Abstract
Yellow head virus (YHV) is a pathogen of the black tiger shrimp (Penaeus monodon) and, with gill-associated virus (GAV), is one of two known invertebrate nidoviruses. We describe sequences of the large replicase gene (ORF1a) and 5′- and 3′-terminal UTRs, completing the 26,662 nt sequence of the YHV genome. ORF1a (12,219 nt) encodes a ∼462,662 Da polypeptide containing a putative 3C-like protease and a putative papain-like protease with the canonical C/H catalytic dyad and α + β fold. The read-through pp1ab polyprotein contains putative uridylate-specific endoribonuclease and ribose-2′-O-methyl transferase domains, and an exonuclease domain incorporating unusual dual Zn2+-binding fingers. Upstream of ORF1a, the 71 nt 5′-UTR shares 82.4% identity with the 68 nt 5′-UTR of GAV. The 677 nt 3′-terminal region contains a single 60 nt ORF, commencing 298 nt downstream of ORF3, that is identical to N-terminal coding region of the 249 nt GAV ORF4. Northern blots using RNA from YHV-infected shrimp and probes directed at ORF1a, ORF1b, ORF2 and ORF3 identified a nested set of 3′-coterminal RNAs comprising the full-length genomic RNA and two sub-genomic (sg) mRNAs. Intergenic sequences upstream of ORF2 and ORF3 share high identity with GAV, particularly in the conserved domains predicted to mediate sgmRNA transcription.
Keywords: YHV, Genome sequence, Transcription, Shrimp, Penaeus monodon
1. Introduction
Yellow head virus (YHV) is a highly virulent pathogen of the black tiger shrimp (Penaeus monodon). It can cause rapidly accumulating mortalities in aquaculture ponds, resulting in total crop loss within several days of the first signs of disease. Yellow head disease first emerged in central Thailand in 1990 (Limsuwan, 1991). It has since been reported in most major shrimp producing countries in Asia and remains a disease of serious concern globally (Walker et al., 2001, OIE, 2007). YHV is now known to be one of several closely related genotypes. These include gill-associated virus (GAV), a less virulent virus that was first detected in Australia in association with a condition named mid-crop mortality syndrome, and at least four other genotypes identified only in healthy P. monodon shrimp from Southeast Asia, India and East Africa (Walker et al., 2001, Wijegoonawardane et al., 2004, Wijegoonawardane et al., 2008).
YHV is a positive-sense (+) ssRNA virus that is currently classified with GAV as a member of the species Gill-associated virus, genus Okavirus, family Roniviridae in the order Nidovirales (Walker et al., 2005). The enveloped virions are rod-shaped (approximately 70 nm × 180 nm) with prominent surface projections and an internal helical nucleocapsid (Wongteerasupaya et al., 1995, Nadala et al., 1997). Virions contain three structural proteins comprising two transmembrane glycoproteins (gp116 and gp64) and a nucleoprotein (p20) (Jitrapakdee et al., 2003, Sittidilokratna et al., 2006). For GAV, the 26,235 nt genome has been shown to be 3′-polyadenylated, and contain five open reading frames (ORFs) bounded by 5′- and 3′-UTRs. ORF1a and ORF1b are expressed from genome-length mRNA and overlap (Cowley et al., 2000, Cowley et al., 2002). ORF1a encodes a long polyprotein (pp1a) that includes a 3C-like protease domain. ORF1b is translated only following a −1 ribosomal frame-shift at a predicted pseudoknot structure in the ORF overlap region and encodes enzymes of the replication complex (Cowley et al., 2000, Ziebuhr et al., 2003). GAV ORF2 and ORF3 are expressed from sub-genomic (sg)mRNAs and encode the nucleoprotein and the virion envelope glycoprotein, respectively (Cowley and Walker, 2002, Cowley et al., 2004). Downstream of ORF3 is a 638 nt region which includes a 249-nt open reading frame (ORF4) that has been reported to be expressed at low level in GAV-infected cells (Dhar et al., 2004).
The YHV genome is less well characterized. ORF1b has been sequenced and shown to encompass elements homologous to the GAV ribosomal frame-shift site. It shares 80.5% overall nucleotide sequence identity with GAV ORF1b and encodes a polypeptide containing recognised replicase functional domains (Sittidilokratna et al., 2002, Snijder et al., 2003). ORF2 has been shown to encode the YHV nucleoprotein and shares 79.0% nucleotide sequence identity with the cognate GAV gene (Sittidilokratna et al., 2006). YHV glycoproteins gp116 and gp64 are encoded in ORF3 and have been shown to be generated by post-translational proteolytic cleavage of a precursor pp3 polyprotein (Jitrapakdee et al., 2003).
In this paper, we report sequences of the 12,219 nt ORF1a gene and the 5′- and 3′-UTRs that complete the YHV genome. We identify for the first time in roniviruses a papain-like cysteine protease in ORF1a, confirm an unusual dual Zn2+-binding exonuclease in ORF1b, and show that the short open reading frame corresponding to GAV ORF4 is severely truncated and unlikely to be functional in YHV. We also clearly demonstrate that, like GAV and other nidoviruses, the YHV transcription strategy employs a nested set of genomic and sub-genomic RNAs.
2. Materials and methods
2.1. Source of virus
The YHV isolate used in this study was obtained from moribund P. monodon shrimp showing signs of yellow head disease that were collected from a farm in Chachoengsao Province, Thailand, in July 1998 (Sittidilokratna et al., 2002). A reference stock of the virus was prepared following experimental passage in P. monodon by methods described previously and stored at −80 °C in aliquots until required (Sittidilokratna et al., 2002, Sittidilokratna et al., 2006).
2.2. RT-PCR amplification and nucleotide sequencing
Genomic RNA was isolated from purified YHV particles using Trizol reagent (Invitrogen) and used as the template for cDNA synthesis and PCR amplification as described previously (Sittidilokratna et al., 2002). The GAV ORF1a sequence (GenBank accession number AF227196) was initially used to design 10 sets of PCR primers to amplify overlapping regions covering the entire ORF1a gene. Four of the primer sets amplified DNA products which were gel purified and sequenced. Sequences obtained from these products and the 5′-terminal genome region amplified by 5′-RACE (see below) were then used to design four sets of YHV-specific primers (Table 1 ) to amplify products spanning the remaining regions of ORF1a. The Superscript One-Step RT-PCR kit (Life Technologies) was used according to the manufacturer's recommended procedure with minor modifications. Reactions (25 μl) contained the kit reagents plus 0.5 μg YHV RNA, 0.4 μM primers and 8 U RNasin (Promega). To amplify DNA products larger than 3 kb, additional (1 μl) Elongase Enzyme mix (Invitrogen) was added to the reaction. RT-PCR was performed in a PerkinElmer 2400 thermal cycler using the conditions 50 °C/30 min and 94 °C/2 min for reverse transcription and enzyme inactivation followed by 35 cycles of 94 °C/20 s, 55 °C/30 s and 68 °C/210 s, and a final extension of 72 °C/10 min. DNA products obtained were gel purified and sequenced directly using each of the PCR primers. Additional primers designed to sequences obtained were used to extend the sequences in both directions and confirm that obtained in other direction.
Table 1.
