Abstract
The small envelope (E) protein of porcine reproductive and respiratory syndrome virus (PRRSV) is known to possess the properties of an ion-channel protein, and in the present study we show that the PRRSV E protein is N-terminal myristoylated. The PRRSV E protein contains the consensus motif of 1MGxxxS6 for myristoylation, and in the presence of 2-hydroxymyristic acid, the virus titer decreased by 2.5 log TCID50 and the level of viral RNA was reduced significantly. When the glycine at position 2 was mutated to alanine (G2A) using an infectious cDNA clone, a viable virus was recoverable and a mutant PRRSV was obtained. The titers of G2A mutant virus were 2.0 × 104 and 1.0 × 106 TCID50/ml for ‘passage-2’ and ‘passage-3’ viruses, respectively, in PAM cells, and these titers were significantly lower than those of wild-type PRRSV. When treated with the myristoylation inhibitor, the G2A mutant virus was resistant to the drug. The data show that the PRRSV E protein myristoylation is non-essential for PRRSV infectivity but promotes the growth of the virus.
Keywords: PRRS, E protein, Infectious clone, Myristoylation, Fatty acid acylation
Porcine reproductive and respiratory syndrome virus (PRRSV) is a small, enveloped, positive-strand RNA virus, belonging to the family Arteriviridae in the order Nidovirales that also includes Coronaviridae and Roniviridae (Cavanagh, 1997, Cowley et al., 2000, González et al., 2003, Smits et al., 2003). Other members of the Arteriviridae family include lactate dehydrogenase-elevating virus (LDV) of mice, equine arteritis virus (EAV), and simian hemorrhagic fever virus (SHFV) (Cavanagh, 1997). The PRRSV genome is approximately 15 kb in size and organized to code for two large non-structural polyproteins 1a and 1a/1b in the 5-terminal 12 kb region and 7 structural proteins in the 3-terminal 3 kb region; GP2 (glycoprotein 2), E (small envelope), GP3, GP4, GP5, M (membrane), and N (nucleocapsid) proteins in order from the 5′ end (Meulenberg et al., 1993, Snijder and Meulenberg, 1998, Wootton et al., 2000). Based on the antigenic and genetic differences, PRRSV isolates are divided into two distinct genotypes: North American genotype and European genotype.
Of the structural proteins, N associates with the viral RNA and makes up the viral capsid. Besides N, six other membrane-associated proteins constitute the virion envelope. GP5 and M proteins form a disulfide-linked heterodimer (Mardassi et al., 1996), which has been shown to be essential for infectivity in LDV and EAV (Faaberg et al., 1995, Snijder et al., 2003). GP2, GP3, and GP4 are minor glycoproteins and form a disulfide-linked heterotrimer, and the heterotrimerization has also been shown to be essential for infectivity in EAV (Wieringa et al., 2003a, Wieringa et al., 2003b). For PRRSV, co-expression of GP2/E, GP3, and GP4 resulted in transport of these proteins from the ER through the Golgi complex followed by their release into the culture medium, suggesting a critical role of the heteromultimeric complex for virus maturation (Wissink et al., 2005).
The E protein is translated from the small internal open reading frame (ORF) within ORF2, starting from the +6 nucleotide position in mRNA2, and comprised of 73 and 70 amino acids for the North American and European types, respectively. The E protein is non-glycosylated and intracellular membrane-associated (Snijder et al., 1999, Wu et al., 2001), and for EAV and PRRSV, has been shown to be essential for virus replication. The E protein is highly hydrophobic but contains a cluster of basic amino acids in the hydrophilic C-terminal region. Although the E protein of North American genotype PRRSV contains two cysteine residues at positions 49 and 54, a study has shown that E is unable to form a disulfide-linked homodimers. Nevertheless, E protein of PRRSV exists as a multimer which is probably mediated by ionic interaction (Lee and Yoo, 2005). In contrast, E protein of EAV does not contain cysteine residues and seems to exist as a monomer. A recent study has shown that the PRRSV E protein is likely an ion-channel protein embedded in the viral envelope and may facilitate the uncoating of virus and release of the genome into the cytoplasm (Lee and Yoo, 2006). The E protein has also been predicted to contain a putative N-terminal N-myristoylation site (Snijder et al., 1999, Wu et al., 2001). However, this aspect has not experimentally been demonstrated, and in the present study, we explored the E protein myristoylation using the reverse genetics approach.
