Abstract
A national control program against bovine respiratory syncytial virus (BRSV) and bovine coronavirus (BCV) was launched in Norway in 2016. A key strategy in the program is to test for presence of antibodies and protect test-negative herds from infection. Because these viruses are endemic, the rate of re-introduction can be high, and a disease-free status will become more uncertain as time from testing elapses. The aim of this study was to estimate the probability of freedom (PostPFree) from BRSV and BCV antibodies over time by use of bulk tank milk (BTM) antibody-testing, geographic information and animal movement data, and to validate the herd-level estimates against subsequent BTM testing.
BTM samples were collected from 1148 study herds in West Norway in 2013 and 2016, and these were analyzed for BRSV and BCV antibodies. PostPFree was calculated for herds that were negative in 2013/2014, and updated periodically with new probabilities every three months. Input variables were test sensitivity, the probability of introduction through animal purchase and local transmission. Probability of introduction through animal purchase was calculated by using real animal movement data and herd prevalence in the region of the source herd. The PostPFree from the final three months in 2015 was compared to BTM test results from March 2016 using a Wilcoxon Rank Sum Test.
The probability of freedom was generally high for test-negative herds immediately after testing, reflecting the high sensitivity of the tests. It did however, decrease with time since testing, and was greatly affected by purchase of livestock. When comparing the median PostPFree for the final three months to the test results in 2016, it was significantly lower (p < 0.01) for test positive herds. Furthermore, there was a large difference in the proportion of test positive herds between the first and fourth quartile of PostPFree. The results show that PostPFree provides a better estimate of herd-level BTM status for both BRSV and BCV than what can be achieved by relying solely on the previous test-result.
Keywords: BRSV, BCV, BCoV, Disease control, BRD, Cattle, Bovine respiratory disease, Bulk tank milk, Animal movements
1. Introduction
Bovine respiratory syncytial virus (BRSV) and bovine coronavirus (BCV) are widespread infectious agents, present in cattle populations around the world, including the Norwegian dairy population (Valarcher and Taylor, 2007; Gulliksen et al., 2009; Boileau and Kapil, 2010). BRSV causes respiratory disease, mostly in young animals, but can affect cattle of all ages (Valarcher and Taylor, 2007). Clinical signs vary from none to severe (Valarcher and Taylor, 2007). BCV is responsible for diarrhea in calves, and for respiratory disease and contagious diarrhea in adult cattle (winter dysentery) (Boileau and Kapil, 2010). These infections lead to increased use of antibiotics due to common secondary bacterial infections, they reduce animal welfare and the associated economic losses can be considerable (Larsen, 2000; Boileau and Kapil, 2010). In 2016, a national control program against BRSV and BCV was launched in Norway, as the first country in the world. The program is conducted as a joint initiative amongst producer organizations, and participation is voluntary. In early 2016, bulk tank milk (BTM) was collected from the majority of Norwegian dairy herds and analyzed for BRSV and BCV antibodies. In a previous study, dairy herds in two counties on the west coast of Norway had also been sampled and tested three years earlier (Toftaker et al., 2016).
A key strategy of the control program is to protect uninfected herds by imposing restrictions on livestock trade. A negative herd status based on BTM lasts for one year after testing, regardless of the degree of contact with other herds. In a previous Norwegian study, it was shown that spread of BRSV between herds was rapid i.e. the elimination rates and introduction rates were high (Klem et al., 2013). Transmission dynamics for BCV has not yet been investigated in Norway, although one study describes a regional outbreak of winter dysentery (Toftaker et al., 2017). Studies from Sweden have shown that recent BCV infection is common, indicating that the infection is easily transmitted (Beaudeau et al., 2010; Ohlson et al., 2013). Due to the constant risk of virus introduction, the assumption that a negative status is valid for a long time is questionable. Several factors can affect the risk of change in status. Purchase of livestock is a well-known route of introduction of infectious agents, and herds that frequently purchase animals are likely at a higher risk of seroconversion (Elvander, 1996; Frössling et al., 2012; Toftaker et al., 2016). In addition to purchase of animals, previous studies have shown that location and herd size are important risk factors for BRSV- and/or BCV antibody positivity (Ohlson et al., 2010b; Toftaker et al., 2016)
Demonstration of freedom from different diseases at the national level is important for international trade purposes, and the use of scenario-tree models has recently provided a more advanced and flexible approach to these calculations (Martin et al., 2007a). More et al. (2013) applied this methodology at herd level within the Irish control program for Johne’s disease. They included information on livestock trade along with test results to calculate probability of freedom from Johne’s disease in test-negative herds. In Norway, information on location of herds, herd size and livestock trade are available from central directories. It was hypothesized that this information could be used along with test results to provide updated estimates of herd probability of freedom from antibodies reflecting the status more accurately than previous BTM test results alone. Estimating a time-varying probability of freedom could potentially form a tool for risk assessment in livestock trade or provide the basis for a risk-based approach to sampling.
The aim of this study was to develop a method for a frequently updated estimate of probability of freedom (PostPFree) from BRSV- and BCV antibodies at the herd level, based on information from BTM testing, geographic location and animal movement data.
