, slides were observed with a Nikon TE 2000 fluorescence microscope. All specimens were tested in duplicate with positives control (specimens of HEV infected swine) and negative control (specimens of normal control piglets).
2.4.4. Serum liver chemistry profile
The activities of ALT, AST, and levels of TBIL in sera were measured with an automated biochemistry analyzer (Olympus 2700, Japan).
2.4.5. Histopathological examination
Tissues fixed in 10% neutral buffered formalin were sectioned at 7 μm and stained with hematoxylin and eosin. The tissue sections were examined and compared with normal controls.
3. Results
3.1. Construction of RdRp recombinant plasmids
The 747 bp RdRp gene of HEV (4114–4830 nt) was amplified by RT-nPCR and introduced into the pcDNA3.1-eGFP vector with the restriction sites of BamHI/BstXI, to yield pcDNA3.1-RdRp-eGFP. The recombinant plasmid was identified by digestion and sequenced.
3.2. Suppression of HEV by siRNAs in A549 cell
3.2.1. Reduction of HEV mRNA
The suppression of RdRp in mRNA level was observed by fluorescence microscopy at 48 h post-transfection (Fig. 1A) and the decrease in HEV mRNA was determined at 48 h post-inoculation by Real-Time qPCR (Fig. 1B). The relative reduction of mRNA in the silenced cells and unsilenced cells was calculated according to the Pfaffl method (Pfaffl, 2001). The transcription level of RdRp mRNA was decreased about 32-fold, 4-fold, 8-fold and 1.319-fold in cells transfected with siRNA-RdRp-1, siRNA-RdRp-2, siRNA-RdRp-3 and siRNA-RdRp-4, respectively. However, no significant changes were noted in cells transfected with scrambled siRNA (Fig. 1B). The mRNA level of GAPDH was not significantly changed in either non-transfected or transfected cells.
Fig. 1.
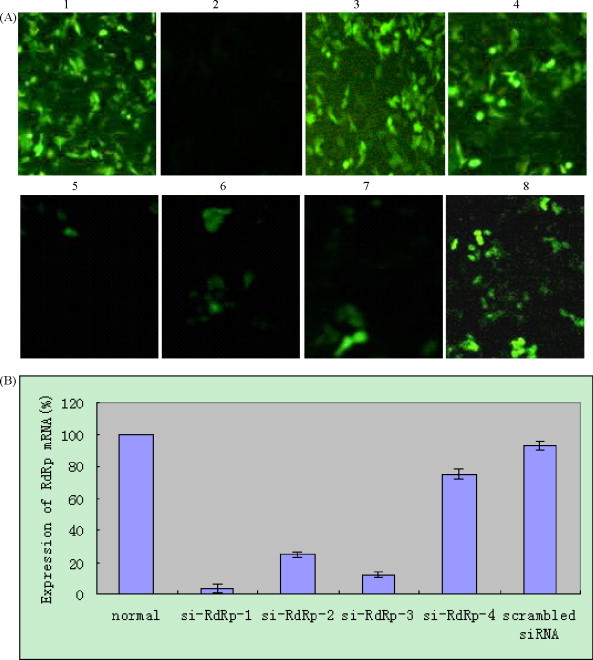
Transfection of pcDNA3.1-RdRp-eGFP with siRNAs in A549 cells. The pcDNA3.1-RdRp-eGFP plasmid was transfected into A549 cells with various siRNAs 48 h before observation by fluorescence microscopy. (A) 1: pcDNA3.1-RdRp-eGFP plasmid transfection; 2: pcDNA3.1 plasmid transfection; 3: pcDNA3.1-eGFP plasmid transfection; 4: pcDNA3.1-RdRp-eGFP plasmid and scrambled siRNA co-transfection; 5–8: pcDNA3.1-RdRp-eGFP plasmid was separately co-transfected with siRNA-RdRp-1, siRNA-RdRp-2, siRNA-RdRp-3 and siRNA-RdRp-4. Pictures were taken at 48 h post-transfection with a Nikon TE2000 microscope system. (B) HEV RdRp mRNA expression was decreased by HEV specific siRNAs detected by RT-qPCR.