The N-terminal myristoylation refers to the linkage of myristic acid (C14:0) via an amide bond to the N-terminal glycine residue of a protein in some cellular, viral, and also a few bacterial proteins (Wilcox et al., 1987, James and Olson, 1990, Boutin, 1997, Nimchuk et al., 2000). The reaction is catalyzed by myristoyl-CoA:N-myristoyltransferase (NMT) (Wilcox et al., 1987). In general, N-myristoylation is an irreversible protein modification that occurs co-translationally following removal of the initiator methionine by a cellular methionylaminopeptidase (Towler et al., 1987, Wolven et al., 1997). N-myristoylation may also occur post-translationally as for the case of pro-apoptotic protein BID where proteolytic cleavage by caspase 8 reveals a hidden myristoylation motif (Zha et al., 2000). Myristoylation has been identified for several viral proteins including VP4 of poliovirus (Chow et al., 1987), Gag protein of simian immunodeficiency virus (SIV) (Henderson et al., 1988), Nef protein of HIV (Harris et al., 1992), pre-S1 of hepatitis B virus (Persing et al., 1987), VP2 of SV40 (Streuli and Griffin, 1987) to name a few, and all of these proteins contain the conserved sequence motif of 1MGxxxS/T, where ‘x’ is any amino acid (Fig. 1 ). Interestingly, E proteins of all PRRSV isolates have been found to contain the conserved myristoylation sequence motif and this motif is also found in E proteins of EAV, LDV and SHFV (Fig. 1), suggesting the importance of E protein myristoylation.
Fig. 1.
The consensus sequence motif for N-terminal glycine myristoylation in viral proteins. E proteins of PRRSV contain the consensus motif of 1MGxxxS6. The myristoylation motif is also found as 1MGxxxS/T in E proteins of equine arteritis virus (EAV), lactate dehydrogenase-elevating virus (LDV), and simian hemorrhagic fever virus (SHFV). SIV, simian immunodeficiency virus; HIV, human immunodeficiency virus; SV40, simian virus 40.
To examine the possibility of E protein myristoylation, we first determined the effects of a myristoylation inhibitor on PRRSV replication. 2-Hydroxymyristic acid (2-hydroxymyristate, HMA; Sigma–Aldrich) is a chemical known to specifically prevent myristoylation without interfering palmitoylation of an acylated protein (Galbiati et al., 1996). HMA is metabolically activated in cells to CoA-thioester, which then competitively inhibits NMT activity. At 0.1 mM concentration, HMA has been shown to reduce the growth of a variety of viruses including herpes simplex virus and arenavirus for up to three orders of magnitude (Harper et al., 1993, Perez et al., 2004). Porcine alveolar macrophages (PAMs) were grown in RPMI-1640 (Invitrogen) containing 10% fetal bovine serum (HyClone, Logan UT), and titrated for cytotoxicity at concentrations of 0, 0.1, 0.5, 1.0, and 2.0 mM of HMA. 1.0 mM or higher concentration of HMA was found to be toxic for PAM cells, and thus 0.1 mM was chosen for subsequent studies. PAMs were infected with 5–10 MOI (multiplicity of infection) with a North American type PRRSV (strain P129). Four hours post-infection, the culture supernatant was replaced with a fresh medium containing 0.1 mM of HMA and the cells were further incubated for an additional 20 h. Cell culture supernatant was collected for virus titration, and viral RNA was also extracted from the supernatant for real-time RT-PCR. In the presence of HMA, titers of PRRSV were reduced by 2.5 log TCID50 and the reduction was statistically significant (P < 0.05) (Fig. 2A). To confirm the reduction of infectivity in the supernatant, real-time RT-PCR was conducted using RNA extracted from the culture supernatant. The level of viral RNA also decreased significantly when compared with that of the untreated cells (Fig. 2B). These results suggest that PRRSV is myristoylated, and since E is the only protein containing the myristoylation motif among PRRSV proteins (data not shown), the E protein is likely the myristoylated protein in PRRSV.
