Abstract
A reovirus was isolated from Operophtera brumata (ObRV) and its parasitoid wasp Phobocampe tempestiva. Each of the 10 dsRNA genome segments of ObRV was sequenced and shown to contain a single open reading frame (ORF). Conserved motifs ([+ve] 5′-AAATAAA … G/TAGGTT-3′) were found at the termini of each segment, with the exception of Seg-6 and Seg-8, where the 5′ termini were 5′-AACAAA…-3′. The putative proteins encoded by each segment were compared with those of other members of the family Reoviridae. Phylogenetic comparisons to published sequences for the RNA-dependent RNA polymerase genes from other reoviruses indicated that ObRV is most closely related to members of the genus Cypovirus. However, unlike the cypoviruses, ObRV has a double-layered capsid structure. When the protein encoded by ObRV Seg-10 was expressed (by inserting the open reading frame into a baculovirus expression vector) no ‘occlusion bodies’ were observed in the recombinant baculovirus infected insect cell cultures. This suggests that unlike the cypoviruses, Seg-10 of ObRV does not contain a polyhedrin gene. Further phylogenetic comparisons also identified relationships between Seg-2 and Seg-10 of ObRV, and genes of Diadromus pulchellus Idnoreovirus 1 (DpIRV1), suggesting that ObRV represents a new species from the genus Idnoreovirus.
Keywords: Reovirus, Winter moth, Operophtera brumata, Protein expression
1. Introduction
Numerous DNA and RNA viral pathogens can infect insect species, including baculoviruses (Cory and Myers, 2003, Fuxa, 2004), entomopoxviruses (Arif and Kurstak, 1991), ascoviruses (Federici et al., 2005), polydnaviruses (Kroemer and Webb, 2004) and reoviruses (Mertens et al., 2005a). The Reoviridae is a large family containing 15 recognised genera of double-stranded RNA (dsRNA) viruses. These include several insect transmitted viruses that cause important diseases of animals or plants, which are classified within the genera Orbivirus, Seadornavirus and Coltivirus, or Fijivirus, Oryzavirus and Phytoreovirus, respectively (Mertens et al., 2005a). However, many of the reoviruses can only infect invertebrates and are classified within the genera Cypovirus (Green et al., 2007, Mertens et al., 2005b, Shapiro et al., 2005), Idnoreovirus (Mertens et al., 2005c) or Dinovernavirus (Attoui et al., 2005). At least 14 other reoviruses that infect insects, crustaceans or arachnids also remain unclassified (Renault et al., 2005), reflecting differences from the recognised genera in their morphology and genome features.
Five reoviruses have previously been isolated from species of Hymenoptera, which appear to be non-pathogenic in their wasp hosts (Renault et al., 2005). Three of these, Microplitis croceipes reovirus-like particles (McRVLP) (Hamm et al., 1994), Hyposoter exiguae idnoreovirus (HeIRV) (Mertens et al., 2005c, Stoltz and Makkay, 2000), and Opius concolor reovirus-like particles (OpRVLP) (Jacas et al., 1997), use their wasp hosts as vectors, and do not appear to be involved in lepidopteran host defence suppression (Renault et al., 2005). All three of these viruses were found in association with other viruses which facilitate the pathogenicity of the parasitoid against the wasp's lepidopteran host. For example, Stoltz and Makkay (2000) observed HeIRV and a DNA polydnavirus, H. exiguae polydnavirus (HePDV) replicating within the same cells of the parasitoid H. exiguae. A fourth virus, Diadromus pulchellus idnoreovirus-1 (DpIRV-1), has a commensal relationship with its host D. pulchellus, and is detected in most populations of this wasp species. DpIRV-1 is always associated with D. pulchellus ascovirus-4 (DpAV-4), which is indispensable for suppressing the immune system of the parasitoid's host. Although DpRV-1 has not been shown to directly increase the parasitic success of D. pulchellus, it may retard the replication of DpAV-4 within the lepidopteran host cells. This prevents rapid cell lysis, thereby enabling normal development of the wasp larva (Renault et al., 2005). The fifth virus, D. pulchellus idnoreovirus-2 (DpIRV-2; Renault et al., 2003), has a mutualistic relationship with D. pulchellus. Its injection by parasitoid females during oviposition is essential for suppressing the immune system of the parasitised lepidopteran host (Renault et al., 2005).
The winter moth, Operophtera brumata L., is a polyphagous univoltine pest. Out-break populations of O. brumata larvae have recently been observed on heather moorland on Orkney, in northern Scotland (Graham et al., 2004, Graham et al., 2006, Kerslake et al., 1996). Three reoviruses were isolated from populations of winter moth on Orkney, which were identified on the basis of their dsRNA genome profile after gel electrophoresis (Graham et al., 2006). Sequence analysis confirmed two of these viruses as belonging to the genus Cypovirus (Graham et al., 2007). However, a third virus, that was initially identified as O. brumata reovirus (ObRV), was isolated from both O. brumata and an ichneumonid parasitoid wasp Phobocampe tempestiva, that is known to attack the winter moth.
