Abstract
Murine-adapted porcine epidemic diarrhea virus (PEDV), MK-p10, shows high neurovirulence and increased fusion activity compared with a non-adapted MK strain. MK-p10 S protein had four mutations relative to the original virus S, and one of these (H → R at position 1381, H1381R) in the cytoplasmic tail (CT) was suggested to be responsible for the increased fusion activity. To explore this, we examined fusion activity using recombinant S proteins. We expressed and compared the fusion activity of MK-p10 S, S with the H1381R mutation, S with the three other mutations that were not thought to be involved in high fusion activity, and the original S protein. The MK-p10 and MK-H1381R S proteins induced larger cell fusions than others. We also examined the distribution of these S proteins; the MK-p10 and MK-H1381R S proteins were transported onto the cell surface more efficiently than others. These findings suggest that the H1381R mutation is responsible for enhanced fusion activity, which may be attributed to the efficient transfer of S onto the cell surface. H1381 is a component of the KxHxx motif in the CT region, which is a retrieval signal of the S protein for the endoplasmic reticulum–Golgi intermediate compartment (ERGIC). Loss of this motif could allow for the efficient transfer of S proteins from ERGIC onto the cell surface and subsequent increased fusion activity.
Abbreviations: CoV, coronavirus; CT, cytoplasmic tail; ERGIC, endoplasmic reticulum–Golgi intermediate compartment; DMEM, Dulbecco's modified Eagle's medium; HR, heptad repeat; PEDV, porcine epidemic diarrhea virus; PBS, phosphate-buffered saline; S, spike; SARS, severe acute respiratory syndrome; TPB, tryptose phosphate broth; TM, transmembrane
Keywords: Coronavirus, Porcine epidemic diarrhea virus (PEDV), Spike protein, Fusion, Cytoplasmic retrieval signal
Porcine epidemic diarrhea virus (PEDV) is a causative agent for pig diarrhea and induces inappetence and weight loss in adult pigs, while it can induce lethal infection in piglets (Pensaert and de Bouck, 1978). PEDV is a group I coronavirus (CoV), enveloped by a viral membrane. Membrane fusion with the host cell is an important event allowing injection of the viral RNA genome into the host cell. CoV membrane fusion is mediated by the spike (S) protein, which consists of two functional subunits: the N-terminal S1 subunit, responsible for receptor binding activity, and the C-terminal transmembrane (TM) S2 subunit, responsible for fusion activity. The S2 subunit can be further structurally divided into three distinct domains: a large ectodomain, a TM anchoring region, and a cytoplasmic tail (CT) region. The role of the ectodomain in fusion activity has been well documented; this domain contains a protease cleavage site, a putative fusion peptide, and two heptad repeat (HR) regions, which have been shown to be the principal players in membrane fusion (Bosch et al., 2003, Li et al., 2006, Matsuyama and Taguchi, 2009, Tripet et al., 2004, Watanabe et al., 2008). On the other hand, the role(s) of the TM and CT regions in fusion activity is not well understood, although a few studies have reported that the aromatic domain of the TM region (Howard et al., 2008, Jeetendra et al., 2003, Sainz et al., 2005) and the cysteine-rich domain of the CT region have regulatory effects on fusogenic activity (Broer et al., 2006, Chang et al., 2000). Another important feature of the CT of the S protein is localization in the endoplasmic reticulum–Golgi intermediate compartment (ERGIC), for which a signal composed of KxHxx or KKxx motif is believed to be critical; this dibasic motif of the S protein of SARS-CoV, and infectious bronchitis virus (IBV) was responsible for retrieving the S protein in the ERGIC, and the lack of this motif resulted in increased surface expression of S protein (Lontok et al., 2004, McBride et al., 2007). The region of S protein located in the ERGIC facilitates an efficient interaction of with M and E proteins in the ERGIC, which is critical for virus budding into those intracellular vesicles. Although some S proteins are transported onto the cell surface, M and E proteins are restricted to those vesicles and are not transported to the cell plasma membrane (Corse and Machamer, 2000, Kapke et al., 1988, Weisz et al., 1993). Thus, maturation of CoVs by budding takes place only into the ERGIC, not from the plasma membrane.
