Abstract
The Rhabdoviridae is one of the most ecologically diverse families of RNA viruses with members infecting a wide range of organisms including placental mammals, marsupials, birds, reptiles, fish, insects and plants. The availability of complete nucleotide sequences for an increasing number of rhabdoviruses has revealed that their ecological diversity is reflected in the diversity and complexity of their genomes. The five canonical rhabdovirus structural protein genes (N, P, M, G and L) that are shared by all rhabdoviruses are overprinted, overlapped and interspersed with a multitude of novel and diverse accessory genes. Although not essential for replication in cell culture, several of these genes have been shown to have roles associated with pathogenesis and apoptosis in animals, and cell-to-cell movement in plants. Others appear to be secreted or have the characteristics of membrane-anchored glycoproteins or viroporins. However, most encode proteins of unknown function that are unrelated to any other known proteins. Understanding the roles of these accessory genes and the strategies by which rhabdoviruses use them to engage, divert and re-direct cellular processes will not only present opportunities to develop new anti-viral therapies but may also reveal aspects of cellar function that have broader significance in biology, agriculture and medicine.
Keywords: Rhabdovirus, Accessory gene, Vesiculovirus, Ephemerovirus, Lyssavirus, Novirhabdovirus, Cytorhabdovirus, Nucleorhabdovirus, Insect-transmitted rhabdovirus
1. Introduction
The Rhabdoviridae is arguably the most diverse family of RNA viruses. Rhabdoviruses have been isolated from a wide range of organisms including placental mammals, marsupials, birds, reptiles, fish, insects and plants (Kuzmin et al., 2009). Their ecology includes marine, freshwater and terrestrial environments. Transmission can be via insect vectors in which they replicate, by direct horizontal transfer via plant sap (wounds or grafts), aerosol, animal bite or immersion in infected water, or by vertical transovarial passage. The Rhabdoviridae is one of only three virus families that include members that infect plants, vertebrates and invertebrates. Replication can occur in the cytoplasm or the nucleus of infected cells. Infection can be either acute or persistent and rhabdoviruses can cause infections that may be unapparent or result in disease that can vary from being mild to severe or even fatal. Over 200 rhabdoviruses have been identified to date and most of them are poorly described (Dietzgen et al., 2012).
For many years, knowledge of rhabdovirus molecular biology has been based primarily on extensive studies of vesicular stomatitis virus (VSV) and rabies virus (RABV). These rhabdoviruses, share very similar and relatively simple genome organisations with only five structural protein genes (N, P, M, G and L) that are transcribed sequentially from the (–) RNA genome as monocistronic mRNAs. Although some variations are now evident, such as the long non-coding region in the RABV G gene (designated pseudogene ψ) (Tordo et al., 1986) and the small alternative open reading frames (ORFs) in the VSV P gene (C and C′) (Peluso et al., 1996), these viruses have generally been regarded as the rhabdovirus archetypes and as very suitable models for studying virus–host interactions. However, it is becoming evident that the ecological diversity displayed by rhabdoviruses is also apparent in the complexity of their genome organisation and in the increasing array of additional proteins which they encode. Although the functions of many of these proteins are yet unknown, preservation of the five canonical structural protein genes (3′-N-P-M-G-L-5′ in negative polarity) by all known rhabdoviruses suggests that the additional proteins may not be essential for virus replication in culture and are likely to have functions associated with enhancing replication efficiency, blocking host innate immune defences, modulating cellular signalling pathways, re-directing normal cellular functions, and allowing effective cell-to-cell transmission. This expectation has been supported by limited studies to date on some of these proteins. By analogy with other RNA viruses with relatively complex genome organisations (e.g., coronaviruses, lentiviruses, paramyxoviruses), we have therefore adopted the term ‘accessory genes’ to describe the ORFs encoding these novel and potentially informative proteins.
2. The Rhabdoviridae
Rhabdoviruses are non-segmented (–) ssRNA viruses that, together with paramyxoviruses, filoviruses and bornaviruses, are classified in the order Mononegavirales. The family Rhabdoviridae currently comprises six genera containing 46 species, five species unassigned to a genus and more than 150 viruses that have not been formally classified (Dietzgen et al., 2012). Several likely species and two proposed new genera (Sigmavirus and Tibrovirus) are currently under review for classification, and a number of other viruses have been identified as rhabdoviruses by morphology and/or serology but have not been further characterised.
The genus Lyssavirus includes Rabies virus and the rabies-related viruses Mokola virus, Lagos bat virus, Duvenhage virus, European bat lyssavirus 1, European bat lyssavirus 2 and Australian bat lyssavirus, Irkut virus, Khujand virus, Aravan virus and West Caucasian bat virus as currently designated species. