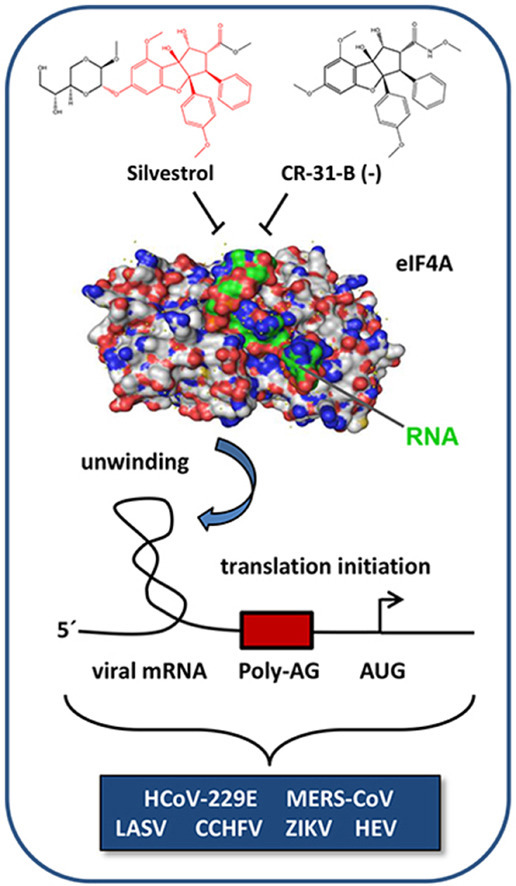
Abstract
Rocaglates, a class of natural compounds isolated from plants of the genus Aglaia, are potent inhibitors of translation initiation. They are proposed to form stacking interactions with polypurine sequences in the 5′-untranslated region (UTR) of selected mRNAs, thereby clamping the RNA substrate onto eIF4A and causing inhibition of the translation initiation complex. Since virus replication relies on the host translation machinery, it is not surprising that the rocaglate Silvestrol has broad-spectrum antiviral activity. Unfortunately, synthesis of Silvestrol is sophisticated and time-consuming, thus hampering the prospects for further antiviral drug development. Here, we present the less complex structured synthetic rocaglate CR-31-B (−) as a novel compound with potent broad-spectrum antiviral activity in primary cells and in an ex vivo bronchial epithelial cell system. CR-31-B (−) inhibited the replication of corona-, Zika-, Lassa-, Crimean Congo hemorrhagic fever viruses and, to a lesser extent, hepatitis E virus (HEV) at non-cytotoxic low nanomolar concentrations. Since HEV has a polypurine-free 5′-UTR that folds into a stable hairpin structure, we hypothesized that RNA clamping by Silvestrol and its derivatives may also occur in a polypurine-independent but structure-dependent manner. Interestingly, the HEV 5′-UTR conferred sensitivity towards Silvestrol but not to CR-31-B (−). However, if an exposed polypurine stretch was introduced into the HEV 5′-UTR, CR-31-B (−) became an active inhibitor comparable to Silvestrol. Moreover, thermodynamic destabilization of the HEV 5′-UTR led to reduced translational inhibition by Silvestrol, suggesting differences between rocaglates in their mode of action, most probably by engaging Silvestrol's additional dioxane moiety.
Keywords: Silvestrol, CR-31-B, Antiviral activity, eIF4A, Translation initiation, Rocaglates
Graphical abstract
1. Introduction
The DEAD-box RNA helicase eIF4A, which is part of the heterotrimeric translation initiation complex eIF4F, unwinds RNA secondary structures in 5′-UTRs of selected mRNAs to enable binding of the 43S preinitiation complex (PIC) (Bhat et al., 2015; Wolfe et al., 2014). In cells, eIF4A has a critical role in the translation of protooncogenic mRNAs with complex-structured 5′-UTRs (Wolfe et al., 2014; Rubio et al., 2014). Viral mRNAs also contain structured 5′-UTRs (Madhugiri et al., 2015; Schlereth et al., 2016), suggesting that viral protein synthesis may also be eIF4A-dependent. Indeed, we have shown that the specific eIF4A inhibitor Silvestrol, a plant-derived rocaglate, has broad-spectrum antiviral activity at non-cytotoxic concentrations in a low nanomolar range (Müller et al., 2018a). Silvestrol inhibits the replication of RNA viruses representing different virus families, such as Ebola- (EBOV), corona- (CoV), Zika- (ZIKV), Chikungunya- (CHIKV) and hepatitis E (HEV) viruses (Müller et al., 2018a; Biedenkopf et al., 2017; Elgner et al., 2018; Glitscher et al., 2018; Henss et al., 2018). Notably, Silvestrol showed good bioavailability (Sarahdi et al., 2011), in vivo antiviral activity (Todt et al., 2018) and remarkably low cytotoxicity in primary cells, thus opening a broad therapeutic window for future applications. To date, however, chemical synthesis of Silvestrol remains difficult and alternative strategies are highly desirable. An example of a structurally less complex natural rocaglate with antiviral activity is RocA (Too et al., 2018), lacking the dioxane moiety of Silvestrol (Pan et al., 2014).
Here, we investigated the antiviral activity of the synthetic RocA analog CR-31-B (−) (Wolfe et al., 2014, Fig. 1 ) and compared its activity with that of Silvestrol. CR-31-B (−) showed similar antiviral activity against HCoV-229E, MERS-CoV, ZIKV, LASV, CCHFV and, to a lesser extent, HEV, thus identifying CR-31-B (−) as another rocaglate with potent broad-spectrum antiviral activity. However, we also found differential activities for these two compounds, depending on the viral 5′-UTR used for the translation inhibition assays. As shown for RocA, clamping of RNA substrates onto eIF4A by CR-31-B (−) requires polypurine sequences (Iwasaki et al., 2019), whereas Silvestrol can also clamp polypurine-free viral RNA substrates, provided that a stable hairpin structure is present. The recently published crystal structure of human eIF4A in complex with a polypurine RNA substrate and RocA (Iwasaki et al., 2019) explains the observed polypurine dependency of RocA because only purines can efficient stack on the phenyl rings of RocA. Although, further structure-based information is missing, we suggest that the dioxane moiety of Silvestrol can mediate polypurine-free RNA clamping onto eIF4A.