In our previous report, the murine-adapted variant of PEDV, MK-p10, which was isolated by 10 sequential passages through suckling mice brains, showed higher virulence for suckling mice when inoculated intracerebrally (Shirato et al., 2010). One interesting features of MK-p10 was increased fusogenic activity compared with the original MK strain. The S protein of MK-p10 had four amino acid mutations, and one of these (H → R at position 1381, H1381R) in the CT region seemed to be involved in the increased fusiogenic activity because fusion enhancement of S protein was coincident with H1381R mutation. The histidine at a position 1381 (H1381) is a constituent of the KxHxx motif. Thus, there is a possibility that the lack of this motif caused by the H1381R mutation enhanced surface expression of MK-p10 S protein, resulting in increased fusion activity. There is another possibility that the mutation reduces virion budding into the ERGIC because reduced localization of S protein at ERGIC decreases the chance of its interaction with M and E proteins at the ERGIC. In this study, to determine the mechanisms of the increased fusogenic activity of the MK-p10 S protein, we generated several recombinant S mutants and analyzed their fusion activities. Our study showed that H1381R mutation damaged the KxHxx motif, allowing for the efficient transport of the S proteins from the ERGIC onto the cell surface, inducing increased fusion activity. The present study in combination with our previous study [17] that MK-p10 multiplies more-or-less similarly to the original virus suggest that the localization of the S at the ERGIC is not critical for virus replication.
We have reported that the S protein of MK-p10 had four amino acid mutations, as depicted in Fig. 1a and the replication kinetics of MK and MK-p10 in Vero cell culture was not different, being identical to our previous report [17]. Therefore, these mutations seem not to affect the efficiency of virus replication. On the other hand, one of these mutations (H → R at position 1381, H1381R) in the CT was suggested to be involved in the increased fusiogenic activity (Shirato et al., 2010). However, the involvement of other amino acids or viral proteins other than S in this activity could not be excluded. To clarify the role of the H1381R mutation of S protein in the increased fusogenicity, S genes were subcloned into pTargeT vector (Promega, Fitchburg, WI, USA) and mutations were inserted using specific primer sets and KOD-Plus Mutagenesis Kit (Toyobo, Osaka, Japan) (Fig. 1a). Fusion activities were analyzed using an S protein expression system on a Vero cells culture. MK-H1381R-S was generated by the introduction of an H1381R mutation into the original S protein, while MK-p10-R1381H-S by the introduction of R1381H mutation into MK-p10. Vero cells were transfected with each S plasmid using DMRIE-C reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's protocol. After 3 days of incubation, cells were treated with trypsin (1.25 μg/mL; Sigma, St. Louis, MO, USA) with DMEM (Sigma) containing 10% tryptose phosphate broth (TPB), and cell fusions were estimated by incubation