ObRV is a non-occluded reovirus which has double-shelled virions ∼55 nm in diameter. Virus isolated from O. brumata and P. tempestiva contained a genome composed of 10 segments of double-stranded RNA, with a total size of approximately 25.3 kb (Graham et al., 2006). We describe the complete sequence analysis of genome segments 1–10 (Seg-1 to Seg-10) of ObRV. For further characterisation and a comparison with known cypoviruses, the protein encoded by the open reading frame (ORF) of Seg-10, was expressed in insect cells using a recombinant-baculovirus vector-system.
2. Materials and methods
2.1. Isolation of viral dsRNA
O. brumata larvae were collected on Calluna vulgaris heather at 15 sites on the Orkney Isles (Graham et al., 2004). P. tempestiva were collected as they emerged from parasitised O. brumata larvae. Purification of non-occluded virus particles was achieved using a method modified from Stoltz and Makkay (2000). Approximately 1 g of pooled insect tissue was homogenised manually in 1 ml of sterile TE buffer (10 mM Tris, 0.1 mM EDTA, pH 8.0). Insect debris was removed by centrifugation at 500 × g for 10 min. The supernatant was loaded onto a two-step sucrose density gradient (50% sucrose [w/w] and 10% sucrose [w/w]) and centrifuged at 60,000 × g for 90 min (SW41 rotor, Beckman). The virus band was collected at the density interface and dialysed in TE buffer for 4 h. The sample was further pelleted at 15,000 × g for 5 min to remove cell debris. The supernatant was collected and centrifuged at 60,000 × g for 1 h. This virus pellet was resuspended in 200 μl TE buffer overnight at 4 °C and stored at −20 °C. Nucleic acid was purified using the DNeasy Tissue Kit (Qiagen). dsRNA was finally eluted in 150 μl of TE into sterile 1.5 ml microtubes, and stored at −20 °C.
2.2. cDNA synthesis, amplification by PCR, cloning and sequencing of segmented reovirus genome
The full-length genome of O. brumata reovirus was cloned using the FLAC method described previously (Maan et al., 2007, Shapiro et al., 2005). Briefly, an anchor-primer 5′P-GAC CTC TGA GGA TTC TAA AC/iSp9/TCC AGT TTA GAA TCC-OH 3′, (iSp9 is a C9 spacer) was ligated to purified viral genome dsRNA using T4 RNA ligase (Promega). The reaction was incubated at 8 °C overnight, heat-denatured and added to a reverse transcription (RT) reaction (AMV reverse transcriptase, Promega; 37 °C for 50 min). A PCR was subsequently performed, with reactions (100 μl) containing the following components: 1 μl (5 U) enzyme (ExTaq, TaKara), 4 μl dNTPs (300 mM), 10 μl 10× buffer (25 mM TAPs [pH 9.3], 50 mM KCl, 2 mM MgCl2, 1 mM DTT), 3 mM primer (5′-GGG ATC CAG TTT AGA ATC CTC AGA GGT C-3′), and 1 μl of cDNA (an initial 20 μl volume of cDNA was added to 200 μl water before 1 μl was used in PCR). Conditions were as follows: (i) 94 °C for 5 min, 1 cycle; (ii) 94 °C for 20 s, 62 °C for 20 s, 68 °C for 4.5 min, 26 cycles; (iii) 70 °C for 10 min, 1 cycle. The product was examined using 1% agarose gel electrophoresis, bands excised, ligated into pGEM-T Easy vector (Promega) and transfected into electrocompetent DH10B cells (Invitrogen). Selected clones were sequenced with M13 primers using a 3730 DNA Analyzer (ABI). Initial sequence data were used to design primers for subsequent direct sequencing of the viral segments. While several segments were sequenced from multiple clones, most were done using both clone and PCR DNA as the sequencing templates.
2.3. Analysis of sequence data
Sequence data was assembled and analysed using PreGap4 (Staden), Gap4 (Staden), and BioEdit (Hall, 1999). Phylogenetic analysis was carried out using ClustalW (Thompson et al., 1994) and Mega3.1 (Kumar et al., 2001) packages. Nucleotide and deduced amino acid sequences were compared with data available in Genbank using the NCBI's online BLAST program (http://www.ncbi.nlm.nih.gov/blast) or a local BLAST program implemented in the DNATools package (version 5.2.018, S.W. Rasmussen: Valby Data Center, Denmark) using a local database constructed from all available sequences of the Reoviridae. Hydrophobicity of segment 10 was analysed as described by Kyte and Doolittle (1982) using BioEdit. Protein motifs were examined using PROSITE (Hulo et al., 2004).
2.4. Construction of the transfer vector, and expression of ObRV S10 protein
Oligonucleotides were designed to amplify a product comprising the complete ORF of genome segment 10 for ObRV (ObRVseg10Forf, 5′CCT GGC CGA ATT GTG AGT A3′ and ObRVseg10Rorf, 5′GGC TCC ATT TCG CAT ACA TC3′). The subsequent product was purified (PCR Purification Kit, Qiagen), ligated into pGEM-T vector and used to transform E. coli (DH5α) (Invitrogen) via heat shock. M13 forward and reverse primers were used to read across the insert to ensure the correct sequence was present. The cloned ORF was ligated into the transfer vector pBacPaK8 (Clontech), using the compatible restriction sites SacI and BglII to derive pBacPAK8.RVS10. Cloned ORF inserts were examined for the correct sequence using primers Bac1 (5′ACC ATC TCG CAA ATA AAT AAG3′) and Bac2 (5′ACA ACG CAC AGA ATC TAG CG3′).