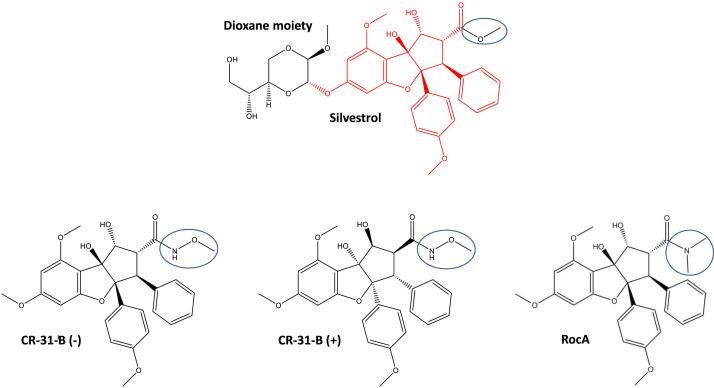
Fig. 1.
Structures of the rocaglates Silvestrol, CR-31-B (−), CR-31-B (+) and RocA. The characteristic cyclopenta[b]benzofurane structure found in all rocaglates is indicated in red in the Silvestrol structure. The dioxane moiety that is only found in Silvestrol is shown on the left side in black. Blue ring: variable region in rocaglates.
2. Material and methods
2.1. Cell culture
Human fetal lung fibroblasts (MRC-5), human lung carcinoma cells (A549) and murine hepatocytes (Hepa1-6 cells) were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% or 5% fetal bovine serum (FBS), 100 U/ml penicillin, and 100 μg/ml streptomycin at 37 °C in an atmosphere containing 5% CO2.
2.2. Reagents
Silvestrol was obtained from the Sarawak Biodiversity Centre (Kuching; North-Borneo, Malaysia; purity >99%). A 6 mM stock solution was prepared in DMSO (sterile-filtered; Roth) and diluted in DMEM. Control cells were treated with corresponding DMSO dilutions lacking Silvestrol. CR-31-B (−) and CR-31-B (+) (Wolfe et al., 2014) were dissolved in DMSO at a concentration of 10 mM and stored at −20 °C.
2.3. Cell toxicity
Cell growth and viability of murine hepatocytes and MRC-5 cells in the presence of the respective compounds was determined by ATPlite assay (PerkinElmer) or the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazoliumbromide (MTT) method as described previously (Günther et al., 2004; Müller et al., 2018a). Cell viability of A549 cells persistently infected with HEV was determined using the PrestoBlue Cell viability reagent (Thermo Fisher Scientific) after treatment with the substances in the respective concentrations for 72 h.
2.4. Human airway epithelial cells
Cryopreserved normal human bronchial epithelial (NHBE) cells were obtained from Lonza. Undifferentiated cells were seeded on transwell plates (Corning Costar) coated with collagen IV (Invitrogen) and grown in a mixture of DMEM (Invitrogen) and BEGM (Lonza) supplemented with retinoic acid (75 nM). Fresh medium was added regularly after 2 days. After reaching confluence, the cells were cultivated under air-liquid conditions for 4 additional weeks for full differentiation into pseudostratified human airway epithelia. Medium from the basolateral compartment was renewed every 2–3 days and the apical surface was washed every week with PBS (Invitrogen).
2.5. Viruses
High-titer stocks of HCoV-229E (NCBI accession number NC_002645) and MERS-CoV (EMC/2012; NC_019843) were produced using Huh-7 cells. High-titer stocks of CCHFV strain Afg-09 2990 (Ölschläger et al., 2011) and LASV strain Ba366 (Lecompte et al., 2006) were produced in Vero E6 cells. The ZIKV strain 976 Uganda (U) was kindly provided by the European Virus Archive. Persistently HEV-infected cells (gt3c strain 47832c; GenBank ID KC618403.1) were previously generated (Johne et al., 2014).
2.6. Antiviral activity
To determine the antiviral activity of CR-31-B and Silvestrol, MRC-5 cells or murine hepatocytes were inoculated with the respective virus at a multiplicity of infection (MOI) of 0.1 or 0.01 at 33 °C (HCoV-229E) or 37 °C (MERS-CoV, LASV, CCHFV). After 1 h, the inoculum was removed and cells were incubated with fresh medium containing the inhibitor at increasing concentrations. Supernatants were collected at 24 h post infection (hpi; HCoV-229E, MERS-CoV) or 3 dpi (LASV, CCHF) and virus titers were analyzed by virus plaque assay (Müller et al., 2018b) or immunofocus assay as described before (Günther et al., 2004). To calculate EC50 values, the virus titer determined for virus-infected cells treated with DMSO only was set to 100% and titers obtained for treated cells were normalized to this value. EC50 values were calculated by non-linear regression analysis using GraphPad Prism 6.0 (GraphPad Software).
For the infection of the primary human airway epithelial cells, the apical surface was washed 3 times with PBS and cells were infected with HCoV-229E (MOI = 3). After 1 h the inoculum was removed and the medium in the basal compartment was replaced with medium containing the indicated inhibitor concentrations. At the indicated time points, the apical surface of the cells was incubated with PBS for 15 min and virus titers in the supernatants were determined by virus plaque assay. Effects on HEV were analyzed using persistently HEV-infected A549 cells. Treatments with the compounds were started 24 h post seeding and cell culture supernatants were analyzed after 72 h via qRT-PCR.
2.7. Western blot analysis
To analyze viral protein accumulation, MRC-5 cells were infected with HCoV-229E at an MOI of 1. After inoculation, the supernatant was replaced with DMEM supplemented with antibiotics and the indicated concentrations of CR-31-B enantiomers. At 24 hpi, cell lysates were prepared and viral proteins were analyzed by Western blotting as described previously (Müller et al., 2018a).
2.8. qRT-PCR of HCoV-229E RNA or extracellular HEV RNA
MRC-5 cells were infected at an MOI of 1 and incubated for 24 h with the indicated inhibitor concentrations. Then, total cellular RNA was isolated using the RNeasy kit (Qiagen) and qRT-PCR was performed using 5 ng RNA and the Luna Universal Probe One-Step RT-qPCR Kit (NEB). Sequences of primers used to amplify genomic and total viral RNA, respectively, and GAPDH mRNA are available upon request. For the analysis of relative fold viral RNA expression in regard to inhibitor treatment the delta-delta Ct method (using GAPDH as reference) was used (Livak and Schmittgen et al., 2001). Isolation and quantification of extracellular HEV RNA was performed as described before (Glitscher et al., 2018).
2.9. Dual luciferase constructs
All constructs are based on the commercially available plasmid pFR_HCV_xb (Addgene) and were produced using PCR-based site-directed mutagenesis. Primers were designed using SnapGene 4.1.9 (GSL Biotech LLC). Primer sequences are available upon request. The respective 5′-UTRs were cloned downstream of the HSV-TK promotor directly followed by the firefly luciferase gene, an HCV IRES and the Renilla luciferase gene. The total length of the analyzed 5′-UTRs, including single-stranded and double-stranded regions, ranges from 25 bp to 292 bp ((AG)15/(AC)15: 30 bp; HEVgt3c: 25 bp; HEVgt3c-G4C: 25 bp; HEVgt3c-G4CC6A: 25 bp; HEVgt3c-Purine: 25 bp; HCoV-229E: 292 bp; VP30: 221 bp; VP35: 97 bp; VP35-HP only: 22 bp; VP35-HP + (AG)5: 32 bp).
