Highlights
► SARS-CoV accessory proteins 6 and 9b interaction is shown for the first time. ► In vivo pull down experiments and proteomic approaches have been used for the identification of the interaction between these SARS-CoV proteins. ► Co-localization of proteins 6 and 9b in SARS-CoV infected cells reinforced this newly described interaction.
Keywords: SARS coronavirus, Protein 6, Protein 9b, Protein–protein interactions, LC–MS
Abstract
The 3′proximal one-third of the severe acute respiratory syndrome coronavirus (SARS-CoV) genome encodes the structural proteins and eight accessory proteins, including 3a, 3b, 6, 7a, 7b, 8a, 8b and 9b, varying in length from 39 to 274 aa which do not share significant homology with viral proteins of known coronaviruses. The SARS-CoV protein 6 is 63 amino acids in length and has been previously involved in virus pathogenicity and replication. To further analyze this functions, the interaction of SARS-CoV protein 6 with other viral and/or cellular factors has been analyzed during SARS-CoV infective cycle. Protein 6 immunoprecipitation from extracts of SARS-CoV infected cells and mass spectrometry analysis revealed an interaction of viral proteins 6 and 9b in biologically relevant conditions. This interaction has been reinforced by co-localization of both proteins in the cytoplasm of SARS-CoV infected cells.
Severe acute respiratory syndrome (SARS) has affected more than 8000 individuals and caused more than 800 deaths in 26 countries since the first case emerged in China in November 2002. The etiological agent of this disease was found to be a previously unknown coronavirus (SARS-CoV) (Drosten et al., 2003, Fouchier et al., 2003, Ksiazek et al., 2003, Lee et al., 2003, Peiris et al., 2003, Rota et al., 2003). In the last years SARS-CoV like viruses have been found circulating in bats from several continents (Drexler et al., 2010, Lau et al., 2005, Quan et al., 2010, Rihtaric et al., 2010) and bats have been described as putative reservoirs of SARS-CoV (Calisher et al., 2006, Li et al., 2005). Thus, the possibility of SARS reoccurrence remains.
Coronaviruses are a family of enveloped viruses with an infectious single-stranded positive-sense RNA genome of ∼30 kb. The SARS-CoV genome organization is similar to that of other coronaviruses. The 5′-proximal two-thirds of the genome encode gene 1, essentially involved in viral RNA synthesis, whereas the 3′-proximal one-third of the genome encodes the structural proteins (spike, S; envelope, E; membrane, M and nucleocapsid, N) and eight accessory proteins (3a, 3b, 6, 7a, 7b, 8a, 8b and 9b) varying in length from 39 to 274 aa, which do not share significant homology with viral proteins of known coronaviruses (Narayanan et al., 2008). Some of these accessory proteins, 3a, 6, 7a, 7b and 9b have been described as structural proteins (Huang et al., 2006, Huang et al., 2007, Ito et al., 2005, Schaecher et al., 2007, Xu et al., 2009).
Although the function of many of the accessory proteins remains unclear, SARS-CoV protein 6 is one of the best characterized accessory proteins. SARS-CoV protein 6 is 63 amino acids in length, its mRNA is present in SARS-CoV infected cells (Snijder et al., 2003) and a minimal transcription regulatory sequence is located upstream of the gene 6 open reading frame (ORF6). Evidence for the presence of protein 6 in clinical specimens has been provided (Chan et al., 2005), and antibodies against its C-terminus have been detected in SARS patients sera (Chow et al., 2006). Protein 6 has been found to localize to the rough endoplasmic reticulum (ER) and Golgi apparatus in transfected cells and in a vesicle-associated intracellular distribution in SARS-CoV infected cells (Geng et al., 2005, Gunalan et al., 2011, Kumar et al., 2007, Pewe et al., 2005). Analysis of a recombinant mouse hepatitis virus (MHV) encoding SARS-CoV protein 6 has demonstrated that it enhances virulence of an attenuated murine coronavirus (Pewe et al., 2005). Protein 6 co-immunoprecipitates with viral RNAs and accelerates replication of a mouse coronavirus (Tangudu et al., 2007). Previous data have shown the intracellular membrane localization of protein 6 in recombinant MHV infected cells (Pewe et al., 2005) and suggested its possible role in the membrane-associated events of coronavirus replication cycle (Tangudu et al., 2007), including viral RNA synthesis (Gosert et al., 2002, Stertz et al., 2007), viral membrane protein synthesis, and virus assembly and secretion (de Haan and Rottier, 2005). More recently protein 6 has been shown to induce membrane rearrangement and we demonstrated that it is required for optimal replication of SARS-CoV (Zhao et al., 2009, Zhou et al., 2010). Furthermore, it has been shown that protein 6 inhibits IFN-β synthesis and signaling (Kopecky-Bromberg et al., 2007) and may act as a cell death inducer (Ye et al., 2010). The amino acid sequence of protein 6 and its previously described transmembrane domain (Zhou et al., 2010) is shown in Fig. 1A.
Fig. 1.

Amino acid sequence and transmembrane domains of SARS-coronavirus proteins 6 and 9b. (A) Amino acid sequence of protein 6 is shown in black. The transmembrane domain is highlighted in gray. (B) Amino acid sequence of protein 9b. The amino acids involved in the hydrophobic lipid-binding tunnel are highlighted in gray.