3.2.2. Detection of HEV infected A549 cells by Western blot
To analyze the suppression efficiency at the protein level, A549 cells were collected for Western blot analysis at 72 h post-inoculation. The inoculated cells expressed the expected ORF2 protein band (71 kDa, Fig. 2A-1), while the cells pre-transfected with HEV specific siRNAs showed protein bands of decreased intensity (Fig. 2A-2–5). The bands reacted with anti-HEV RdRp antibody. Western blot analysis showed that the HEV ORF2 protein expression was significantly reduced in cells pre-transfected with HEV specific siRNAs, especially in siRNA-RdRp-1 (Fig. 2A-2), compared with cells that were not transfected with siRNAs before inoculation. GAPDH was used as a loading control in this experiment (Fig. 2B).
Fig. 2.
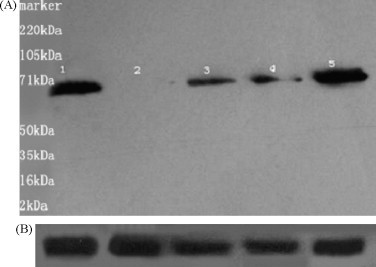
HEV protein expression inhibited by siRNAs 72 h post-inoculation. Western blotting was performed on equal amounts of protein harvested from A549 cells 72 h post-inoculation. (A) 1 line: HEV inoculation alone; 2–5 lines: pre-transfected siRNAs (siRNAs-RdRp-1–4, respectively). (B) GAPDH used as a loading control.
3.3. Inhibition of HEV infection by shRNA in piglets
3.3.1. Clinical evaluation and gross and microscopic lesions
Piglets in Group 2 (inoculated HEV), Group 6 (pre-injected with shRNA-RdRp-4) and Group 7 (pre-injected with shRNA-scrambled) developed severe hepatitis with icteric viral hepatitis or diarrhea, which was consistent with previous studies (Aggarwal and Krawczynski, 2000, Feagins et al., 2008). The piglets pre-injected with shRNA-RdRp-2 and shRNA-RdRp-3 showed a mild viral hepatitis. However, no symptoms were observed in piglets injected with shRNA-RdRp-1 expression plasmids.
3.3.2. Anti-HEV antibody determination by ELISA
Elevated anti-HEV IgM was detected in all the infected piglets in Group 2 and Group 7, and in one member of the Group 6. However, none of the piglets injected with HEV specific shRNA-RdRp-1, shRNA-RdRp-2 or shRNA-RdRp-3 expression plasmids showed anti-HEV IgM. Anti-HEV IgG was not detected in any of the piglets in this study.
3.3.3. HEV antigens in piglets inhibited by shRNAs
HEV RNA was detected in feces in Group 2 (HEV inoculated) from 5 days post-inoculation until human euthanization, which was consistent with previous studies (Feagins et al., 2008, Kasorndorkbua et al., 2004). HEV RNA was also detected in Group 7 (pre-injected with shRNA-scrambled) and in one member of Group 6 (pre-injected shRNA-HEV-4) in feces on the same day. There was no HEV RNA detected in Group 3 (pre-injected shRNA-HEV-1) throughout the entire study (Table 2). The presence of HEV antigens were observed by indirect immunofluorescence in the liver, spleen, and kidney (Fig. 3A-1–3) in the inoculated group (Group 2), but no signs were observed in the normal control group. HEV antigens in the liver, spleen, and kidney were obviously reduced by the shRNA-RdRp-1 expression plasmid (Fig. 3B-1–3).
Fig. 3.
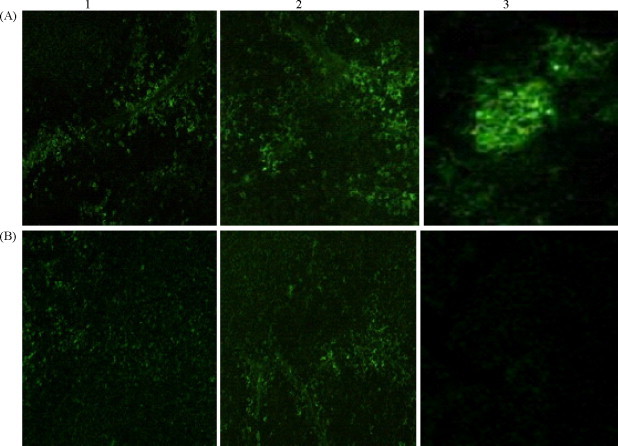
HEV antigens in tissues of piglets were obviously reduced by shRNA-RdRp-1. (A) 1–3: HEV antigens distributed in the liver (1), spleen (2) and kidney (3) of HEV infected piglets (Group 2) 7 days post-inoculation, observed by indirect immunofluorescence. (B) 1–3: HEV antigens distributed in the liver (1), spleen (2) and kidney (3) of HEV infected piglets (Group 3) 7 days post-inoculation. Piglets were injected with shRNA-RdRp-1 expression plasmid 24 h before HEV inoculation.