Fig. 2.
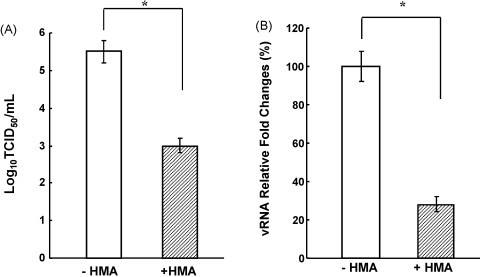
Inhibition of PRRSV replication in the presence of 0.1 mM 2-hydroxymyristic acid (HMA). PAM cells were infected with PRRSV (strain P129) and incubated with RPMI-1640 containing 10% fetal bovine serum. At 4 h post-infection, the supernatant was replaced with a fresh medium containing 0.1 mM HMA. The culture supernatant was harvested at 24 h post-infection and subjected to virus titration by TCID50 assay (A) and to real-time PCR for ORF7 using a primer set of N-Fwd (5′-AATAACAACGGCAAGCAGCAG-3′) and N-Rev (5′-CCTCTGGACTGGTTTTGCTGA-3′) (B) −HMA, absence of 2-hydroxymyristate; +HMA, presence of 2-hydroxymyristate. Experiments were conducted in duplicate and repeated three times. The data were compared using analysis of variance and t-test, and shown as mean ± standard error. Stars indicate a statistical value of p < 0.05.
To confirm this finding, the potential myristoylation motif was mutated using an infectious cDNA clone for PRRSV and the impact of HMA on the mutant virus was examined. The GGG codon for Gly at position 2 of the E protein was mutated to GCT for Ala using the full-length infectious clone (Lee and Yoo, 2005), using the QuikChange® II XL site-directed mutagenesis kit (Stratagene, La Jolla, CA) and the following primer set: P129-E-G2A-Fwd (5′-GAATTGAAATGAAATGGCTTCTATA CAAAGCCTCT-3′) and P129-E-G2A-Rev (5′-AGAGGCTTTGTATAGAAGCCATT TCATTTCAATTC-3′), where underlined letters indicate codons for mutagenized amino acid. PCR products were used to transform Escherichia coli and appropriate clones were screened based on the SmaI digestion pattern of the plasmid followed by sequencing for verification of the mutation. Five clones were independently obtained from two different mutagenesis experiments. MARC-145 cells at 70% confluency seeded in 35-mm-diameter dishes were transfected with 2 μg of the wild-type infectious clone (P129-WT) or myristoylation-mutagenized clone (P129-E-G2A), and incubated for 5 days. PRRSV-specific cytopathic effects (CPE) appeared for P129-WT transfected cells by 4 days post-transfection and the CPE became prominent by 5 days post-transfection. On the contrary, CPE was not readily visible in P129-E-G2A transfected cells. The culture supernatants were harvested at 5 days post-transfection and designated ‘passage-1’ virus. The ‘passage-1’ virus was used to inoculate fresh MARC-145 cells and the 5-day harvest was designated ‘passage-2.’ The ‘passage-3’ virus was prepared in the same way as for ‘passage-2.’ Each passage virus was aliquoted and stored at −80 °C until use.