of an additional 15-h. As expected, cells expressing the original S protein showed very small cell fusions, as revealed by microscopic observation. In contrast, the cells expressing MK-p10 S protein showed large cell fusions (Fig. 1b; MK vs MK-p10). Moreover, the cells expressing MK-p10-R1381H S showed slightly larger cell fusions, while the cells expressing MK-H1381R-S displayed apparently larger cell fusions, similar in size to those of the MK-p10 S protein (Fig. 1b; MK-p10-R1381H vs MK-p10). The mean diameter of cell fusions of MK-H1381R-S and MK-p10-S was statistically larger than those of MK and MK-p10-R1381H [340 ± 142 (MK-H1381R) and 297 ± 89 (MK-p10) vs 154 ± 52 (MK) and 214 ± 81 (MK-p10-R1381H) μm, respectively; n = 50; p < 0.0001]. To quantify the fusiogenic activity of each S protein, we used a luciferase reporter fusion assay, as described by Branigan et al. (2005), with positive control plasmids (pFR-luc and pBD-NFκB) from a Mammalian Two-Hybrid Assay Kit (Stratagene, La Jolla, CA, USA) and a ONE-Glo Luciferase Assay System (Promega) using COS7 cells with some modifications. Briefly, S protein plasmid and pFR-luc were co-transfected into COS7 cells using the TransIT-COS reagent (Mirus, Madison, WI, USA) and other COS7 cells were transfected with pBD-NFκB plasmid. After 24 h of incubation, cells that had been transfected with pBD-NFκB were detached and added into S protein plasmid-transfected cells with fresh medium. After an additional 36 h of incubation, cells were washed with PBS and DMEM containing 1.5% carboxymethyl cellulose and 5 μg/mL of trypsin and incubated at 37 °C. After 8 h of incubation, luminescence was measured with a ONE-Glo Luciferase Assay System following the manufacturer's instructions, and was expressed as the relative luminescence level/10,000 copies of S protein mRNA quantitated by specific real-time PCR. As shown in Fig. 2 , the relative luminescence values of MK-p10-R1381H-S were slightly higher than those of MK-S (MK-p10-R1381H vs MK; n = 8, p < 0.05), while those of MK-p10 and MK-H1381R-S were significantly higher (MK-p10 and MK-H1381R vs MK; n = 8, p < 0.01). In both methods, MK-p10-R1381H-S showed slightly increased fusion activity, indicating that these mutations contributed weakly to fusogenicity. These results collectively indicated that the H1381R mutation in the CT of S protein was a major determinant of the increased fusogenicity.
Fig. 1.
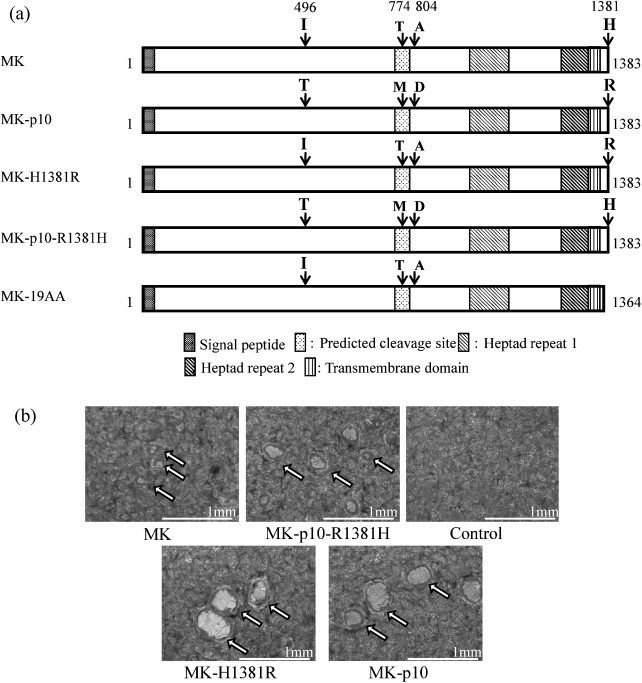
Cell fusion induced by recombinant S proteins of PEDV MK and MK-p10. (a) A schematic presentation of amino acid substitutions in the S protein of MK-p10 compared with the parental MK S protein and of constructed plasmids. (b) Cell fusion induced by recombinant PEDV S proteins. The S protein expression plasmids were transfected into Vero cells using the DMRIE-C reagent. After 3 days of incubation, cells were washed with PBS, added to DMEM containing 10% TPB and 1.25 μg/mL of trypsin, and incubated for 15 h. These cells were fixed and stained with PBS containing 20% formalin and 0.1% crystal violet and observed by microscopy. Fused cells are indicated by white arrows.
Fig. 2.