3.3.4. The activities of liver enzymes
The sera were separated and enzyme activities were characterized using an automated biochemistry analyzer. The infected piglets showed a significant elevation in ALT activity, 2.125 times higher than the upper limits of normal controls (∼40), the activity of AST was 1.825 times higher than the upper limits (∼40), and the level of TBIL was 3.396 times higher than the upper limits (∼1.06) of normal controls. The activities of ALT (P = 0.0023), AST (P = 0.0021) and TBIL (P = 0.0018) were significantly decreased in piglets injected with the shRNA-RdRp-1 expression plasmid compared with HEV infected group (Fig. 4 ).
Fig. 4.
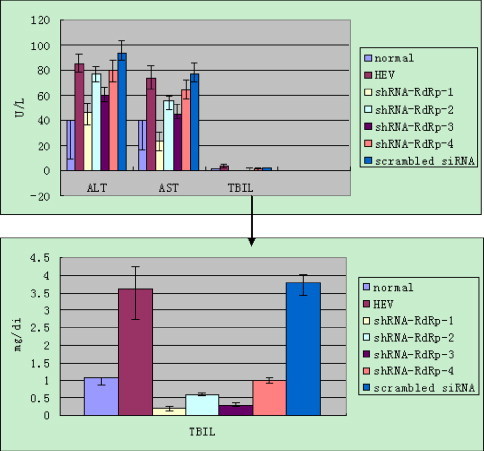
The enzyme activities of AST, ALT and in level of TBIL in piglets 7 days post-inoculation. The normal value presents the upper limits with negative deviation. The data represent the average of three experiments, with the standard deviation indicated by error bars (±S.D.).
3.3.5. Histopathological changes
The hepatic injuries caused by HEV infection were observed in all of the infected piglets. Enlarged livers were filled with inflammatory exudates, hepatocyte necrosis, and liver hemorrhages (Fig. 5A-2). The enlarged spleen displayed multiple hemorrhages, focal necrosis, and lymphocyte proliferation (Fig. 5B-2). The degrees of hepatic and splenetic injuries were clearly attenuated in piglets injected with the shRNA-RdRp-1 expression plasmid (Fig. 5A-4 and B-4). The shRNA-RdRp-2, shRNA-RdRp-3 or shRNA-RdRp-4 expression plasmids showed some protective effects as well (Fig. 5A-5, -6 and -7, B-5, -6 and -7). However, piglets injected with shRNA-scrambled expression plasmid showed no increased protection (Fig. 5A-3 and B-3). There was no injury in the livers or spleens in the normal control group (Fig. 5A-1 and B-1).
Fig. 5.
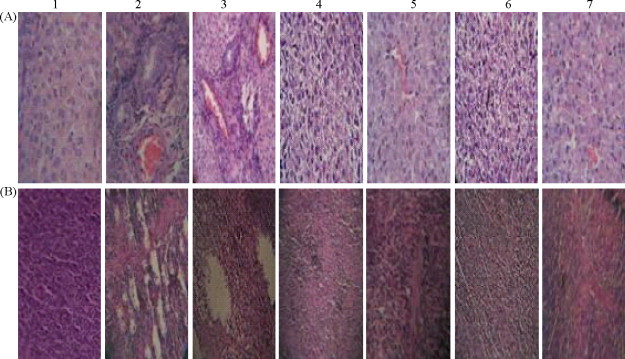
Histopathological changes in piglets 7 days post-inoculation. (A) 1: livers of piglets in the normal control group, ×400; 2: livers of piglets inoculated with HEV, ×400; 3: livers of piglets injected with shRNA-scrambled expression plasmid 24 h pre-inoculation, ×200; 4–7: livers of piglets separately injected with shRNA-RdRp-1–4 expression plasmids 24 h pre-inoculation, ×400. (B) 1: spleens of piglets in normal control group, ×200; 2: spleens of piglets inoculated with HEV, ×200; 3: spleens of piglets injected with shRNA-scrambled expression plasmid 24 h pre-inoculation, ×200; 4–7: spleens of piglets separately injected with shRNA-RdRp-1–4 expression plasmids 24 h pre-inoculation, ×200. Tissues were stained with hematoxylin and eosin.