2. Materials and methods
2.1. Study area and study population
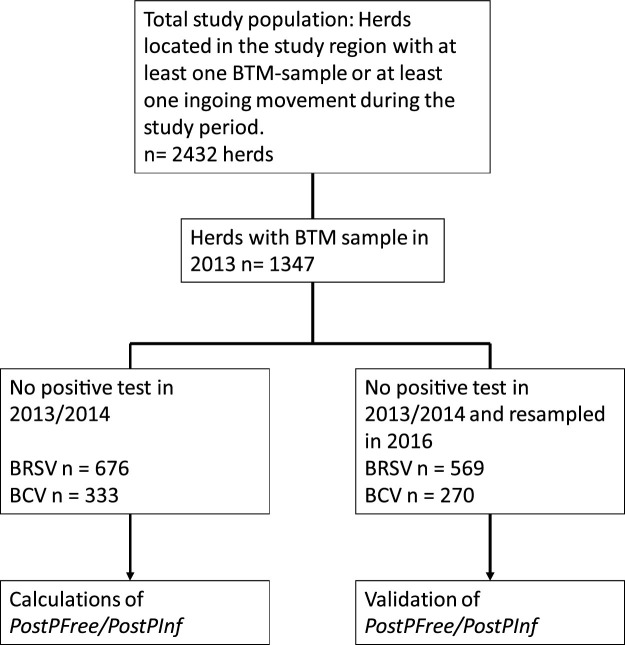
The study area was two neighboring counties on the west coast of Norway. The southern county; “Sogn og Fjordane” and the northern county; “Møre og Romsdal”. Herds located in the study region were included if they had either at least one ingoing animal movement or contributed with at least one BTM sample during January 2013 to March 2016. We had no information on herds without movements or BTM samples; hence, the total cattle population in the study region was not known. A flowchart was made to describe the different subsets of herds used for the different analyses (Fig. 1 ).
Fig. 1.
Flow-chart outlining the study sample and subsets of herds included in different calculations in a study estimating the probability of freedom from BRSV- and BCV antibodies in dairy herds located in two counties in western Norway during the period January 2013–March 2016.
2.2. Sampling and analysis of BTM
During December 2012 to June 2013, BTM samples were collected from 1347 herds (out of 1854 herds delivering milk in 2013) in the study area as part of a cross-sectional risk factor study (Toftaker et al., 2016). For the PostPFree calculations, BTM samples collected in December 2012 were assigned to the first time period i.e. the first three months of 2013. Some of the test-negative herds were resampled the following year (n = 275, February 2014–August 2014). Finally, 1148 herds also had a BTM sample collected in March 2016 as part of the national BRSV/BCV control program. All BTM samples were collected by the milk truck driver in conjunction with milk collection and cooled at a temperature of 2–4 °C until received at the laboratory (TINE Mastitis Laboratory, Molde, Norway) where samples were frozen between −18 and −20 °C until the time of analysis. The 2013 and 2014 samples were analyzed in the Norwegian laboratory, whereas the 2016 samples were shipped over-night to a laboratory in Ireland (Enfer Scientific, Naas, Ireland).
BTM samples collected in 2012–2014 were tested for antibodies against BRSV and BCV using the SVANOVIR® BRSV-Ab and SVANOVIR® BCV-Ab, respectively. Samples were analyzed following the manufacturer’s instructions as described by Toftaker et al. (2016). A cut-off value of 10 percent positivity (PP) was used for both tests, according to the test manual (Svanova, 2018a,b). From 2016, all samples were analyzed with the new MVD-Enferplex BCV/BRSV multiplex, hereafter referred to as the multiplex. This test detects BRSV and BCV antibodies simultaneously using a panel of two recombinant proteins and two synthetic peptides for BRSV (BRSV-A -D) along with one recombinant protein (BCV-A) for BCV, as antigens. A positive test response results in chemiluminescence, captured by an imaging system, and measured in relative light units (RLU) by the Quansys Q view software (v 1.5.4.7). Antigens were combined in a parallel reading, i.e. the test was considered positive when the RLU-value of at least one antigen was above the cut-off. The applied cut-off values for the four different BRSV-antigens were: 2000 for BRSV-A, 4000 for BRSV-B, 7000 for BRSV-C and 1700 for BRSV-D. For BCV-A a cut-off value of 10,000 was used. The sensitivity (Se) of the multiplex was set to 0.94 for BRSV and 0.995 BCV. The Se was set to 0.998 for the SVANOVIR® BRSV-Ab and 0.999 for SVANOVIR® BCV-Ab. Test parameters at the applied cut-off values were based on a diagnostic test evaluation study, evaluating the multiplex along with the SVANOVIR® BRSV-Ab and SVANOVIR® BCV for BTM (Toftaker et al., 2018).
All the tests detect antibodies, not the antigen itself, consequently we will in the present study use “positive” when referring to animals, herds or regions as having BRSV and/or BCV antibodies. Furthermore, all input variables in the probability model relates to antibodies, hence, the calculated probabilities relate to presences of antibodies, and not necessarily infection or presence of virus.
2.3. Data sources and software
The Norwegian food safety authority provided data on cattle movements (The Norwegian Livestock registry). In the current study, animal movements refer to movements where there is a change of owner, for which reporting is mandatory. Information about herd size was retrieved from the Norwegian dairy herd recording system (NDHRS) which in 2011 included 98% of Norwegian dairy herds (Espetvedt et al., 2013). BTM test results were provided by the largest producer organisation, TINE SA, and information on location of herds (coordinates, EUREF89/WGS 1984 UTM-32) was provided by the Norwegian Agriculture Agency. All data management, calculations and analyses were performed using Stata (Stata SE/14; Stata Corp., College Station, TX).