2.10. Dual luciferase reporter assay
The dual luciferase reporter assay was done as described previously (Müller et al., 2018a). All experiments were performed in at least three independent replicates.
3. Results
3.1. Antiviral activity of CR-31-B (−) against coronaviruses in vitro
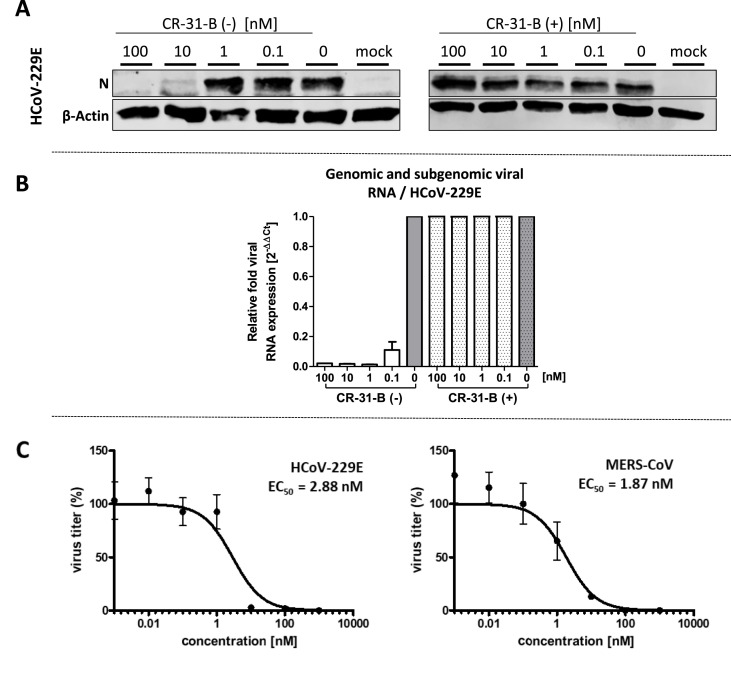
We have recently shown that Silvestrol can efficiently inhibit viral protein synthesis in MERS-CoV- and HCoV-229E-infected MRC-5 cells (Müller et al., 2018a). Here, we investigated the antiviral potential of CR-31-B, a synthetic rocaglate lacking the dioxane moiety of Silvestrol (Fig. 1). MRC-5 cells were infected with HCoV-229E at an MOI of 1 and the effects of the CR-31-B (−)- and (+)-enantiomers on viral protein synthesis were analyzed at 24 hpi. Coronavirus nucleoprotein (N) levels were found to be strongly reduced in the presence of the (−)-enantiomer at concentrations ≥ 10 nM, while CR-31-B (+) had no detectable effect (Fig. 2 A). Similarly, the genomic and subgenomic RNA levels of HCoV-229E were reduced in the presence of subnanomolar concentrations of CR-31-B (−) (Fig. 2B). In line with this, the formation of viral replication/transcription complexes was reduced in the presence of CR-31-B (−) as shown by immunofluorescence analysis using antibodies directed against nonstructural protein 8 (nsp8) and double-stranded RNA (dsRNA; Fig. S1).
Fig. 2.
Antiviral activities of the synthetic rocaglate CR-31-B (−) against HCoV-229E and MERS-CoV. (A) Western blot analysis of HCoV-229E N protein accumulation (top panel) in cells treated with different enantiomers of CR-31-B. β-Actin (lower panel) was used as a loading control. (B) Total (genomic and subgenomic) viral RNA produced in HCoV-229E-infected MRC-5 cells treated with the two enantiomers of CR-31-B. Relative changes in viral RNA levels were determined by RT-qPCR. The data were normalized to infected but untreated cells as well as GAPDH mRNA using the comparative ΔΔCt method. (C) HCoV-229E and MERS-CoV titers in supernatants collected from infected MRC-5 cells (MOI = 0,1) at 24 hpi. Cells were treated with CR-31-B (−/+) as indicated. Data from three independent experiments were used to calculate EC50 values (2.88 nM for HCoV-229E- and 1.87 nM for MERS-CoV).
Next, we analyzed the effects of CR-31-B (−) on the production of coronavirus progeny using HCoV-229E- and MERS-CoV-infected MRC-5 cells. For both viruses, CR-31-B (−) was revealed to reduce viral titers efficiently, with EC50 values of 2.88 nM for HCoV-229E and 1.87 nM for MERS-CoV (Fig. 2C). At a concentration of 100 nM CR-31-B (−), the MERS-CoV titer was reduced by about 5 log levels (Suppl. Fig. S2A). Cytotoxicity tests using MRC-5 cells revealed that CR-31-B (−) and CR-31-B (+) caused a slight reduction of cell viability by 10–25% and 10%, respectively, if the cells were incubated for 24 h with concentrations of up to 5 μM of the respective compound (Suppl. Fig. S2B), indicating low cytotoxicities for both compounds with selectivity indices of >1000 (see Table 1 ). Even if the cells were treated for extended periods of time (48 and 72 h) using concentrations of up to 1 μM, no major cytotoxic effects were observed for CR-31-B (Fig. S2C). We further determined the CC50 values of a racemic mixture of CR-31-B using a range of human liver and skin carcinoma cell lines as well as primary human dermal fibroblasts (HDF). As expected, CC50 values in all analyzed cancer cell lines (7 liver and 3 skin carcinoma cell lines) were found to be in the low nanomolar range whereas the CC50 in primary HDF was calculated at ~ 500 nM (Fig. S3). Taken together, our data demonstrate low cytotoxicity of CR-31-B (−) in primary cells compared to the known cytotoxic effects in fast growing cancer cell lines.
Table 1.
