Abstract
This paper characterises a virulent strain (CB/05) of canine coronavirus (CCoV) isolated from the internal organs of pups that had died of a systemic disease without evidence of other common canine pathogens. High viral RNA titres were detected in the internal organs by a real-time RT-PCR assay specific for CCoV type II. Sequence analysis of the 3′ end (8.7 kb) of the genomic RNA of strain CB/05 revealed conserved structural as well as non-structural proteins, with the exception of a truncated form of non-structural protein 3b. The exceptional form was due to a 38-nucleotide deletion and a frame shift in ORF3b that introduced an early stop codon. By phylogenetic analysis of the structural proteins, the spike (S) protein was found to cluster with feline coronavirus type II strain 79-1683, whereas, the envelope (E), membrane (M) and nucleocapsid (N) proteins segregated together with the reference strain Purdue of transmissible gastroenteritis virus of swine.
Keywords: Pantropic canine coronavirus, Systemic disease, Genetic analysis
1. Introduction
Canine coronavirus (CCoV) is an enveloped, single-stranded RNA virus responsible for mild to severe enteritis in dogs. CCoV belongs to group I coronaviruses within the family Coronaviridae, along with feline coronaviruses (FCoVs) types I and II, transmissible gastroenteritis virus (TGEV) of swine, porcine respiratory coronavirus (PRCoV), porcine epidemic diarrhoea virus (PEDV) and human coronavirus 229E (HCoV-229E) (Enjuanes et al., 2000). The main non-structural protein of CCoV is the replicase, which is encoded by two large open, partially overlapping, reading frames (ORFs), ORF1a and ORF1b that encompass the 5′ two-thirds of the genomic RNA. The structural proteins comprise the S, E, M and N proteins encoded by ORFs 2, 4, 5 and 6, respectively (Enjuanes et al., 2000). The region between the S and M genes (ORFs 3a, 3b, 3c and 4) comprises four accessory genes that encode non-structural (nsp3a, nsp3b, nsp3c) as well as structural (E) proteins. The very 3′ end of the CCoV RNA is occupied by an untranslated region with a poly-A tail, preceded by two ORFs (7a and 7b) that encode two non-structural proteins (De Groot et al., 1988, Garwes et al., 1989, Horsburgh et al., 1992, Tung et al., 1992, Vennema et al., 1992a, Vennema et al., 1992b).
To date, two different CCoV genotypes are known, which have been designated CCoVs type I and type II on the basis of their genetic relatedness to FCoVs type I and type II, respectively (Pratelli et al., 2003a, Pratelli et al., 2003b). Molecular methods have been established to differentiate those two genotypes (Decaro et al., 2005c), which have been shown to cause enteric infections in dogs (Pratelli et al., 2004). CCoV type I has not been adapted to in-vitro growth and differs from CCoV type II with a higher genetic relatedness to FCoV type I. There also is an additional ORF, ORF3, located between ORFs 2 and 3a (N. Decaro, unpublished data).
CCoV is generally recognised as the aetiological agent of self-limiting infections of the small intestine, which can lead to clinical signs typical of gastroenteritis, i.e., inappetence, diarrhoea and vomiting. Fatal disease commonly occurs as a consequence of mixed infections with CCoV together with canine parvovirus type 2 (CPV-2) (Pratelli et al., 1999), canine adenovirus type 1 (CAdV-1) (Pratelli et al., 2001, Decaro et al., 2006) or canine distemper virus (CDV) (Decaro et al., 2004a). Systemic disease has not previously been ascribed to CCoV infection alone, although the virus has been isolated previously from several tissues (tonsils, lungs and liver) of pups infected experimentally (Tennant et al., 1991).
Recently, a highly virulent CCoV strain (CB/05) was described (Buonavoglia et al., 2006) and it was shown to have been responsible for an outbreak of fatal, systemic disease in pups. In the present study, we report the molecular characterisation of CCoV strain CB/05.
2. Methods
2.1. Clinical case
In May 2005, a severe outbreak of a fatal, systemic disease affected seven dogs housed in a pet shop in the Apulia region of Italy (Buonavoglia et al., 2006). Clinical signs were first observed in four pups that comprised three Miniature Pinschers and one Cocker Spaniel, 45 and 53 days of age, respectively. Signs in the index case consisted of fever (39.5–40 °C), lethargy, inappetance, vomiting, haemorrhagic diarrhoea, leukopenia (WBC counts < 3000 cells/μl) and neurological signs (ataxia, seizures), followed by death within 2 days after the onset of the symptoms. Similar signs were observed a few days later in two additional Miniature Pinschers (117/05-A, 117/05-B) and in a 56 day-old Pekinese pup (117/05-C) that died rapidly. The bodies of those three pups were submitted for laboratory investigation. Necropsy examination revealed severe gross lesions in lungs, liver, spleen, and kidneys. Extensive sub-acute bronchopneumonia was evidenced both in the cranial and caudal lobes, while coalescing degeneration was present within the livers and spleens; also zones of haemorrhagic infarction were observed in the kidneys.