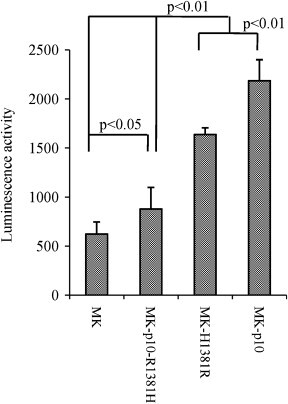
Quantitation of fusion activity of PEDV S protein. One group of COS7 cells was transfected with PEDV S plasmids and pBD-NFκB control plasmid, and another group was transfected with pFR-luc reporter plasmids for 24 h of incubation. These cells were mixed and incubated for 36 h. CMC-DMEM containing 5 μg/mL of trypsin was then added into each well and incubated for 8 h. Cells were treated with ONE-Glo Luciferase Assay System reagent following the manufacturer's protocol, and luminescence values were measured. The luminescence obtained was represented as the relative value of luminescence/10,000 copies of PEDV S mRNA (n = 8).
The S proteins of many CoVs have been reported to be cleaved by various proteases to generate the S2 subunit, which was essential for virus–cell entry and cell–cell fusion activity (Matsuyama and Taguchi, 2002, Taguchi and Shimazaki, 2000). Thus, the protease sensitivity of S proteins of PEDV was expected to affect their fusogenic activity. To evaluate the protease sensitivity of S proteins, we examined the cleavability of the S protein from MK and MK-p10 with trypsin. Viruses collected from virus-infected cells cultured without trypsin were treated with 1–25 μg/mL of trypsin at room temperature for 5 min, and the S protein was analyzed by Western blotting, using an S-protein specific antibody, which recognized the 1366-80 amino acid residue of the PEDV S protein. As shown in Fig. 3 , both S proteins were cleaved by trypsin and the S protein cleavage pattern was not significantly different between MK and MK-p10. This cleavage pattern was also observed similarly in S protein expressed on cells and treated with trypsin before cell collection (data not shown). These suggest the S protein of MK and MK-p10 were cleaved by trypsin in a similar fashion.
Fig. 3.
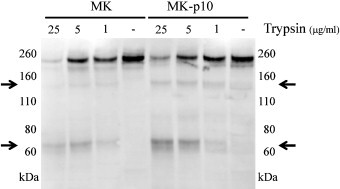
Treatment of the S protein with trypsin. Western blot analysis of PEDV S protein after trypsin treatment. Vero cells were infected with PEDV and cultured for 3 days in the absence of trypsin. These cells were collected and treated with the indicated concentration of trypsin at room temperature for 5 min. After trypsin treatment, samples were separated with 10% SDS-PAGE gel electrophoresis, and PEDV S protein was detected with a specific rabbit polyclonal antibody.
Because the H1381R mutation caused the loss of the KxHxx motif, which is known to be an S protein retrieval signal in the ERGIC (Table 1 ), we hypothesized that S proteins with H1381R mutation could be transferred efficiently from the ERGIC onto the cell surface and enhance their surface expression, which could finally result in increased fusogenicity. Thus, the cells expressing MK-S, MK-H1381R, and MK-p10 S proteins were examined by immunofluorescence under permeabilized and non-permeabilized conditions. We also examined the cells expressing MK-19aa, in which the C-terminal 19 amino acids of its S protein are deleted, completely losing the KxHxx motif. The S protein that has this deletion showed increased fusogenicity and incorporation of VSV-pseudotyped virus in SARS-CoV (Fukushi et al., 2005, Petit et al., 2005). As shown in Fig. 4a, each S protein was distributed in a similar fashion in permeabilized cells (Fig. 4a, left panel), while it seemed different in non-permeabilized cells; although MK-S was rarely seen on the cell surface, MK-H1381R-S, MK-p10-S, and MK-19aa were widely distributed there (MK-H1381R, MK-p10 and MK-19aa vs MK; Fig. 4a, right panel). The areas of S protein expression in non-permeabilized cells were calculated and normalized with the data obtained from empty vector-transfected control. The amount of surface-expressed S protein was represented as the relative levels (%) as compared to those of the area of intracellular-expressed S protein (Fig. 4b). This result also showed that the S protein with a H1381R mutation was significantly more efficiently expressed on the cell surface than was the S protein without the mutation. This indicated that high distribution on the cell surface was attributable to the H1381R mutation and loss of the KxHxx motif. These results suggested that enhanced surface expression caused increased fusogenic activity.