CC50 and EC50 values determined for Silvestrol and CR-31-B (−)-treated cells that were mock infected (CC50) or infected with the indicated viruses (EC50). SI =Selectivity Index. Experiments were done in biological triplicates.
| cell type | virus | compound | CC50 [nM] | EC50 [nM] | SI |
|---|---|---|---|---|---|
| MRC-5 | HCoV-229E | Silvestrol | >10000 | 3 | >3300 |
| CR-31-B (−) | >5000 | 2.88 | >1736 | ||
| MERS-CoV | Silvestrol | >10000 | 1.3 | >7690 | |
| CR-31-B (−) | >5000 | 1.87 | >2674 | ||
| murine hepatocytes | LASV | Silvestrol | >5000 | 50.73 | >99 |
| CR-31-B (−) | >5000 | 36.11 | >139 | ||
| CCHFV | Silvestrol | >5000 | 28.53 | >175 | |
| CR-31-B (−) | >5000 | 20.04 | >250 | ||
| A549 | ZIKV | Silvestrol | 9.42 | 1.08 | 8.8 |
| CR-31-B (−) | 19.34 | 1.13 | 17.1 |
3.2. Antiviral activity of Silvestrol and CR-31-B (−) in a human bronchial epithelial cell system
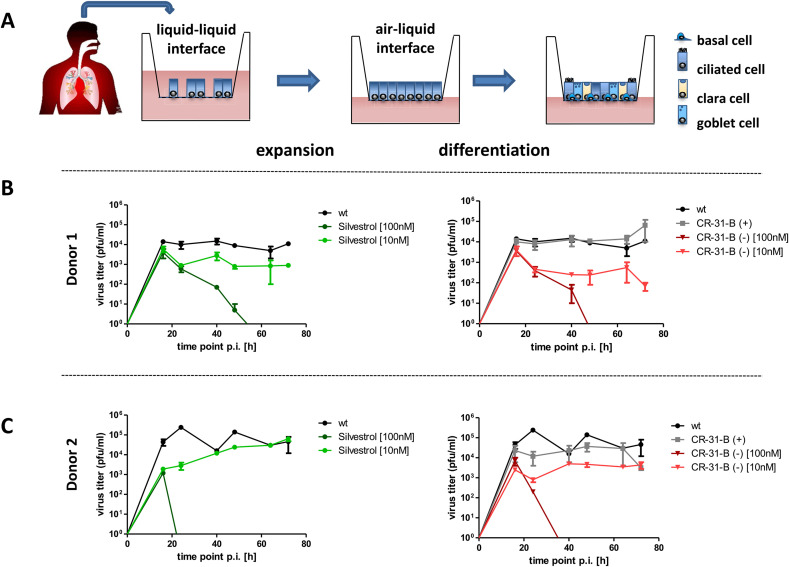
For further evaluation of the antiviral potential of Silvestrol and CR-31-B (−) in a relevant ex vivo system for respiratory viruses, we used 3D primary human airway epithelial cell cultures. Normal human bronchial epithelial (NHBE) cells were differentiated under air/liquid conditions to pseudostratified (columnar) epithelia. The epithelium was then infected with HCoV-229E to mimic viral infection of the different cell types in human airways in the presence of inhibitor or solvent control (Fig. 3 A). At a concentration of 10 nM, CR-31-B (−) reduced the virus titer in the supernatant by about 1.5 orders of magnitude using cells from two different donors, which was similar to the antiviral effect observed for 10 nM Silvestrol in this ex vivo model. At 100 nM, both compounds reduced infectious virus production to undetectable levels (Fig. 3B + C), whereas 100 nM CR-31-B (+) did not significantly reduce viral replication.
Fig. 3.
Comparison of antiviral effects of CR-31-B (−) and Silvestrol using human bronchial epithelial cells infected with HCoV-229E. (A) Human bronchial epithelial cells were cultivated and differentiated at an air-liquid interface into different airway epithelial cell types (basal, ciliated, clara and goblet cells) und used to assess antiviral effects of the respective compounds. (B + C) HCoV-229E titers in cell culture supernatants collected at the indicated time points p.i. Cells obtained from two different donors were infected and treated with CR-31-B (−), Silvestrol (100/10 nM) or Cr-31-B (+) (100 nM), respectively.
3.3. Comparison of broad-spectrum antiviral activities of Silvestrol and CR-31-B (−)
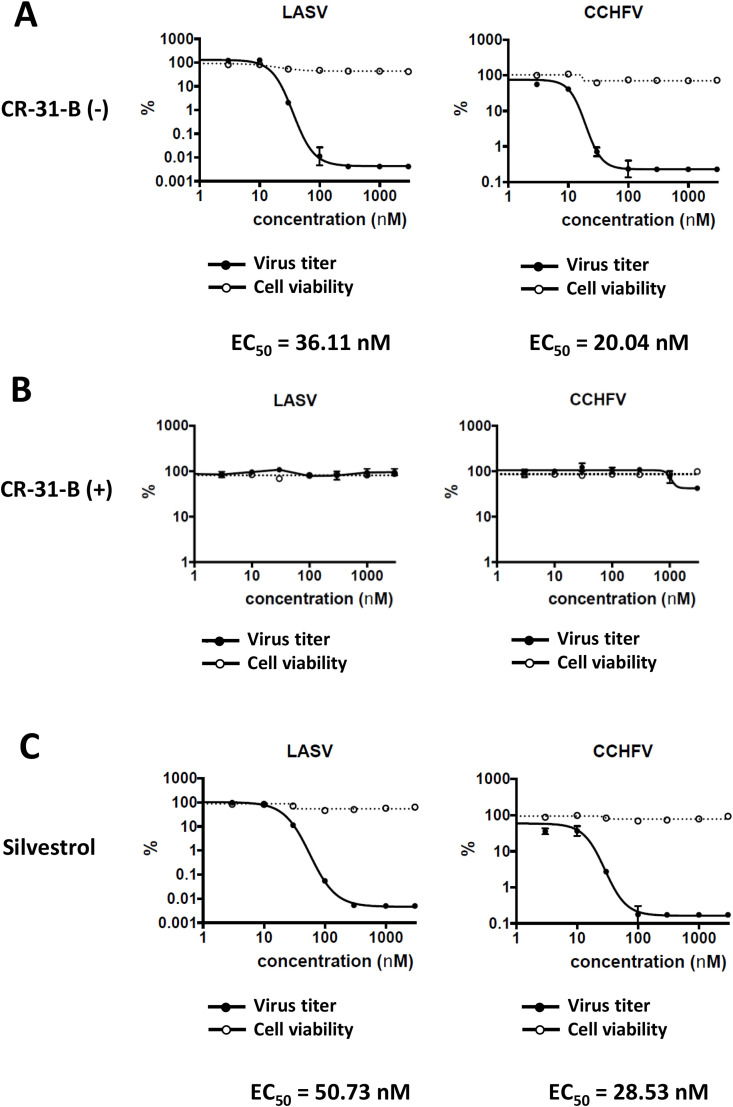
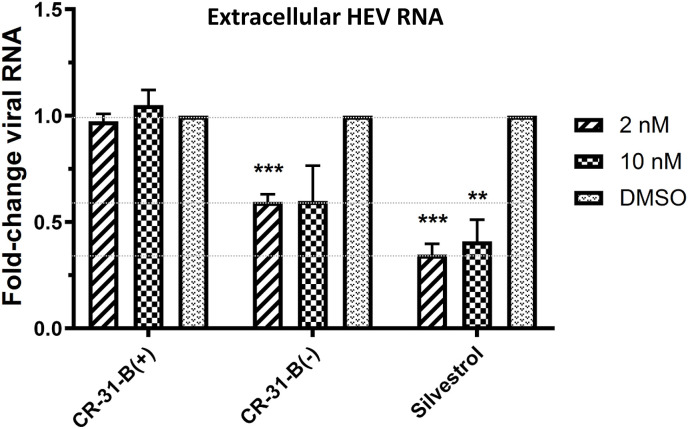
We have recently shown that Silvestrol inhibits ZIKV replication in the human lung epithelial cell line A549 and in primary human hepatocytes (Elgner et al., 2018). In the present study, we found that also CR-31-B (−) causes a strong reduction of ZIKV RNA levels with a calculated EC50 value of 1.13 nM (Table 1; Suppl. Fig. S4). To assess potential broad-spectrum antiviral activities of CR-31-B (−) and Silvestrol against other highly pathogenic emerging viruses, we analyzed their effects in primary murine hepatocytes infected with LASV or CCHFV (Fig. 4 ). The data revealed that CR-31-B (−) and Silvestrol have potent antiviral activities with EC50 values between ~20 and 50 nM with no detectable cytotoxicity at concentrations of