PCR primers used to amplify YHV ORF1a segments
| Primer name | Sequence (5′–3′) | Product (kb) |
|---|---|---|
| 2S001 | ACTGTTCTGTCTGCCGTCAGA | 3.8 |
| 2A018 | ACTGTCTGTACGGTGTGAGA | |
| 2S022 | CACACTTCTCCTCACAGTCA | 3.3 |
| 2A004a | CATGTGTAGACGGGATGTTG | |
| 2S013 | CACGGCATCGAACAATCTGG | 3.9 |
| 2A021 | GTCATTCGTGCTAGCAAGAG | |
| 2S010 | GCTCAACTATGTCACCGGTG | 3.2 |
| 2A008 | GGTAGCGGTTTACTGGAAGA | |
The sequence of the YHV genome from the 3′-end of ORF1b gene to the 3′-polyA tail was obtained from a 6.7 kb RT-PCR product amplified using an ORF1b-specific primer (5′-GATCGGGGTACCTAAGCTTATGCTATCGACCTA-3′) in combination with a primer (5′-TCTAGAGGATCCCCGGTACCTTTTTTTTTTTTTTTTTTTT-3′) designed to extend from the polyA tail. The amplification and partial sequence analysis of this product has been described previously (Jitrapakdee et al., 2003, Sittidilokratna et al., 2006). The entire sequence of the 6.7 kb product was determined by primer walking in both directions.
2.3. 5′-RACE
The 5′-terminal sequence of the YHV genome was determined by 5′-RACE (Dumas et al., 1991) using the complementary anchor-PCR primers 156 and 2668 as described previously (Walker et al., 1994, Cowley et al., 2002). Briefly, cDNA was synthesized using ∼2 μg RNA isolated from purified YHV, 150 ng either anti-sense primer GAV94 (5′-CGTGGTGTGATCATAGTCCTT-3′) or GAV95 (5′-CTAAGCTCTGGAGGTTGATCAT-3′) designed to sequences located near to the GAV genome 5′-terminus and 200 U SuperScript II RT (Invitrogen) according to the manufacturer's protocol. After incubation at 42 °C for 1 h, RNA was digested with 1.5 U RNase H at 37 °C for 1 h and excess primer was removed using a Sephadex S400-HR column (Pharmacia). Primer 156 (0.2 μg) that had been 5′-phosphorylated using [γ-32P]ATP and T4 polynucleotide kinase (Promega) was ligated to 3′-cDNA ends using T4 RNA ligase (New England Biolabs). Excess primer was removed using a S400-HR column and the cDNA was amplified by PCR using primer 2668 in combination with primer GAV96 (5′-TCTGACGGCAGACAGAACAGT-3′) designed to a GAV sequence upstream of primers GAV94 and GAV95, Taq DNA polymerase and buffer (Promega) containing 2.5 mM MgCl2 and 40 cycles of 95 °C/30 s, 54 °C/30 s and 72 °C/45 s. The ∼200 bp PCR products were purified using QIAquick columns, cloned into pGEM-T vector and sequenced.
2.4. Sequence analysis
Nucleotide sequencing was performed at the Bio Service Unit, National Center for Genetic Engineering and Biotechnology, National Science and Technology Development Agency (NSTDA), Thailand. Overlapping contigs were compiled using SeqEd 1.0.3 software (ABI, 1992). ORF Finder and BLAST 2.0 (Altschul et al., 1997) accessed at http://www.ncbi.nlm.nih.gov were used to identify open reading frames and in database sequence similarity searches. Amino acid sequences were translated and analyzed using EMBOSS Transeq and pI/MW Tool software available at http://au.expasy.org. Hydrophobic domains were predicted by the TMPred method (Hofmann and Stoffel, 1993) available at http://www.EMBnet.ch. ClustalX 1.8 was used for pair-wise and multiple sequence alignments (Thompson et al., 1997). Secondary structure predictions were conducted using the PHDsec program accessed via the PredictProtein server (Rost and Sander, 1993, Rost et al., 2004) at http://www.predictprotein.org.
2.5. RNA extraction and northern blot hybridization
Lymphoid organ tissue from specific pathogen-free (SPF) P. monodon was kindly provided by Prof. Chu Fang Lo, National Taiwan University, Taipei. Lymphoid organ tissue was also obtained from ∼20 g P. monodon injected into the third abdominal segment with 100 μl YHV inoculum (Sittidilokratna et al., 2002) and sampled at 24, 36, 48 and 60 h post-infection. Lymphoid organ tissue was collected into RNAlater (Ambion) that was placed at 4 °C overnight before being stored at −20 °C. Total RNA was isolated from lymphoid organ tissue using Trizol reagent (Invitrogen), resuspended in DEPC-treated water and stored at −80 °C until required. Only those RNA preparations with an OD260/280 ratio >1.5 were used for northern blot hybridization.
Lymphoid organ total RNA (4 μg) from uninfected and YHV-infected shrimp was denatured in formamide–formaldehyde solution (60% formamide, 4 M formaldehyde, 2 mM EDTA, 40 mM HEPES, pH 7.4) at 70 °C for 10 min and resolved by electrophoresis at 50 ± 2 °C in a 1.2% agarose-TAE gel as described by Almeida et al. (2000). The denatured RNA was then transferred to a 0.45-μm Biodyne® B membrane (Pall Corporation) using 50 mM NaOH and downward capillary flow (Ingelbrecht et al., 1998). After washing briefly in 2× SSC, membranes were pre-hybridized in UltraHyb solution (Ambion) in a shaking water-bath at 42 °C for 3 h. Digoxygenin (DIG)-labeled DNA probes were generated by PCR using the primers shown in Table 2 and the DIG-DNA Labeling Mix (Roche) as described by Cowley et al. (2002) but with a modified cycling conditions of 94 °C/5 min, 35 cycles of 94 °C/15 s, 55 °C/30 s and 72 °C/30 s, and final extension of 72 °C/7 min. DIG-labeled probes were denatured at 100 °C for 10 min and then incubated with membranes at 42 °C in a shaking water-bath for 16–18 h. Following hybridization, membranes were washed individually twice at 42 °C for 15 min in 2 × SSC, 0.1% SDS and twice for 15 min in 0.5 × SSC, 0.1% SDS. DIG-labeled DNA probe hybridized to RNA was detected by chemiluminescence using 1:2000 anti-DIG-alkaline phosphatase Fab fragment and 1:100 CDP-Star reagent (Roche) according to the manufacturer's instructions.
Table 2.