2.2. Screening for canine pathogens
Using standardised methods, samples of brain, lung, liver, spleen, kidney, mesenteric lymph nodes and intestinal contents were examined for the major canine bacterial and viral pathogens, e.g., Bordetella bronchiseptica, Pasteurella multocida, Leptospira interrogans (Gravekamp et al., 1993), reoviruses (Leary et al., 2002, Decaro et al., 2005a), rotaviruses (Gouvea et al., 1994), caliciviruses (Jiang et al., 1999, Marsilio et al., 2005), CPV-2 (Decaro et al., 2005b), canine adenoviruses (Hu et al., 2001), CDV (Elia et al., 2006), canid herpesvirus (Schulze and Baumgartner, 1998), CCoV (Decaro et al., 2004b).
2.3. Virus isolation
Intestinal contents and tissue samples were homogenised (10%, w/v) in Dulbecco's minimal essential medium (D-MEM), together with antibiotics (penicillin 5000 IU/ml, streptomycin 2500 μg/ml, amphotericin B 10 μg/ml), and inoculated into cell cultures.
The following cell lines were used for virus isolation attempts: canine fibroma (A-72), Madin Darby canine kidney (MDCK), Walter Reed canine cells (WRCC), Crandell feline kidney (CrFK), and felis catus whole foetus (fcwf). The cells were grown in D-MEM supplemented with 10% foetal calf serum (FCS). When the monolayers were confluent, the medium was removed and the cells were washed two times with FCS-free medium and inoculated with clarified tissue homogenates. After an adsorption period of 60 min at 37 °C, the fluid portion of each inoculated cell culture was replaced with fresh serum-free medium. Infected cells were monitored daily for the occurrence of cytopathic effects (CPE) and, after 5 days of incubation, the inoculated cells were tested for CCoV antigen by an immunofluorescence (IF) assay using a monoclonal antibody targeting the N protein. Each sample was passaged three times prior to being judged negative for CCoV.
2.4. Typing and quantitation of CCoV
RNAs extracted with the commercial kits QIAamp® Viral RNA Mini Kit (Qiagen S.p.A., Milan, Italy) from the intestinal contents, and QIAamp® RNeasy Mini Kit (Qiagen S.p.A.) from the tissue samples, were subjected to differential real-time RT-PCR assays for identification and quantitation of CCoV type I and type II (Decaro et al., 2005c). Reverse transcription was carried out using GeneAmp® RNA PCR (Applied Biosystems, Applera Italia, Monza, Italy), following the manufacturer's recommendations. Real-time PCR for both genotype-specific assays was performed in a 50 μl-reaction mixture containing 25 μl of IQ™ Supermix (Bio-Rad Laboratories Srl), 600 nM of primers CCoVI-F (CGTTAGTGCACTTGGAAGAAGCT)/CCoVI-R (ACCAGCCATTTTAAATCCTTCA) or CCoVII-F (TAGTGCATTAGGAAGAAGCT)/CCoVII-R (AGCAATTTTGAACCCTTC), 200 nM of probe CCoVI-Pb (FAM-CCTCTTGAAGGTACACCAA-TAMRA) or CCoVII-Pb (FAM-CCTCTTGAAGGTGTGCC-TAMRA) and 20 μl of c-DNA. The thermal profile consisted of activation of iTaq DNA polymerase at 95 °C for 10 min, followed by 45 cycles of denaturation at 95 °C for 15 s, annealing at 53 °C (CCoV type I-specific assay) or 48 °C (CCoV type II-specific assay) for 30 s and extension at 60 °C for 1 min.
2.5. Sequence and phylogenetic analyses of the virulent CCoV strain
The 3′ end of the genome of strain CB/05 was amplified using SuperScript™ One-Step RT-PCR for Long Templates (Life Technologies, Invitrogen, Milan, Italy), according to the manufacturer's instructions. Seven partially overlapping fragments encompassing the ORFs 2 (S gene), 3a, 3b, 3c, 4 (E gene), 5 (M gene), 6 (N gene) 7a and 7b were amplified using primer pairs El-Ins1 (GGATTACTAARGADKGGTAAGTTGC)/S2 (CAACTTCACTTGAAGCAACA), SIIF (GCATTAGTAGTTGAAAACACAGC)/SIIR (TCAAGTCTGGCACCCATTG), S3 (GAGACTTTCAACCAATTAGC)/UCD6R (TTAATGAATGTGAACTTTTTCAATAGG), SM1F (CAGTTTGAAAGTTATGAACCTATTGA)/M6 (TCCCTGAGAGGACATTTAGA), MNIF (CAACAGATGCAAGAACTGAYAA)/MNIR (TTAGTTCGTTACCTCATCAATAATCTC), 7abF (TGATCCTAAGACTGGACAATTCC/7abR (AAATCTAGCATTGCCAAATCAAA).