up to 10 μM (Fig. 4A–C). Virus titers showed a ~4-log drop for LASV- and a 3-log drop for CCHFV-infected cells (Fig. 4A,C). Furthermore, we analyzed if CR-31-B (−) inhibits HEV replication (Fig. 5 ). Persistently HEV-infected A549 cells were treated with different concentrations of Silvestrol and the two CR-31-B enantiomers. At 72 hpi the extracellular viral RNA levels, which correlate with released viral particles, were analyzed using quantitative RT-PCR. In line with our previous results (Glitscher et al., 2018), we confirmed the antiviral activity of Silvestrol, while the antiviral effect of CR-31-B (−) against HEV was found to be slightly weaker at the low nanomolar concentrations used in this assay (Fig. 5). However, some cytotoxicity of Silvestrol was already observed at a concentration of 2 nM whereas CR-31-B was not cytotoxic at this concentration, yet at 10 nM some cytotoxicity of CR-31-B (−) could be observed (~35% reduced viability) in persistently HEV-infected A549 cells (Fig. S5).
Fig. 4.
CR-31-B (−) and Silvestrol inhibit LASV and CCHFV replication in primary murine hepatocytes with comparable efficiencies in a concentration range between 20 and 50 nM. (A) Potent antiviral activity of CR-31-B (−) against LASV and CCHFV without cytotoxicity in murine hepatocytes. (B) No antiviral effects of CR-31-B (+) up to a concentration of 5 μM. (C) Potent antiviral activity of Silvestrol against LASV and CCHFV without cytotoxicity in murine hepatocytes.
Fig. 5.
CR-31-B (−) and Silvestrol reduce the levels of extracellular HEV RNA at low nanomolar concentrations. qRT-PCR measurement of extracellular HEV RNA of CR-31-B (+), CR-31-B (−) and Silvestrol treated, persistently HEV-infected cells. All data are referred to the DMSO control.
3.4. Analyses of the 5′-UTR-mediated inhibitory activities of Silvestrol and CR-31-B (−)
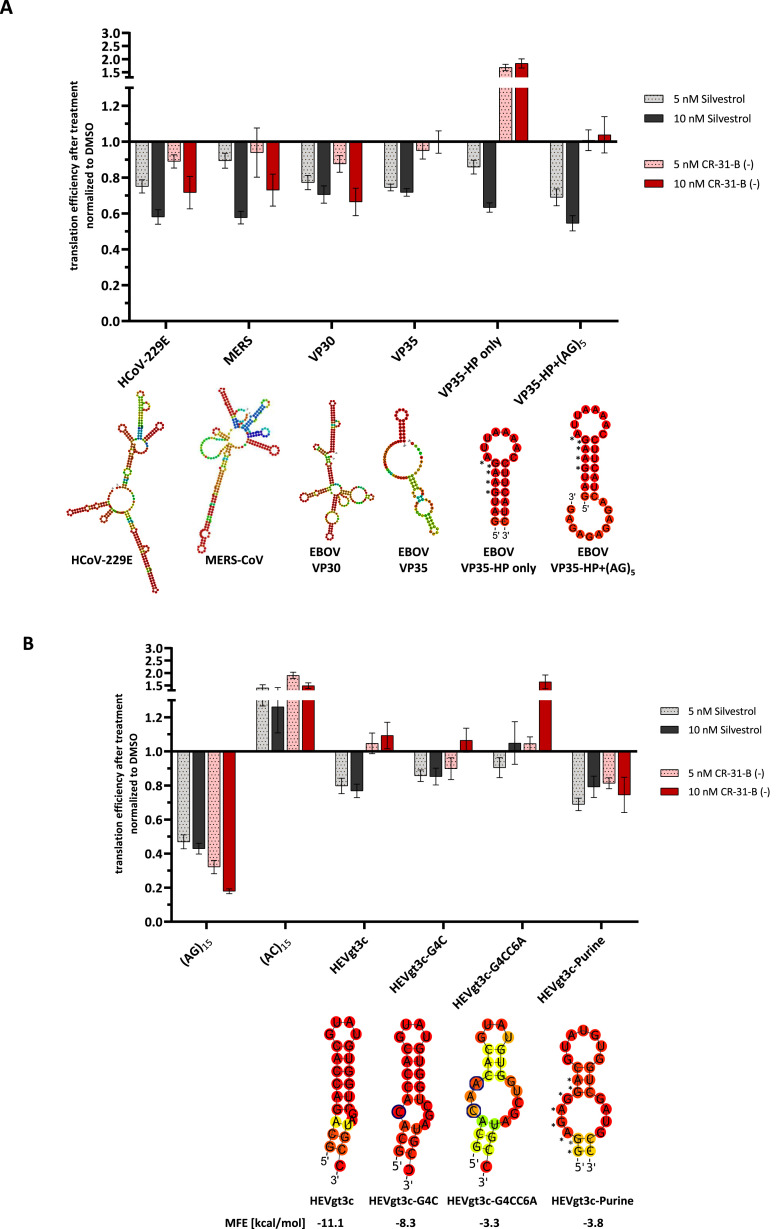
To gain more mechanistic insights into the effects of Silvestrol and CR-31-B (−) on translation initiation and RNA clamping, we compared the inhibitory effects of the two rocaglates on different viral 5′-UTRs in a dual luciferase reporter assay (Fig. S6; Müller et al., 2018a). The 5′-UTRs of HCoV-229E, MERS-CoV and EBOV VP30 mRNA were found to be similarly sensitive to translation inhibition by Silvestrol and CR-31-B (−) (Fig. 6 A). In contrast, the 5′-UTR of EBOV VP35 was insensitive to CR-31-B (−) but sensitive to Silvestrol. Interestingly, the VP30 and VP35 mRNAs both carry a likely inaccessible pentapurine stretch in their 5′-terminal stem structures (Fig. 6A). However, in the VP30 5′-UTR, this pentapurine stretch is followed by a decapurine stretch that seems to be unstructured and thus predicted to enable RNA clamping (Iwasaki et al., 2019). Such a second purine stretch is absent in the VP35 5′-UTR. The 5′-terminal hairpin of VP35 alone was sufficient to mediate translation inhibition by Silvestrol. Surprisingly, this hairpin increased the reporter activity about twofold in the presence of CR-31-B (−) (Fig. 6A, VP35-HP only). This increase was reduced back to baseline levels if an additional (AG)5-polypurine stretch was inserted at the 3′-end of the VP35 hairpin. Also, this insertion resulted in a slightly increased translation inhibition by Silvestrol, indicating that polypurine sequences strengthen the inhibitory effects of rocaglates on eIF4A (Fig. 6A, VP35-HP+(AG)5).