PCR primers used to amplify DIG-labeled DNA probes
| Target ORF | Primer | Sequence | Probe length | Genome position |
|---|---|---|---|---|
| ORF1a | 2S026 | 5′-GCCAAATCGACTACGATAGAT-3′ | 346 | G3640-T3985 |
| 2A018 | 5′-TCTCACACCGTACAGACAGT-3′ | |||
| ORF1b | 2S28 | 5′-AAGGTACCGACAACAAGAGCC-3′ | 354 | A18156-G18509 |
| 2A34 | 5′-ACCATCCCATCTGGTACACAG-3′ | |||
| ORF2 | NS6 | 5′-ATGCCTCGTCGTCGCCTACC-3′ | 366 | A20490-T20855 |
| NA10 | 5′-ATCGTTCCCGATCCTTCCCTT 3′ | |||
| ORF3 | YS7 | 5′-GTTACAGTAGATCAACGTTGC-3′ | 313 | G22299-T22611 |
| YA4 | 5′-TGCATCCACACGTCACGATT-3′ | |||
3. Results
3.1. Nucleotide sequence of the YHV genome
The complete nucleotide sequence of the YHV genome (Chachoengsao 1998 isolate) was compiled from ORF1a and 5′- and 3′-terminal sequences described here and ORF1b, ORF2, ORF3 and intergenic sequences reported previously (Sittidilokratna et al., 2002, Sittidilokratna et al., 2006, Jitrapakdee et al., 2003). The (+) ssRNA genome is 26,662 nt in length excluding the polyA tail (GenBank accession number EU487200). The genome organization is illustrated in Fig. 1 . The 71 nt 5′-UTR shares 82.4% nucleotide sequence identity with the 68 nt 5′-UTR of GAV, including a 5′-terminal stretch of 20 identical nucleotides (Fig. 2a). The 12,219 nt ORF1a overlaps ORF1b by 37 nt, terminating at a site 30 nt downstream of a ‘slippery’ sequence (AAAUUUU) at which a −1 ribosomal frame-shift is predicted to allow read-through translation of ORF1b to generate the large pp1ab replicase polyprotein (Sittidilokratna et al., 2002). Overall identity between ORF1a sequences of YHV and GAV is 79.0%. The sequences of ORF1b (7887 nt) encoding the replicase component of the pp1ab replicase, ORF2 (441 nt) encoding the nucleoprotein, ORF3 (4998 nt) encoding the membrane glycoproteins, and the intervening UTRs have been described in detail elsewhere (Sittidilokratna et al., 2002, Sittidilokratna et al., 2006, Jitrapakdee et al., 2003).
Fig. 1.
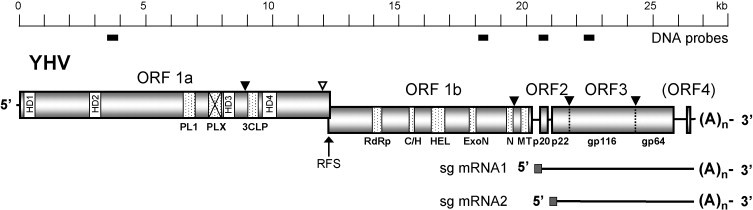
Schematic representation of the 26,662 nt polyadenylated genome of YHV and transcribed (sub-genomic) sgmRNA1 and sgmRNA2. Functional domains in ORF1a: hydrophobic regions (HD1-HD4), 3C-like protease (3CLP), papain-like protease (PLP1) and a domain with homology to PLP1 but lacking the canonical α + β fold of papain-like proteases (PLPX). Functional domains in ORF1b: RNA polymerase (RdRp), cysteine- and histidine-rich domain (C/H), helicase (HEL), exoribonuclease (ExoN), uridylate-specific endoribonuclease (N) and ribose-O-methyl transferase (MT). ORF2 encodes the nucleoprotein (p20). ORF3 encodes a polyprotein (pp3) is processed post-translation to generate envelope glycoproteins (gp116 and gp64) and an N-terminal triple-membrane-spanning fragment of unknown function (p22). The ribosomal frame-shift site (RFS) allows read-through translation of polyprotein pp1ab from ORF1a and ORF1b. Known (▾) and likely (▿) sites of proteolytic cleavage of expressed polyproteins (pp1a, pp1ab and pp3) are indicated. The locations of DNA probes used for northern blot hybridizations are indicated below the scale bar.
Fig. 2.

ClustalX alignments of YHV and GAV genomic sequences and deduced amino acid sequences. (A) 5′-terminal regions up to the commencement of ORF1a and (B) 3′-terminal regions from the start of ORF4 to the polyA tail. Potential initiation and termination codons are indicated in bold face and underlined. Multiple termination codons in all reading frames beyond the point of truncation of YHV ORF4 are indicated. Identical nucleotides in aligned sequences are indicated (*).
The 3′-terminal region of the YHV genome from the ORF3 termination codon to the polyA tail is 677 nt in length. The region contains a single open reading frame of 60 nt that commences 298 nt downstream of the ORF3 termination codon and displays 100% nucleotide sequence identity with the first 60 nt of the putative 249 nt ORF4 in GAV. Downstream of the termination codon of this severely truncated YHV open reading frame, a relatively high level of nucleotide identity (79.5%) is maintained with the GAV ORF4 sequence. However, the YHV sequence is punctuated by a series of additions, deletions and substitutions that introduce multiple termination codons in all three reading frames (Fig. 2b). The 3′-terminal region of the YHV genome, downstream of the sequence corresponding to ORF4 in GAV, comprises 131 nt and shares 92.4% sequence identity with the 129 nt terminal region of GAV, including perfect identity of the 18 nucleotides immediately preceding the polyA tail.
In GAV, the intergenic regions upstream of ORF2 and ORF3 contain a stretch of 16 identical nucleotides that includes a common sequence motif (5′-ACAACCU) corresponding to the 5′-terminus of sub-genomic (sg)mRNAs 1 and 2 (Cowley et al., 2002). As shown in Fig. 3 , there is a high level of nucleotide sequence conservation between YHV and GAV in these intergenic regions and the sgmRNA 5′-terminal sequence motif is invariant. In GAV, the region upstream of putative ORF4 contains a similar sequence including a motif corresponding to the sgmRNA 5′-terminal sequence in which the 5′-terminal position is corrupted by an A > U transversion (i.e., 5′-UCAACCU) (Cowley et al., 2002). In YHV, this region shares almost 100% nucleotide sequence identity with GAV but the sgmRNA 5′-terminal sequence motif upstream of the truncated ORF4 is corrupted by an A > G transition (i.e., GCAACCU), rather than the A > U transversion (Fig. 3).
Fig. 3.
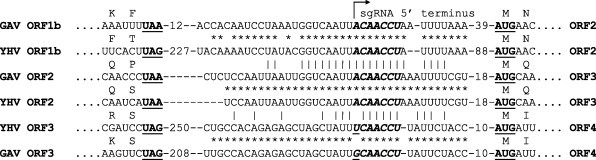
Conserved sequences in the intergenic regions upstream of ORF2, ORF3 and ORF4 in YHV and GAV. The initiation and termination codons of flanking ORFs are indicated in bold face and underlined. The putative 5′-terminal sequence (5′ ACAACC…) of ORF2 and ORF3 sgmRNA are indicated in bold italic face. Base substitutions in similar sequences in the intergenic region upstream of ORF4 are underlined. Identical nucleotides in aligned cognate YHV and GAV intergenic sequences (*) and aligned sequences of the different intergenic regions (|) are indicated.
3.2. Deduced amino acid sequences of YHV pp1a and pp1ab polyproteins
YHV ORF1a encodes a polypeptide (pp1a) of 4072 amino acids with a calculated molecular mass of 462,662.48 Da. Assuming a ribosomal frame-shift occurs at the conserved slippage site identified in GAV within the ORF1a/ORF1b sequence overlap (Cowley et al., 2000, Sittidilokratna et al., 2002), translation through YHV ORF1b would result in an extended polyprotein (pp1ab) of 6688 amino acids with a molecular mass of 762,017.85 Da. There is a high level of overall amino acid sequence identity between YHV and GAV pp1a (82.4% identity) and pp1ab (84.9% identity), with sequence variations distributed relatively evenly throughout each polyprotein. YHV pp1a is 15 amino acids longer than GAV pp1a due to insertions of stretches of up to 3 amino acids that occur primarily in the N-terminal third of the protein and a C-terminal deletion of 3 amino acids beyond the ribosomal frame-shift site. YHV pp1ab is also 15 amino acids longer than GAV pp1ab due to an insertion of an additional stretch of 3 amino acids (Sittidilokratna et al., 2002).