The PCR-amplified products were sequenced by Genome Express (Meylan, France) and the obtained sequences were assembled and analysed using the BioEdit software package (Hall, 1999) and the NCBI's (http://www.ncbi.nlm.nih.gov) and EMBL's (http://www.ebi.ac.uk) analysis tools. GenBank accession number DQ112226 was assigned to the sequenced 8.7-kb fragment. ORFs contained in the amplified genomic region were determined either with the ORF Finder tool of NCBI or on the basis of the similarity to known coronavirus proteins. The ORFs identified in this manner were translated and the predicted amino acid (aa) sequences were saved as individual files for further analyses. Phylogenetic and molecular evolutionary analyses were conducted using Mega3 (Kumar et al., 2004). Phylogenetic trees, based on the S, E, M and N proteins of CCoV strain CB/05 were elaborated using both parsimony and neighbor-joining methods, supplying a statistical support with bootstrapping over 100 replicates.
3. Results
3.1. CCoV detection and isolation from tissue samples of dogs infected naturally
The bodies of the dead pups tested negative by all of the screening assays; however, real-time RT-PCR for CCoV detected viral RNA in the intestinal contents and, unexpectedly, in all the tissue samples examined. By genotype-specific real-time RT-PCR assays, both CCoV genotypes were detected in the intestinal contents. Viral loads ranged from 4.23 × 104 to 8.99 × 103 RNA copies/μl and from 9.24 × 104 to 6.12 × 105 RNA copies/μl of template for CCoV type I and type II, respectively. On the other hand, only CCoV type II was detected in the parenchymatous organs, with RNA titres ranging from 9.52 × 102 (pup 117/05-B, brain) to 6.87 × 106 RNA copy numbers/μl of template (pup 117/05-C, spleen) (Table 1 ). Sequence analysis showed a 100% nucleotide (nt) identity among the real-time RT-PCR products amplified from the samples of the three pups (data not shown).
Table 1.
CCoV RNA titres in the organs of dogs infected with strain CB/05
| Tissue/sample | 117/05-A |
117/05-B |
117/05-C |
|||
|---|---|---|---|---|---|---|
| RRT-PCR | VI | RRT-PCR | VI | RRT-PCR | VI | |
| Faeces | 6.12 × 105 | + | 9.24 × 104 | + | 3.17 × 105 | + |
| Lung | 1.08 × 106 | + | 2.74 × 104 | + | 2.32 × 106 | + |
| Spleen | 4.46 × 106 | + | 1.20 × 105 | + | 6.87 × 106 | + |
| Liver | 9.02 × 104 | + | 5.98 × 104 | + | 3.12 × 105 | + |
| Kidney | 7.54 × 105 | + | 1.40 × 104 | + | 2.53 × 105 | + |
| Mesenteric lymph node | 8.09 × 104 | + | 2.01 × 104 | + | 5.17 × 104 | + |
| Brain | 5.23 × 103 | − | 9.52 × 102 | − | 1.25 × 104 | − |
RRT-PCR, real-time RT-PCR. Results are expressed as CCoV type II RNA copy number/μl of template. VI, virus isolation. Results are expressed as positive (+) or negative (−).
CCoV type II was successfully isolated in cell cultures from all tissue samples, except the brain. Viral titres of the brain samples were found to be very low by type-specific real-time RT-PCR (Table 1). Only A-72 and fcwf cells were found to be permissive to viral infection; in contrast, VERO, MDCK and CrFK cells were negative by the IF test for CCoV after three passages. Virus isolation was achieved at the first passage from the spleen and lungs of pup 117/05-C (strain CB/05). The CPE was characterised by cell rounding and lysis of the infected monolayers. Viral titres reached at the third passage on A-72 and fcwf cells were 105.75 and 103.25 TCID50/50 μl of viral suspension, respectively.
3.2. Sequence analysis and phylogeny
The nt sequence of a 8.7-kb genomic region of strain CB/05 was determined by amplification and sequencing of overlapping fragments, including the full length of the spike (S), 3a, 3b, 3c, envelope (E), membrane (M), nucleoprotein (N), 7a and 7b genes. The inferred amino acid (aa) sequences were compared to the analogous sequences available in the online databases. The spike protein was 1454 aa in length and, by analysis with the NetNglyc server (http://www.cbs.dtu.dk/services/NetNGlyc/), was found to contain 31 potential N-glycosylation sites, matching the best identity (95%) to FCoV type II strain 79-1683 (Table 2 ). By phylogenetic analysis, two divergent CCoV clusters could be identified, with strain CB/05 segregating with type II CCoVs and FCoVs (Fig. 1a). Several residues encountered in the N-terminus of the S protein were unique to strain CB/05, including Ile-4, Val-8, Ala-18, Asp-34, Pro-73, Ala-116, Asn-125, His-129, Thr-143, Arg-154, His-197, Asp-255, and Gly-291, Ala-407, Thr-459. In order to verify whether the same substitutions were shared by other strains circulating in Italy, the 5′ end of the S gene, encoding for the N-terminal 500 amino acids of the S protein, was amplified from five type II CCoV strains recovered from recent field outbreaks of enteritis in dogs from different regions of Italy. By sequence analysis, a 94–98% identity was found in the deduced amino acid sequences (data not shown). All of the N-terminus residues initially found unique to strain CB/05 were shared by the CCoV strains currently circulating in Italy, with the exception of residues Pro-73, Asn-125 and Ala-407 that were present only in strain CB/05. An aa substitution (Asp to His) at position 125 also was displayed by the virulent strain BGF10.
Table 2.