Fig. 6.
Comparison of the inhibitory effects of CR-31-B (−) and Silvestrol on reporter gene expression constructs containing different viral 5′-UTRs. (A) Effects of 5 and 10 nM Silvestrol or CR-31-B (−) on reporter gene expression in the context of 5′-UTRs from coronaviruses HCoV-229E and MERS-CoV as well as EBOV VP30 and VP35. The VP35 5′-terminal hairpin and the VP35 hairpin with (AG)5 extensions were also analyzed. The predicted RNA secondary structures of the indicated 5′-UTRs are shown. Asterisks mark the positions of purines as part of the polypurine stretch in the VP35 hairpin. (B) The 5′-UTR of HEV and derivatives thereof were analyzed towards their sensitivity against 5 and 10 nM Silvestrol and CR-31-B (−) treatment in a dual luciferase assay. (AG)15 and (AC)15 sequences were used as positive and negative controls, respectively. Predicted RNA secondary structures of the HEV 5′-UTRs are shown. The reporter gene expression data were normalized to the transfection efficiencies and the corresponding DMSO controls. Blue circles indicate the mutated nucleotides in HEVgt3c-G4C and HEVgt3c-G4CC6A. Asterisks mark the positions of purines as part of the polypurine stretch in HEVgt3c-Purine. Standard errors of the mean of at least three independent experiments are shown. MFE = minimal free energy (kcal/mol).
To confirm the idea of polypurines being required for stable stacking interactions with rocaglates, we constructed 5′-UTRs consisting of a 30-nt long (AG)15 or an unstructured (AC)15 sequence as negative control. As presumed, Silvestrol inhibited translation of the (AG)15 construct by ~55% and CR-31-B (−) even by ~80% at concentrations of 10 nM (Fig. 6B). Remarkably, the presence of the (AC)15 sequence caused a 1.5-fold induction of luciferase activity with both compounds (Fig. 6B), indicating that the helicase activity of eIF4A was dispensable in this case.
Interestingly, the 5′-UTR of HEV (HEVgt3c) lacks any polypurine sequence element, but it is predicted to form a stable RNA hairpin structure which, most likely, requires unwinding during translation initiation. As shown before, HEV replication can be inhibited by Silvestrol (Glitscher et al., 2018) and, to a slightly lesser extent, by CR-31-B (−) (Fig. 5). Therefore, we asked if the polypurine-free 5′-UTR of HEV is also sensitive to Silvestrol or CR-31-B (−) treatment. At a concentration of 10 nM Silvestrol, the 5′-UTR of HEV mediated reduced luciferase activity, demonstrating that Silvestrol can indeed clamp this viral RNA onto eIF4A. Importantly, this was not the case for CR-31-B (−) treatment (Fig. 6B, HEVgt3c). Moreover, this indicated that the dioxane moiety of Silvestrol may play a critical role in clamping structured RNAs onto eIF4A in a polypurine-independent manner. To test this assumption, the HEV 5′-UTR hairpin structure was thermodynamically destabilized by disrupting one or two base pairs in the stem (Fig. 6B, HEVgt3c-G4C and HEVgt3c-G4CC6A). These changes led to a gradual loss of the inhibitory effect of Silvestrol. To further analyze the relevance of polypurine stretches in viral 5′-UTRs, the sequence in the HEV 5′-UTR was changed at the 5′-end from 5′-GCAGACCA… into 5′-GGAGAGGA... (Fig. 6B, HEVgt3c-Purine), thereby introducing a stretch of 8 consecutive purines. Although the thermodynamic stability of the hairpin structure was reduced by these sequence changes, the HEVgt3c-Purine 5′-UTR became now sensitive to CR-31-B (−) and Silvestrol treatment (Fig. 6B, HEVgt3c-Purine).
3.5. Modelling of Silvestrol onto the surface structure of the human eIF4A-polypurine RNA complex
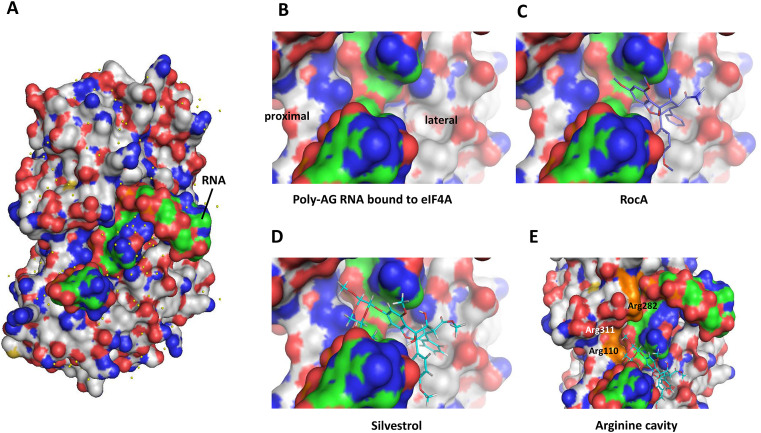
Our reporter assay results indicated differences in the mode of action between Silvestrol and rocaglates lacking the dioxane moiety. Since no structural data from co-crystallization of Silvestrol, RNA and eIF4A are available, we modeled the structure of Silvestrol into the published eIF4A-polyAG structure surface (Iwasaki et al., 2019). By comparing the localization of RocA and Silvestrol, we found that the dioxane moiety of Silvestrol can completely cross the surface of the bound RNA substrate, thus enabling Silvestrol to make additional contacts e.g. via H-bonds to arginines in eIF4A which cannot be formed in the absence of the dioxane moiety (Fig. 7 ). This suggests that Silvestrol is able to clamp RNA without an absolute requirement for stacking interactions with purine bases because stabilizing interactions with eIF4A at the proximal side of the bound RNA may be sufficient for locking the RNA on the helicase.