Nidovirus polyproteins pp1a and pp1ab (also known as the replicase proteins) are processed post-translation to generate elements of the viral replication complex (Ziebuhr et al., 2000). Autolytic cleavage of the polyproteins occurs via a 3C-like ‘main’ protease (named after the picornavirus 3C proteases) located within the C-terminal third of pp1a, and one or more paralogous papain-like ‘accessory’ proteases located within the N-terminal half of pp1a. Analysis of the YHV pp1a sequence identified a 3C-like protease domain between I2866 and G3006. This domain shares a high level of amino acid sequence identity (87.9%) with the corresponding domain described previously for GAV (Cowley et al., 2000), including a core of key residues that are conserved across coronaviruses and arteriviruses (Fig. 4A). As reported previously for GAV, the YHV 3C-like protease employs the H/C catalytic dyad that is characteristic of coronaviruses, rather than the H/S dyad of the arteriviruses and toroviruses (Fig. 4A). Two proteolytic cleavage sites identified previously in GAV pp1a and pp1ab are highly conserved in the YHV sequence. These are located at 2838LVTHE↓VNTGN2847 and 6455KVNHE↓LYHVA6464 in pp1ab and conform to the proposed substrate consensus sequence, VxHE↓(L,V) (Ziebuhr et al., 2003).
Fig. 4.

Amino acid sequence alignments of conserved functional domains in nidovirus pp1a and pp1ab polyproteins. (A) 3C-like protease (3CLP) domain; (B) uridylate-specific endoribonuclease (XendoU) domain; (C) exonuclease (ExoN) domain; (D) ribose-2′-O-methyl transferase (2′-O-MT) domain. The alignments include cognate sequences in the roniviruses YHV and GAV, SARS coronavirus (SARS), porcine reproductive and respiratory syndrome arterivirus (PRRSV), bovine torovirus (BToV) and the white bream bafnivirus (WBV). Absolutely conserved (*) and similar (: or .) amino acids are indicated according to the similarity groups defined in ClustalX. YHV amino acids identical in at least three of the aligned sequences are highlighted by light shadowing. Conserved cysteine and histidine residues predicted to form Zn2+-binding fingers in the ExoN domain are indicated (+) with dark shadowing. Alignments were generated using ClustalX and adjusted to optimize sequence similarities.
In overall amino acid sequence, nidovirus accessory protease domains are poorly conserved (Ziebuhr et al., 2000). However, they share similarities with papain-like cysteine proteases of other positive-sense RNA viruses that include variations of a characteristic α + β fold and a C/H catalytic dyad in which the Cys residue is usually followed immediately by a large aromatic residue (Tyr or Trp). In coronaviruses and arteriviruses, the accessory proteases also feature, within the framework of the papain-like fold, a Zn2+-binding finger that is essential for proteolytic activity and appears also to function as a transcription factor in sgmRNA synthesis (Herold et al., 1999, Tijms et al., 2001, Ziebuhr, 2005). Scanning of the YHV pp1a sequence identified four regions in which a CY/W motif is followed by H at a suitable distance (70–180 amino acids) downstream. However, only one of the four possible sites (C2102/H2281) is predicted to adopt a secondary structure that features a α-helix domain downstream of the catalytic Cys residue, followed by a β-sheet domain that incorporates the catalytic His residue (Fig. 5A). The sequence of this domain (designated YHV PLP1) is only moderately conserved in GAV pp1a (73.7% identity). However, the C/H catalytic dyad is preserved and a similar α + β fold is predicted. The domain is similar in length to the PLP1 and PLP2 domains of coronavirus pp1a polyproteins but, as illustrated in Fig. 5A for the PLP2 domain of mouse hepatitis virus (MHV), amino acid sequence similarity with papain-like proteases is weak and, significantly, like those of toroviruses, the domain lacks a Zn2+-binding finger. Approximately 120 amino acids downstream of YHV PLP1, and immediately upstream of hydrophobic domain (HD3) flanking the 3CL protease, there is a second region (C2461/H2607) that features the (CY…H) motif and shares striking amino acid sequence homology (50.8% similarity; 17.8% identity) with the PLP1 domain (Fig. 5B). However, in both YHV and GAV, this region (designated PLPX) lacks both the Zn2+-binding finger and the canonical α + β fold.
Fig. 5.
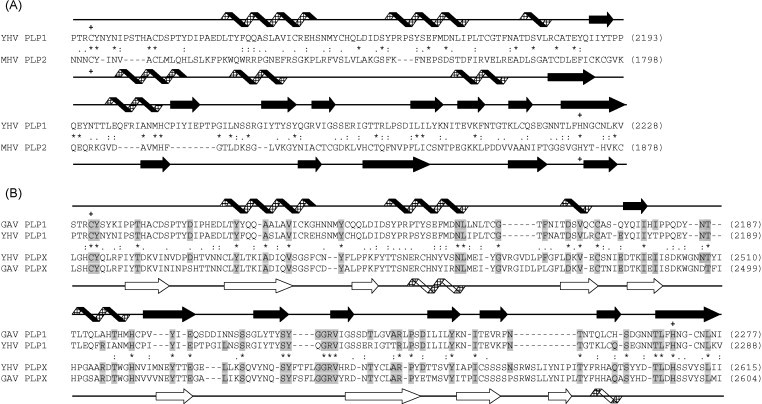
Amino acid sequence alignments of papain-like protease domains. (A) Alignment of YHV PLP1 domain with the PLP2 domain of coronavirus mouse hepatitis virus (MHV). (B) Alignment of YHV and GAV PLP1 domains with a downstream region of pp1a (PLPX) in which there is some recognizable similarity that may be indicative of sequence duplication. Amino acids identical in at least three of the sequences are indicated by light shadowing. The predicted C/H catalytic dyads in YHV and GAV PLP1 and MHV PLP2 are indicated (+). Absolutely conserved (*) and similar (: or .) amino acids are indicated according to the similarity groups defined in ClustalX. Secondary structure fold predictions generated by using the PHDsec program are indicated according to the assignment of α-helices (helix) or β-sheets (block arrow). Unfilled helicies and block arrows emphasize the deviation of PLPX domains from the papain-like α + β fold.
Other conserved catalytic domains identified in nidovirus pp1a polyproteins were not evident in either YHV or GAV. These include the ADP-ribose 1″-phosphatase (ADRP) domain that is adjacent to PLP domains in coronaviruses and toroviruses (Putics et al., 2006, Gorbelenya et al., 2006) and has recently also been reported in the fish bafnivirus (white bream virus) (Schultze et al., 2006), and the cyclic phosphodiesterase domain that has been identified in the C-terminal region of pp1a in toroviruses (Snijder et al., 1991, Draker et al., 2006). Analysis of the YHV and GAV pp1a sequences did identify four conserved hydrophobic domains (HD1–HD4). Hydrophobic domains contain transmembrane helixes that appear to anchor the nidovirus replication complex to intracellular membranes (Prentice et al., 2004). HD1 is located within the first 150 amino acids of pp1a and does not appear to occur in other nidoviruses; HD2 is located in the N-terminal third of the polyprotein, well upstream of the protease domains; HD3 and HD4 flank the 3C-like protease domain and correspond to those identified in other nidoviruses (Fig. 1).