Amino acid identity in the structural proteins of group I coronavirus reference strains to strain CB/05
| Coronavirus strain | GenBank accession no. | Amino acid identity (%) to isolate CB/05 |
|||
|---|---|---|---|---|---|
| S | E | M | N | ||
| CCoV II-Insavc1 | D13096 | 92 | 95 | 90 | 92 |
| CCoV II-BGF10 | AY342160 | 92 | 87 | 89 | 92 |
| CCoV I-Elmo/02 | AY307020 (S), AY426983 (E), ND (M, N) | 44 | 85 | 87 | 89 |
| CCoV I-23/03 | AY307021 (S), AY426984 (E) AY548235 (M, N) | 44 | 86 | 86 | 89 |
| FCoV II-79-1146 | NC_007025 | 94 | 78 | 84 | 77 |
| FCoV II-79-1683 | X80799 (S), FCY13921 (E, M, N) | 95 | 97 | 87 | 79 |
| FCoV I-KU-2 | D32044 (S), AAB47501 (M) AB086881 (N) | 43 | NA | 82 | 76 |
| FCoV I-Black | AB088223 (S), AB086903 (M, N), | 42 | NA | 82 | 77 |
| FCoV I-UCD1 | AB088222 (S), AB086902 (M, N) | 43 | NA | 84 | 77 |
| TGEV-Purdue | NC_002306 | 80 | 96 | 94 | 95 |
ND, sequence not deposited. NA, sequence not available.
Fig. 1.
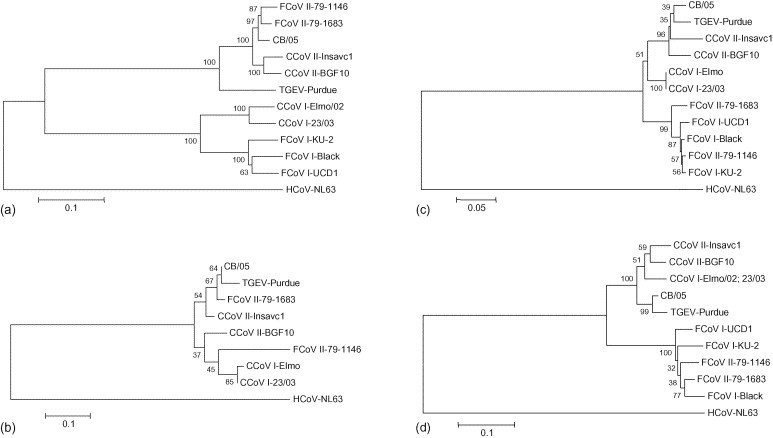
Neighbor-joining trees based on the S (a), E, (b), M (c) and N (d) proteins of group I coronaviruses. Accession numbers of the strains used for phylogeny are reported in Table 2. The trees are rooted on the distantly related group I coronavirus HCoV-NL63 (accession no. NC_005831) and drawn to scale. A statistical support was provided by bootstrapping over 100 replicates. The scale bars represent the numbers of amino acid substitutions.
The envelope protein was 82 aa long and had the highest identity (97%) to FCoV type II strain 79-1683 (Table 2), in whose cluster it was found to fall (Fig. 1b). Due to a lack of data from the 3′ end of the CCoV genome in the genes encoding for non-structural proteins, comparison of strain CB/05 was possible only with CCoV type II strains Insavc-1 (Horsburgh et al., 1992) and BGF10 (Sanchez-Morgado et al., 2004) and with CCoV type I strains Elmo/02 and 23/03 (N. Decaro, unpublished data). While the non-structural protein (nsp) 3a had the same length as the other type I and type II CCoVs (78 aa), the nsp3b was 22-aa long (49-aa shorter than expected) due to the presence of a 38-nt deletion at position 4704 and to a frame shift in the sequence downstream from the deletion that introduced an early stop codon (Fig. 2 ). Genetic analysis of enteric CCoV strains circulating in Italy revealed that the deletion in ORF3b was unique to CB/05 strain (data not shown). The nsp3c (244 aa) was 6 aa shorter and 79 aa longer than the cognate proteins of the enteropathogen strain BGF10 and the attenuated strain Insavc-1, respectively.
Fig. 2.
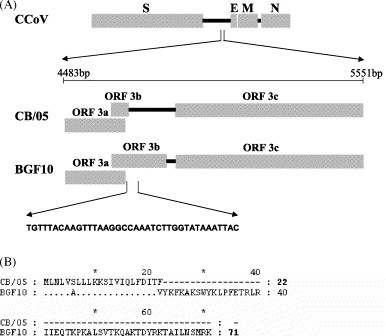
(A) Schematic representation of the 3′ end of the viral genome of CCoV strains CB/05 and BGF10. The 38-nt deletion in ORF3b of strain CB/05 is reported. (B) Comparison between the amino acid sequences of nsp3b of strains CB/05 and BGF10.