2.4. Animal movements
All recorded animal movements where the destination herd was located in the study area were included. Duplicate records, i.e. movements where animal ID, source county, destination herd and movement date where identical, were reduced to single records (n = 8237). Records of movements where the same animal was moved back and forth between the same two herds, or to two different recipient herds, on the same day, were omitted (n = 179). Records where the source county or the source herd was missing, and could not be retrieved from other variables, were also omitted (n = 56). After editing, the dataset included records of 45,208 movements to 1802 destination herds located in the study region.
2.5. Probability of freedom
PFree was calculated for all herds starting the study period with negative BTM test results in 2013 (and, if tested, in 2014). This was done separately for each virus. The probability of freedom was updated periodically according to the chosen time period; every three months.
The framework presented here is based on a combination of concepts from the following studies; a) scenario-tree modelling of freedom from disease using multiple sources of data presented by Martin et al. (2007a, 2007b), b) calculations of probability of disease freedom on herd level in the Irish control program for Johne´s disease by More et al. (2013) and c) a novel method to identify herds with an increased probability of disease due to animal trade developed by Frössling et al. (2014). The probability of freedom was calculated for each herd using the following Eqs. (1), (2), (3), (4), (5):
First, the probability of introducing at least one positive animal, PIntroTrade, to the destination herd was calculated for each unique combination sd of source herd s and destination herd d for each time period:
| (1) |
where P(D+)a was the within-herd prevalence in the source herd, set to 0.5 (i.e. a 50–50 probability of infection/freedom) for all herds, and n was the number of animals purchased from the source herd.
The total probability of introduction from all animal purchases within each time period t was calculated for each destination herd:
| (2) |
where P(D+)h is the probability that the source herd is antibody positive at the herd level. As an estimate of P(D+)h the herd prevalence in the county of the source herd, based on the national BTM screening was used.
As virus can be introduced, not only through purchase of livestock, but also by indirect transfer, we included a factor for probability of indirect transmission; PIntroLocal. This factor was estimated using the proportion of herds that were negative at the first sampling (2013) and positive at the last sampling (2016), in the group that did not purchase animals, hereafter designated closed herds. This was done separately for the two viruses and for the two counties as we knew that the prevalence, and likely the infectious pressure, was higher in the northern county (Toftaker et al., 2016). In addition, herd size was taken into account as several studies have found an association between herd size and seropositivity (Norström et al., 2000; Solís-Calderón et al., 2007; Ohlson et al., 2010b; Toftaker et al., 2016). In the study by Toftaker et al. (2016) conducted in the same region, the odds of testing positive increased with 12% across the inter quartile range of herd size. The effect of herd size was the same for both viruses. Based on this, we divided the study herds into two groups with median herd size as cut-point and assigned a value of PIntroLocal 12% higher in the “large” compared to the small herds. In summary, this resulted in four categories of PIntroLocal for each virus based on herd size below or above median, and which county the herd was located in (north/south). The total probability of introduction through animal purchase and by indirect transmission for each time period t was then calculated:
| (3) |
The prior probability of infection at time t, PriorPInft, was estimated as follows:
| (4) |
For the first time period, the prior probability of infection (PriorPInf) was set to 0.5, resembling testing a herd with unknown status, i.e. no prior information on herd prevalence in the region available and an equal probability of being positive and negative. PriorPInf was then calculated for each time period by taking the posterior probability of infection from the previous time period (PostPInft-1) and adding the probability of introduction during time period t calculated from Eq. (3), and adjusting for the possibility that the herd might already have been antibody positive but undetected, at the end of the previous time period (t − 1).
After each three month period, an updated probability of freedom (PostPFree) was calculated using Bayes theorem as described by Martin et al. (2007b):
| (5) |
The probability of infection (PostPInf) was the complement to PostPFree. The change in PostPFree over time was visualized for two example herds in a line plot.
2.6. Sensitivity analysis
Due to the uncertainty of the local factor, a sensitivity analysis was performed, using 50% lower and 50% higher values of PIntroLocal, and assessing the effect on the outcome; PostPFree.
2.7. Model evaluation
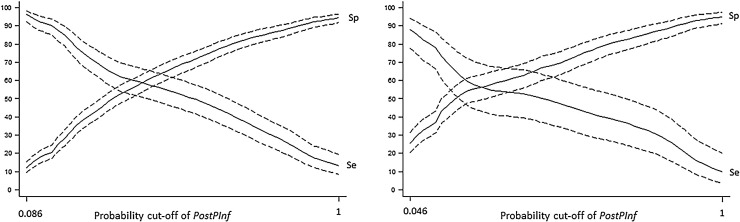
To assess the usefulness of the developed method, the PostPFree calculations for the final three month period was compared to the results from BTM testing in 2016, using a Wilcoxon Rank Sum Test. Bar charts were made showing the proportion of test positive herds in each quartile of PostPFree. The accuracy of the PostPInf was explored by treating it as a diagnostic test, comparing the PostPInf results to the 2016 BTM test-results (used as gold standard). A smoothed line plot of Se and Sp versus probability cut-off of PostPInf was made, and the Se and Sp at different cut-offs of PostPInf were tabulated (results not shown).