4. Discussion
Hepatitis E infection is now of considerably concern, as its antibody is now found globally. HEV is one of the most important causes of acute clinical hepatitis in adults in developing countries (Tei et al., 2003). The nonstructural polyprotein RdRp of virus is involved in the earliest stages of viral replication (Arbuthnot and Thompson, 2008, DeMarini et al., 2003, Zhong et al., 2000, Brockway et al., 2003). Because of this, RdRp may prove to be the best target and best timed strategy for suppression of HEV replication by RNA interference. As an antiviral therapy tool, RNA interference has great promise (Kumar et al., 2008, Lupberger et al., 2008, Ni et al., 2005) and represents a realistic approach for suppressing recalcitrant viral infections such as those induced by HEV.
In the current study, HEV-specific siRNAs effectively and specifically inhibited the expression of HEV RdRp gene in A549 cells. Cells transfected with HEV specific siRNA-RdRp-1 were capable of preventing infection of host cells by HEV. These in vitro findings indicated that this protocol may be an effective antiviral strategy for HEV infection.
The further demonstration of an equally efficient in vivo suppression of HEV infection in piglets following injection with HEV-specific shRNA expression plasmids is particularly encouraging. In Group 3 piglets, which were injected with the shRNA-RdRp-1 expression plasmid, no HEV RNA was detected in the group throughout the entire study. In addition, HEV antigens were obviously reduced in the liver, spleen, and kidney of these piglets, as were activities of ALT, AST and the level of TBIL. This clear reduction in in vivo infection symptoms is a strong indication of the potential usefulness of the RNAi approach as an alternative to the relatively unsuccessful vaccination approaches that have been attempted previously.
In conclusion, the findings of the present study strongly suggest that HEV-specific siRNA (siRNA-RdRp-1) is capable of inhibiting the HEV expression and replication both in vitro in A549 cells and in vivo in piglets. Thus, RNAi looks extremely promising as a potential antiviral therapy for suppression of HEV infection.
Acknowledgements
We thank Shengbo Zheng, Xubin Zhang, Weijun Yang and Xinyong Qi for their generous help. This work was supported by Key Project of Shanghai Science and Technology committee of China under Grant No. 063919121 and the Major Special Science and Technological programme of Department of Agriculture in China under Grant No. 2008ZX08006-004.
Contributor Information
Fen Huang, Email: huangfen6789@163.com.
Xiuguo Hua, Email: hxg@sjtu.edu.cn.
Shixing Yang, Email: yangshixing@sjtu.edu.cn.
Congli Yuan, Email: ycl@sjtu.edu.cn.
Wen Zhang, Email: z0216wen@sjtu.edu.cn.