The interaction of protein 6 with other viral proteins has been previously described, although most of these interactions are limited to two-hybrid or cotransfection assays (Pan et al., 2008, von Brunn et al., 2007). Only the co-localization of protein 6 with nsp8 has been described in SARS-CoV infected cells (Kumar et al., 2007).
The protein 9b is synthesized from an alternative reading frame of the N gene in SARS-CoV and is 98 amino acids in length. Antibodies against protein 9b have been found in patients, demonstrating that it is produced during infection (Qiu et al., 2005). Expression of truncated 9b recombinant proteins in mammalian cells has previously revealed that protein 9b binds to cellular membranes and appears to be associated with intracellular vesicular structures, indicating a possible role in virus assembly via membrane association (Meier et al., 2006). Accordingly, protein 9b has been described as a virion associated protein (Xu et al., 2009), although its function remains to be fully described. The crystal structure of protein 9b indicates that this is an unusual membrane binding protein with a long hydrophobic lipid-binding tunnel (Meier et al., 2006). The amino acid sequence of protein 9b and the amino acids involved in this central hydrophobic cavity that binds lipid molecules are highlighted in Fig. 1B.
To elucidate protein 6-mediated function and to analyze any possible interaction of SARS-CoV protein 6 with other cellular and/or viral factors during the SARS-CoV viral infective cycle, a proteomic approach was carried out in SARS-CoV infected cells, which may offer more biologically relevant results than other methods and assays performed previously. Rabbit anti-protein 6 antibody and a pre-immune rabbit antibody were used for immunoprecipitation of mock and SARS-CoV infected cells. Vero E6 cells were SARS-CoV infected at an MOI of 8 or mock-infected as a control. All work was done by triplicate in biosafety level 3 containment facilities by personnel wearing positive-pressure air-purifying respirators (HEPA AirMate; 3M, Saint Paul, MN). To select the appropriate time post-infection to perform the interaction assays and to corroborate its subcellular localization during infection, a time course expression of protein 6 was monitored by Western blot and immunofluorescence assays at high multiplicity of infection (Fig. 2 ). Expression kinetic of protein 6 was monitored at various times post-infection in total cell extracts of SARS-CoV or mock infected cells by Western blot with rat anti-protein 6 antibody (Zhao et al., 2009). A specific band of 7 kDa was observed in SARS-CoV infected cell extracts (Fig. 2A, anti-protein 6) and this band was not observed in mock-infected extracts. SARS-CoV infection was corroborated monitoring nucleoprotein (N) expression (Imgenex), which appears as a double band of approximately 46 kDa (Fig. 2A, anti-nucleoprotein). Protein 6 was detected in SARS-CoV infected cells at least from 4 hours post-infection (hpi) (Fig. 2A, lane 2) and high levels of protein 6 were detected at 17 hpi (Fig. 2A, lane 4). To analyze the subcellular location of protein 6, rabbit anti-PDI as endoplasmic reticulum (ER) marker (Santa Cruz Biotechnology) (Fig. 2B, in green), and rat anti-protein 6 (Fig. 2B, in red) antibodies were used for confocal microscopy analysis. This assay revealed that protein 6 partially localized to the ER in SARS-CoV infected cells (Fig. 2B, merge). Thus, it has been shown that protein 6 is expressed at high levels from 17 hpi during the SARS-CoV infection cycle and that this protein indeed localized in the cytoplasm and partially to the ER of SARS-CoV infected cells at 17 hpi.
Fig. 2.
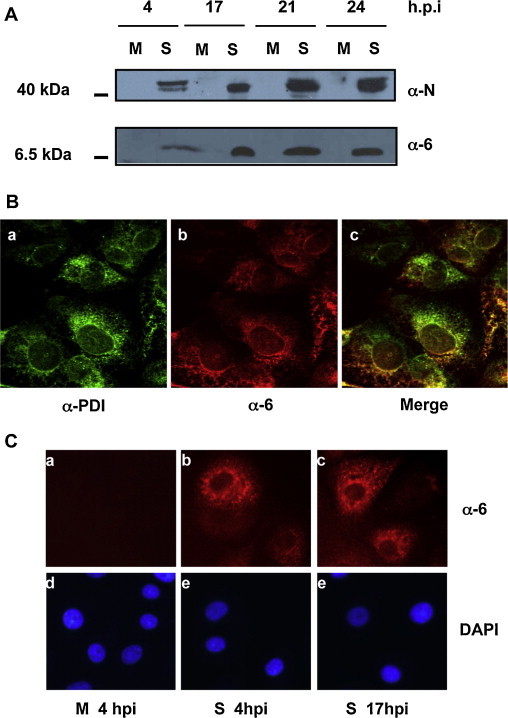
SARS-CoV protein 6 expression and subcellular localization. Vero E6 cells were infected with SARS-CoV (S) or mock-infected (M). At the hours post-infection indicated in the figure (h.p.i), cells were either fixed for immunofluorescence or total cells extracts were obtained in Laemmli buffer. (A) Total cell extracts were separated by SDS-PAGE and expression of nucleoprotein (N) and protein 6 (6) was analyzed by Western blot. (B) SARS-CoV infected cell cultures were fixed at 17 h.p.i. Confocal immunofluorescence of protein 6 and ER was developed with anti-PDI rabbit antibody (ER marker) and anti-protein 6 rat antibody. Data were visualized with Alexa Fluor 488-conjugated anti-rabbit (green) and TxRed-conjugated anti-rat (red) antibodies. Co-localization of protein 6 in the ER is shown in yellow (merge). (C) Protein 6 expression was detected by immunofluorescence using an anti-protein 6 rabbit antibody and Tx-Red-conjugated anti-rabbit antibody (a–c). DAPI (blue) was used for cellular nuclei staining (d–f). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.)