The membrane protein was 262 aa in length and contained 3 N-glycosylation sites. The highest identity (94%) was obtained against TGEV strain Purdue (Table 2), in whose cluster the virulent CCoV was found to fall by phylogeny (Fig. 1c). In the N protein, which was 383 aa long, strain CB/05 was found to be closely related to TGEV strain Purdue both by sequence analysis (95% of aa identity, Table 2) and phylogeny (Fig. 1d). Nsp7a was 101 aa long and had one N-glycosylation site, whereas, nsp7b had a length of 213 aa and no N-glycosylation sites, unlike to other type I and type II CCoVs that displayed one N-glycosylation site. However, by sequence analysis, both nsp7a and nsp7b were found closely related to the analogous proteins of strains BGF (96% and 79% of aa identity, respectively) and Insavc-1 (95% and 79% of aa identity, respectively).
4. Discussion
The emergence of a pantropic variant of CCoV, reported in the present study, is paradigmatic of the evolution of coronaviruses. The ORF2 of PRCoV has a 200-aa deletion in the N-terminus with respect to TGEV, from which it presumably had arisen. Most likely, this deletion is responsible for the change in the viral pathobiology (Vaughn et al., 1994). Nevertheless, minor amino acid differences in the sequence of the spike protein have been shown to change the virulence of even very closely related TGEV isolates (Sanchez et al., 1999). The enteric biotype of FCoV, feline enteric coronavirus (FECV), causes persistent infections of the intestinal mucosa that may lead to point mutations in the S gene and/or deletions in the group-specific genes 3c, 7b (Vennema et al., 1998) or 7a (Kennedy et al., 2001). Those mutations are considered responsible for changes in the tropism of the virus, which acquired the ability to infect monocytes/macrophages and to cause a systemic, fatal disease of cats (feline infectious peritonitis, FIP). An FIPV mutant lacking the group-specific genes 3abc or 7ab resulted in live attenuated vaccines protective against FIP (Haijema et al., 2004). Similar drastic shifts of tissue tropism have been observed with murine coronaviruses (Haspel et al., 1978). Adaptation to humans of the recently recognised SARS-associated coronavirus (SARS-CoV) appears to be related to minor genome mutations, consisting of a 29-nt deletion in the genome of a wild-mammal coronavirus, that resulted in the translation of two different ORFs, 10 and 11, instead of the single ORF10 (Guan et al., 2003).
The association of strain CB/05 to a severe, fatal disease of dogs, together with the isolation of the virus from organs with extensive lesions, strongly suggests that CCoV also may have changed its tropism and acquired the ability to spread from the enteric tract to the internal organs.
CCoV strain CB/05 displayed intact structural and non-structural proteins, with a spike protein closely related to that of other type II CCoVs and FCoVs, mostly of FCoV type II strain 79-1683 (95% of aa identity). In contrast to reference type II CCoVs and FCoVs, several point mutations were found in the N-terminus of the S protein of strain CB/05. However, most of these amino acid changes were not associated to increased pathogenicity or change in tropism since the same substitutions were observed in enteric CCoVs recovered from recent outbreaks of canine gastroenteritis. Only residues Pro-73, Asn-125 and Ala-407 were unique to strain CB/05, although a substitution at position 125 (Asp to His instead Asn) was found in the virulent strain BGF10.
A conspicuous change observed in the CB/05 genome was the truncated form of nsp3b, but whether the deletion in the ORF3b is involved in the change of virulence should be assessed by using reverse genetics systems similar to those recently used for FIPV (Haijema et al., 2003). Further studies also are required to verify whether the truncated protein is translated. Recently, a 61–64-conspicuous nucleotide deletion in the intergenic S-3a region was tentatively associated to increased virulence of the enteric CCoV strain BGF10 (Sanchez-Morgado et al., 2004). Similar mutations also have been shown to influence the level of nsp3b translation in TGEV (McGoldrick et al., 1999). However, the same or more extended deletions have been found in strain CB/05 as well as in almost all non-virulent strains circulating in Italy (C. Buonavoglia, unpublished data). Moreover, in strain BGF10 a long nsp3b protein (79-aa longer than the 171-aa protein of strain Insavc-1) is present, in contrast with the truncated protein predicted for the virulent strain CB/05.
Two cases of fatal coronavirus disease in pups without evidence of co-infection by CPV-2 have been reported by Evermann et al. (2005). CCoV infection was demonstrated by immunohistochemistry on gut sections and electron microscopy of intestinal contents. Although CPV-2 was not identified in those cases, the gross lesions and histopathological changes were suggestive of CPV infection. It is possible, however, that the moderate depletion and necrosis of lymphoid tissues (thymus, spleen, lymph nodes and gut-associated lymphoid tissues) in both pups could be related solely to a pantropic CCoV infection. However, this hypothesis cannot be supported since investigations for CCoV were not carried out on the internal organs.
The growth characteristics of strain CB/05 were evaluated by using cell lines commonly used for isolation of canine viral pathogens. Viral growth occurred only on canine A-72 and feline fcwf cells, with higher titres found in the former cell line. Despite the high similarity to FCoV strain 79-1683 in the S protein, responsible for viral attachment on cell receptors (Godet et al., 1994, Kubo et al., 1994), strain CB/05 did not grow on feline CrFK cells. This finding is in contrast with observations on other type II CCoV and FCoV strains which are easily adapted to CrFK cells (Binn et al., 1974, Pedersen et al., 1984).