References
- Abreu C. Viral hepatitis in travellers. Acta Med. Port. 2007;20:557–566. [PubMed] [Google Scholar]
- Aggarwal R., Krawczynski K. Hepatitis E: an overview and recent advances in clinical and laboratory research. J. Gastroenterol. Hepatol. 2000;15:9–20. doi: 10.1046/j.1440-1746.2000.02006.x. [DOI] [PubMed] [Google Scholar]
- Agrawal S., Gupta D., Panda S.K. The 3′ end of hepatitis E virus (HEV) genome binds specifically to the virus RNA-dependent RNA polymerase (RdRp) Virology. 2001;282:87–101. doi: 10.1006/viro.2000.0819. [DOI] [PubMed] [Google Scholar]
- Ansari I.H., Nanda S.K., Durgapal H., Agrawal S., Mohanty S.K., Gupta D., Jameel S., Panda S.K. Cloning, sequencing, and expression of the hepatitis E virus (HEV) nonstructural open reading frame 1 (ORF1) J. Med. Virol. 2000;60:275–283. [PubMed] [Google Scholar]
- Arankalle V.A., Joshi M.V., Kulkarni A.M., Gandhe S.S., Chobe L.P., Rautmare S.S., Mishra A.C., Padbidri V.S. Prevalence of anti-hepatitis E virus antibodies in different Indian animal species. J. Viral. Hepat. 2001;8:223–227. doi: 10.1046/j.1365-2893.2001.00290.x. [DOI] [PubMed] [Google Scholar]
- Arbuthnot P., Thompson L.J. Harnessing the RNA interference pathway to advance treatment and prevention of hepatocellular carcinoma. World J. Gastroenterol. 2008;14:1670–1681. doi: 10.3748/wjg.14.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockway S.M., Clay C.T., Lu X.T., Denison M.R. Characterization of the expression, intracellular localization, and replication complex association of the putative mouse hepatitis virus RNA-dependent RNA polymerase. J. Virol. 2003;77:10515–10527. doi: 10.1128/JVI.77.19.10515-10527.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMarini D.J., Johnston V.K., Konduri M., Gutshall L.L., Sarisky R.T. Intracellular hepatitis C virus RNA-dependent RNA polymerase activity. J. Virol. Methods. 2003;113:65–68. doi: 10.1016/s0166-0934(03)00226-x. [DOI] [PubMed] [Google Scholar]
- Feagins A.R., Opriessnig T., Huang Y.W., Halbur P.G., Meng X.J. Cross-species infection of specific-pathogen-free pigs by a genotype 4 strain of human hepatitis E virus. J. Med. Virol. 2008;80:1379–1386. doi: 10.1002/jmv.21223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner M.B., Luciw P.A. Macaque models of human infectious disease. ILAR. J. 2008;49:220–255. doi: 10.1093/ilar.49.2.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitlin L., Myers C.P., Andino R. Short interfering RNA confers intracellular antiviral immunity in human cells. Nature. 2002;418:430–434. doi: 10.1038/nature00873. [DOI] [PubMed] [Google Scholar]
- Halbur P.G., Kasorndorkbua C., Gilbert C., Guenette D., Potters M.B., Purcell R.H., Emerson S.U., Toth T.E., Meng X.J. Comparative pathogenesis of infection of pigs with hepatitis E viruses recovered from a pig and a human. J. Clin. Microbiol. 2001;39:918–923. doi: 10.1128/JCM.39.3.918-923.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh S.Y., Meng X.J., Wu Y.H., Liu S.T., Tam A.W., Lin D.Y., Liaw Y.F. Identity of a novel swine hepatitis E virus in Taiwan forming a monophyletic group with Taiwan isolates of human hepatitis E virus. J. Clin. Microbiol. 1999;37:3828–3834. doi: 10.1128/jcm.37.12.3828-3834.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R., Li D., Wei S., Li Q., Yuan X., Geng L., Li X., Liu M. Cell culture of sporadic hepatitis E virus in China. Clin. Diagn. Lab. Immunol. 1999;6:729–733. doi: 10.1128/cdli.6.5.729-733.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F.F., Sun Z.F., Emerson S.U., Purcell R.H., Shivaprasad H.L., Pierson F.W., Toth T.E., Meng X.J. Determination and analysis of the complete genomic sequence of avian hepatitis E virus (avian HEV) and attempts to infect rhesus monkeys with avian HEV. J. Gen. Virol. 2004;85:1609–1618. doi: 10.1099/vir.0.79841-0. [DOI] [PubMed] [Google Scholar]