To perform interaction studies, a rabbit antibody specific for protein 6 was generated using the same methodology used to generate rat-anti-protein 6 antibody (Zhao et al., 2009). To corroborate rabbit anti-protein 6 antibody specificity, immunofluorescence analysis was carried out with rabbit-anti-protein 6 antibody at several times post infection (Fig. 2C, boxes a, b and c, in red). DAPI staining was used as a control to localize the cell nucleus (Fig. 2C, boxes d, e and f, in blue). The rabbit antibody showed a specific signal only in infected cells and the subcellular distribution pattern of the signal is similar to that observed for protein 6 (Fig. 2B and Geng et al., 2005, Pewe et al., 2005), indicating that the antibody specifically recognized the protein 6.
To identify viral and cellular proteins that interact with protein 6, immune matrices were prepared by overnight incubation of protein A-agarose with either anti-protein 6 or pre-immune rabbit sera. Soluble extracts from mock or SARS-CoV Vero E6 infected cells were prepared at 17 hpi in TNE-1% NP-40 buffer (100 mM NaCl, 5 mM EDTA, 50 mM Tris–HCl, 1% Nonidet P-40, pH 7.5) containing protease inhibitors (Roche). The matrices were incubated with the soluble extracts for 3 h and washed in the same buffer using decreasing detergent concentration. Final equilibration in 50 mM ammonium bicarbonate was required for further trypsin digestion and mass spectrometric analysis. An aliquot of the bound material was eluted in Laemmli buffer and processed for Western blot using rat-anti-protein 6 antibody to corroborate specific protein 6 immunoprecipitation (Fig. 3A). The rabbit-anti-protein 6 antibody specifically immunoprecipitated protein 6 present in SARS-CoV infected cells (Fig. 3A, lane 6), as no protein was detected with this antibody in non-infected cell extracts (Fig. 3A, lane 5), and no protein was detected using a pre-immune sera for immunoprecipitation of SARS-CoV infected cells (Fig. 3A, lane 4). The complexities of these protein samples were appreciated in a silver-stained gel image (Fig. 3B). An overwhelming amount of protein A and IgGs was detected in the immunoprecipitated samples (Fig. 3B, lines 4–7).
Fig. 3.
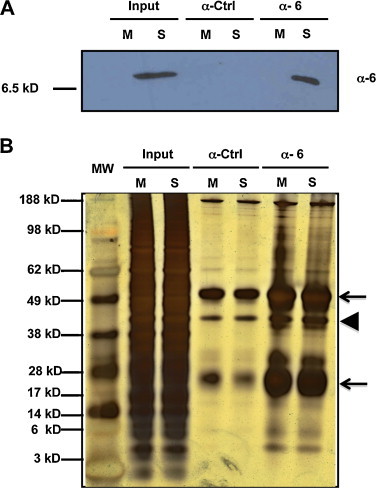
Immunoprecipitation of protein 6 in SARS-CoV infected cells. Immune matrices were prepared by incubation of protein A-Sepahrose with either anti-protein 6 (anti-protein 6) or preimmune rabbit sera (anti-control). These matrices were incubated with soluble extracts from SARS-CoV infected (S) or mock-infected cells (M). (A) After washing, aliquots of total extracts (Input) or the immunoprecipitates were analyzed by Western-blot using anti-protein 6 rat antibodies. (B) The complexity of the total extracts (Input) and immunoprecipitated samples was analyzed by silver-staining. The arrows show the IgG and the arrowhead shows the protein A.
The remaining aliquots of resin-bound immunoprecipitated proteins were digested with 1 μg of modified porcine trypsin (Sequence grade, Promega) for 1 h at 37 °C under shaking conditions (1300 rpm). The reactions were stopped by adding acetic acid and the tryptic peptides were analyzed by LC–MS on an Esquire HCT Ultra ion-trap (Bruker-Daltoniks, Bremen, Germany) mass spectrometer following described procedures (Calvo et al., 2005). Despite the high amount of protein A in comparison with other proteins, a peptide was detected within the chromatogram in the SARS-CoV infected-sample immunoprecipitated with specific anti-protein 6 antibody (retention time at 34.1 min, not shown), but not in the control sample immunoprecipitated with a pre-immune sera. The comprehensive analysis of the corresponding MS/MS spectrum (Fig. 4A) unambiguously identified the sequence LGSQLSLSMAR of the SARS-CoV protein 9b, with a Mascot score of 68, signifying individual ions scores > 57 identity or extensive homology (p < 0.05). This sequence represented 11% of protein coverage, a value which lies within described common values due to the short sequence of protein 9b. We attempted to use an anti-9b antiserum in Western blot to asses co-IP of 9b protein. However, this technique was not sensitive enough to detect 9b protein under these experimental conditions. To improve the obtained results by MS, a new set of samples was analyzed by LC–MS with a more sensitive scan mode (Single Ion Monitoring, SIM), which only analyses the indicated masses. Thus the selected 9b protein-derived ions can be detected and fragmented more efficiently. Masses at m/z 581.8 and 725.3, corresponding to doubly-charged peptides LGSQLSLSMAR and AFQSTPIVVQMTK respectively, were monitored along the gradient. Both ions were properly detected and analyzed in the anti-6 immunoprecipitated infected samples but were not detected in the control samples immunoprecipitated with a pre-immune sera (Fig. 4B). All these data indicated that proteins 6 and 9b interact in vivo.