Table 1.
Localization motifs in the cytoplasmic tail region of S protein.
| Virus | Carboxyl terminal sequences |
|---|---|
| MK | |
| MK-p10 |
TM, transmembrane; Underline, YxxΦ motif; Double underline, KxHxx motif.
Fig. 4.
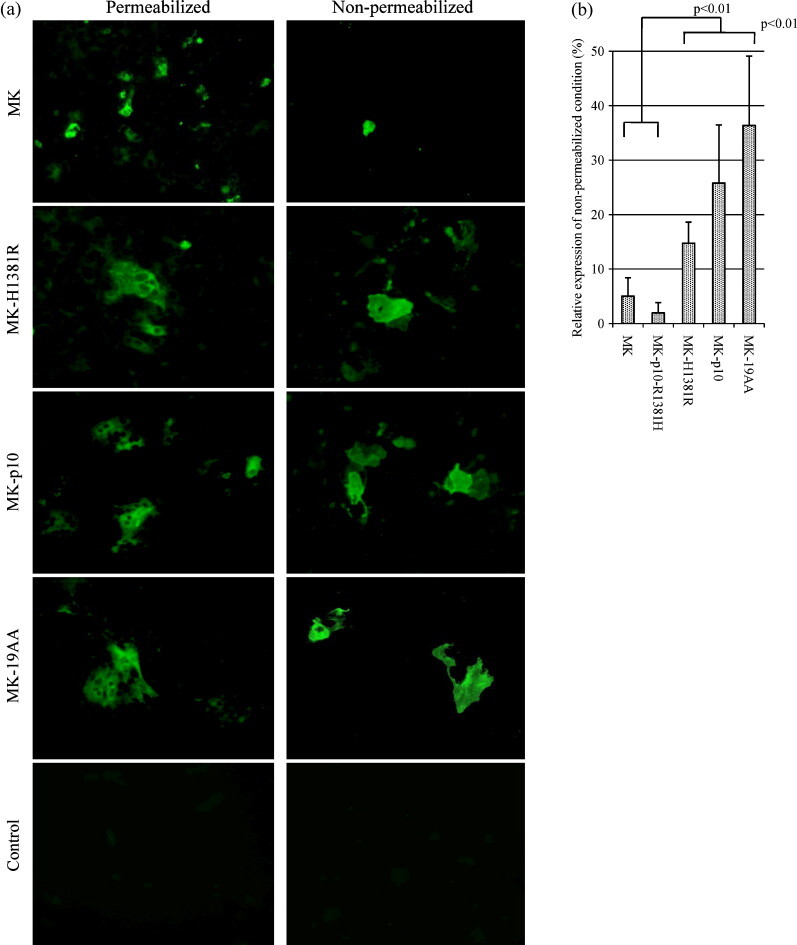
(a) Detection of recombinant PEDV S protein by immunofluorescence. The plasmids containing each of the PEDV S genes were transfected into Vero cell suspensions with DIMRIE-C reagent. After 3 days of incubation, cells were fixed with PBS containing 2% formaldehyde for 12 min (permeabilized) or with PBS containing 2.5% formaldehyde and 0.5% triton-100 for 20 min (non-permeabilized), and S protein was stained with PEDV-immunized pig serum (magnification 200×). (b) Quantitation of S protein expression on the cell surface. PEDV S proteins were expressed on Vero cell cultures and fixed and stained as described in Fig. 4a. After immunostaining, cell images were digitally captured, and the area of S protein expression was calculated with VH-H1A5 analyzer (KEYENCE, Osaka, Japan) and normalized with the data from empty vector-transfected control. The amount of S protein expression on the cell surface was represented as the relative value (%) (value of non-permeabilized condition/value of permeabilized condition) (n = 5).