Spodoptera frugiperda-21 cells (Sf-21) were inoculated with a sample containing 500 ng of pBacPAK8.RVS10 DNA, 100 ng of flashBac™ DNA (Oxford Expression Technologies) and an equal volume of lipofectin (King and Possee, 1992). Cells were incubated at 28 °C. The virus stock was amplified in secondary cultures of the same cells and the infectivity determined by plaque assay (King and Possee, 1992). Cell cultures (106 cells per 35 mm dish) were inoculated with virus (10 pfu per cell), incubated at 28 °C and photographed at intervals. At 72 h p.i., the cells were harvested and proteins were analysed by SDS-PAGE, running samples for 16 h in 12% polyacrylamide gels. Following electrophoresis, proteins were stained with Coomassie brilliant blue and photographed.
3. Results
3.1. Sequence determination and analysis of ObRV
The full sequences of ObRV Seg-1 to Seg-10 were determined. The complete genome is 25,194 bp long and the corresponding sequences have been deposited in the Genbank database (see Table 1 for accession numbers). Each genome segment contains a single ORF, starting with the codon ‘AUG’. Analysis of the non-coding regions (NCRs) showed that all of the segments share seven conserved nucleotides at the 5′ ends and five conserved nucleotides at the 3′ ends, except for S6 and S8 (Table 1).
Table 1.
The accession numbers and conserved terminal sequences for ObRV isolated from winter moth
| Segment name | Accession number | Segment length | Encoded protein length | Mass | 5’ non-coding region |
3’ non-coding region |
||
|---|---|---|---|---|---|---|---|---|
| Length | Terminal | Terminal | Length | |||||
| S1 | DQ192235 | 4170 | 1358 | 155.7 | 32 | AAATAAAACCC | CCGATGAGGTT | 61 |
| S2 | DQ192236 | 3780 | 1207 | 137 | 77 | AAATAAATCTC | TTGGGTAGGTT | 79 |
| S3 | DQ192237 | 3595 | 1161 | 133 | 15 | AAATAAAACCC | TGATGGAGGTT | 94 |
| S4 | DQ192238 | 3362 | 1091 | 122.7 | 14 | AAATAAATCC | TTGTGTAGGTT | 72 |
| S5 | DQ192239 | 2106 | 620 | 69.2 | 30 | AAATAAAACCC | TTGGGTAGGTT | 213 |
| S6 | DQ192240 | 1935 | 594 | 68.4 | 62 | AACAAATCTCC | GTTGGTAGGTT | 88 |
| S7 | DQ192241 | 1606 | 499 | 57.1 | 17 | AAATAAAATCT | GAGGTAGGTT | 89 |
| S8 | DQ192242 | 1584 | 467 | 51.6 | 98 | AACAAAAACCC | TCATGTAGGTT | 82 |
| S9 | DQ192243 | 1547 | 437 | 49.6 | 76 | AAATAAAACTC | TAGTGTAGGTT | 156 |
| S10 | DQ192244 | 1509 | 467 | 53.6 | 21 | AAATAAAACCC | TGATGGAGGTT | 84 |
Predicted protein sizes are shown, as are the non-coding region sequence lengths. The bold letters indicate the conserved terminal sequences and the two italic bold sequences indicate different conserved sequence terminal.
Seg-1 of ObRV is 4170 nt in length (Table 1), with a single large ORF between nt 33 and 4109 (stop codon TGA), coding for a predicted protein of 1358 aa (155.7 kDa), which is identified as VP1. BLASTp searches showed VP1 to be homologous to the RNA-dependant RNA polymerases (RdRp) from Lymantria dispar CPV-14 (21% identity, e-value = 1 × 10−23); Fiji Disease Virus (24% identity, e-value = 9 × 10−19); Mal de Rio Cuarto virus (24% identity, e-value = 1 × 10−19); Bombyx mori CPV-1 (20% identity, e-value = 2 × 10−15); Cryphonectria parasitica reovirus (23% identity, e-value = 2 × 10−14); O. brumata CPV-19 (21% identity, e-value 2 × 10−14); and Colorado tick fever Virus (27% identity, e-value = 1 × 10−13); indicating that VP1 is the RdRp of ObRV. A leucine-zipper motif was found between aa 1310 and 1331, which has been suggested as a DNA-binding structure (Landschulz et al., 1988).
Seg 2 of ObRV is 3780 nt in length (Table 1), with an ORF between 78 and 3701 nt (stop codon TGA) coding for a predicted protein of 1207 aa (137 kDa) that is identified as VP2. The use of local BLASTp identified a cognate protein from DpIRV-1 (encoded by the 4.230 kb genome segment: accession number X82049), with 20% amino acid identity (42% aa similarity) over the full-length of the protein.