Fig. 4.
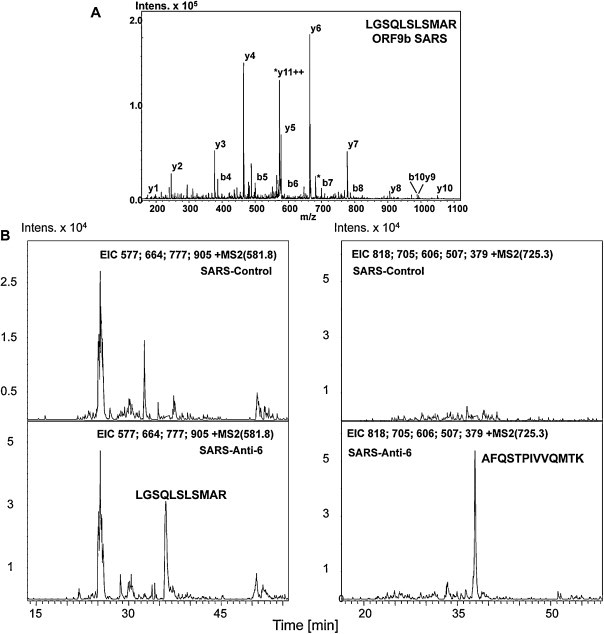
Identification of SARS-CoV interacting 9b protein by LC–MS. (A) MS/MS spectrum from the doubly-charged ion at m/z 581.8 Da spanning the sequence LGSQLSLSMAR, and corresponding to protein 9b from SARS-CoV. Figure displays the main fragmentation series (y-carboxy and b-amino). Water loss is marked with an (*). (B) Extracted Ion Chromatogram (EIC) of the SIM experiments monitoring the doubly-charged ions at m/z 581.8 (LGSQLSLSMAR, left) and 725.3 (AFQSTPIVVQMTK, right). Upper and lower panels shows respectively the results from control and anti-6-immunoprecipited SARS-infected cells preparations.
We detected for the first time the interaction of protein 6 with protein 9b using pull-down experiments during SARS-CoV infection. Previously, it has been described that SARS-CoV protein 6 interacts with karyopherin α2 and viral nsp8, nsp3 and 7b proteins (Frieman et al., 2007, Kumar et al., 2007, von Brunn et al., 2007), that have not been identified in this work. Because different techniques within this and the cited articles had been used, different results might emerge. Several SARS-CoV viral protein–protein interaction studies have shown different results depending on the technology used to perform them. Thus, two-hybrid assays carried out in mammalian cells have shown only partially overlapping results with those shown in yeast two-hybrid assays (Imbert et al., 2008, Pan et al., 2008, von Brunn et al., 2007). Surprisingly, even two different studies performing similar yeast two hybrid assays showed no overlapping interactions (Imbert et al., 2008, von Brunn et al., 2007). The presence of high levels of protein A in this work could hamper the detection of other previously described SARS-CoV protein 6-interacting proteins.
To corroborate the interaction between proteins 6 and 9b, confocal immunofluorescence assays were performed (Fig. 5 ). Mock and SARS-CoV infected Vero E6 cells were washed and fixed at 17 hpi with 10% paraformaldehyde for 20 min, and processed for immunofluorescence following described procedures (Garaigorta et al., 2005). Cells were incubated with previously described rat and mouse antibodies specific for 6 and 9b proteins, respectively (Xu et al., 2009, Zhao et al., 2009). The preparations were mounted in Prolong reagent and analyzed by confocal microscopy using a Leica TCS SP5 laser scanning system. Images were acquired sequentially every 0.5 μm employing LAS AF 2.6.0 software (Leica Microsystems). To assess the extent of co-localization of fluorescence signals, quantitative analysis was performed using the same program. A minimum of 44 cells were analyzed and all of them showed an overlap coefficient >0.64, indicating a partial co-localization of both proteins. Three representative quantifications are shown in Fig. 5 (Fig. 5, mask and co-localization).
Fig. 5.