As described above, the H1381R mutation increased the transport of S protein on cell surface. This suggests to us that virions with the H1381R mutation contains less S than those containing the KxHxx motif because it is expected that sufficient S protein without retrieval domain is not located in the ERGIC. To examine this hypothesis, the rate of S protein comparative to nucleocapsid (N) protein incorporated in virion was estimated. PEDVs were infected into Vero/TMPRSS2 cells, which was constitutively expressed the transmembrane serine protease TMPRSS2 (Shirogane et al., 2008) and was able to increase the virion release into culture medium in PEDV infection (Shirato et al., 2011). After 2 days of incubation, the culture medium were collected, ultracentrifuged, and analyzed by SDS-PAGE and WB analysis (Fig. 5 ). The band densitometries were analyzed with MultiGauge software (Fujifilm, Tokyo, Japan) and the S/N rate were calculated. As shown in Fig. 5, the S/N rates were 0.13 ± 0.06 (MK) and 0.16 ± 0.05 (MK-p10), respectively (n = 5), and there was no statistical difference. This suggests that the amount of S protein incorporated into virions is not different between MK and MK-p10. This may be attributed to the over-production of S proteins in cells and thus plenty amounts of S protein remains in ERGIC even if some of those are transported to the plasma membrane. Similar ratio of S protein to N protein found on MK and MK-p10 virions could also explain why MK and MK-p10 grows equally in cultured cells as well as in the mouse as shown in our previous report [17]. In addition, this result also suggests that the retrieval motif is not a prerequisite for the multiplication strategy of PEDV. To determine whether this is a general feature of Coronaviridae, analysis using mutant viruses with mutations in this region of other coronaviruses by reverse genetics should be conducted.
Fig. 5.
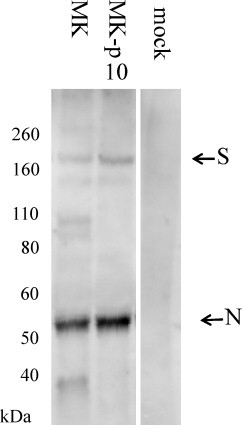
Estimation of amounts of S protein expressed on released virions of MK and MK-p10. PEDVs were infected to the Vero/TMPRSS2 cells. After 2 days of incubation, culture medium was collected and ultracentrifuged (35,000 rpm, 4 °C, 1.5 h), and separated with SDS PAGE. The S protein (200 kDa) and N protein (58 kDa) were detected by Western blot analysis using PEDV-immunized pig serum. N protein was also examined to normalize the amounts of virions.
Recently, it has been reported that the CT of CoV S protein played a regulatory role in intracellular trafficking or its localization in the ERGIC, and several motifs were identified (Fukushi et al., 2005, Lontok et al., 2004, McBride et al., 2007, Schwegmann-Wessels et al., 2004). Schwegmann-Wessels et al. (2004) and Tan reported that the S protein of CoV had a tyrosine-dependent motif (YXXΦ, where Φ is a hydrophobic amino acid) in the CT region and that this motif was essential for the intracellular localization of S protein (Fukushi et al., 2005, Schwegmann-Wessels et al., 2004). Lontok et al. and McBride et al. reported that a dibasic motif (KxHxx-COOH or KKxx-COOH) in some CoVs, such as SARS-CoV and IBV, was responsible for retrieving the S protein in the ERGIC and that the lack of this motif resulted in increased surface expression of the S protein (Lontok et al., 2004, McBride et al., 2007). The S protein CT region of the group 1 CoV PEDV had both “YEAF” as the tyrosine-dependent motif at positions 1374-1377 and “KVHVQ” as a dibasic motif at positions 1379–1383 (Table 1). In this study, MK-p10 S protein, which lost the KxHxx motif due to H1381R mutation, but retained the YXXΦ motif, showed enhanced cell surface expression and subsequent increased fusogenicity, suggesting that the intracellular localization function of the YXXΦ motif alone seems to be insufficient to retrieve S protein in the ERGIC.
As described in a previous report, MK-p10 is more virulent to suckling mice than the original MK strain. To analyze the difference in virulence, we must first identify the differences between entire genomes and examine the effect of each of those mutations on virulence. To do this, establishment of reverse genetics in PEDV is critical. Also, to clarify how MK-p10 virus exhibits higher neurovirulence for suckling mice, the study is in progress to see the distribution of viral antigens in the brain by histopathological and immunohistological methods.