3. Results
3.1. Study population
The dataset consisted of 2432 beef and dairy herds located in “Sogn og Fjordane” and “Møre og Romsdal” counties. A BTM result from 2013 was available for 1347 herds, of which 275 had a follow up sample in 2014. Of the 1347 herds, 676 and 333 did not have antibodies against BRSV and BCV (in 2013 or 2014), respectively, and were used for probability of freedom calculations. Of the 1347 herds sampled in 2013, 1148 also had a BTM sample in 2016 of which 569 and 270 were initially negative for BRSV and BCV, respectively, and were used for validation of PostPFree/PostPInf. For an overview of study sample and subset of herds used in different calculations, see Fig. 1.
3.2. BTM results
At the first sampling in 2013, 622 out of 1347 sampled herds were BRSV-antibody positive and 973 were BCV-antibody positive, i.e. a proportion of test positive of 46.2% for BRSV and 72.2% for BCV as previously reported (Toftaker et al., 2016). The national control program started in March 2016, resulting in BTM samples from 1565 herds in the study area. On this final screening, 688 herds (44.0%) were antibody positive for BRSV and 1210 herds (77.3%) were antibody positive for BCV. Of the initially negative herds that were also sampled in 2016, 178 (29%) had changed status for BRSV and 89 (29%) for BCV. An overview of counts and proportions of test outcomes are presented in Table 1 .
Table 1.
Overview of BRSV- and BCV antibody test result for bulk tank milk samples in 2013 and 2016, in a study estimating the probability of freedom from BRSV- and BCV antibodies in dairy herds located in two counties in western Norway.
| Year | BRSV+ | BRSV− | BCV+ | BCV− | ||||
|---|---|---|---|---|---|---|---|---|
| n (%) | n (%) | n (%) | n (%) | |||||
| 2013 n = 1347 |
622 (46.2) | 725 (53.8) | 973 (72.2) | 374 (27.8) | ||||
| 2016 n = 1565 |
688 (44.0) | 877 (56.0) | 1210 (77.3) | 355 (22.7) | ||||
| 2013/2016 | +/+ | +/− | −/+ | −/− | +/+ | +/− | −/+ | −/− |
| n = 1148 | 334 (29.1) | 200 (17.4) | 178 (15.5) | 436 (38.0) | 724 (63.0) | 120 (10.5) | 89 (7.8) | 215 (18.7) |
3.3. Local transmission factor
3.3.1. BRSV
Of the closed herds (n = 384), 104 herds were initially test-negative for BRSV in each county. When retested in 2016, 21 (20%) of the initially negative herds had changed status in the southern county, and 36 (35%) in the northern county.
3.3.2. BCV
For BCV, 60 herds were initially test-negative in the northern county, and 66 in the southern county in the group that did not purchase animals. When retested in 2016, 16 (27%) and seven (11%) herds went from negative to positive in the northern- and southern county, respectively. The resulting local transmission rate, PIntroLocal, per three month time period for each virus is presented in Table 2 .
Table 2.
Local transmission rate, PIntroLocal, per three month time period in the four different categories of herds, in a study estimating the probability of freedom from BRSV- and BCV antibodies in dairy herds located in two counties in western Norway. PIntroLocal was estimated from the proportion of herds that went from antibody- negative to positive during the study period (2013–2016) and did not purchase livestock.
| Herd size |
PIntroLocal |
|||
|---|---|---|---|---|
| BRSV |
BCV |
|||
| Northern county | Southern county | Northern county | Southern county | |
| Small herds | 0.025 | 0.015 | 0.019 | 0.0078 |
| Large herds | 0.028 | 0.016 | 0.022 | 0.0087 |
3.4. Probability of freedom
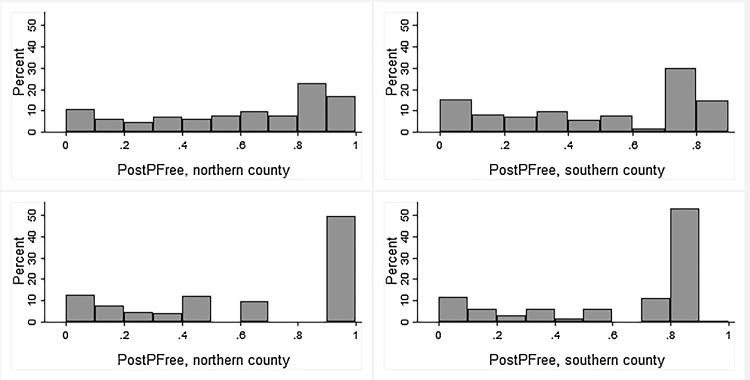
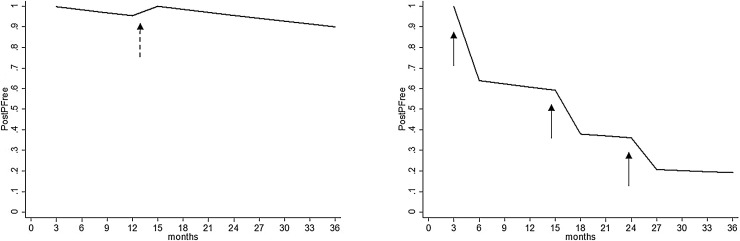
PostPFree was high after the initial negative tests for both viruses. The median PostPFree in the 12th, i.e. the last, time period was 0.62 (range 0–0.91) for BRSV and 0.80 (range 0–0.95) for BCV. The distribution of PostPFree in time period twelve is shown by county in Fig. 2 . Purchase of animals greatly affected the PostPFree for both agents, resulting in different slopes for herds that purchased animals compared to closed herds, as illustrated by two example herds, in Fig. 3 .
Fig. 2.