Fig. 7.
Possible binding mode of Silvestrol and RNA clamping using structure-based comparative modeling of RocA or Silvestrol onto a 10-mer polyAG RNA bound to the surface of human eIF4A. (A) Surface of human eIF4A with a bound polyAG 10mer (adapted from Iwasaki et al., 2019). (B-E) Zoom in to show the binding region of RocA and Silvestrol. The dioxane moiety of Silvestrol is able to cross the RNA stretch to make additional contacts with proximal positioned arginine residues in eIF4A. Pymol was used for graphical illustration. eIF4A: grey; RNA: green; RocA: purple; Silvestrol; cyan.
4. Discussion
We have identified the synthetic rocaglate CR-31-B (−) as a novel potent antiviral compound with broad-spectrum activity against HCoV-229E, MERS-CoV, LASV, CCHFV, HEV and ZIKV. Moreover the antiviral potential of Silvestrol and CR-31-B (−) was compared in an ex vivo human airway epithelial cell system under air/liquid conditions. These differentiated airway epithelial cells were used as a model for the primary airway defense barrier against inhaled pathogens, mimicking the natural situation in the infected host. This system provides a pseudostratified organization of basal, ciliated, goblet and other less common types of cells and plays a crucial role in maintaining airway homeostasis by regulating innate and acquired immunity through the production of a wide range of cytokines as well as chemokines (Davies, 2014). In this relevant ex vivo system, we could confirm the antiviral effect of CR-31-B (−) as well as Silvestrol against HCoV-229E replication.
CR-31-B (−) has a potent antiviral activity similar to that of the more complex-structured Silvestrol. It is active at low nanomolar concentrations with low cytotoxicity in primary human cells while it has a higher cytotoxicity in cancer cell lines (see Fig. S3). Thus, CR-31-B (−) opens a broad therapeutic window for the treatment of viral infections and qualifies as an interesting synthetic rocaglate for further in vivo evaluations. Even though the antiviral potential of CR-31-B (−) is similar to Silvestrol, we identified substantial mechanistic differences between the two compounds as detailed below.
RNAs generally bind to eIF4A in a sequence- and structure-independent manner via their phosphate backbone. Thus, if no RNA clamping by rocaglates occurs, active eIF4A retains its ability to unwind secondary structures in the bound substrate RNA. In our reporter assays, Silvestrol was able to clamp polypurine-free stable hairpin structures onto eIF4A, whereas CR-31-B (−) required an accessible polypurine-sequence in close proximity to the 5′-terminal hairpin. This conclusion is supported by data showing that translation of the reporter construct containing the EBOV VP35 5′-UTR is insensitive to CR-31-B (−) while it is sensitive to Silvestrol. Reduced hairpin stability correlated with a loss in Silvestrol sensitivity, suggesting that thermodynamic stability of hairpin structures is of critical importance. This is also supported by our observation that introduction of an unstructured (AC)15 sequence rendered the reporter construct insensitive to both Silvestrol and CR-31-B (−), whereas an (AG)15 polypurine sequence, which might be able to form a stable hairpin structure by making non-Watson-Crick G-A basepair contacts, strongly require the helicase activity of eIF4A. Remodeling the structure of Silvestrol onto the surface of the published eIF4A-RNA structure revealed that Silvestrol is in principle capable to contact eIF4A via phenylalanine (Phe163) at the lateral (Iwasaki et al., 2019) and via arginines at the proximal site of the bound substrate RNA (Fig. 7). This leads us to suggest that Silvestrol may be able to clamp any RNA sequence onto eIF4A. Our results (Fig. 6B) indicate that an unstructured sequence of sufficient length, e.g. the (AC)15 sequence does not require the helicase activity of eIF4A to allow 43S PIC binding as a prerequisite for translation initiation, whereas a stable hairpin structure likely prevents 43S PIC binding and thus needs to be unwound.
The published crystal structure of human eIF4A in complex with a polypurine RNA substrate and RocA (Iwasaki et al., 2019) helps to explain why polypurine sequences are required to clamp RNA onto eIF4A by RocA. Stacking interactions between two phenyl rings of RocA and two consecutive purines in the substrate RNA are stable enough for RNA clamping onto eIF4A, whereas less stable interactions with pyrimidines are insufficient (Iwasaki et al., 2019). As a consequence, a stable hairpin structure without a polypurine stretch may not be bound efficiently by RocA or CR-31-B (−) due to the lack of the dioxane moiety. To date, there is no structural information on the exact binding mode of Silvestrol onto eIF4A. We therefore initiated co-crystallization studies with human eIF4A, poly-AG RNA and Silvestrol to identify the exact position of the dioxane moiety.
Surprisingly, we observed some antiviral activity of CR-31-B (−) in HEV-producing A549 cells at a non-cytotoxic concentration of 2 nM even though the compound did not inhibit reporter translation in the presence of a polypurine-free HEVgt3c 5′-UTR. We propose an indirect antiviral effect of CR-31-B (−) that could be caused by inhibition of eIF4A-dependent cellular mRNAs of unknown identity.
Importantly, the effects of CR-31-B in vivo are comparable to Silvestrol. In a xenograft mouse model, both compounds reduced tumor growth efficiently (Wolfe et al., 2014). CR-31-B treatment did not change body weight or the number of blood cells and no toxicity could be observed in the gastrointestinal tract of mice. Moreover, serum levels of aminotransferases (ALT and AST), albumin, total bilirubin as well as creatinine were not significantly changed two weeks after cessation of treatment, indicating that CR-31-B has a favorable toxicity profile in vivo (Wolfe et al., 2014). So far, no published data on the pharmacokinetic properties of CR-31-B (−) are available. It was already shown that daily intraperitoneal injection of CR-31-B (−) works well in mice (Wolfe et al., 2014; Chan et al., 2019) while the systemic availability of rocaglates following oral application seems to be limited (Saradhi et al., 2011). Based on this and our own data, we conclude that CR-31-B (−) may represent an interesting alternative to the broad-spectrum antiviral Silvestrol, which remains to be confirmed in appropriate in vivo studies to evaluate and compare the antiviral potential, toxicity and pharmacokinetics profiles of the two compounds.