Since the CPE was shown much slower for G2A mutant virus, titers of the virus were determined for ‘passage-2’ and ‘passage-3’ in both MARC-145 cells (Fig. 3A) and PAMs (Fig. 3B) by microtitration infectivity assay and recorded as 50% tissue culture infectious dose per milliliter (TCID50/ml). In MARC-145 cells, P129-E-G2A grew to a titer of only 3.2 × 101 and 3.2 × 102 TCID50/ml for ‘passage-2’ and ‘passage-3,’ whereas the titers of wild-type PRRSV were 1.4 × 104 and 2.5 × 105 TCID50/ml (Fig. 3A), respectively. These titers are considered low even for wild-type PRRSV and it is because these viruses were freshly reconstituted from infectious clones and required subsequent passages in cell culture for higher titers. Since PAMs appeared to be more sensitive for PRRSV than MARC-145 cells, titration was also conducted in PAMs (Fig. 3B). As expected, the titers were higher in PAMs for ‘passage-2’ and ‘passage-3’ of P129-WT with 4 × 106 and 3.2 × 108 TCID50/ml, respectively. On the contrary, P129-E-G2A titers remained low with 2.0 × 104 and 1.0 × 106 TCID50/ml for ‘passage-2’ and ‘passage-3,’ respectively, and these titers were significantly lower than those of wild-type PRRSV (Fig. 3B). All five clones of P129-E-G2A that were recovered independently from two different experiments showed identical virological properties and titers (data not shown), confirming that the virological phenotype of P129-E-G2A was indeed due to the Gly-2 mutation of E.
Fig. 3.

Titers of P129-E-G2A and P129-WT at ‘passage-2’ and ‘passage-3’ in MARC-145 and PAMs. Experiments were conducted in duplicate and repeated three times. The data were compared using analysis of variance and t-test, and shown as mean ± standard error.
To further characterize the myristoylation mutant, MARC-145 cells were infected with ‘passage-2’ of P129-E-G2A and stained for viral proteins using N-specific and Nsp2/3-specific antibodies. MARC-145 cells were seeded on microscope coverslips and grown to 70% confluence. The cells were infected with ‘passage-2’ of P129-E-G2A at an MOI of 0.1, and at 2 days post-infection, cells were fixed and co-stained with MAb SDOW17 for N and a rabbit serum for Nsp2/3 followed by staining with respective secondary antibodies (Molecular Probes; Eugene, OR). Both N and Nsp2/3 proteins were clearly visible in the same cell indicating the expression of both structural and non-structural proteins in P129-E-G2A infected cells (Fig. 4A). The virological property of P129-E-G2A was then examined by plaque assays using the ‘passage-2’ virus. The plaque morphology of P129-E-G2A appeared to be a pin-hole type and much smaller than the wild-type plaques (Fig. 4B). When PAMs were infected with P129-E-G2A, CPE and cell death were evident by 2 days post-infection (Fig. 4C). Data taken from the IFA, plaque assay, and infectivity assay, a conclusion was made that the G2A mutation in the E protein was not essential for PRRSV infectivity and a viable virus could be generated.
Fig. 4.

Characterization of the myristoylation knock-out virus P129-E-G2A. (A) Immunofluorescence of MARC-145 cells infected with the ‘passage-2’ virus for 2 days. Cells were stained with N (green)-specific MAb SDOW17 and Nsp2/3 (red)-specific rabbit antiserum, followed by staining with goat anti-mouse antibody conjugated with Alexa green, and goat anti-rabbit antibody conjugated with Texas red, respectively. (B) Plaque morphologies of P129-E-G2A and P129-WT. MARC-145 cells were infected with dilutions of ‘passage-2’ virus for 1 h, overlaid with 1% agarose, and incubated for 6 days until plaques developed. Plaques were stained by 1% crystal violet. (C) CPE in PAM cells infected with ‘passage-2’ of P129-E-G2A. CPE is evident in PAMs infected with the P129-E-G2A mutant or P129-WT virus. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.)
Since HMA inhibited the replication of wild-type PRRSV (Fig. 2) and because the inhibitory effect was most likely due to the inhibition of myristoylation of E, it is conceivable that HMA would have minimal effects on replication of the myristoylation-negative virus. The HMA inhibition assay was conducted as described above. As expected, titers of P129-E-G2A remained unchanged in the presence or absence of HMA (Fig. 5A), and similarly, the level of viral RNA synthesis was also unchanged (Fig. 5B). This is because P129-E-G2A mutant lacks the myristoylation site in the E protein and thus the myristoylation-negative PRRSV becomes resistant to myristoylation inhibitor. These data demonstrate that the E protein myristoylation is dispensable for virus infectivity but promotes PRRSV replication.
Fig. 5.