The distribution of the estimated herd level probability of freedom (PostPFree) from BRSV antibodies (top panel) and BCV antibodies (bottom panel), by county, in the final three-month time period (time period 12) before subsequent testing. Calculations were based on BTM antibody testing, herd location and animal movement data during the period January 2013–December 2016, and were performed for n = 676 (BRSV) and n = 333 (BCV) dairy herds in two counties in western Norway during the period January 2013–December 2016. All herds had a negative test result at inclusion.
Fig. 3.
Herd level probability of freedom (PostPFree) from BRSV antibodies over 36 months for two example herds both starting with a negative test. The herd to the left has no purchases, but a second bulk tank milk test indicated by a dashed arrow, whereas the herd to the right has purchased livestock on several occasions indicated by solid arrows. Calculations were based on BTM antibody testing, herd location and animal movement data during the period January 2013–December 2016. PostPFree was updated every three months.
3.5. Sensitivity analysis
For BRSV, reducing the value of PIntroLocal by 50% gave a mean increase in PostPFree of 10.6% (SD 4.6%), and increasing the value of PIntroLocal gave a mean decrease in PostPFree of 9.6% (SD 3.8%). For BCV, reducing the value of PIntroLocal by 50% gave a mean increase in PostPFree of 5.4% (SD 3.3%), and increasing the value of PIntroLocal by 50% gave a mean decrease in PostPFree of 5.0% (SD 2.9%).
3.6. Model evaluation
The Wilcoxon Rank Sum test showed a significant (p < 0.01) difference in PostPFree between BTM positive and BTM negative herds in 2016. This was true for both BRSV and BCV.
3.6.1. BRSV
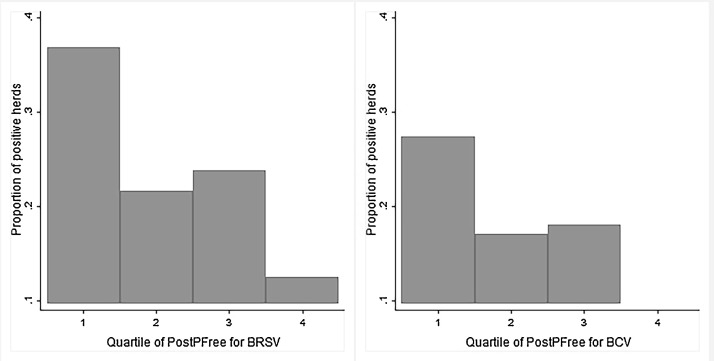
When assessing PostPInf as a diagnostic test, the Se decreased with increasing cut-off, and when 0.25 was used as cut-off, the Se for detecting herds that were BTM positive in 2016 was 0.76 (95% CI: 0.68–0.82). In a practical sense this means that a recommended retesting at this value would capture an estimated 76% of the positive herds i.e. herds that are misclassified as negative based solely on the previous BTM test. No herds had PostPInf < 0.05 (PostPFree > 0.95) at the end of the study period, but at the lowest estimated value, PostPInf < 0.086, two out of 15 herds (13%) were test positive. The proportion of test positive herds in each quartile of PostPFree is illustrated in Fig. 4 , and the Se and Sp of PostPInf is illustrated in Fig. 5 .
Fig. 4.
Proportion of test positive herds in each quartile of the herd level probability of freedom (PostPFree) from BRSV- (left panel, n = 676) and BCV (right panel, n = 333) antibodies in the last of 12 (three month) time periods in dairy herds located in two counties in western Norway.
Fig. 5.
Relative sensitivity and specificity of PostPInf (the probability of antibody positivity) for BRSV (left panel, n = 569) and BCV (right panel, n = 270)) in the last three month time period before subsequent bulk tank milk (BTM) testing versus cut-off value, when the subsequent BTM antibody-test was used as gold standard. Estimation of PostPInf was based on BTM antibody testing, herd location and animal movement data during the period January 2013–December 2016.
3.6.2. BCV
For BCV the Se decreased with increasing cut-off as for BRSV, however when using a cut-off value of 0.25 for PostPInf the Se for detecting BTM positive herds in 2016 was only 0.55 (95% CI 0.42–0.68). At a cut-off value of 0.1 the Se was 0.83 (95% CI 0.71-0.92). At PostPInf < 0.05, two out of 19 herds (10%) were test positive. The proportion of test positive herds in each quartile of PostPFree is illustrated in Fig. 4, and the Se and Sp of PostPInf is illustrated in Fig. 5.
4. Discussion
This study shows that the PostPFree of BRSV and BCV can be used as an updated measure of the probability of freedom from antibodies at the herd level. For both infections, PostPFree of a test-negative herd was high immediately after a negative test, reflecting the high sensitivity of the tests, but gradually decreased with time. It is intuitive that the confidence of freedom from infection will decrease with time since sampling, as long as there is a risk of introduction. The advantage of our approach is that it offers a quantification of this decrease in confidence, through the regularly updated PostPFree resulting in herd specific slopes over time based on purchase of livestock, location of the herd and herd size (Fig. 3). Based on our calculations, the effect of the local factor was small compared to the effect of purchasing livestock, which had a large impact on the probability of freedom. Large differences in PostPFree were observed in the study herds at the end of the study period, depending on to which extent the individual herd had purchased animals. When herds that were test negative in 2013 were retested in 2016, 29% had changed antibody status to positive, and even though this proportion was likely lower after only one year (when retesting is required), this indicated that, in many cases, inferring a herd’s current status from an old BTM sample is problematic. Because most herds in the present study were not retested until 2016, a validation before this point was not possible. Consequently, we could only assess the overall performance of the method across three years, and not assess any variations between years. If implemented in the ongoing control program a continuous evaluation of the tool would be advisable so that adjustments can be made accordingly.