In this study, we have analysed a pantropic variant of CCoV associated with a fatal disease of dogs characterised by leukopenia, gastroenteritis and severe lesions in the major organs. Our findings suggest that the novel pantropic CCoV variant can cause a fatal disease of dogs, whose clinical and diagnostic features should be recognised by veterinarians since the clinical course may resemble CPV-2 infection. In contrast, the gross lesions of this new CCoV disease are very similar to those observed in dogs with infectious canine hepatitis caused by CAdV-1.
Further investigation would provide new insights into the molecular mechanisms responsible for the change in viral pathobiology and into the pathogenic and immunological aspects of the pantropic CCoV infection.
Acknowledgements
This study was supported by grants of Ministry of Education, University and Research to N.D. (PRIN 2005, project “Il coronavirus del cane: aspetti molecolari e patogenetici”) and of Ministry of Health to M.T. (Ricerca corrente 2003, project “Studio del coronavirus del cane come modello animale per le ricombinazioni genetiche dei coronavirus”). The authors are grateful to the undergraduate students Giuseppe Grandolfo and Angela Segreto for their excellent assistance with part of the experimental work.
Footnotes
The DDBJ/EMBL/GenBank accession number of the sequence reported in this paper is DQ112226.
References
- Binn, L.N., Lazar, E.C., Keenan, K.P., Huxsoll, D.L., Marchwicki, B.S., Strano, A.J., 1974. Recovery and characterization of a coronavirus from military dogs with diarrhea. Proceedings of the 78th Ann Mtg USAHA, pp. 359–366. [PubMed]
- Buonavoglia C., Decaro N., Martella V., Elia G., Campolo M., Desario C., Castagnaro M., Tempesta M. Canine coronavirus highly pathogenic for dogs. Emerg. Infect. Dis. 2006;12:492–494. doi: 10.3201/eid1203.050839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Groot R.J., Andeweg A.C., Horzinek M.C., Spaan W.J. Sequence analysis of the 3′-end of the feline coronavirus FIPV 79-1146 genome: comparison with the genome of porcine coronavirus TGEV reveals large insertions. Virology. 1988;167:370–376. doi: 10.1016/0042-6822(88)90097-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro N., Camero M., Greco G., Zizzo N., Elia G., Campolo M., Pratelli A., Buonavoglia C. Canine distemper and related diseases: report of a severe outbreak in a kennel. New Microbiol. 2004;27:177–181. [PubMed] [Google Scholar]
- Decaro N., Pratelli A., Campolo M., Elia G., Martella V., Tempesta M., Buonavoglia C. Quantitation of canine coronavirus RNA in the faeces of dogs by TaqMan RT-PCR. J. Virol. Meth. 2004;119:145–150. doi: 10.1016/j.jviromet.2004.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro N., Campolo M., Desario C., Ricci D., Camero M., Lorusso E., Elia G., Lavazza A., Martella V., Buonavoglia C. Virological and molecular characterization of a mammalian orthoreovirus type 3 strain isolated from a dog in Italy. Vet. Microbiol. 2005;109:19–27. doi: 10.1016/j.vetmic.2005.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro N., Elia G., Martella V., Desario C., Campolo M., Di Trani L., Tarsitano E., Tempesta M., Buonavoglia C. A real-time PCR assay for rapid detection and quantitation of canine parvovirus type 2 DNA in the feces of dogs. Vet. Microbiol. 2005;105:19–28. doi: 10.1016/j.vetmic.2004.09.018. [DOI] [PubMed] [Google Scholar]
- Decaro N., Martella V., Ricci D., Elia G., Desario C., Campolo M., Cavaliere N., Di Trani L., Tempesta M., Buonavoglia C. Genotype-specific fluorogenic RT-PCR assays for the detection and quantitation of canine coronavirus type I and type II RNA in faecal samples of dogs. J. Virol. Meth. 2005;130:72–78. doi: 10.1016/j.jviromet.2005.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro, N., Campolo, M., Elia, G., Buonavoglia, D., Colaianni, M.L., Lorusso, A., Mari, V., Buonavoglia, C., 2006. Infectious canine hepatitis: an “old” disease reemerging in Italy. Res. Vet. Sci., Epub ahead of print, doi:10.1016/j.rvsc.2006.11.009. [DOI] [PMC free article] [PubMed]
- Elia G., Decaro N., Martella V., Cirone F., Lucente M.S., Lorusso E., Di Trani L., Buonavoglia C. Detection of canine distemper virus in dogs by real-time RT-PCR. J. Virol. Meth. 2006;136:171–176. doi: 10.1016/j.jviromet.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Enjuanes L., Brian D., Cavanagh D., Holmes K., Lai M.M.C., Laude H., Masters P., Rottier P., Siddell S., Spaan W.J.M., Taguchi F., Talbot P. Family Coronaviridae. In: van Regenmortel M.H.V., Fauquet C.M., Bishop D.H.L., Carstens E.B., Estes M.K., Lemon S.M., Maniloff J., Mayo M.A., McGeoch D.J., Pringle C.R., Wickner R.B., editors. Virus Taxonomy, Classification and Nomenclature of Viruses. Academic Press; New York: 2000. pp. 835–849. [Google Scholar]