Seg-3 of ObRV is 3595 nt long, with a single ORF between nt 16 and 3501 (stop codon TAA: Table 1), coding for VP3 which is 1161 aa long (133 kDa). Tentative homology was detected, between aa 582 and 766, with Fijivirus segment 4 (27% identity, e-value = 0.007). The use of the local BLAST program also identified aa identities to partial sequences of the proteins encoded by Seg-3 of rice dwarf virus (RDV—genus Phytoreovirus) (29% identity, 55% similarity) 55% similarity: amino acids 1–70 of RDV P3), and African horse sickness virus (AHSV—genus Orbivirus) (31% identity, 47% similarity: amino acids 149–220 of AHSV VP3). Both of these proteins form the subcore shell (T2 layer, made of 120 copies) of their respective virus particles. The protein encoded by Seg-3 of ObRV may therefore also form the sub-core layer of the virus. Further analysis predicted the existence of three leucine zipper motifs at aa 446–467, 878–899 and 885–906.
Seg-4 of ObRV is 3362 nt long, with a single large ORF between nt 15 and 3290 (Table 1: stop codon TAG). The encoded protein (VP4) has a predicted size of 1091 aa (122.7 kDa). A local BLAST search showed 32% aa identity (46% similarity) to a partial sequence of RDV P2 (encoded by Seg-2) (aa 879–980 of RDV: compared to aa 809–900 of ObRV). The P2 of RDV is an outer coat protein that is essential for vector transmission.
Seg-5 of ObRV consists of 2106 nt (Table 1), with a single ORF between nt 31 and 1893 (stop codon TAA) coding for a VP5 protein that is 620 aa long (69.2 kDa). No homology was found with other virus sequences. The only tentative homology detected was between aa 85 and 218, with ‘copper amine oxidase’ of the common garden pea Pisum sativum (29% identity, e-value = 3.5).
Seg-6 of ObRV consists of 1935 nt (Table 1), with a single ORF between nt 63 and 1847 (stop codon TAG), coding for a VP6 protein that consist of 594 aa (68.4 kDa). Partial sequences of VP6 matched with VP6 of rice ragged stunt virus (RRSV—genus Oryzavirus) (aa 357–435 of RRSV, compared to aa 348–417 of ObRV: aa identity 26%, aa similarity 47%). Amino acid identity of 41% (e-value = 5.6) was also detected with aa 379 and 417 of the protein encoded by Seg-9 of Oat Sterile Dwarf Virus (genus Fijivirus).
Seg-7 of ObRV is 1606 nt long (Table 1), with a single long ORF between nt 18 and 1517 (stop codon TAA), which codes for the VP7 protein, with a predicted length of 499 aa (57.1 kDa). BLASTp searches showed no homology with the proteins of any virus previously sequenced. Tentative homology was seen between aa 339 and 442 with Trypanosoma brucei hypothetical protein Tb106k154000 (32% identity, e-value = 2.1).
Seg-8 of ObRV is 1584 nt long, with a single ORF between nt 99 and 1502 (Table 1) coding for a predicted VP8 protein that is 467 aa long (51.6 kDa). Tentative homology was observed between 52 and 119 aa with the Apicomplexan Theileria parva ABC transporter protein (31% identity, e-value = 1.4).
Seg-9 of ObRV is 1547 nt long, with a single large ORF between nt 77 and 1390 (Table 1), coding for a predicted VP9 protein that is 437 aa long (49.6 kDa). A local BLAST search identified a partial match with aa 85–150 of VP6 of Colorado tick fever virus, CTFV (29% identity, 49% similarity).
Seg-10 of ObRV is 1509 nt long, with a single large ORF between nt 22 and 1425 (stop codon TAG, Table 1), coding for VP10 with a predicted length of 467 aa (53.6 kDa). Tentative homology was detected between aa 298 and 395 of VP10, with the surface antigen D15 from the bacterium Ralstoni metallidurans CH34 (29% identity, e-value = 2.4). A local BLAST analysis showed a match over the whole length of the protein with an unassigned protein of DpIRV-1 (accession number X82045) with aa identity of 20% and a number of conserved motifs.
The hydrophobicity of ObRV VP10 showed no direct similarity to hydrophobicity plots or secondary structures obtained for the polyhedrin proteins of different cypoviruses (Echeverry et al., 1997). An analysis of the predicted amino acid sequence indicated that, unlike the cypovirus polyhedrin proteins, VP10 of ObRV is not especially rich in tyrosine (4.49%), aromatic residues (phenylalanine, tryptophane), or basic amino acids (lysine and arginine). However, it was particularly rich in leucine (10.04%), which is an amino acid periodically involved in signalling motifs (Hurley, 2003), which could be a factor in cell necrosis-inducing activities (Sawada et al., 2000).
3.2. Phylogenetic analysis
The RdRp is the most conserved reovirus protein and can be used for phylogenetic comparisons across the family as a whole. A tree was constructed using RdRp sequences from representatives of the different genera of the family Reoviridae, including that of ObRV (Fig. 1 ). Unfortunately, no RdRp sequences are available for any of the idnoreoviruses. ObRV is clearly distinct from the other reoviruses where the RdRp sequence is known.
Fig. 1.