The estimated PIntroLocal was smaller for BCV than for BRSV. This was expected, as previous studies have indicated that the relative importance of purchase of livestock is higher for BCV than for BRSV (Frössling et al., 2012; Toftaker et al., 2016). The reason for the low estimated PIntroLocal was that few of the initially negative, closed herds seroconverted during the study period, 27% for BRSV and 18% for BCV. When herds purchasing animals also were included, 29% of the previously negative herds changed status to positive for each of the viruses. This is within the same range as in a Swedish study where between 11.1 and 66.7% of different categories of study herds went from BCV antibody negative to positive during a three-year period, when herd classification where based on pooled samples of primiparous cows (Ohlson et al., 2013). Only two herds had a negative BTM test. Even though the total study period was the same as in the current study, some herds did not become negative until after the study had started, thus the time at risk for each herd differed. Compared to our results for BRSV, Klem et al. (2013) found a considerably higher introduction rate (42%) over a period of only six months in a previous Norwegian study. However, the latter study differed from the present in two important aspects; it used a random sample of herds from the national dairy population, and herd classification was based on serum samples from a group of young stock. The difference in introduction rates could therefore be due to regional differences in disease occurrence and dynamics, and/or it might reflect that BTM negative herds represent a low risk stratum of the population. A negative BTM test means that the herd has likely been free from circulating virus for a long time, as animals continue to produce antibodies several years after infection (Alenius et al., 1991; Tråvén et al., 2001; Klem et al., 2014). If a herd has managed to stay free from infection for many years, it might have certain characteristics that makes it likely to remain free. PIntroLocal was used as a parameter for transmission through other routes than officially recorded animal movements. Indirect transfer via fomites is likely the most important factor, however, direct animal contact is possible e.g. on shared pastures, or if animals are temporarily moved (without change in ownership).
The estimation of PIntroLocal in the present study was based on a small sample size, and support from literature was scarce. The sensitivity analysis showed that the change in output (PostPFree) was moderate when PIntroLocal was increased or decreased with 50%, suggesting that if the true rate of local transmission is very different from the estimated local factor, this could affect the predictive ability of the model. It seems likely that local differences in prevalence and geographically dependent risk factors such as herd density might cause important differences in PIntroLocal. Differences in the importance of local transmission should therefore be investigated for different regions if the presented framework is to be applied at a national scale.
Currently, the control program is moving towards classification of herds based on pooled individual serum or milk samples, but these test results were not yet available for research purposes. The presented framework could be extended to encompass herd classification based on individual samples. This would include estimation of herd Se for the different types of sampling strategies, as described by More et al. (2013) for Johne’s disease in Ireland and recently by Ågren (2017) for Salmonella surveillance in Sweden. When individual samples are used for herd classification the time span reflected by a positive test, in terms of time of exposure to virus, is shorter compared to using BTM. The length of the time span will depend on the age of the tested animals, i.e. young stock will reflect a shorter time period than primiparous cows. There might also be differences between categories of herds based on other factors, such as biosecurity level, production type, and herd size. The herd size in the study region is smaller than the national average (Anonymous, 2017), hence herds categorized as “large” in the present study, are small even in a Norwegian context. A different cut-off between large and small herds, or more categories of herd size might be appropriate for application at a larger scale.
As mentioned, PostPFree relates to presence of antibodies and not necessarily presence of virus. Ideally, one would prefer to use a test detecting the antigen itself in order to achieve a herd’s true infection status; however, this is demanding to do on a large scale, and antibody testing is commonly used (Hägglund et al., 2006; Beaudeau et al., 2010; Ohlson et al., 2010a). Animal purchase might mean introducing an antibody positive animal and not necessarily introducing virus. As Norwegian herds are small the purchase of a single antibody positive lactating cow will likely suffice to produce a positive BTM test. Because we used BTM testing as the “gold standard” the herd would be classified as a “true positive” in the validation. Altogether, it is important to keep in mind that serologic classification in general as well as the output of our model (PostPFree) likely overestimates the proportion of herds in which there is actual virus circulation. Therefore, the estimated PostPFree from antibodies is likely lower than the true probability of freedom from circulating virus. However, the consequences of a positive test result in the control program is the same, regardless of why there are test-positive animals in a herd.
In the present study, the within-herd prevalence was set to 0.5 (50%) for all source herds. There are likely variations in within-herd prevalences depending on time since outbreak, and an increase in seroprevalence with age has previously been shown (Bidokhti et al., 2009). In a previous Norwegian study, Klem et al. (2013) reported a mean within-herd prevalence for BRSV of 55% based on serology of young stock (>6 months age), and Gulliksen et al. (2009) found a mean within-herd prevalence of 50% and 39% for BCV and BRSV, respectively, at calf level when calves with maternal antibodies were included. Studies sampling across age groups are lacking, hence the validity of the assumption of a 50% prevalence is hard to asses. Ideally, studies investigating the range of within-herd-prevalences should be performed.