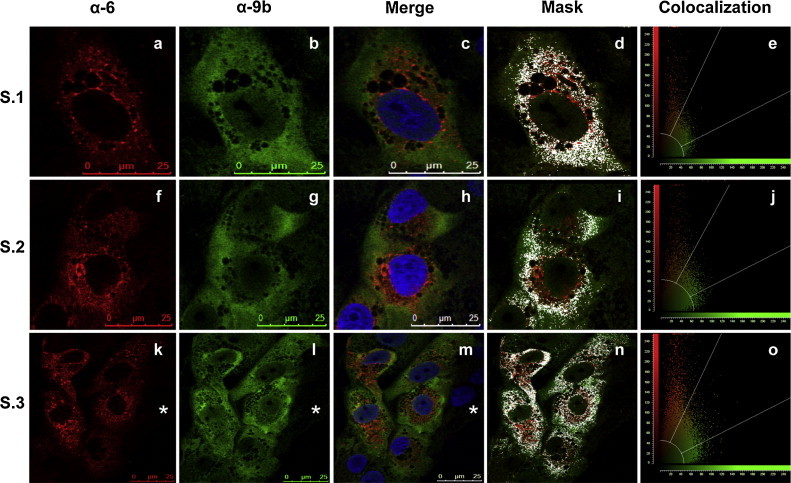
Proteins 6 and 9b partially co-localize in the cytoplasm of SARS-CoV infected cells. Vero E6 cell cultures were SARS-CoV infected and were fixed and analyzed by confocal immunofluorescence. Three representative examples are shown (S.1–S.2 and S.3). SARS-CoV protein 6 was detected with rat anti-protein 6 antibody and visualized with Tx-Red-conjugated anti-rat antibody (a, f, k). SARS-CoV protein 9b was detected with mouse anti-protein 9b antibody and visualized with FITC-conjugated anti-mouse antibody (b, g, i). Merge images show the localization of proteins 6, 9b, and DAPI stained nucleus in blue (c, h, m). The masks show the partial co-localization of proteins 6 and 9b in the cytoplasm of SARS-CoV infected cells (d, i, n). The graphics show the quantification of co-localization signal (e, j, o). The asterisk shows a non-infected cell, as a control. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.)
A cytoplasmic partial co-localization of proteins 6 and 9b during SARS-CoV infection was shown (Fig. 5, mask), which indicates that the two proteins are less than about 200 nm apart, reinforcing the notion that proteins 6 and 9b may interact in vivo. The expression of protein 9b in SARS-CoV infected cells was observed partially localized both in the cytoplasm and in the nucleus, in agreement with the described subcellular localization of recombinant 9b protein in transfected cells (Moshynskyy et al., 2007, von Brunn et al., 2007) and in SARS-CoV infected cells at similar hpi (Sharma et al., 2011) (Fig. 5, merge). The co-immunoprecipitation and partial cytosolic co-localization showed by the overlap coefficient of proteins 6 and 9b at 17 hpi is in agreement with the time kinetics shown for protein 9b during infection (Sharma et al., 2011). To corroborate that there is no cross-signal between the different channels used in the immunofluorescence assays, a non-infected cell is shown in Fig. 5 (Fig. 5, S.3 anti-6, anti-9b and merge).
We describe here an interaction of two viral proteins in vivo in SARS-CoV infected cells, where the conditions were the most likely to be biologically relevant. However, the possible existence of an additional factor mediating the interaction between proteins 6 and 9b during the infection remains to be elucidated, and could explain the lack of this interaction in the previous analysis of SARS-CoV protein–protein interactions (Pan et al., 2008, von Brunn et al., 2007). Interestingly, similar mass spectrometric-based studies herein performed have already been described for other viral–viral or viral–host protein interactions (Kang et al., 2006, Mayer et al., 2007) and for the detection of specific proteins in highly complex protein mixtures (Calvo et al., 2005, Wolf et al., 2004, Zhang et al., 2005).
SARS-CoV is the most pathogenic human CoV known (Weiss and Navas-Martin, 2005) and it encodes a set of non essential accessory proteins such as 3a, 3b, 6, 7a, 7b, 8a, 8b and 9b. These proteins, such as 3b and 6 counteract host defenses (Frieman et al., 2007, Kopecky-Bromberg et al., 2007) contributing to the high virulence of the virus. To identify new protein-mediated viral functions and intraviral protein–protein interactions, a proteomic approach using SARS-CoV 6 protein as a bait was used. We describe a novel interaction of SARS-CoV protein 6 with viral protein 9b in vivo. We also confirm a partial co-localization of both proteins in the cytoplasm of SARS-CoV infected cells by confocal microscopy, further reinforcing this conclusion. Nevertheless, further studies will be necessary to determine if this is a direct interaction or if there is any other viral/cellular protein mediating this interaction.
The interaction between these two proteins supports a possible role of protein 6 in SARS-CoV replication, which has been suggested by co-immunoprecipitation of protein 6 with viral RNAs (Pewe et al., 2005, Tangudu et al., 2007). Additional data have also suggested the implication of protein 6 in replication, as protein 6 is located in the ER and replication takes place at the double membranes from ER (Stertz et al., 2007) and we have later demonstrated the requirement of protein 6 for optimal replication of SARS-CoV (Zhao et al., 2009). Protein 9b is also a small protein, which may interact with several SARS-CoV viral proteins involved in viral RNA replication, such as nsp3N, nsp3C, nsp5, nsp7, nsp12, nsp13, nsp14, nsp15, 7a, 7b, nsp14 and nsp8 (von Brunn et al., 2007), which also interacts with protein 6 (Kumar et al., 2007). Furthermore, it has been demonstrated the interaction between protein 6 and nsp8, a second RdRp uniquely encoded by the SARS-CoV (Imbert et al., 2006). All these data could indicate an nsp8, or an RNA mediated interaction between protein 6 and 9b, but further studies will be necessary to clearly assess the interaction between these proteins.