Conserved domains in the ORF1b region of YHV pp1ab described previously include RdRP, helicase and Zn2+-binding domains, and conserved NTP-binding motifs A and B (Sittidilokratna et al., 2002). Sequence alignments shown in Fig. 4 also indicate that, downstream of the helicase domain, YHV pp1ab contains exonuclease (ExoN), uridylate-specific endoribonuclease (XendoU) and ribose-O-methyl transferase (2′-O-MT) domains that have recently been identified in GAV and other nidoviruses (Gorbelenya et al., 2006, Cowley and Walker, 2008). YHV and GAV share a high level of amino acid sequence identity in these domains and feature amino acids residues that have been reported to be conserved in other nidovirus pp1ab polyproteins. However, the ExoN domains of YHV and GAV are significantly larger than those of other nidoviruses due to a sequence insertion between conserved sequence blocks II and III that appears to provide a second Zn2+-binding finger (Fig. 4C). As observed previously, the second Zn-finger is absent from the ExoN domains of all nidoviruses for which pp1ab sequence is currently available, including the fish bafnivirus, white bream virus (Snijder et al., 2003).
3.3. Identification of YHV mRNAs
Total RNA was extracted from the lymphoid organs of normal (SPF) P. monodon shrimp and, at 3 d post-infection, from shrimp infected experimentally with YHV. The RNA was denatured in formamide–formaldehyde solution, resolved by agarose gel electrophoresis, blotted onto a nylon membrane, and hybridized with DIG-labeled probes targeted to regions in ORF1a, ORF1b, ORF2 and ORF3 (see Fig. 1). As shown in Fig. 6 , the probes targeting ORF1a and ORF1b each reacted with a discrete band of infected lymphoid organ RNA with a significantly larger mass than the 9 kb RNA marker. The ORF2 probe also reacted with the >9 kb RNA as well as a smaller discrete RNA of ∼6.5 kb. The ORF3 probe reacted with both the >9 kb and ∼6.5 kb RNAs, as well as a slightly smaller discrete RNA with a mass of ∼6.0 kb. No RNA from uninfected shrimp reacted with any of the probes. A probe targeted to ORF4 was also tested but no evidence was obtained of a ∼400–700-nt RNA initiating in the upstream UTR (data not shown).
Fig. 6.

Northern blot detection of YHV genomic and sub-genomic RNAs in infected shrimp cells. Total RNA was extracted from lymphoid organs of uninfected (u) or YHV-infected (i) shrimp, resolved by ‘hot’ agarose gel electrophoresis, transferred to positively charged nylon membranes and hybridized with DIG-labeled DNA probes corresponding to sequences in ORF1a, ORF1b, ORF2 or ORF3, as shown. RNAs hybridizing with DIG-labeled probes were detected by chemiluminescence using anti-DIG-alkaline phosphatase Fab fragment and CDP-Star reagent. The locations of RNA ladder markers are indicated and arrows indicate the locations of >9.0 kb YHV genomic RNA (gRNA), ∼6.5 kb sub-genomic mRNA (sgmRNA1) and ∼6.0 kb sub-genomic mRNA (sgmRNA2).
4. Discussion
We report here sequences that complete the 26,662 nt genome of YHV. We also show that YHV transcribes 3′-coterminal genomic and sub-genomic RNAs in infected shrimp cells. Together with previously published data, the analyses indicate that, like closely related GAV, the YHV genome organization and transcription strategy are consistent with its classification in the order Nidovirales (Cowley et al., 2000, Cowley et al., 2002, Sittidilokratna et al., 2002, Sittidilokratna et al., 2006, Jitrapakdee et al., 2003). Features shared with coronaviruses, toroviruses, arteriviruses and bafniviruses (Spaan et al., 2005, Schultze et al., 2006) include a polyadenylated (+) ssRNA genome, overlapping reading frames encoding elements of the viral replicase, a −1 ribosomal frame-shift site followed by a predicted RNA pseudoknot structure in the overlap region, a characteristic ‘SDD’ sequence motif at the polymerase active site, and the expression of the replicase and virion structural proteins from a nested set of 3′-coterminal genomic and sub-genomic mRNAs. We also show that the replicase functional domains in YHV are arranged similarly to those of other nidoviruses and include putative papain-like protease (PLP) and 3C-like protease (3CLP) domains in pp1a, and putative helicase, exonuclease (ExoN), uridylate-specific endoribonuclease (XendoU) and ribose-O-methyl transferase (2′-O-MT) domains in the downstream region of pp1ab. Other domains not identified in YHV include the ADRP that occurs coronaviruses, toroviruses and bafniviruses (Snijder et al., 2003, Draker et al., 2006, Putics et al., 2006, Gorbelenya et al., 2006, Schultze et al., 2006) and the cyclic phosphodiesterase that has been identified in pp1a of toroviruses and in the ORF2 product of some coronaviruses (Snijder et al., 1991). It is not yet clear whether these domains are totally absent from YHV pp1a or have diverged beyond the limits of recognizable sequence homology.
Nidovirus proteases encoded in pp1a and pp1ab include a ‘main’ 3C-like protease that processes the key downstream replicase domains, including the RdRp and helicase enzymes, and one or more papain-like ‘accessory’ proteases that recognize sites in the N-terminal half of the replicase polyproteins (Ziebuhr et al., 2000). YHV pp1a appears to contain a single PLP domain (PLP1) which was identified on the basis of the conserved C/H catalytic dyad and structural similarity to the characteristic α + β fold of other papain-like proteases (Herold et al., 1999). The catalytic dyad and fold structure are each preserved in the corresponding region of GAV pp1a in which there is relatively high amino acid sequence identity with YHV (74.7%, data not shown). Coronaviruses, arteriviruses and toroviruses may encode up to three papain-like cysteine proteases and it has been suggested that, during the course of nidovirus evolution, these domains may have been duplicated (Ziebuhr et al., 2000). The arteriviruses papain-like protease domains are located near the N-terminus of pp1a (den Boon et al., 1995). In coronaviruses, PLP domains are located in the central region of pp1a and are almost twice as long. Each adopts the canonical α + β fold and the C/H catalytic dyad is separated by a relatively short sequence similar to other viral papain-like proteases (Herold et al., 1999, Ziebuhr et al., 2000). Coronavirus and arterivirus PLPs commonly include a Zn2+-binding finger that connects the α and β domains of the papain-like fold and appears to have a role in regulating transcription (Herold et al., 1999, Tijms et al., 2001). In group 1 coronaviruses (e.g., HCoV-229E) and group 2a coronaviruses (e.g., MHV), there are two PLP domains (Ziebuhr et al., 2000). In group 3 coronaviruses (e.g., IBV) two domains are evident but the first (PLP1) is not active, and in SARS coronavirus, only a single PLP domain (PLP2) is present (Snijder et al., 2003). Toroviruses (e.g., EToV) also encode only a single short PLP domain that, although cysteine-rich, appears to lack a typical Zn2+-binding finger (Draker et al., 2006). In YHV and GAV, the size and location of the single PLP domain (PLP1) in pp1a is similar to coronaviruses but, like toroviruses, it lacks the central Zn2+-binding finger. A second downstream domain (PLPX) also features the (CY…H) motif but lacks both the canonical α + β fold and the Zn2+-binding finger (Fig. 5B) and is therefore unlikely to be functional as a papain-like protease. Nevertheless, the region is similar in size and sequence to the PLP1 domain and may have arisen by duplication of an ancestral PLP.