The prior probability of infection, PriorPInf, was set to 0.5 for the first time period. This is a conservative estimate as it assumes no useful prior information about infection status (Martin et al., 2007b). However, the high Se of the BTM antibody tests will entail a high probability of freedom immediately after testing even if the prior probability is low. The model is therefore robust regarding choice of prior in this case.
Fixed values were used for all parameters in the present study. A stochastic approach is possible, and could potentially capture some of the uncertainty in the probability of disease. However, the aim of the present study was not to simulate disease spread, but to introduce a herd-specific measure as a decision support tool in the ongoing control program.
The model evaluation suggested that PostPFree is a useful tool for updated herd probability of freedom. When comparing PostPFree to the BTM result from 2016 (Wilcoxon Rank Sum Test), there was significant difference (p < 0.01) between groups for both models (BRSV and BCV), suggesting a benefit of using PostPFree instead of relying on the previous BTM result alone. Another indication of the usefulness of PostPFree was the clear differences in proportion of test positive herds between the first and fourth quartile of PostPFree as shown in Fig. 4. When assessing PostPInf as a diagnostic test we showed how many herds would be correctly classified at different cut-off values of PostPInf. In a practical setting, this is equal to the expected proportion of true positive herds that is detected if retesting is recommended at a certain value of PostPInf (PostPFree). If used on close to real time data, one could decide on a cut-off, and have an alarm when PostPFree drops below this value. This could enable timely intervention and a more risk-based approach to sampling and re-testing of herds. The relative Se (cut-off PostPInf >0.25) was lower for BCV than for BRSV suggesting that a more stringent cut-off might be appropriate for BCV if used for targeted sampling. In addition to test strategy purposes, the PostPFree could be used to classify herds in more than two categories, thus providing a more updated input for risk assessment prior to livestock purchase.
In conclusion, estimation of the probability of freedom for individual herds over time, based on the framework presented in this study, gave considerable variation in values among study herds even when they had equal starting points, i.e. negative test results. Validation against subsequent BTM sampling indicated a benefit of using PostPFree for an updated probability of a herd's antibody status instead of relying solely on a previous BTM test result.
Acknowledgements
The authors gratefully acknowledge The Norwegian Food Safety authorities for providing data on animal movements and Norwegian Agriculture Agency for access to data on location of herds. This project was a collaboration between The Norwegian University of Life Sciences and TINE SA, and was funded by Regional Research Funds, Region West (project No 257074).
References
- Ågren E. Publisher, Uppsala; 2017. Salmonella in Swedish Cattle.http://urn.kb.se/resolve?urn=urn:nbn:se:slu:epsilon-e-4007 Available from: (Accessed 14 January 2018) [Google Scholar]
- Alenius S., Niskanen R., Juntti N., Larsson B. Bovine coronavirus as the causative agent of winter dysentery: serological evidence. Acta Vet. Scand. 1991;32:163–170. doi: 10.1186/BF03546976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anonymous . TINE advisory service; 2017. Statistikksamling 2016.https://medlem.tine.no/aktuelt/nyheter/hk-statistikker/statistikksamling-2016 [Google Scholar]
- Beaudeau F., Ohlson A., Emanuelson U. Associations between bovine coronavirus and bovine respiratory syncytial virus infections and animal performance in Swedish dairy herds. J. Dairy Sci. 2010;93:1523–1533. doi: 10.3168/jds.2009-2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidokhti M.R.M., Tråvén M., Fall N., Emanuelson U., Alenius S. Reduced likelihood of bovine coronavirus and bovine respiratory syncytial virus infection on organic compared to conventional farms. Vet. J. 2009;182 doi: 10.1016/j.tvjl.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boileau M.J., Kapil S. Bovine coronavirus associated syndromes. Vet. Clin. N. Am. Food Anim. Pract. 2010;26:123–146. doi: 10.1016/j.cvfa.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elvander M. Severe respiratory disease in dairy cows caused by infection with bovine respiratory syncytial virus. Vet. Rec. 1996;138:101–105. doi: 10.1136/vr.138.5.101. [DOI] [PubMed] [Google Scholar]
- Espetvedt M.N., Reksen O., Rintakoski S., Østerås O. Data quality in the Norwegian dairy herd recording system: agreement between the national database and disease recording on farm. J. Dairy Sci. 2013;96:2271–2282. doi: 10.3168/jds.2012-6143. [DOI] [PubMed] [Google Scholar]
- Frössling J., Ohlson A., Björkman C., Håkansson N., Nöremark M. Application of network analysis parameters in risk-based surveillance - examples based on cattle trade data and bovine infections in Sweden. Prev. Vet. Med. 2012;105:202–208. doi: 10.1016/j.prevetmed.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frössling J., Nusinovici S., Nöremark M., Widgren S., Lindberg A. A novel method to identify herds with an increased probability of disease introduction due to animal trade. Prev. Vet. Med. 2014;117:367–374. doi: 10.1016/j.prevetmed.2014.07.013. [DOI] [PubMed] [Google Scholar]