Acknowledgements
We thank Silvia Gutierrez and Juan C. González Armas from the CNB, CSIC and CNM, ISCIII respectively, for confocal microscopy assistance. We are gratefully to Dr. B. Sun by kindly providing anti-9b antibody. We thank Dr. I. Casas and Dr. F. Pozo for scientific advises. This work was supported by the European Community Frame VI, DISSECT PROJECT, SP22-CT-2004-511060.
References
- Calisher C.H., Childs J.E., Field H.E., Holmes K.V., Schountz T. Bats: important reservoir hosts of emerging viruses. Clinical Microbiology Reviews. 2006;19(3):531–545. doi: 10.1128/CMR.00017-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo E., Escors D., Lopez J.A., Gonzalez J.M., Alvarez A., Arza E., Enjuanes L. Phosphorylation and subcellular localization of transmissible gastroenteritis virus nucleocapsid protein in infected cells. Journal of General Virology. 2005;86(Pt 8):2255–2267. doi: 10.1099/vir.0.80975-0. [DOI] [PubMed] [Google Scholar]
- Chan W.S., Wu C., Chow S.C., Cheung T., To K.F., Leung W.K., Chan P.K., Lee K.C., Ng H.K., Au D.M., Lo A.W. Coronaviral hypothetical and structural proteins were found in the intestinal surface enterocytes and pneumocytes of severe acute respiratory syndrome (SARS) Modern Pathology. 2005;18(11):1432–1439. doi: 10.1038/modpathol.3800439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow S.C., Ho C.Y., Tam T.T., Wu C., Cheung T., Chan P.K., Ng M.H., Hui P.K., Ng H.K., Au D.M., Lo A.W. Specific epitopes of the structural and hypothetical proteins elicit variable humoral responses in SARS patients. Journal of Clinical Pathology. 2006;59(5):468–476. doi: 10.1136/jcp.2005.029868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan C.A., Rottier P.J. Molecular interactions in the assembly of coronaviruses. Advances in Virus Research. 2005;64:165–230. doi: 10.1016/S0065-3527(05)64006-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler J.F., Gloza-Rausch F., Glende J., Corman V.M., Muth D., Goettsche M., Seebens A., Niedrig M., Pfefferle S., Yordanov S., Zhelyazkov L., Hermanns U., Vallo P., Lukashev A., Muller M.A., Deng H., Herrler G., Drosten C. Genomic characterization of severe acute respiratory syndrome-related coronavirus in European bats and classification of coronaviruses based on partial RNA-dependent RNA polymerase gene sequences. Journal of Virology. 2010;84(21):11336–11349. doi: 10.1128/JVI.00650-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., Berger A., Burguiere A.M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.C., Muller S., Rickerts V., Sturmer M., Vieth S., Klenk H.D., Osterhaus A.D., Schmitz H., Doerr H.W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. New England Journal of Medicine. 2003;348(20):1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- Fouchier R.A., Kuiken T., Schutten M., van Amerongen G., van Doornum G.J., van den Hoogen B.G., Peiris M., Lim W., Stohr K., Osterhaus A.D. Aetiology: Koch's postulates fulfilled for SARS virus. Nature. 2003;423(6937):240. doi: 10.1038/423240a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieman M., Yount B., Heise M., Kopecky-Bromberg S.A., Palese P., Baric R.S. SARS-CoV ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rER/Golgi membrane. Journal of Virology. 2007;81(18):9812–9823. doi: 10.1128/JVI.01012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garaigorta U., Falcon A.M., Ortin J. Genetic analysis of influenza virus NS1 gene: a temperature-sensitive mutant shows defective formation of virus particles. Journal of Virology. 2005;79(24):15246–15257. doi: 10.1128/JVI.79.24.15246-15257.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng H., Liu Y.M., Chan W.S., Lo A.W., Au D.M., Waye M.M., Ho Y.Y. The putative protein 6 of the severe acute respiratory syndrome-associated coronavirus: expression and functional characterization. FEBS Letters. 2005;579(30):6763–6768. doi: 10.1016/j.febslet.2005.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosert R., Kanjanahaluethai A., Egger D., Bienz K., Baker S.C. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. Journal of Virology. 2002;76(8):3697–3708. doi: 10.1128/JVI.76.8.3697-3708.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunalan V., Mirazimi A., Tan Y.J. A putative diacidic motif in the SARS-CoV ORF6 protein influences its subcellular localization and suppression of expression of co-transfected expression constructs. BMC Research Notes. 2011;4:446. doi: 10.1186/1756-0500-4-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Ito N., Tseng C.T., Makino S. Severe acute respiratory syndrome coronavirus 7a accessory protein is a viral structural protein. Journal of Virology. 2006;80(15):7287–7294. doi: 10.1128/JVI.00414-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Peters C.J., Makino S. Severe acute respiratory syndrome coronavirus accessory protein 6 is a virion-associated protein and is released from 6 protein-expressing cells. Journal of Virology. 2007;81(10):5423–5426. doi: 10.1128/JVI.02307-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbert I., Guillemot J.C., Bourhis J.M., Bussetta C., Coutard B., Egloff M.P., Ferron F., Gorbalenya A.E., Canard B. A second, non-canonical RNA-dependent RNA polymerase in SARS coronavirus. EMBO Journal. 