Another unusual feature of the YHV replicase is the presence of two Zn2+-binding fingers in the exonuclease (ExoN) domain of pp1ab (Gorbelenya et al., 2006; Fig. 4C). In coronaviruses and toroviruses, the ExoN domain displays homology to conserved catalytic residues of DEDD superfamily RNase homologues in yeast and bacteria but varies by the insertion of a single Zn2+-binding finger between conserved sequence blocks I and II (Snijder et al., 2003). In YHV (and GAV), this Zn2+-binding structure is preserved and a second Zn-finger lies between conserved blocks II and III. The DEDD superfamily of exoribonucleases is a subset of a much larger superfamily that includes the proofreading domains of many DNA polymerases as well as other DNA exonucleases (Bernad et al., 1989, Moser et al., 1997, Zuo and Deutscher, 2001). It has been suggested that nidovirus ExoN may also be involved in proofreading, allowing these large RNA genomes to avoid error catastrophe during replication (Snijder et al., 2003). Domains including dual Zn-finger motifs have been identified as essential to the exonuclease activity of some eukaryote and archaea DNA polymerases that appear to be involved in recognition and repair of DNA damage (Dua et al., 1998, Shen et al., 2003).
Northern blot detection of YHV RNA transcripts in infected shrimp cells identified three RNAs that were similar to those reported previously for GAV (Cowley et al., 2002) and appear to correspond to a nested set of full-length genomic mRNA (>9.0 kb band) and two sgmRNAs (∼6.5 and ∼6.0 kb bands) extending from conserved transcription regulatory sequences (TRS) immediately upstream of ORF2 and ORF3, respectively (Fig. 2). The sgmRNAs are slightly larger than those reported for GAV (∼6.0 and ∼5.5 kb) and this is consistent with the larger size of the YHV genome. As reported previously for GAV (Cowley et al., 2002), there is no evidence of a smaller YHV transcript from the corrupted TRS immediately upstream of the severely truncated ORF4.
YHV and GAV are each currently classified as the same virus species (Gill-associated virus) in the genus Okavirus, family Roniviridae, order Nidovirales (Walker et al., 2005). However, a consideration of the relative virulence, geographic distribution and host range, and the genome sequence data reported here, raises issues with respect to the possible taxonomic assignment of YHV as a separate virus species. YHV is one of the most significant pathogens of farmed shrimp, causing a disease that develops rapidly and often results in total crop loss within several days (Flegel et al., 1995). These characteristics distinguish YHV from GAV which, although associated mortalities in P. monodon in Australia, occurs commonly in healthy shrimp in Australasia and Asia and is far less virulent (∼106-fold by LD50) in experimental bioassays (Sittidilokratna and Walker, unpublished data). YHV and GAV share a similar genome organization (5′-ORF1a-ORF1b-ORF2-ORF3-(ORF4)-polyA-3′) and common strategies for gene expression that employ a nested set of genomic and sub-genomic mRNAs, a ribosomal frame-shift in the ORF1a/ORF1b overlap and proteolytic processing of polyproteins generated from both the ORF1a/ORF1b replicase and ORF3 glycoprotein genes. The level of nucleotide sequence identity between the genomes is ∼79% overall and varies in the range ∼74% in ORF3 to ∼82% in ORF1b. Amino acid sequence identity varies from approximately 73% in gp116 to 84% in pp1ab. Excluding the polyA tail, the YHV genome (26,662 nt) is somewhat larger than the GAV genome (26,235 nt), due to sequence insertions that occur primarily in several large blocks. Two of these are located in untranslated regions between ORF1b and ORF2 (259 nt insertion) and immediately following ORF3 (41 nt insertion). There are also multiple blocks of inserted sequence within ORF3 (76 nt total insertion) in the region encoding the hypervariable N-terminal domain of YHV gp116 and these significantly increase its calculated molecular mass (101.5 kDa prior to glycosylation), compared to GAV gp116 (98.1 kDa). Other deletions are distributed throughout the coding regions but occur primarily in ORF1a as single or triple codon deletions that maintain the reading frame. Relative to YHV, there are very few sequence insertions in the GAV genome, suggesting it may be more stable, perhaps reflecting a stable longer term association with its natural host (P. monodon) in which low-level benign infections commonly occur at high prevalence (Walker et al., 2001).
The 3′-terminal region between ORF3 and the polyA tail also displays several significant distinguishing features. In GAV, this 638 nt region contains an open reading frame (ORF4) encoding a 83 aa (9.6 kDa) protein that has been reported to be expressed at low levels in GAV-infected shrimp tissues (Dhar et al., 2004). In YHV, the corresponding 677 nt 3′-terminal region contains a severely truncated open reading frame encoding a polypeptide of only 20 aa. Although this product shares perfect amino acid sequence identity with the N-terminus of the GAV ORF4 protein, it is unlikely to be expressed as a functional protein. This is consistent with the observation reported for GAV (Cowley et al., 2002) that no discrete transcript is generated from the corrupted TRS immediately upstream of ORF4, suggesting that expression occurs either by internal initiation from sgmRNA2 or from variable-length RNAs transcribed in low abundance from multiple sites in the upstream UTR (Cowley et al. unpublished data). However, the existence of a TRS-like sequence upstream of this region in both YHV and GAV, and the high level of sequence homology within ORF4 and downstream of the point of truncation, indicate that this 3′-region of the ronivirus genome is a genetic resource in active evolutionary transition.
Virus species demarcation is a complex task that is based on the concept, endorsed by the International Committee on Taxonomy of Viruses (ICTV), of a species as ‘a polythetic class of viruses that constitute a replicating lineage and occupy a particular ecological niche’, and requires a holistic consideration of molecular and biological properties (van Regenmortel, 2000). On the basis of nucleic acid or amino acid sequence identity alone, the data presented here indicate that YHV and GAV fulfill species demarcation criteria applied to such diverse RNA viruses as potyviruses (<85% whole genome identity) and geminiviruses (<85–90% identical over genome component A) infecting plants, and lyssaviruses (<86% replicase identity) and flaviviruses (<88% whole genome identity) infecting animals. The significant difference in overall genome size (427 nt) due primarily to large insertions/deletions in non-coding regions is also consistent with a view that YHV and GAV represent distinct genetic lineages. Although they share the same natural host (Penaeus monodon shrimp) and a similar susceptible host range, YHV and GAV differ significantly in virulence and pathogenicity, and have natural distributions that are geographically remote (i.e., Asia and Australia, respectively). Nevertheless, YHV and GAV represent only two of several distinct genetic lineages of closely related viruses infecting P. monodon amongst which there is evidence for natural genetic recombination, possibly as a consequence of co-infections resulting from the translocation and co-habitation of infected broodstock and larvae for aquaculture (Walker et al., 2001, Wijegoonawardane et al., 2004, Wijegoonawardane et al., 2008). This common practice is likely to confound any speciation established through past natural geographic isolation. It appears, therefore, that despite molecular and biological evidence to the contrary, the current taxonomic assignment of YHV and GAV as the same species should be maintained until a clearer picture of this complex of viruses emerges.