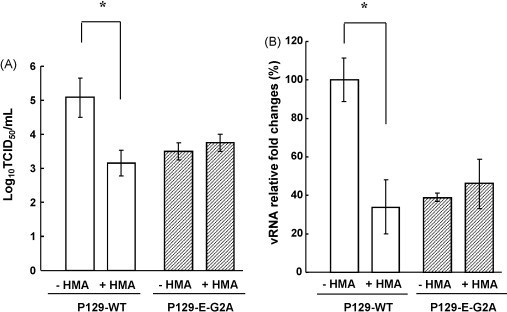
Resistance of the myristoylation knock-out mutant virus P129-E-G2A to 0.1 mM 2-hydroxymyristate (HMA). Cell culture supernatants were harvested at 24 h post-infection in the presence of HMA and subjected TCID50 assay (A) and real-time RT-PCR (B) for ORF7 gene to measure the infectivity and the amount of viral RNA in the supernatant. −HMA, absence of 2-hydroxymyristate; +HMA, presence of 2-hydroxymyristate. Experiments were conducted in duplicate and repeated three times. The data were compared using analysis of variance and t-test, and shown as mean ± standard error. Stars indicate a statistical value of p < 0.05.
Myristoylated proteins exhibit diverse cellular functions. Some myristoylated proteins are involved in signaling processes while others promote membrane association or protein–protein interactions. The hydrophobic nature of myristoylation allows for reversible membrane interactions in concern with additional factors on the protein for membrane targeting and attachments, including palmitoylation of cysteine residues, clusters of positively charged amino acids, phospholipid-binding domains, transmembrane regions, and protein–protein interactions. Both specific targeting and membrane affinity are achieved through harmonized effects of these factors, for example, dual acylation of a protein by myristoylation and palmitoylation often directs the protein to lipid rafts (Zacharias et al., 2002). In viruses, the myristoyl moiety not only targets to membranes or specific lipids (Hearps and Jans, 2007, Saad et al., 2006), but can also be involved in direct interactions with other proteins (Hayashi et al., 2002) or in the conformational switches that affect tertiary and quaternary structures of a protein (Liemann et al., 2002). For E protein of PRRSV, the myristoylation may attribute to membrane association and protein interactions rather than signaling processes.
During the course of our study, Thaa et al. (2009) reported that the E protein homolog of EAV was indeed a myristoylated protein at the glycine residing at position 2, and showed that the E protein myristoylation contributed to EAV infectivity. This study is consistent with our finding that the PRRSV E protein is also myristoylated and the myristoylation is a non-essential modification for virus infectivity. A considerable inhibition of EAV infectivity was observed at a concentration of 0.1 mM HMA, and similarly, 0.1 mM HMA negatively affected PRRSV replication in our study.
The E protein is an ion-channel protein for SARS (severe acute respiratory syndrome) coronavirus (Madan et al., 2005, Liao et al., 2004, Wilson et al., 2004). The PRRSV E protein also contains similar functional properties to those of SARS coronavirus E protein but differs in their structures (Lee and Yoo, 2006). For ion-channel proteins, multimerization is a common property. The PRRSV E protein contains two well-conserved cysteine residues for the North American genotype but these residues are not conserved in LDV, EAV, and SHFV, including the European genotype PRRSV. Additionally, the PRRSV E protein has been shown not to form a disulfide-linked homodimer and therefore does not undergo cysteine-linked multimerization for pore formation (Lee and Yoo, 2006). Consequently, the cysteine residues in E have been shown to be non-essential for PRRSV infectivity (Lee and Yoo, 2005). However, by cross-linking studies, the PRRSV E protein has been shown to exist as non-covalently linked homo-multimers in virus-infected cell, suggesting that the physical basis for pore formation of E to function as an ion-channel is likely a non-covalent, ionic interaction between monomeric E proteins. The PRRSV E protein is highly hydrophobic and contains a cluster of basic, positively charged amino acids in the hydrophilic C-terminal region. Thus, it is predicted that the major role of myristoylation for PRRSV E is to enhance its membrane association and protein interactions. This will facilitate the stable pore formation of E on the viral membrane and permit stable ion influx and subsequent pH alterations in the virion interior, which in turn will lead to efficient uncoating processes of the viral capsid. This premise is supported by the findings in the present study that the myristoylation knock-out mutant virus was viable but its replication was severely affected. Coronavirus E proteins resemble PRRSV E proteins in many aspects as an ion-channel protein (Wilson et al., 2004, Wilson et al., 2006). Coronavirus E proteins, however, do not contain the myristoylation motif. Similarly, toroviruses and roniviruses, other member viruses in the Nidovirales order, do not appear to contain the myristoylation motif in their homologs. Thus, it is conceivable that the E protein myristoylation is a common and unique feature to the family Arteriviridae in the order Nidovirales.