2006;25(20):4933–4942. doi: 10.1038/sj.emboj.7601368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbert I., Snijder E.J., Dimitrova M., Guillemot J.C., Lecine P., Canard B. The SARS-Coronavirus PLnc domain of nsp3 as a replication/transcription scaffolding protein. Virus Research. 2008;133(2):136–148. doi: 10.1016/j.virusres.2007.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito N., Mossel E.C., Narayanan K., Popov V.L., Huang C., Inoue T., Peters C.J., Makino S. Severe acute respiratory syndrome coronavirus 3a protein is a viral structural protein. Journal of Virology. 2005;79(5):3182–3186. doi: 10.1128/JVI.79.5.3182-3186.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S., Hawkridge A.M., Johnson K.L., Muddiman D.C., Prevelige P.E., Jr. Identification of subunit-subunit interactions in bacteriophage P22 procapsids by chemical cross-linking and mass spectrometry. Journal of Proteome Research. 2006;5(2):370–377. doi: 10.1021/pr050356f. [DOI] [PubMed] [Google Scholar]
- Kopecky-Bromberg S.A., Martinez-Sobrido L., Frieman M., Baric R.A., Palese P. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. Journal of Virology. 2007;81(2):548–557. doi: 10.1128/JVI.01782-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A.E., Humphrey C.D., Shieh W.J., Guarner J., Paddock C.D., Rota P., Fields B., DeRisi J., Yang J.Y., Cox N., Hughes J.M., LeDuc J.W., Bellini W.J., Anderson L.J. A novel coronavirus associated with severe acute respiratory syndrome. New England Journal of Medicine. 2003;348(20):1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- Kumar P., Gunalan V., Liu B., Chow V.T., Druce J., Birch C., Catton M., Fielding B.C., Tan Y.J., Lal S.K. The nonstructural protein 8 (nsp8) of the SARS coronavirus interacts with its ORF6 accessory protein. Virology. 2007;366(2):293–303. doi: 10.1016/j.virol.2007.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.K., Woo P.C., Li K.S., Huang Y., Tsoi H.W., Wong B.H., Wong S.S., Leung S.Y., Chan K.H., Yuen K.Y. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(39):14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.K., Tso E.Y., Chau T.N., Tsang O.T., Choi K.W., Lai T.S. Asymptomatic severe acute respiratory syndrome-associated coronavirus infection. Emerging Infectious Diseases. 2003;9(11):1491–1492. doi: 10.3201/eid0911.030401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H., Wang H., Crameri G., Hu Z., Zhang H., Zhang J., McEachern J., Field H., Daszak P., Eaton B.T., Zhang S., Wang L.F. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310(5748):676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- Mayer D., Molawi K., Martinez-Sobrido L., Ghanem A., Thomas S., Baginsky S., Grossmann J., Garcia-Sastre A., Schwemmle M. Identification of cellular interaction partners of the influenza virus ribonucleoprotein complex and polymerase complex using proteomic-based approaches. Journal of Proteome Research. 2007;6(2):672–682. doi: 10.1021/pr060432u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier C., Aricescu A.R., Assenberg R., Aplin R.T., Gilbert R.J., Grimes J.M., Stuart D.I. The crystal structure of ORF-9b, a lipid binding protein from the SARS coronavirus. Structure. 2006;14(7):1157–1165. doi: 10.1016/j.str.2006.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moshynskyy I., Viswanathan S., Vasilenko N., Lobanov V., Petric M., Babiuk L.A., Zakhartchouk A.N. Intracellular localization of the SARS coronavirus protein 9b: evidence of active export from the nucleus. Virus Research. 2007;127(1):116–121. doi: 10.1016/j.virusres.2007.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan K., Huang C., Makino S. SARS coronavirus accessory proteins. Virus Research. 2008;133(1):113–121. doi: 10.1016/j.virusres.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J., Peng X., Gao Y., Li Z., Lu X., Chen Y., Ishaq M., Liu D., Dediego M.L., Enjuanes L., Guo D. Genome-wide analysis of protein–protein interactions and involvement of viral proteins in SARS-CoV replication. PLoS ONE. 2008;3(10):e3299. doi: 10.1371/journal.pone.0003299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris J.S., Lai S.T., Poon L.L., Guan Y., Yam L.Y., Lim W., Nicholls J., Yee W.K., Yan W.W., Cheung M.T., Cheng V.C., Chan K.H., Tsang D.N., Yung R.W., Ng T.K., Yuen K.Y. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361(9366):1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pewe L., Zhou H., Netland J., Tangudu C., Olivares H., Shi L., Look D., Gallagher T., Perlman S. A severe acute respiratory syndrome-associated coronavirus-specific protein enhances virulence of an attenuated murine coronavirus. Journal of Virology. 2005;79(17):11335–11342. doi: 10.1128/JVI.79.17.11335-11342.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu M., Shi Y., Guo Z., Chen Z., He R., Chen R., Zhou D., Dai E., Wang X., Si B., Song Y., Li J., Yang L., Wang J., Wang H., Pang X., Zhai J., Du Z., Liu Y., Zhang Y., Li L., Sun B., Yang R. Antibody responses to individual proteins of SARS coronavirus and their neutralization activities. Microbes and Infection. 