Acknowledgments
We thank Miyuki Kawase for excellent technical assistance. This work was supported by a Grant-in-Aid for Scientific Research (B; No. 19390135) and the strategic Research Base Development Program for private Universities 2008 from the Ministry of Education, Culture, Sports, Science and Technology, Japan. We thank Dr. Tetsuo Nunoya and Dr. Koutarou Tsuchiya (The Nippon Institute for Biological Science, Tokyo, Japan) for helpful discussion.
References
- Bosch B.J., van der Zee R., de Haan C.A., Rottier P.J. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 2003;77(16):8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branigan P.J., Liu C., Day N.D., Gutshall L.L., Sarisky R.T., Del Vecchio A.M. Use of a novel cell-based fusion reporter assay to explore the host range of human respiratory syncytial virus F protein. Virol. J. 2005;2:54. doi: 10.1186/1743-422X-2-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broer R., Boson B., Spaan W., Cosset F.L., Corver J. Important role for the transmembrane domain of severe acute respiratory syndrome coronavirus spike protein during entry. J. Virol. 2006;80(3):1302–1310. doi: 10.1128/JVI.80.3.1302-1310.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang K.W., Sheng Y., Gombold J.L. Coronavirus-induced membrane fusion requires the cysteine-rich domain in the spike protein. Virology. 2000;269(1):212–224. doi: 10.1006/viro.2000.0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corse E., Machamer C.E. Infectious bronchitis virus E protein is targeted to the Golgi complex and directs release of virus-like particles. J. Virol. 2000;74(9):4319–4326. doi: 10.1128/jvi.74.9.4319-4326.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushi S., Mizutani T., Saijo M., Matsuyama S., Miyajima N., Taguchi F., Itamura S., Kurane I., Morikawa S. Vesicular stomatitis virus pseudotyped with severe acute respiratory syndrome coronavirus spike protein. J. Gen. Virol. 2005;86(Pt 8):2269–2274. doi: 10.1099/vir.0.80955-0. [DOI] [PubMed] [Google Scholar]
- Howard M.W., Travanty E.A., Jeffers S.A., Smith M.K., Wennier S.T., Thackray L.B., Holmes K.V. Aromatic amino acids in the juxtamembrane domain of severe acute respiratory syndrome coronavirus spike glycoprotein are important for receptor-dependent virus entry and cell–cell fusion. J. Virol. 2008;82(6):2883–2894. doi: 10.1128/JVI.01805-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeetendra E., Ghosh K., Odell D., Li J., Ghosh H.P., Whitt M.A. The membrane-proximal region of vesicular stomatitis virus glycoprotein G ectodomain is critical for fusion and virus infectivity. J. Virol. 2003;77(23):12807–12818. doi: 10.1128/JVI.77.23.12807-12818.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapke P.A., Tung F.Y., Hogue B.G., Brian D.A., Woods R.D., Wesley R. The amino-terminal signal peptide on the porcine transmissible gastroenteritis coronavirus matrix protein is not an absolute requirement for membrane translocation and glycosylation. Virology. 1988;165(2):367–376. doi: 10.1016/0042-6822(88)90581-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Berardi M., Li W., Farzan M., Dormitzer P.R., Harrison S.C. Conformational states of the severe acute respiratory syndrome coronavirus spike protein ectodomain. J. Virol. 2006;80(14):6794–6800. doi: 10.1128/JVI.02744-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lontok E., Corse E., Machamer C.E. Intracellular targeting signals contribute to localization of coronavirus spike proteins near the virus assembly site. J. Virol. 2004;78(11):5913–5922. doi: 10.1128/JVI.78.11.5913-5922.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S., Taguchi F. Communication between S1N330 and a region in S2 of murine coronavirus spike protein is important for virus entry into cells expressing CEACAM1b receptor. Virology. 2002;295(1):160–171. doi: 10.1006/viro.2002.