- Gulliksen S.M., Jor E., Lie K.I., Løken T., Åkerstedt J., Østerås O. Respiratory infections in Norwegian dairy calves. J. Dairy Sci. 2009;92:5139–5146. doi: 10.3168/jds.2009-2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hägglund S., Svensson C., Emanuelson U., Valarcher J.F., Alenius S. Dynamics of virus infections involved in the bovine respiratory disease complex in Swedish dairy herds. Vet. J. 2006;172 doi: 10.1016/j.tvjl.2005.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klem T.B., Gulliksen S.M., Lie K.I., Løken T., Østerås O., Stokstad M. Bovine respiratory syncytial virus: infection dynamics within and between herds. Vet. Rec. 2013 doi: 10.1136/vr.101936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klem T.B., Tollersrud T., Østerås O., Stokstad M. Association between the level of antibodies in bulk tank milk and bovine respiratory syncytial virus exposure in the herd. Vet. Rec. 2014;175:47. doi: 10.1136/vr.102403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen L.E. Bovine respiratory syncytial virus (BRSV): a review. Acta Vet. Scand. 2000;41:1–24. doi: 10.1186/BF03549652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin P.A., Cameron A.R., Barfod K., Sergeant E.S., Greiner M. Demonstrating freedom from disease using multiple complex data sources 2: case study--classical swine fever in Denmark. Prev. Vet. Med. 2007;79:98–115. doi: 10.1016/j.prevetmed.2006.09.007. [DOI] [PubMed] [Google Scholar]
- Martin P.A., Cameron A.R., Greiner M. Demonstrating freedom from disease using multiple complex data sources 1: a new methodology based on scenario trees. Prev. Vet. Med. 2007;79:71–97. doi: 10.1016/j.prevetmed.2006.09.008. [DOI] [PubMed] [Google Scholar]
- More S.J., Sergeant E.S.G., Strain S., Cashman W., Kenny K., Graham D. The effect of alternative testing strategies and bio-exclusion practices on Johne’s disease risk in test-negative herds. J. Dairy Sci. 2013;96:1581–1590. doi: 10.3168/jds.2012-5918. [DOI] [PubMed] [Google Scholar]
- Norström M., Skjerve E., Jarp J. Risk factors for epidemic respiratory disease in Norwegian cattle herds. Prev. Vet. Med. 2000;44:87–96. doi: 10.1016/s0167-5877(99)00113-0. [DOI] [PubMed] [Google Scholar]
- Ohlson A., Emanuelson U., Traven M., Alenius S. The relationship between antibody status to bovine corona virus and bovine respiratory syncytial virus and disease incidence, reproduction and herd characteristics in dairy herds. Acta Vet. Scand. 2010;52(37) doi: 10.1186/1751-0147-52-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlson A., Heuer C., Lockhart C., Tråvén M., Emanuelson U., Alenius S. Risk factors for seropositivity to bovine coronavirus and bovine respiratory syncytial virus in dairy herds. Vet. Rec. 2010;167:201–206. doi: 10.1136/vr.c4119. [DOI] [PubMed] [Google Scholar]
- Ohlson A., Alenius S., Tråvén M., Emanuelson U. A longitudinal study of the dynamics of bovine corona virus and respiratory syncytial virus infections in dairy herds. Vet. J. 2013;197:395–400. doi: 10.1016/j.tvjl.2013.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solís-Calderón J.J., Segura-Correa J.C., Aguilar-Romero F., Segura-Correa V.M. Detection of antibodies and risk factors for infection with bovine respiratory syncytial virus and parainfluenza virus-3 in beef cattle of Yucatan, Mexico. Prev. Vet. Med. 2007;82:102–110. doi: 10.1016/j.prevetmed.2007.05.013. [DOI] [PubMed] [Google Scholar]
- Svanova. Svanova Manual. Bovine Coronavirus Antibodytest. Boehringer Ingelheim Svanova, Uppsala, Sweden. Available from: http://www.svanova.com/content/dam/internet/ah/svanova/dk_EN/documents/Kit%20inserts/Insert%20BCV-Ab%2019-2400-00_08.pdf. (Accessed 12 April 2016).
- Svanova. Svanova Manual. Bovine Respiratory Syncytial Virus Antibody Test. Boehringer Ingelheim Svanova, Uppsala, Sweden. Available from: http://www.svanova.com/content/dam/internet/ah/svanova/dk_EN/documents/Kit%20inserts/Insert%20BRSV-Ab%2019-2500-00_09.pdf. (Accessed 12 April 2016).
- Toftaker I., Sanchez J., Stokstad M., Nødtvedt A. Bovine respiratory syncytial virus and bovine coronavirus antibodies in bulk tank milk − risk factors and spatial analysis. Prev. Vet. Med. 2016;133:73–83. doi: 10.1016/j.prevetmed.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toftaker I., Holmøy I., Nødtvedt A., Østerås O., Stokstad M. A cohort study of the effect of winter dysentery on herd-level milk production. J. Dairy Sci. 2017;100:6483–6493. doi: 10.3168/jds.2017-12605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toftaker I., Toft N., Stokstad M., Sølverød L., Harkiss G., Watt N., O’ Brien A., Nødtvedt A. Evaluation of a multiplex immunoassay for bovine respiratory syncytial virus and bovine coronavirus antibodies in bulk tank milk against two indirect ELISAs using latent class analysis. Prev. Vet. Med. 2018;154:1–8. doi: 10.1016/j.prevetmed.2018.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tråvén M., Näslund K., Linde N., Linde B., Silván A., Fossum C., Hedlund K.O., Larsson B. Experimental reproduction of winter dysentery in lactating cows using BCV: comparison with BCV infection in milk-fed calves. Vet. Microbiol. 2001;81:127–151. doi: 10.1016/S0378-1135(01)00337-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valarcher J.F., Taylor G. Bovine respiratory syncytial virus infection. Vet. Res. 2007;38:153–180. doi: 10.1051/vetres:2006053. [DOI] [PubMed] [Google Scholar]