- Jameel S. Molecular biology and pathogenesis of hepatitis E virus. Expert. Rev. Mol. Med. 1999;1999:1–16. doi: 10.1017/S1462399499001271. [DOI] [PubMed] [Google Scholar]
- Jothikumar N., Cromeans T.L., Robertson B.H., Meng X.J., Hill V.R. A broadly reactive one-step real-time RT-PCR assay for rapid and sensitive detection of hepatitis E virus. J. Virol. Methods. 2006;131:65–71. doi: 10.1016/j.jviromet.2005.07.004. [DOI] [PubMed] [Google Scholar]
- Kasorndorkbua C., Guenette D.K., Huang F.F., Thomas P.J., Meng X.J., Halbur P.G. Routes of transmission of swine hepatitis E virus in pigs. J. Clin. Microbiol. 2004;42:5047–5052. doi: 10.1128/JCM.42.11.5047-5052.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawczynski K., Kamili S., Aggarwal R. Global epidemiology and medical aspects of hepatitis E. Forum (Genova) 2001;11:166–179. [PubMed] [Google Scholar]
- Krawczynski K. Hepatitis E vaccine—ready for prime time? N. Engl. J. Med. 2007;356:949–951. doi: 10.1056/NEJMe068311. [DOI] [PubMed] [Google Scholar]
- Kumar P., Ban H.S., Kim S.S., Wu H., Pearson T., Greiner D.L., Laouar A., Yao J., Haridas V., Habiro K., Yang Y.G., Jeong J.H., Lee K.Y., Kim Y.H., Kim S.W., Peipp M., Fey G.H., Manjunath N., Shultz L.D., Lee S.K., Shankar P. T cell-specific siRNA delivery suppresses HIV-1 infection in humanized mice. Cell. 2008;134:577–586. doi: 10.1016/j.cell.2008.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo F.R., Tanaka T., Takahashi H., Ichiyama K., Hoshino Y., Yamada K., Inoue J., Takahashi M., Okamoto H. Mutational events during the primary propagation and consecutive passages of hepatitis E virus strain JE03-1760F in cell culture. Virus Res. 2008;137:86–96. doi: 10.1016/j.virusres.2008.06.005. [DOI] [PubMed] [Google Scholar]
- Lu A., Zhang H., Zhang X., Wang H., Hu Q., Shen L., Schaffhausen B.S., Hou W., Li L. Attenuation of SARS coronavirus by a short hairpin RNA expression plasmid targeting RNA-dependent RNA polymerase. Virology. 2004;324:84–89. doi: 10.1016/j.virol.2004.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig L.B. RNA silencing and HIV: a hypothesis for the etiology of the severe combined immunodeficiency induced by the virus. Retrovirology. 2008;5:79. doi: 10.1186/1742-4690-5-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupberger J., Brino L., Baumert T.F. RNAi: a powerful tool to unravel hepatitis C virus–host interactions within the infectious life cycle. J. Hepatol. 2008;48:523–525. doi: 10.1016/j.jhep.2007.12.007. [DOI] [PubMed] [Google Scholar]
- McIntyre G.J., Fanning G.C. Design and cloning strategies for constructing shRNA expression vectors. BMC Biotechnol. 2006;6(1) doi: 10.1186/1472-6750-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mushahwar I.K. Hepatitis E virus: molecular virology, clinical features, diagnosis, transmission, epidemiology, and prevention. J. Med. Virol. 2008;80:646–658. doi: 10.1002/jmv.21116. [DOI] [PubMed] [Google Scholar]
- Nanda S.K., Panda S.K., Durgapal H., Jameel S. Detection of the negative strand of hepatitis E virus RNA in the livers of experimentally infected rhesus monkeys: evidence for viral replication. J. Med. Virol. 1994;42:237–240. doi: 10.1002/jmv.1890420306. [DOI] [PubMed] [Google Scholar]
- Ni B., Shi X., Li Y., Gao W., Wang X., Wu Y. Inhibition of replication and infection of severe acute respiratory syndrome-associated coronavirus with plasmid-mediated interference RNA. Antivir. Ther. 2005;10:527–533. [PubMed] [Google Scholar]
- Okamoto H., Takahashi M., Nishizawa T., Usui R., Kobayashi E. Presence of antibodies to hepatitis E virus in Japanese pet cats. Infection. 2004;32:57–58. doi: 10.1007/s15010-004-3078-0. [DOI] [PubMed] [Google Scholar]