Acknowledgements
The authors acknowledge the support of the Australian Centre for International Agricultural Research (ACIAR) and the assistance and advice of Professor Timothy Flegel and the staff of Centex Shrimp, Bangkok, Thailand, in the conduct of this project.
References
- ABI, 1992. SEQED: Sequence Editor, version 1.0.3. ABI, Foster City.
- Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucl. Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida T.A., Pérez J.A., Pinto F.M. Size-fractionation of RNA by hot agarose electrophoresis. Biotechniques. 2000;28:414–416. doi: 10.2144/00283bm04. [DOI] [PubMed] [Google Scholar]
- Bernad A., Blanco L., Lazaro J.M., Martin G., Salas M. A conserved 3′ → 5′ exonuclease active site in prokaryotic and eukaryotic DNA polymerases. Cell. 1989;59:219–228. doi: 10.1016/0092-8674(89)90883-0. [DOI] [PubMed] [Google Scholar]
- Cowley J.A., Cadogan L.C., Spann K.M., Sittidilokratna N., Walker P.J. The gene encoding the nucleocapsid protein of gill-associated nidovirus of Penaeus monodon prawns is located upstream of the glycoprotein gene. J. Virol. 2004;78:8935–8941. doi: 10.1128/JVI.78.16.8935-8941.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowley J.A., Dimmock C.M., Spann K.M., Walker P.J. Gill-associated virus Penaeus monodon prawns: an invertebrate nidovirus with ORF1a and ORF1b gene related arteri- and coronaviruses. J. Gen. Virol. 2000;81:1473–1484. doi: 10.1099/0022-1317-81-6-1473. [DOI] [PubMed] [Google Scholar]
- Cowley J.A., Dimmock C.M., Walker P.J. Gill-associated nidovirus of Penaeus monodon prawns transcribes 3′-coterminal subgenomic RNAs that do not possess 5′-leader sequences. J. Gen. Virol. 2002;83:927–935. doi: 10.1099/0022-1317-83-4-927. [DOI] [PubMed] [Google Scholar]
- Cowley J.A., Walker P.J. The complete sequence of gill-associated virus of Penaeus monodon prawns indicates a gene organisation unique among nidoviruses. Arch. Virol. 2002;147:1977–1987. doi: 10.1007/s00705-002-0847-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowley J.A., Walker P.J. Molecular biology and pathogenesis of roniviruses. In: Perlman S., Gallagher T., Snijder E.J., editors. Nidoviruses. ASM Press; WA: 2008. pp. 361–377. [Google Scholar]
- den Boon A.J., Faaberg K.S., Meulenberg J.J.M., Wassenaar A.L.M., Plagemann P.G.W., Gorbalenya A.E., Snijder E.J. Processing and evolution of the N-terminal region of the arterivirus replicase ORF1a protein: identification of two papainlike cysteine proteases. J. Virol. 1995;69:4500–4505. doi: 10.1128/jvi.69.7.4500-4505.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar A.K., Cowley J.A., Hasson K.W., Walker P.J. Genomic organization, biology, and diagnosis of Taura syndrome virus and yellow head virus of penaeid shrimp. Adv. Virus Res. 2004;63:347–415. doi: 10.1016/S0065-3527(04)63006-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draker R., Roper R.L., Petric M., Tellier R. The complete sequence of the bovine torovirus genome. Virus Res. 2006;115:56–68. doi: 10.1016/j.virusres.2005.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dua R., Levy D.L., Campbell J.L. Role of the putative zinc finger domain of Saccharomyces cerevisiae DNA polymerase ɛ in DNA replication and the S/M checkpoint pathway. J. Biol. Chem. 1998;273:30046–30055. doi: 10.1074/jbc.273.45.30046. [DOI] [PubMed] [Google Scholar]
- Dumas J.B., Edwards M., Delort J., Mallet J. Oligodeoxynucleotide ligation of single-stranded cDNAs: a new tool for cloning 5′ ends of mRNAs and for constructing cDNA libraries by in vitro amplification. Nucl. Acids Res. 1991;19:5227–5232. doi: 10.1093/nar/19.19.5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flegel T.W., Sriurairatant S., Wongteerasupaya C., Boonsaeng V., Panyim S., Withyachumnarnkul B. Progress in characterization of yellow-head virus of Penaeus monodon. In: Browdy C.L., Hopkins J.S., editors. Swimming through Troubled Water, Proceedings of the Special Session on Shrimp Farming, Aquaculture ‘95. World Aquaculture Society; Baton Rouge: 1995. pp. 76–83. [Google Scholar]
- Gorbelenya A.E., Enjuanes L., Ziebuhr J., Snijder E.J. Nidovirales: evolving the largest RNA virus genome. Virus Res. 2006;117:17–37. doi: 10.1016/j.virusres.2006.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold J., Siddell S.G., Gorbalenya A.E. A human RNA viral cysteine proteinase that depends upon a unique Zn2+-binding finger connecting the two domains of a papain-like fold. J. Biol. Chem. 1999;274:14918–14925. doi: 10.1074/jbc.274.21.14918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann K., Stoffel W. TMbase—a database of membrane spanning proteins segments. Biol. Chem. Hoppe-Seyler. 1993;374:166. [Google Scholar]
- Ingelbrecht I.L., Mandelbaum C.I., Mirkov T.E. Highly sensitive northern hybridization using a rapid protocol for downward alkaline blotting of RNA. Biotechniques. 1998;25:420–426. doi: 10.2144/98253st03. [DOI] [PubMed] [Google Scholar]
- Jitrapakdee S., Unajak S., Sittidilokratna N., Hodgson R.A.J., Cowley J.A., Panyim S., Boonsaeng V., Walker P.J. Identification and analysis of gp116 and gp64 structural glycoproteins of yellow head nidovirus of Penaeus monodon shrimp. J. Gen. Virol. 2003;84:863–873. doi: 10.1099/vir.0.18811-0. [DOI] [PubMed] [Google Scholar]
- Limsuwan C. Tamsetakit Co., Ltd.; Bangkok (in Thai): 1991. Handbook for Cultivation of Black Tiger Prawns. [Google Scholar]
- Moser M.J., Holley W.R., Chatterjee A., Mian I.S. The proofreading domain of Escherichia coli DNA polymerase I and other DNA and/or RNA exonuclease domains. Nucl. Acids Res. 1997;25:5110–5118. doi: 10.1093/nar/25.24.5110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadala E.C.B., Tapay L.M., Loh P.C. Yellow-head virus: a rhabdovirus-like pathogen of penaeid shrimp. Dis. Aquat. Org. 1997;31:141–146. [Google Scholar]
- OIE, 2007. Aquatic Animal Health Code, 10th ed. World Animal Health Organization, Paris.