In conclusion, our results confirm that CR-31-B (−) has potent broad-spectrum antiviral activity similar to that of Silvestrol (Table 1). Moreover, polypurine sequences in the 5′-UTR were found to be required for rocaglate-dependent clamping onto eIF4A if the dioxane moiety of Silvestrol is missing as shown for the synthetic CR-31-B (−) compound. Our data also suggest that Silvestrol retains its ability to clamp RNA substrates containing a stable hairpin structure in cases where polypurine sequences are not accessible (see the VP35 5′-UTR) or absent (see the HEV 5′-UTR).
Acknowledgements
We like to thank Nadja Karl for technical assistance and Dr. Janis Müller and Dr. Serghei Glinca for their assistance in structural modeling of the human eIF4A-RNA-Silvestrol complex.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.antiviral.2020.104706.
Disclosure statement
The authors declare no conflict of interest.
Funding
The work was supported by the LOEWE Center DRUID (projects A2, B2 and D3 to A.G., J.Z. and E.H.), the German Center for Infection Research (DZIF), partner sites Giessen and Hamburg, Germany (TTU Emerging Infections, to J.Z. and L.O.), Deutsche Forschungsgemeinschaft (SFB 1021 ‘RNA viruses: RNA metabolism, pathogenesis and host response’; projects A01 and A02, to J.Z. and R.K.H., respectively; CRU KFO309, project P3 to J.Z.), and the Leibniz Gesellschaft (Leibniz Best Minds, to L.O.)
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Bhat M., Robichaud N., Hulea L., et al. Targeting the translation machinery in cancer. Nat. Rev. Drug Discov. 2015;14:261–278. doi: 10.1038/nrd4505. (submitted for publication) [DOI] [PubMed] [Google Scholar]
- Biedenkopf N., Lange-Grünweller K., Schulte F.W., et al. The natural compound silvestrol is a potent inhibitor of Ebola virus replication. Antivir. Res. 2017;137:76–81. doi: 10.1016/j.antiviral.2016.11.011. [DOI] [PubMed] [Google Scholar]
- Chan K., Robert F., Oertlin C., et al. eIF4A supports an oncogenic translation program in pancreatic ductal adenocarcinoma. Nat. Commun. 2019;10:5151. doi: 10.1038/s41467-019-13086-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies D.E. Epithelial barrier function and immunity in asthma. Ann. Am. Thorac. Soc. 2014;11(Suppl. 5):S244–S251. doi: 10.1513/AnnalsATS.201407-304AW. (submitted for publication) [DOI] [PubMed] [Google Scholar]
- Elgner F., Sabino C., Basic M., et al. Inhibition of Zika virus replication by silvestrol. Viruses. 2018;10:E149. doi: 10.3390/v10040149. pii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glitscher M., Himmelsbach K., Woytinek K., et al. Inhibition of hepatitis E virus spread by the natural compound silvestrol. Viruses. 2018;10:E301. doi: 10.3390/v10060301. pii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günther S., Asper M., Röser C., et al. Application of real-time PCR for testing antiviral compounds against Lassa virus, SARS coronavirus and Ebola virus in vitro. Antivir. Res. 2004;63:209–215. doi: 10.1016/j.antiviral.2004.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henss L., Scholz T., Grünweller A., Schnierle B.S. Silvestrol inhibits Chikungunya virus replication. Viruses. 2018;10:E592. doi: 10.3390/v10110592. pii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki S., Iwasaki W., Takahashi M., et al. The translation inhibitor rocaglamide targets a bimolecular cavity between eIF4A and polypurine RNA. Mol. Cell. 2019;73:738–748. doi: 10.1016/j.molcel.2018.11.026. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johne R., Reetz J., Ulrich R.G., et al. An ORF1-rearranged hepatitis E virus derived from a chronically infected patient efficiently replicates in cell culture. J. Viral Hepat. 2014;21:447–456. doi: 10.1111/jvh.12157. [DOI] [PubMed] [Google Scholar]
- Lecompte E., Fichet-Calvet E., Daffis S., et al. Mastomys natalensis and Lassa fever, west Africa. Emerg. Infect. Dis. 2006;12:1971–1974. doi: 10.3201/eid1212.060812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Madhugiri R., Fricke M., Marz M., Ziebuhr J. Coronavirus cis-acting RNA elements. Adv. Virus Res. 2016;96:127–163. doi: 10.1016/bs.aivir.2016.08.007. (submitted for publication) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller C., Schulte F.W., Lange-Grünweller K., et al. Broad-spectrum antiviral activity of the eIF4A inhibitor silvestrol against corona- and picornaviruses. Antivir. Res. 2018;150:123–129. doi: 10.1016/j.antiviral.2017.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller C., Hardt M., Schwudke D., et al. Inhibition of cytosolic phospholipase A2α impairs an early step of coronavirus replication in cell culture. J. Virol. 2018;92 doi: 10.1128/JVI.01463-17. pii: e01463-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ölschläger S., Gabriel M., Schmidt-Chanasit J., et al. Complete sequence and phylogenetic characterisation of Crimean-Congo hemorrhagic fever virus from Afghanistan. J. Clin. Virol. 2011;50:90–92. doi: 10.1016/j.jcv.2010.09.018. [DOI] [PubMed] [Google Scholar]
- Pan L., Woodard J.L., Lucas D.M., et al. Rocaglamide, silvestrol and structurally related bioactive compounds from Aglaia species. Nat. Prod. Rep. 2014;31:924–939. doi: 10.1039/c4np00006d. (submitted for publication) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio C.A., Weisburd B., Holderfield M., et al. Transcriptome-wide characterization of the eIF4A signature highlights plasticity in translation regulation. Genome Biol. 2014;15(10):476. doi: 10.1186/s13059-014-0476-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saradhi U.V., Gupta S.V., Chiu M., et al. Characterization of silvestrol pharmacokinetics in mice using liquid chromatography-tandem mass spectrometry. AAPS J. 2011;13:347–356. doi: 10.1208/s12248-011-9273-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlereth J., Grünweller A., Biedenkopf N., et al. RNA binding specificity of Ebola virus transcription factor VP30. RNA Biol. 2016;13:783–798. doi: 10.1080/15476286.2016.1194160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todt D., Moeller N., Praditya D., et al. The natural compound silvestrol inhibits hepatitis E virus (HEV) replication in vitro and in vivo. Antivir. Res. 2018;157:151–158. doi: 10.1016/j.antiviral.2018.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Too I.H.K., Bonne I., Tan E.L., et al. Prohibitin plays a critical role in Enterovirus 71 neuropathogenesis. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1006778. e1006778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe A.L., Singh K., Zhong Y., et al. RNA G-quadruplexes cause eIF4A-dependent oncogene translation in cancer. Nature. 2014;513:65–70. doi: 10.1038/nature13485. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.