Acknowledgments
This study was supported by the National Research Initiatives of the US Department of Agriculture Cooperative State Research Education and Extension Service, grant number 2008-35204-04634 awarded to DY.
References
- Boutin J.A. Myristoylation. Cell. Signal. 1997;9:15–35. doi: 10.1016/s0898-6568(96)00100-3. [DOI] [PubMed] [Google Scholar]
- Cavanagh D. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch. Virol. 1997;142:629–633. [PubMed] [Google Scholar]
- Chow M., Newman J.F., Filman D., Hogle J.M., Rowlands D.J., Brown F. Myristylation of picornavirus capsid protein VP4 and its structural significance. Nature. 1987;327:482–486. doi: 10.1038/327482a0. [DOI] [PubMed] [Google Scholar]
- Cowley J.A., Dimmock C.M., Spann K.M., Walker P.J. Gill-associated virus of Penaeus monodon prawns: an invertebrate virus with ORF1a and ORF1b genes related to arteri- and coronaviruses. J. Gen. Virol. 2000;81:1473–1484. doi: 10.1099/0022-1317-81-6-1473. [DOI] [PubMed] [Google Scholar]
- Faaberg K.S., Even C., Palmer G.A., Plagemann P.G. Disulfide bonds between two envelope proteins of lactate dehydrogenase-elevating virus are essential for viral infectivity. J. Virol. 1995;69:613–617. doi: 10.1128/jvi.69.1.613-617.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbiati F., Guzzi F., Magee A.I., Milligan G., Parenti M. Chemical inhibition of myristoylation of the G-protein Gi1 alpha by 2-hydroxymyristate does not interfere with its palmitoylation or membrane association. Evidence that palmitoylation, but not myristoylation, regulates membrane attachment. Biochem. J. 1996;313:717–720. doi: 10.1042/bj3130717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González J.M., Gomez-Puertas P., Cavanagh D., Gorbalenya A.E., Enjuanes L. A comparative sequence analysis to revise the current taxonomy of the family Coronaviridae. Arch. Virol. 2003;148:2207–2235. doi: 10.1007/s00705-003-0162-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper D.R., Gilbert R.L., Blunt C., McIlhinney R.A. Inhibition of varicella-zoster virus replication by an inhibitor of protein myristoylation. J. Gen. Virol. 1993;74:1181–1184. doi: 10.1099/0022-1317-74-6-1181. [DOI] [PubMed] [Google Scholar]
- Harris M., Hislop S., Patsilinacos P., Neil J.C. In vivo derived HIV-1 nef gene products are heterogeneous and lack detectable nucleotide binding activity. AIDS Res. Hum. Retroviruses. 1992;8:537–543. doi: 10.1089/aid.1992.8.537. [DOI] [PubMed] [Google Scholar]
- Hayashi N., Matsubara M., Jinbo Y., Titani K., Izumi Y., Matsushima N. Nef of HIV-1 interacts directly with calciumbound calmodulin. Protein Sci. 2002;11:529–537. doi: 10.1110/ps.23702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hearps A.C., Jans D.A. Regulating the functions of the HIV-1 matrix protein. AIDS Res. Hum. Retroviruses. 2007;23:341–346. doi: 10.1089/aid.2006.0108. [DOI] [PubMed] [Google Scholar]
- Henderson L.E., Benveniste R.E., Sowder R., Copeland T.D., Schultz A.M., Oroszlan S. Molecular characterization of gag proteins from simian immunodeficiency virus (SIVMne) J. Virol. 1988;62:2587–2595. doi: 10.1128/jvi.62.8.2587-2595.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James G., Olson E.N. Fatty acylated proteins as components of intracellular signaling pathways. Biochemistry. 1990;29:2623–2634. doi: 10.1021/bi00463a001. [DOI] [PubMed] [Google Scholar]
- Lee C., Yoo D. Cysteine residues of the porcine reproductive and respiratory syndrome virus small envelope protein are non-essential for virus infectivity. J. Gen. Virol. 2005;86:3091–3096. doi: 10.1099/vir.0.81160-0. [DOI] [PubMed] [Google Scholar]