- Prentice E., McAuliffe J., Lu X., Subbarao K., Denison M.R. Identification and characterization of severe acute respiratory syndrome coronavirus replicase proteins. J. Virol. 2004;78:9977–9986. doi: 10.1128/JVI.78.18.9977-9986.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putics A., Gorbalenya A.E., Ziebuhr J. Identification of protease and ADP-ribose 1″-monophosphatase activities associated with transmissible gastroenteritis virus non-structural protein 3. J. Gen. Virol. 2006;87:651–656. doi: 10.1099/vir.0.81596-0. [DOI] [PubMed] [Google Scholar]
- Rost B., Sander C. Improved prediction of protein secondary structure by use of sequence profiles and neural networks. Proc. Natl. Acad. Sci. U.S.A. 1993;90:7558–7562. doi: 10.1073/pnas.90.16.7558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost B., Yachdav G., Liu J. The PredictProtein server. Nucl. Acids Res. 2004;32(Web Server issue):W321–W326. doi: 10.1093/nar/gkh377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultze H., Ulferts R., Schelle B., Bayer S., Granzow H., Hoffmann B., Mettenleiter T.C., Ziebuhr J. Characterization of White bream virus reveals a novel genetic cluster of nidoviruses. J. Virol. 2006;80:11598–11609. doi: 10.1128/JVI.01758-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Tang X.F., Matsui I. Subunit interaction and regulation of activity through terminal domains of the family D polymerase from Pyrococcus horikoshii. J. Biol. Chem. 2003;278:21247–21257. doi: 10.1074/jbc.M212286200. [DOI] [PubMed] [Google Scholar]
- Sittidilokratna N., Hodgson R.A.J., Cowley J.A., Jitrapakdee S., Boonsaeng V., Panyim S., Walker P.J. Complete ORF1b-gene sequence indicates yellow head virus is an invertebrate nidovirus. Dis. Aquat. Org. 2002;50:87–93. doi: 10.3354/dao050087. [DOI] [PubMed] [Google Scholar]
- Sittidilokratna N., Phetchampai N., Boonsaeng V., Walker P.J. Structural and antigenic analysis of the yellow head virus nucleocapsid protein p20. Virus Res. 2006;116:21–29. doi: 10.1016/j.virusres.2005.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., den Boon J.A., Horzinek M.C., Spaan W.J.M. Comparison of the genome organization of toro- and coronaviruses: evidence for two nonhomologous RNA recombination events during Berne virus evolution. Virology. 1991;180:448–452. doi: 10.1016/0042-6822(91)90056-H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Bredenbeek P.J., Dobbe J.C., Thiel V., Ziebuhr J., Poon L.L.M., Guan Y., Rozanov M., Spaan W.J.M., Gorbalenya A.E. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 2003;331:991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaan, W.J.M., Cavanagh, D., de Groot, R.J., Enjuanes, L., Gorbalenya, A.E., Snijder, E.J., Walker, P.J., 2005. Order Nidovirales. In: Fauquet, C.M., Mayo, M.A., Maniloff, J., Desselberger, U., Ball, L.A. (Eds.), Virus Taxonomy, VIIIth Report of the ICTV. Elsevier/Academic Press, London, pp. 937–945.
- Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucl. Acids Res. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijms M.A., van Dinten L.C., Gorbalenya A.E., Snijder E.J. A zinc finger-containing papain-like protease couples subgenomic mRNA synthesis to genomic translation in a positive-stranded RNA virus. Proc. Natl. Acad. Sci. U.S.A. 2001;98:1889–1894. doi: 10.1073/pnas.041390398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Regenmortel, M.H.V., 2000. Introduction to the species concept in virus taxonomy. In: van Regenmortel, M.H.V., Fauquet, C.M., Bishop, D.H.L., Carstens, E.B., Estes, M.K., Lemon, S.M., Maniloff, J., Mayo, M.A., McGeoch, D.J., Pringle, C.R., Wickner, R.B. (Eds.), Virus Taxonomy, VIIth Report of the ICTV. Academic Press, San Diego, pp. 3–16.
- Walker, P.J., Cowley, J.A., Spann, K.M., Hodgson, R.A.J., Hall, M.R., Withyachumnarnkul, B., 2001. Yellow head complex viruses: transmission cycles and topographical distribution in the Asia-Pacific region. In: Browdy, C.L., Jory, D.E. (Eds.), The New Wave: Proceedings of the Special Session on Sustainable Shrimp Culture, Aquaculture 2001. The World Aquaculture Society, Baton Rouge, pp. 227–237.
- Walker, P.J., Bonami, J.R., Boonsaeng, V., Chang, P.S., Cowley, J.A., Enjuanes, L., Flegel, T.W., Lightner, D.V., Loh, P.C., Snijder, E.J., Tang, K., 2005. Family Roniviridae. In: Fauquet, C.M., Mayo, M.A., Maniloff, J., Desselberger, U., Ball, L.A. (Eds.), Virus Taxonomy, VIIIth Report of the ICTV. Elsevier/Academic Press, London, pp. 973–977.
- Walker P.J., Wang Y., Cowley J.A., McWilliam S.M., Prehaud C.J.N. Structural and antigenic analysis of the nucleoprotein of bovine ephemeral fever rhabdovirus. J. Gen. Virol. 1994;75:1889–1899. doi: 10.1099/0022-1317-75-8-1889. [DOI] [PubMed] [Google Scholar]
- Wijegoonawardane P.K.M., Cowley J.A., Phan T., Hodgson R.A.J., Nielsen L., Kiatpathomchai W., Walker P.J. Genetic diversity in the yellow head nidovirus complex. Virology. 2008 doi: 10.1016/j.virol.2008.07.005. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijegoonawardane P., Cowley J.A., Kiatpathomchai W., Nielsen L., Walker P.J. Phylogenetic analysis and evidence of genetic recombination among six genotypes of yellow head complex viruses from Penaeus monodon. Proceedings of the Seventh Asian Fisheries Forum; Penang, Malaysia; 2004. p. 210. (Book of Abstracts) [Google Scholar]
- Wongteerasupaya C., Vickers J., Sriurairatana S., Nash G.L., Akarajamorn A., Boonsaeng V., Panyim S., Tassanakorn A., Withyachumnarnkul B., Flegel T.W. A non-occluded, systemic baculovirus that occurs in cells of ectodermal and mesodermal origin and causes high mortality in the black tiger prawn Penaeus monodon. Dis. Aquat. Org. 1995;21:69–77. [Google Scholar]
- Ziebuhr J. The coronavirus replicase. Curr. Top. Microbiol. Immunol. 2005;287:57–94. doi: 10.1007/3-540-26765-4_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr J., Snidjer E.J., Gorbalenya A.E. Viral-encoded proteases and proteolytic processing in the Nidovirales. J. Gen. Virol. 2000;81:853–879. doi: 10.1099/0022-1317-81-4-853. [DOI] [PubMed] [Google Scholar]
- Ziebuhr J., Bayer S., Cowley J.A., Gorbalenya A.E. The 3C-like proteinase of an invertebrate nidovirus links coronavirus and potyvirus homologs. J. Virol. 2003;77:1415–1426. doi: 10.1128/JVI.77.2.1415-1426.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Y., Deutscher M.P. Exoribonuclease superfamilies: structural analysis and phylogenetic distribution. Nucl. Acids Res. 2001;29:1017–1026. doi: 10.1093/nar/29.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]