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S., Taguchi F. Two-step conformational changes in a coronavirus envelope glycoprotein mediated by receptor binding and proteolysis. J. Virol. 2009;83(21):11133–11141. doi: 10.1128/JVI.00959-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride C.E., Li J., Machamer C.E. The cytoplasmic tail of the severe acute respiratory syndrome coronavirus spike protein contains a novel endoplasmic reticulum retrieval signal that binds COPI and promotes interaction with membrane protein. J. Virol. 2007;81(5):2418–2428. doi: 10.1128/JVI.02146-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pensaert M.B., de Bouck P. A new coronavirus-like particle associated with diarrhea in swine. Arch. Virol. 1978;58(3):243–247. doi: 10.1007/BF01317606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit C.M., Melancon J.M., Chouljenko V.N., Colgrove R., Farzan M., Knipe D.M., Kousoulas K.G. Genetic analysis of the SARS-coronavirus spike glycoprotein functional domains involved in cell-surface expression and cell-to-cell fusion. Virology. 2005;341(2):215–230. doi: 10.1016/j.virol.2005.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainz B., Jr., Rausch J.M., Gallaher W.R., Garry R.F., Wimley W.C. The aromatic domain of the coronavirus class I viral fusion protein induces membrane permeabilization: putative role during viral entry. Biochemistry. 2005;44(3):947–958. doi: 10.1021/bi048515g. [DOI] [PubMed] [Google Scholar]
- Schwegmann-Wessels C., Al-Falah M., Escors D., Wang Z., Zimmer G., Deng H., Enjuanes L., Naim H.Y., Herrler G. A novel sorting signal for intracellular localization is present in the S protein of a porcine coronavirus but absent from severe acute respiratory syndrome-associated coronavirus. J. Biol. Chem. 2004;279(42):43661–43666. doi: 10.1074/jbc.M407233200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirato K., Maejima M., Hirai A., Ami Y., Takeyama N., Tsuchiya K., Kusanagi K., Nunoya T., Taguchi F. Enhanced cell fusion activity in porcine epidemic diarrhea virus adapted to suckling mice. Arch. Virol. 2010;155(12):1989–1995. doi: 10.1007/s00705-010-0790-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirato K., Matsuyama S., Ujike M., Taguchi F. Role of proteases in the release of porcine epidemic diarrhea virus from infected cells. J. Virol. 2011;85(15):7872–7880. doi: 10.1128/JVI.00464-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirogane Y., Takeda M., Iwasaki M., Ishiguro N., Takeuchi H., Nakatsu Y., Tahara M., Kikuta H., Yanagi Y. Efficient multiplication of human metapneumovirus in Vero cells expressing the transmembrane serine protease TMPRSS2. J. Virol. 2008;82(17):8942–8946. doi: 10.1128/JVI.00676-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi F., Shimazaki Y.K. Functional analysis of an epitope in the S2 subunit of the murine coronavirus spike protein: involvement in fusion activity. J. Gen. Virol. 2000;81(Pt 12):2867–2871. doi: 10.1099/0022-1317-81-12-2867. [DOI] [PubMed] [Google Scholar]
- Tripet B., Howard M.W., Jobling M., Holmes R.K., Holmes K.V., Hodges R.S. Structural characterization of the SARS-coronavirus spike S fusion protein core. J. Biol. Chem. 2004;279(20):20836–20849. doi: 10.1074/jbc.M400759200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe R., Matsuyama S., Shirato K., Maejima M., Fukushi S., Morikawa S., Taguchi F. Entry from the cell surface of severe acute respiratory syndrome coronavirus with cleaved S protein as revealed by pseudotype virus bearing cleaved S protein. J. Virol. 2008;82(23):11985–11991. doi: 10.1128/JVI.01412-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisz O.A., Swift A.M., Machamer C.E. Oligomerization of a membrane protein correlates with its retention in the Golgi complex. J. Cell. Biol. 1993;122(6):1185–1196. doi: 10.1083/jcb.122.6.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]