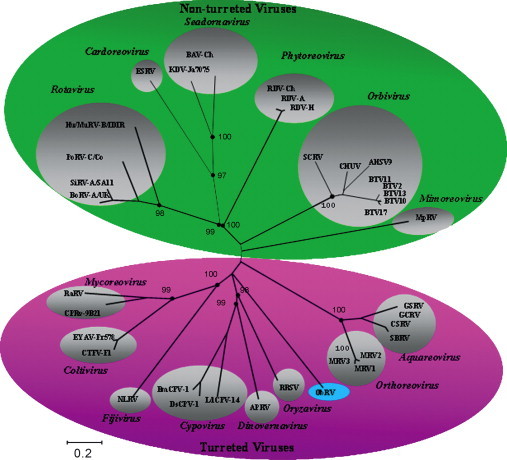
Phylogenetic tree of the Reoviridae family based upon sequences of the RdRp gene. Numbers given at each node correspond to the percentage bootstrap values (for 1000 repetitions). Amino acid identities were calculated for 19 viruses representative of the different genera: Bombyx mori Cypovirus (BmCPV1; accession number AAR88092; 20% identity); Lymantria dispar Cypovirus (LdCPV14; AAK73087; 20%); Operophtera brumata Cypovirus (OpbuCPV19; ABB17221; 21%); Mal de Rio Cuarto virus (MRCV; AAO73182; 21%); Rice black streaked disease virus (RBSDV; NP_620452; 19%); Fiji disease virus (FDV; AAK40249; 21%); Nilaparvata lugens reovirus (NLRV; BAA08542; 21%); Cryphonectria parasitica reovirus (CpRV; AAP45577; 23%); Colorado Tick Fever virus (CTFV; AAG34362; 27%); Eyach virus (EYAV; AAM18342; 26%); Golden Shiner Reovirus (GoCh; AAM92745; 22%); Grass Carp Reovirus (GCRV; NP_938029; 22%); Ndelle virus (NDEV; AAL36027; 20%); Rosellinia Anti-rot virus (RARV; BAC98431; 27%); Rice Ragged Stunt virus (RRSV; NP_620541; 20%); Simian RotaVirus (SirV; P22678; 13%); Rice Dwarf Virus (RDV; AAB18743; 12%); African Horse Sickness Virus (AHSV; AAC40586; 10%); St. Croix River virus (SCRV; AAG34363; 11%).
3.3. Expression of ObRV S10 open-reading frame
VP10 of ObRV was synthesised using a baculovirus expression system. Recombinant-baculoviruses were inoculated onto Sf-21 cells, and virus infection (CPE) observed at 72 h p.i. (Fig. 2 ). However, no polyhedra-like occlusion bodies (OB) were observed as a result of VP10 expression. SDS-PAGE analysis of the baculovirus-infected cell lysates showed the presence of a ∼54 kDa protein (Fig. 2). ObCPV-19 polyhedrin was synthesised as a positive control (as previously described, Graham et al., 2007). The presence of OB was detected at 72 h p.i. (Fig. 2). SDS-PAGE analysis of the infected insect cell lysates confirmed the presence of the ObCPV-19 polyhedrin at ∼29.2 kDa (Fig. 2).
Fig. 2.
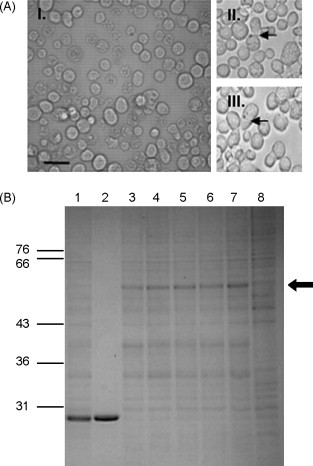
(a) Light microscope images of virus-infected Sf-21 cells. Panel I: recombinant ObRV S10 expression, showing no evidence of polyhedrin structures. Panels II and III: positive control, recombinant OpbuCPV19 polyhedrin expression. Arrows indicate cells showing crystal-like polyhedrin structures. Bar, 20 μm. (b) Expression of the polyhedrin gene of ObRV. Gel stained with Coomassie brilliant blue following electrophoresis at 80 V for 16 h. Lane 1: positive control, recombinant OpbuCPV19 polyhedrin expression ∼30 kDa; lane 2: positive control OpbuCPV19 virus, indicating the size of the polyhedrin protein; lanes 3–7: recombinant virus expressing ObRV S10 protein (∼54 kDa); lane 8: negative control, uninfected Sf-21 cells. The arrow indicates the position of the expressed protein resulting from the recombinant ObRV S10-expressing virus. Molecular sizes (in kDa) are shown to the left of the gel.
4. Discussion
A reovirus (ObRV) was isolated from the lepidopteran O. brumata and the ichneumonid wasp P. tempestiva. ObRV was more abundant in the parasitoid host (100% of wasp adults were infected) than in its lepidopteran host (of which only 11.5% were infected, Graham et al., 2006). Recent studies have suggested that three types of relationship may occur between certain reoviruses and parasitoid wasps; non-pathogenic, commensal viruses (occur and replicate in wasp populations, and may be found in association with other types of viruses); putative mutualistic viruses (found in association with other viruses, but which do not appear to play a major role in host/parasitoid relationships); mutualistic viruses (not associated with other viruses, and contribute significantly to parasitoid success) (Renault et al., 2005). It is difficult to be certain of the exact nature of the relationship of ObRV with P. tempestiva without further study.