2005;7(5-6):882–889. doi: 10.1016/j.micinf.2005.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan P.L., Firth C., Street C., Henriquez J.A., Petrosov A., Tashmukhamedova A., Hutchison S.K., Egholm M., Osinubi M.O., Niezgoda M., Ogunkoya A.B., Briese T., Rupprecht C.E., Lipkin W.I. Identification of a severe acute respiratory syndrome coronavirus-like virus in a leaf-nosed bat in Nigeria. MBio. 2010;1(4) doi: 10.1128/mBio.00208-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rihtaric D., Hostnik P., Steyer A., Grom J., Toplak I. Identification of SARS-like coronaviruses in horseshoe bats (Rhinolophus hipposideros) in Slovenia. Archives of Virology. 2010;155(4):507–514. doi: 10.1007/s00705-010-0612-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.H., Tong S., Tamin A., Lowe L., Frace M., DeRisi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., Gunther S., Osterhaus A.D., Drosten C., Pallansch M.A., Anderson L.J., Bellini W.J. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300(5624):1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- Schaecher S.R., Mackenzie J.M., Pekosz A. The ORF7b protein of severe acute respiratory syndrome coronavirus (SARS-CoV) is expressed in virus-infected cells and incorporated into SARS-CoV particles. Journal of Virology. 2007;81(2):718–731. doi: 10.1128/JVI.01691-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma K., Akerstrom S., Sharma A.K., Chow V.T., Teow S., Abrenica B., Booth S.A., Booth T.F., Mirazimi A., Lal S.K. SARS-CoV 9b protein diffuses into nucleus, undergoes active Crm1 mediated nucleocytoplasmic export and triggers apoptosis when retained in the nucleus. PLoS ONE. 2011;6(5):e19436. doi: 10.1371/journal.pone.0019436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Bredenbeek P.J., Dobbe J.C., Thiel V., Ziebuhr J., Poon L.L., Guan Y., Rozanov M., Spaan W.J., Gorbalenya A.E. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. Journal of Molecular Biology. 2003;331(5):991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stertz S., Reichelt M., Spiegel M., Kuri T., Martinez-Sobrido L., Garcia-Sastre A., Weber F., Kochs G. The intracellular sites of early replication and budding of SARS-coronavirus. Virology. 2007;361(2):304–315. doi: 10.1016/j.virol.2006.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tangudu C., Olivares H., Netland J., Perlman S., Gallagher T. Severe acute respiratory syndrome coronavirus protein 6 accelerates murine coronavirus infections. Journal of Virology. 2007;81(3):1220–1229. doi: 10.1128/JVI.01515-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Brunn A., Teepe C., Simpson J.C., Pepperkok R., Friedel C.C., Zimmer R., Roberts R., Baric R., Haas J. Analysis of intraviral protein–protein interactions of the SARS Coronavirus ORFeome. PLoS ONE. 2007;2:e459. doi: 10.1371/journal.pone.0000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss S.R., Navas-Martin S. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiology and Molecular Biology Reviews. 2005;69(4):635–664. doi: 10.1128/MMBR.69.4.635-664.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf R., Hoffmann T., Rosche F., Demuth H.U. Simultaneous determination of incretin hormones and their truncated forms from human plasma by immunoprecipitation and liquid chromatography–mass spectrometry. Journal of Chromatography B: Analytical Technologies in the Biomedical and Life Sciences. 2004;803(1):91–99. doi: 10.1016/j.jchromb.2003.11.044. [DOI] [PubMed] [Google Scholar]
- Xu K., Zheng B.J., Zeng R., Lu W., Lin Y.P., Xue L., Li L., Yang L.L., Xu C., Dai J., Wang F., Li Q., Dong Q.X., Yang R.F., Wu J.R., Sun B. Severe acute respiratory syndrome coronavirus accessory protein 9b is a virion-associated protein. Virology. 2009;388(2):279–285. doi: 10.1016/j.virol.2009.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Z.D., Wong C.K., Li P., Xie Y. The role of SARS-CoV protein, ORF-6, in the induction of host cell death. Hong Kong Medical Journal. 2010;16(5 Suppl. 4):22–26. [PubMed] [Google Scholar]
- Zhang Y., Wolf-Yadlin A., Ross P.L., Pappin D.J., Rush J., Lauffenburger D.A., White F.M. Time-resolved mass spectrometry of tyrosine phosphorylation sites in the epidermal growth factor receptor signaling network reveals dynamic modules. Molecular and Cellular Proteomics. 2005;4(9):1240–1250. doi: 10.1074/mcp.M500089-MCP200. [DOI] [PubMed] [Google Scholar]
- Zhao J., Falcon A., Zhou H., Netland J., Enjuanes L., Perez Brena P., Perlman S. Severe acute respiratory syndrome coronavirus protein 6 is required for optimal replication. Journal of Virology. 2009;83(5):2368–2373. doi: 10.1128/JVI.02371-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Ferraro D., Zhao J., Hussain S., Shao J., Trujillo J., Netland J., Gallagher T., Perlman S. The N-terminal region of severe acute respiratory syndrome coronavirus protein 6 induces membrane rearrangement and enhances virus replication. Journal of Virology. 2010;84(7):3542–3551. doi: 10.1128/JVI.02570-09. [DOI] [PMC free article] [PubMed] [Google Scholar]