- Evermann J.F., Abbott J.R., Han S. Canine coronavirus-associated puppy mortality without evidence of concurrent canine parvovirus infection. J. Vet. Diagn. Invest. 2005;17:610–614. doi: 10.1177/104063870501700618. [DOI] [PubMed] [Google Scholar]
- Garwes D.J., Stewart F., Britton P. The polypeptide of Mr 14000 of porcine transmissible gastroenteritis virus. gene assignment and intracellular location. J. Gen. Virol. 1989;70:2495–2499. doi: 10.1099/0022-1317-70-9-2495. [DOI] [PubMed] [Google Scholar]
- Godet M., Grosclaude J., Delmas B., Laude H. Major receptor-binding and neutralization determinants are located within the same domain of the transmissible gastroenteritis virus (coronavirus) spike protein. J. Virol. 1994;68:8008–8016. doi: 10.1128/jvi.68.12.8008-8016.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouvea V., Santos N., Timenetsky, Mdo C. Identification of bovine and porcine rotavirus G types by PCR. J. Clin. Microbiol. 1994;32:1338–1340. doi: 10.1128/jcm.32.5.1338-1340.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravekamp C., Van de Kemp H., Franzen M., Carrington D., Schoone G.J., Van Eys G.J., Everard C.O., Hartskeerl R.A., Terpstra W.J. Detection of seven species of pathogenic leptospires by PCR using two sets of primers. J. Gen. Microbiol. 1993;139:1691–1700. doi: 10.1099/00221287-139-8-1691. [DOI] [PubMed] [Google Scholar]
- Guan Y., Zheng B.J., He Y.Q., Liu X.L., Zhuang Z.X., Cheung C.L., Luo S.W., Li P.H., Zhang L.J., Guan Y.J., Butt K.M., Wong K.L., Chan K.W., Lim W., Shortridge K.F., Yuen K.Y., Peiris J.S., Poon L.L. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- Haijema B.J., Volders H., Rottier P.J. Switching species tropism: an effective way to manipulate the feline coronavirus genome. J. Virol. 2003;77:4528–4538. doi: 10.1128/JVI.77.8.4528-4538.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haijema B.J., Volders H., Rottier P.J. Live, attenuated coronavirus vaccines through the directed deletion of group-specific genes provide protection against feline infectious peritonitis. J. Virol. 2004;78:3863–3871. doi: 10.1128/JVI.78.8.3863-3871.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall T.A. BioEdit: a user-friendly biological sequence alignment and analysis program for Windows 95/98/NT. Nucl. Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- Haspel M.V., Lampert P.W., Oldstone M.B. Temperature-sensitive mutants of mouse hepatitis virus produce a high incidence of demyelination. Proc. Natl. Acad. Sci. U.S.A. 1978;75:4033–4036. doi: 10.1073/pnas.75.8.4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu R.L., Huang G., Qiu W., Zhong Z.H., Xia X.Z., Yin Z. Detection and differentiation of CAV-1 and CAV-2 by polymerase chain reaction. Vet. Res. Commun. 2001;25:77–84. doi: 10.1023/a:1006417203856. [DOI] [PubMed] [Google Scholar]
- Horsburgh B.C., Brierley I., Brown T.D. Analysis of a 9.6 kb sequence from the 3′ end of canine coronavirus genomic RNA. J. Gen. Virol. 1992;73:2849–2862. doi: 10.1099/0022-1317-73-11-2849. [DOI] [PubMed] [Google Scholar]
- Jiang X., Huang P.W., Zhong W.M., Farkas T., Cubitt D.W., Matson D.O. Design and evaluation of a primer pair that detects both Norwalk- and Sapporo-like caliciviruses by RT-PCR. J. Virol. Meth. 1999;83:145–154. doi: 10.1016/s0166-0934(99)00114-7. [DOI] [PubMed] [Google Scholar]
- Kennedy M., Boedeker N., Gibbs P., Kania S. Deletions in the 7a ORF of feline coronavirus associated with an epidemic of feline infectious peritonitis. Vet. Microbiol. 2001;81:227–234. doi: 10.1016/S0378-1135(01)00354-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo H., Yamada Y.K., Taguchi F. Localization of neutralizing epitopes and the receptor-binding site within the amino-terminal 330 amino acids of the murine coronavirus spike protein. J. Virol. 1994;68:5403–5410. doi: 10.1128/jvi.68.9.5403-5410.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Tamura K., Nei M. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Leary P.L., Erker J.C., Chalmers M.L., Cruz A.T., Wetzel J.D., Desai S.M., Mushahwar I.K., Dermody T.S. Detection of mammalian reovirus RNA by using reverse transcription-PCR: sequence diversity within the λ3-encoding L1 gene. J. Clin. Microbiol. 2002;40:1368–1375. doi: 10.1128/JCM.40.4.1368-1375.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsilio F., Di Martino B., Decaro N., Buonavoglia C. Nested PCR for the diagnosis of calicivirus infections in the cat. Vet. Microbiol. 2005;105:1–7. doi: 10.1016/j.vetmic.2004.09.017. [DOI] [PubMed] [Google Scholar]