- Lee C., Yoo D. The small envelope protein of porcine reproductive and respiratory syndrome virus possesses ion channel protein-like properties. Virology. 2006;355:30–43. doi: 10.1016/j.virol.2006.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Lescar J., Tam J.P., Liu D.X. Expression of SARS-coronavirus envelope protein in Escherichia coli alters membrane permeability. Biochem. Biophys. Res. Commun. 2004;325:374–380. doi: 10.1016/j.bbrc.2004.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liemann S., Chandran K., Baker T.S., Nibert M.L., Harrison S.C. Structure of the reovirus membranepenetration protein, Mu1, in a complex with its protector protein, Sigma3. Cell. 2002;108:283–295. doi: 10.1016/s0092-8674(02)00612-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madan V., de García M.J., Sanz M.S., Carrasco L. Viroporin activity of murine hepatitis virus E protein. FEBS Lett. 2005;579:3607–3612. doi: 10.1016/j.febslet.2005.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardassi H., Massie B., Dea S. Intracellular synthesis, processing, and transport of proteins encoded by ORFs 5 to 7 of porcine reproductive and respiratory syndrome virus. Virology. 1996;221:98–112. doi: 10.1006/viro.1996.0356. [DOI] [PubMed] [Google Scholar]
- Meulenberg J.J., Hulst M.M., de Meijer E.J., Moonen P.J.M., den Besten A., De Kluyver E.P., Wensvoort G., Moormann R.J. Lelystad virus, the causative agent of porcine epidemic abortion and respiratory syndrome (PEARS), is related to LDV and EAV. Virology. 1993;192:62–72. doi: 10.1006/viro.1993.1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimchuk Z., Marois E., Kjemtrup S., Leister R.T., Katagiri F., Dangl J.L. Eukaryotic fatty acylation drives plasma membrane targeting and enhances function of several type III effector proteins from Pseudomonas syringae. Cell. 2000;101:353–363. doi: 10.1016/s0092-8674(00)80846-6. [DOI] [PubMed] [Google Scholar]
- Perez M., Greenwald D.L., de la Torre J.C. Myristoylation of the RING finger Z protein is essential for arenavirus budding. J. Virol. 2004;78:11443–11448. doi: 10.1128/JVI.78.20.11443-11448.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persing D.H., Varmus H.E., Ganem D. The preS1 protein of hepatitis B virus is acylated at its amino terminus with myristic acid. J. Virol. 1987;61:1672–1677. doi: 10.1128/jvi.61.5.1672-1677.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saad J.S., Miller J., Tai J., Kim A., Ghanam R.H., Summers M.F. Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly. Proc. Natl. Acad. Sci. U.S.A. 2006;103:11364–11369. doi: 10.1073/pnas.0602818103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits S.L., Lavazza A., Matiz K., Horzinek M.C., Koopmans M.P., de Groot R.J. Phylogenetic and evolutionary relationships among torovirus field variants: evidence for multiple intertypic recombination events. J. Virol. 2003;77:9567–9577. doi: 10.1128/JVI.77.17.9567-9577.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Dobbe J.C., Spaan W.J. Heterodimerization of the two major envelope proteins is essential for arterivirus infectivity. J. Virol. 2003;77:97–104. doi: 10.1128/JVI.77.1.97-104.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Meulenberg J.J. The molecular biology of arteriviruses. J. Gen. Virol. 1998;79:961–979. doi: 10.1099/0022-1317-79-5-961. [DOI] [PubMed] [Google Scholar]
- Snijder E.J., van Tol H., Pedersen K.W., Raamsman M.J., de Vries A.A. Identification of a novel structural protein of arteriviruses. J. Virol. 1999;73:6335–6345. doi: 10.1128/jvi.73.8.6335-6345.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streuli C.H., Griffin B.E. Myristic acid is coupled to a structural protein of polyoma virus and SV40. Nature. 1987;326:619–622. doi: 10.1038/326619a0. [DOI] [PubMed] [Google Scholar]