Five hymenopteran reoviruses have been previously described (Renault et al., 2005). An interesting feature of these viruses is that four have never been found alone in their wasp host; they are always associated with another virus type. HeRV is associated with a polydnavirus in the ichneumonid parasitoid H. exiguae; DpRV-1 with an ascovirus (DpAV-4) in the ichneumonid D. pulchellus; McRVLP with a baculovirus (or nudivirus) or polydnavirus in the braconid M. croceipes; OpRVLP with a coronavirus in the braconid O. concolor (Hamm et al., 1994, Rabouille et al., 1994, Jacas et al., 1997, Stoltz and Makkay, 2000). In its hymenopteran host, ObRV was predominantly found alone, apart from two individuals additionally infected with a cypovirus (Graham et al., 2006). The ‘independent’ nature of ObRV suggests a replication strategy similar to that of the other cypoviruses or idnoreoviruses, both of which infect insect species.
The observation of ObRV in seemingly non-parasitised O. brumata indicates the ability of this virus to persist in this host without the need for the wasp (although wasp probing may have occurred without oviposition). The presence of ObRV in adult winter moth also suggests transmission of the virus through lepidopteran life stages (Graham et al., 2006). This shows similarity with infections caused by a cypovirus from D. pulchellus (Renault et al., 2003), which was able to infect and multiply in lepidopteran gut cells after oviposition (Renault et al., 2003), and assisted development of wasp larvae by down-regulating the immune response of the host. Interestingly, ObRV isolated from female wasps appears to contain a genome of 11 segments (Graham et al., 2006). Studies on the hymenopteran D. pulchellus showed that the Idnoreovirus DpIRV-1 isolated from diploid wasps (female or sterile diploid male wasps) also contained an extra genomic segment of dsRNA when compared to virus isolated from haploid wasps (fertile males; Rabouille et al., 1994). This extra segment is thought to provide additional nucleic information, necessary for the development of eggs laid by the female parasitoid in the lepidopteran host (Rabouille et al., 1994). Unfortunately, during the course of this study, insufficient viral dsRNA was available from female P. tempestiva to sequence the 11th segment. As more reoviruses are being isolated from insects, and specifically from parasitic wasps, it is becoming clear that some may play a close and important role in ensuring the success of the parasitoid host, and that complex relationships have developed to increase the fitness of both parasitoid and virus.
All 10 genome segments of ObRV were cloned and sequenced, showing that in each case they contain a single large ORF. Seg-1 encodes a putative RNA-dependent-RNA-polymerase. The dsRNA viruses use a viral RdRp to synthesis mRNA, functioning both as a transcriptase and replicase. The viral mRNA molecules can associate with other viral proteins to form large complexes. The viral RdRps can subsequently use these mRNAs as templates for the synthesis of complementary −ve strands, reforming the dsRNA genome of the virus. Within the family Reoviridae, RdRp aa identity of over 30% groups members within the same genus, while members of distinct genera have aa identities lower than 20% (Attoui et al., 2000, Rao et al., 2003). However, two exceptions to this rule have been discussed: these include aquareoviruses and orthoreoviruses, which clearly have a common ancestor with an aa identity of 42%; and rotavirus B which has only 22% identity to other rotaviruses. Amino acid identity values of between 19 and 27% were observed when comparing ObRV RdRp with the RdRps of the “turreted reoviruses”, while the “non-turreted reoviruses” gave only 11 to 13% aa identities. These data indicate that ObRV does not belong to any of the existing genera of the Reoviridae family for which RdRp sequences are available. A comparison of the ObRV Seg-10 sequence with those in Genbank, generated no matches with other insect viruses.
Although ObRV Seg-10 is 1509 bp in length, some species of cypovirus also have a large Seg-10 > 1 kb. For example, Seg-10 of Choristoneura fumiferana CPV-16 and Antheraea mylitta CPV-4 are ∼1.5 and ∼1.2 kb, respectively, although they only encode proteins of ∼260 aa in length (Echeverry et al., 1997, Sinha-Datta et al., 2005), corresponding to the expected size of their respective polyhedrin proteins. Expression of the ORF confirmed that ObRV Seg-10 encodes a protein with a molecular mass of ∼54 kDa, which did not form occlusion bodies in infected Sf-21 cells. Further analysis on the function of this protein remains to be done. The double-layered capsid, the non-occluded nature of ObRV (Graham et al., 2006), phylogenetic analyses of the RdRp, and the expression analysis of VP10 all indicate that that ObRV was not a member of the genus Cypovirus.However, similarities were detected between VP2 from ObRV and VP2 protein from an Idnoreovirus (DpIRV-1), over the whole length of the molecule. VP10 of ObRV also showed similarities to VP10 (encoded by Seg-10) of DpIRV-1 protein again over the whole length of the molecule. Although the lack of sequence information from this genus (particularly the RdRp), makes it difficult to make further comparisons, the data presented here, together with the detection of an eleventh genome segment in female wasps (Graham et al., 2006), suggest that ObRV represents a new member (a new species) within the genus Idnoreovirus, which contains other non-occluded insect reoviruses. In this case the isolate would belong to the species ‘Idnoreovirus-6’ and should be identified as ‘Operophtera brumata idnoreovirus-6’ (ObIRV-6).
Acknowledgements
This project was funded through a NERC non-thematic Research Studentship allocated to the Centre for Ecology and Hydrology and by a NERC non-thematic grant (NE/D008077/1) awarded to RDP, SMS and RSH.