- McGoldrick A., Lowings J.P., Paton D.J. Characterisation of a recent virulent transmissible gastroenteritis virus from Britain with a deleted ORF 3a. Arch. Virol. 1999;144:763–770. doi: 10.1007/s007050050541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen N.C., Black J.W., Boyle J.F., Evermann J.F., McKeirnan A.J., Ott R.L. Pathogenic differences between various feline coronavirus isolates. Adv. Exp. Med. Biol. 1984;173:365–380. doi: 10.1007/978-1-4615-9373-7_36. [DOI] [PubMed] [Google Scholar]
- Pratelli A., Tempesta M., Roperto F.P., Sagazio P., Carmichael L., Buonavoglia C. Fatal coronavirus infection in puppies following canine parvovirus 2b infection. J. Vet. Diagn. Invest. 1999;11:550–553. doi: 10.1177/104063879901100615. [DOI] [PubMed] [Google Scholar]
- Pratelli A., Martella V., Elia G., Tempesta M., Guarda F., Capucchio M.T., Carmichael L.E., Buonavoglia C. Severe enteric disease in an animal shelter associated with dual infection by canine adenovirus type 1 and canine coronavirus. J. Vet. Med. B. 2001;48:385–392. doi: 10.1046/j.1439-0450.2001.00466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratelli A., Martella V., Decaro N., Tinelli A., Camero M., Cirone F., Elia G., Cavalli A., Corrente M., Greco G., Buonavoglia D., Gentile M., Tempesta M., Buonavoglia C. Genetic diversity of a canine coronavirus detected in pups with diarrhoea in Italy. J. Virol. Meth. 2003;110:9–17. doi: 10.1016/S0166-0934(03)00081-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratelli A., Martella V., Pistello M., Elia G., Decaro N., Buonavoglia D., Camero M., Tempesta M., Buonavoglia C. Identification of coronaviruses in dogs that segregate separately from the canine coronavirus genotype. J. Virol. Meth. 2003;107:213–222. doi: 10.1016/S0166-0934(02)00246-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratelli A., Decaro N., Tinelli A., Martella V., Elia G., Tempesta M., Cirone F., Buonavoglia C. Two genotypes of canine coronavirus simultaneously detected in fecal samples of dogs with diarrhea. J. Clin. Microbiol. 2004;42:1797–1799. doi: 10.1128/JCM.42.4.1797-1799.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez C.M., Izeta A., Sanchez-Morgado J.M., Alonso S., Sola I., Balasch M., Plana-Duran J., Enjuanes L. Targeted recombination demonstrates that the spike gene of transmissible gastroenteritis coronavirus is a determinant of its enteric tropism and virulence. J. Virol. 1999;73:7607–7618. doi: 10.1128/jvi.73.9.7607-7618.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Morgado J.M., Poynter S., Morris T.H. Molecular characterization of a virulent canine coronavirus BGF strain. Virus Res. 2004;104:27–31. doi: 10.1016/j.virusres.2004.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze C., Baumgartner W. Nested polymerase chain reaction and in situ hybridization for diagnosis of canine herpesvirus infection in puppies. Vet. Pathol. 1998;35:209–217. doi: 10.1177/030098589803500306. [DOI] [PubMed] [Google Scholar]
- Tennant B.J., Gaskell R.M., Kelly D.F., Carter S.D., Gaskell C.J. Canine coronavirus infection in the dog following oronasal inoculation. Res. Vet. Sci. 1991;51:11–18. doi: 10.1016/0034-5288(91)90023-H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung F.Y.T., Abraham S., Sethna M., Hung S.L., Sethna P., Hogue B.G., Brian D.A. The 9-kDa hydrophobic protein encoded at the 3′ end of the porcine transmissible gastroenteritis coronavirus genome is membrane-associated. Virology. 1992;186:676–683. doi: 10.1016/0042-6822(92)90034-M. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughn E.M., Halbur P.G., Paul P.S. Three new isolates of porcine respiratory coronavirus with various pathogenicities and spike (S) gene deletions. J. Clin. Microbiol. 1994;32:1809–1812. doi: 10.1128/jcm.32.7.1809-1812.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vennema H., Heijnen L., Rottier P.J., Horzinek M.C., Spaan W.J. A novel glycoprotein of feline infectious peritonitis coronavirus contains a KOEL-like endoplasmic reticulum retention signal. J. Virol. 1992;66:4951–4956. doi: 10.1128/jvi.66.8.4951-4956.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vennema H., Rossen J.W., Wesseling J., Horzinek M.C., Rottier P.J. Genomic organization and expression of the 3′ end of the canine and feline enteric coronaviruses. Virology. 1992;191:134–140. doi: 10.1016/0042-6822(92)90174-N. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vennema H., Poland A., Foley J., Pedersen N.C. Feline infectious peritonitis viruses arise by mutation from endemic feline enteric coronaviruses. Virology. 1998;243:150–157. doi: 10.1006/viro.1998.9045. [DOI] [PMC free article] [PubMed] [Google Scholar]