- Panda S.K., Nanda S.K., Zafrullah M., Ansari I.H., Ozdener M.H., Jameel S. An Indian strain of hepatitis E virus (HEV): cloning, sequence, and expression of structural region and antibody responses in sera from individuals from an area of high-level HEV endemicity. J. Clin. Microbiol. 1995;33:2653–2659. doi: 10.1128/jcm.33.10.2653-2659.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda S.K., Ansari I.H., Durgapal H., Agrawal S., Jameel S. The in vitro-synthesized RNA from a cDNA clone of hepatitis E virus is infectious. J Virol. 2000;74:2430–2437. doi: 10.1128/jvi.74.5.2430-2437.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda S.K., Thakral D., Rehman S. Hepatitis E virus. Rev. Med. Virol. 2007;17:151–180. doi: 10.1002/rmv.522. [DOI] [PubMed] [Google Scholar]
- Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasterk R.A. RNA silencing: the genome's immune system. Science. 2002;296:1263–1265. doi: 10.1126/science.1072148. [DOI] [PubMed] [Google Scholar]
- Saayman S., Barichievy S., Capovilla A., Morris K.V., Arbuthnot P., Weinberg M.S. The efficacy of generating three independent anti-HIV-1 siRNAs from a single U6 RNA Pol III-expressed long hairpin RNA. PLoS ONE. 2008;3:e2602. doi: 10.1371/journal.pone.0002602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabalina S.A., Koonin E.V. Origins and evolution of eukaryotic RNA interference. Trends Ecol. Evol. 2008;23:578–587. doi: 10.1016/j.tree.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha M.P., Scott R.M., Joshi D.M., Mammen M.P., Jr., Thapa G.B., Thapa N., Myint K.S., Fourneau M., Kuschner R.A., Shrestha S.K., David M.P., Seriwatana J., Vaughn D.W., Safary A., Endy T.P., Innis B.L. Safety and efficacy of a recombinant hepatitis E vaccine. N. Engl. J. Med. 2007;356:895–903. doi: 10.1056/NEJMoa061847. [DOI] [PubMed] [Google Scholar]
- Tacke F., Trautwein C. Efficient recombinant hepatitis E virus vaccine: mission accomplished? Hepatology. 2007;46:941–943. doi: 10.1002/hep.21909. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Takahashi M., Kusano E., Okamoto H. Development and evaluation of an efficient cell-culture system for Hepatitis E virus. J. Gen. Virol. 2007;88:903–911. doi: 10.1099/vir.0.82535-0. [DOI] [PubMed] [Google Scholar]
- Tei S., Kitajima N., Takahashi K., Mishiro S. Zoonotic transmission of hepatitis E virus from deer to human beings. Lancet. 2003;362:371–373. doi: 10.1016/S0140-6736(03)14025-1. [DOI] [PubMed] [Google Scholar]
- ter Brake O., Legrand N., von Eije K.J., Centlivre M., Spits H., Weijer K., Blom B., Berkhout B. Evaluation of safety and efficacy of RNAi against HIV-1 in the human immune system (Rag-2(−/−)gammac(−/−)) mouse model. Gene Ther. 2009;16:148–153. doi: 10.1038/gt.2008.124. [DOI] [PubMed] [Google Scholar]
- Vitral C.L., Pinto M.A., Lewis-Ximenez L.L., Khudyakov Y.E., dos Santos D.R., Gaspar A.M. Serological evidence of hepatitis E virus infection in different animal species from the Southeast of Brazil. Mem. Inst. Oswaldo. Cruz. 2005;100:117–122. doi: 10.1590/s0074-02762005000200003. [DOI] [PubMed] [Google Scholar]
- Yarbough P.O. Hepatitis E virus. Advances in HEV biology and HEV vaccine approaches. Intervirology. 1999;42:179–184. doi: 10.1159/000024978. [DOI] [PubMed] [Google Scholar]
- Zhai L., Dai X., Meng J. Hepatitis E virus genotyping based on full-length genome and partial genomic regions. Virus Res. 2006;120:57–69. doi: 10.1016/j.virusres.2006.01.013. [DOI] [PubMed] [Google Scholar]
- Zhong W., Uss A.S., Ferrari E., Lau J.Y., Hong Z. De novo initiation of RNA synthesis by hepatitis C virus nonstructural protein 5B polymerase. J. Virol. 2000;74:2017–2022. doi: 10.1128/jvi.74.4.2017-2022.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]