Footnotes
References
- Arif B.M., Kurstak E. The entomopoxviruses. In: Kurstak E., editor. Viruses of Invertebrates. Marcel Dekker; New York: 1991. [Google Scholar]
- Attoui H., Billoir F., Biagini P., Micco P., de Lamballerie X. Complete sequence determination and genetic analysis of Banna virus and Kadipiro virus: proposal for assignment to a new genus (Seadornavirus) within the family Reoviridae. J. Gen. Virol. 2000;81:1507–1515. doi: 10.1099/0022-1317-81-6-1507. [DOI] [PubMed] [Google Scholar]
- Attoui H., Mohd Jaafar F., Belhouchet M., Biagini P., Cantaloube J.-F., de Micco P., de Lambalerie X. Expansion of family Reoviridae to include nine-segmented dsRNA viruses: isolation and characterisation of a new virus designated Aedes pseudoscutellaris reovirus assigned to a proposed genus (Dinovernavirus) Virology. 2005;343:212–223. doi: 10.1016/j.virol.2005.08.028. [DOI] [PubMed] [Google Scholar]
- Cory J.S., Myers J.H. The ecology and evolution of insect baculoviruses. Annu. Rev. Ecol. Evol. Syst. 2003;34:239–272. [Google Scholar]
- Echeverry F., Bergeron J., Kaupp W., Guertin C., Arella M. Sequence analysis and expression of the polyhedrin gene of Choristoneura fumiferana cytoplasmic polyhedrosis virus (CfCPV) Gene. 1997;198:399–406. doi: 10.1016/s0378-1119(97)00355-7. [DOI] [PubMed] [Google Scholar]
- Federici B.A., Bigot Y., Granados R.R., Hamm J.J., Miller L.K., Newton I., Stasiak K., Vlak J.M. Family Ascoviridae. In: Fauguet C.M., Mayo M.A., Maniloff J., Desselberger U., Ball L.A., editors. Virus Taxonomy. Eighth Report of the International Committee on Taxonomy of Viruses. Elsevier; Amsterdam, The Netherlands: 2005. pp. 247–250. [Google Scholar]
- Fuxa J.R. Ecology of insect nucleopolyhedroviruses. Agric. Ecosystems Environ. 2004;103:27–43. [Google Scholar]
- Graham R.I., Tyne W.I., Possee R.D., Sait S.M., Hails R.S. Genetically variable nucleopolyhedroviruses isolated from spatially separate populations of the winter moth Operophtera brumata (Lepidoptera: Geometridae) in Orkney. J. Invertebr. Pathol. 2004;87:29–38. doi: 10.1016/j.jip.2004.06.002. [DOI] [PubMed] [Google Scholar]
- Graham R.I., Rao S., Possee R.D., Sait S.M., Mertens P.P.C., Hails R.S. Detection and characterisation of three novel species of reovirus (Reoviridae), isolated from geographically separate populations of the winter moth Operophtera brumata (Lepidoptera: Geometridae) on Orkney. J. Invertebr. Pathol. 2006;91:79–87. doi: 10.1016/j.jip.2005.11.003. [DOI] [PubMed] [Google Scholar]
- Graham R.I., Rao S., Hails R.S., Sait S.M., Mertens P.P.C., Possee R.D. Characterisation and sequence analysis of two novel cypoviruses isolated from the winter moth Operophtera brumata (Lepidoptera: Geometridae) Virus Genes. 2007;35:463–471. doi: 10.1007/s11262-007-0113-0. [DOI] [PubMed] [Google Scholar]
- Green T.B., White S., Rao S., Mertens P., Adler P.H., Becnel J.J. Biological and molecular studies of a cypovirus from the black fly Simulium ubiquitum (Diptera: Simuliidae) J. Invertebr. Pathol. 2007;95:26–32. doi: 10.1016/j.jip.2006.10.006. [DOI] [PubMed] [Google Scholar]
- Hall T.A. BioEdit: a user friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- Hamm J.J., Styer E.L., Steiner W.W. Reovirus-like particle in the parasitoid Microplitis croceipes (Hymenoptera: Braconidae) J. Invertebr. Pathol. 1994;63:304–306. [Google Scholar]
- Hulo N., Sigrist C.J.A., Le Saux V., Langendijk-Genevaux P.S., Bordoli L., Gattiker A., De Castro E., Bucher P., Bairoch A. Recent improvements to the PROSITE database. Nucleic Acids Res. 2004;32:134–137. doi: 10.1093/nar/gkh044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley J.H. Leucine in the sky with diamonds. Structure. 2003;11:1192–1193. doi: 10.1016/j.str.2003.09.009. [DOI] [PubMed] [Google Scholar]
- Jacas J.Y., Budia F., Rodriguez-Cerezo E., Vinuela E. Virus-like particles in the poison gland of the parasitic wasp Opius concolor. Ann. Appl. Biol. 1997;130:587–592. doi: 10.1111/j.1744-7348.1997.tb07685.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerslake J.E., Kruuk L.E.B., Hartley S.E., Wooding S.J. Winter moth (Operophtera brumata (Lepidoptera: Geometridae)) outbreaks on Scottish heather moorlands: effects of host plant and parasitoids on larval survival and development. Bull. Entomol. Res. 1996;86:155–164. [Google Scholar]