- Thaa B., Kabatek A., Zevenhoven-Dobbe J.C., Snijder E.J., Herrmann A., Veit M. Myristoylation of the arterivirus E protein: the fatty acid modification is not essential for membrane association but contributes significantly to virus infectivity. J. Gen. Virol. 2009;90:2704–2712. doi: 10.1099/vir.0.011957-0. [DOI] [PubMed] [Google Scholar]
- Towler D.A., Adams S.P., Eubanks S.R., Towery D.S., Jackson-Machelski E., Glaser L., Gordon J.I. Purification and characterization of yeast myristoyl CoA:protein N-myristoyltransferase. Proc. Natl. Acad. Sci. U.S.A. 1987;84:2708–2712. doi: 10.1073/pnas.84.9.2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieringa R., De Vries A.A., Post S.M., Rottier P.J. Intra- and intermolecular disulfide bonds of the GP2b glycoprotein of equine arteritis virus: relevance for virus assembly and infectivity. J. Virol. 2003;77:12996–13004. doi: 10.1128/JVI.77.24.12996-13004.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieringa R., de Vries A.A., Rottier P.J. Formation of disulfidelinked complexes between the three minor envelope glycoproteins (GP2b, GP3, and GP4) of equine arteritis virus. J. Virol. 2003;77:6216–6226. doi: 10.1128/JVI.77.11.6216-6226.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox C., Hu J.S., Olson E.N. Acylation of proteins with myristic acid occurs cotranslationally. Science. 1987;238:1275–1278. doi: 10.1126/science.3685978. [DOI] [PubMed] [Google Scholar]
- Wilson L., Gage P., Ewart G. Hexamethylene amiloride blocks E protein ion channels and inhibits coronavirus replication. Virology. 2006;353:294–306. doi: 10.1016/j.virol.2006.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson L., McKinlay C., Gage P., Ewart G. SARS coronavirus E protein forms cation-selective ion channels. Virology. 2004;330:322–331. doi: 10.1016/j.virol.2004.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissink E.H., Kroese M.V., van Wijk H.A., Rijsewijk F.A.M., Meulenberg J.J., Rottier P.J. Envelope protein requirements for the assembly of infectious virions of porcine reproductive and respiratory syndrome virus. J. Virol. 2005;79:12495–12506. doi: 10.1128/JVI.79.19.12495-12506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolven A., Okamura H., Rosenblatt Y., Resh M.D. Palmitoylation of p59fyn is reversible and sufficient for plasma membrane association. Mol. Biol. Cell. 1997;8:1159–1173. doi: 10.1091/mbc.8.6.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wootton S.K., Yoo D., Rogan D. Full-length sequence of a Canadian porcine reproductive and respiratory syndrome virus (PRRSV) isolate. Arch. Virol. 2000;145:2297–2323. doi: 10.1007/s007050070022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W.H., Fang Y., Farwell R., Steffen-Bien M., Rowland R.R., Christopher-Hennings J., Nelson E.A. A 10-kDa structural protein of porcine reproductive and respiratory syndrome virus encoded by ORF2b. Virology. 2001;287:183–191. doi: 10.1006/viro.2001.1034. [DOI] [PubMed] [Google Scholar]
- Zacharias D.A., Violin J.D., Newton A.C., Tsien R.Y. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 2002;296:913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- Zha J., Weiler S., Oh K.J., Wei M.C., Korsmeyer S.J. Posttranslational N-myristoylation of BID as a molecular switch for targeting mitochondria and apoptosis. Science. 2000;290:1761–1765. doi: 10.1126/science.290.5497.1761. [DOI] [PubMed] [Google Scholar]