- King L.A., Possee R.D. Chapman and Hall; London: 1992. The Baculovirus Expression System: a Laboratory Guide. [Google Scholar]
- Kroemer J.A., Webb B.A. Polydnavirus genes and genomes: emerging gene families and new insights into polydnavirus replication. Annu. Rev. Entomol. 2004;49:431–456. doi: 10.1146/annurev.ento.49.072103.120132. [DOI] [PubMed] [Google Scholar]
- Kumar S., Tamura K., Jakobsen I.B., Nei M. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics. 2001;17:1244–1245. doi: 10.1093/bioinformatics/17.12.1244. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Landschulz W.H., Johnson P.F., McKnight S.L. The leucine zipper: a hypothetical structure common to a new class of DNA-binding proteins. Science. 1988;240:1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- Maan S., Rao S., Maan N.S., Anthony S.J., Attoui H., Samuel A.R., Mertens P.P.C. Rapid cDNA synthesis and sequencing techniques for the genetic study of bluetongue and other dsRNA viruses. J. Virol. Methods. 2007;143:132–139. doi: 10.1016/j.jviromet.2007.02.016. [DOI] [PubMed] [Google Scholar]
- Mertens P.P.C., Duncan R., Attoui H., Dermody T.S. Reoviridae. In: Fauguet C.M., Mayo M.A., Maniloff J., Desselberger U., Ball L.A., editors. Virus Taxonomy. Eighth Report of the International Committee on Taxonomy of Viruses. Elsevier; Amsterdam, The Netherlands: 2005. pp. 447–454. [Google Scholar]
- Mertens P.P.C., Rao S., Zhou H. Cypovirus. In: Fauguet C.M., Mayo M.A., Maniloff J., Desselberger U., Ball L.A., editors. Virus Taxonomy. Eighth Report of the International Committee on Taxonomy of Viruses. Elsevier; Amsterdam, The Netherlands: 2005. pp. 522–533. [Google Scholar]
- Mertens P.P.C., Makkay A., Duncan R., Bergoin M., Dermody T.S. Idnoreovirus, Reoviridae. In: Fauguet C.M., Mayo M.A., Maniloff J., Desselberger U., Ball L.A., editors. Virus Taxonomy. Eighth Report of the International Committee on Taxonomy of Viruses. Elsevier; Amsterdam, The Netherlands: 2005. pp. 517–521. [Google Scholar]
- Rabouille A., Bigot Y., Drezen J.-M., Sizaret P.-Y., Hamelin M.-H., Periquet G. A member of the Reoviridae (DpRV) has a ploidy-specific genomic segment in the wasp Diadromus pulchellus (Hymenoptera) Virology. 1994;205:228–237. doi: 10.1006/viro.1994.1638. [DOI] [PubMed] [Google Scholar]
- Rao S., Carner G.R., Scott S.W., Omura T., Hagiwara K. Comparison of the amino acid sequences of RNA-dependant RNA polymerases of cypoviruses in the family Reoviridae. Arch. Virol. 2003;148:209–219. doi: 10.1007/s00705-002-0923-2. [DOI] [PubMed] [Google Scholar]
- Renault S., Bigot S., Lemesle M., Sizaret P.Y., Bigot Y. The cypovirus Diadromus pulchellus RV-2 is sporadically associated with the endoparasitoid wasp D. pulchellus and modulates the defence mechanisms of pupae of the parasitized leek-moth, Acrolepiopsis assectella. J. Gen. Virol. 2003;84:1799–1807. doi: 10.1099/vir.0.19038-0. [DOI] [PubMed] [Google Scholar]
- Renault S., Stasiak K., Federici B., Bigot Y. Commensal and mutualistic relationships of reoviruses with their parasitoid wasp hosts. J. Insect Physiol. 2005;51:137–148. doi: 10.1016/j.jinsphys.2004.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada T., Hashimoto S., Tohma S., Nishioka Y., Nagai T., Sato T., Ito K., Inoue T., Iwata M., Yamamoto K. Inhibition of l-leucine methyl ester mediated killing of THP-1, a human monocytic cell line, by a new anti-inflammatory drug, T614. Immunopharmacology. 2000;49:285–294. doi: 10.1016/s0162-3109(00)00242-3. [DOI] [PubMed] [Google Scholar]
- Shapiro A., Green T., Rao S., White S., Carner G., Mertens P.P.C., Becnel J.J. Morphological and molecular characterization of a Cypovirus (Reoviridae) from the mosquito Uranotaenia sapphirina (Diptera: Culicidae) J. Virol. 2005;79:9430–9438. doi: 10.1128/JVI.79.15.9430-9438.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha-Datta U., Chavali V.R.M., Ghosh A.K. Molecular cloning and characterisation of Antheraea mylitta cytoplasmic polyhedrosis virus polyhedrin gene and its variant forms. Biochem. Biophys. Res. Commun. 2005;332:710–718. doi: 10.1016/j.bbrc.2005.05.011. [DOI] [PubMed] [Google Scholar]
- Stoltz D., Makkay A. Co-replication of a reovirus and a polydnavirus in the ichneumonid parasitoid Hyposoter exiguae. Virology. 2000;278:266–275. doi: 10.1006/viro.2000.0652. [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]


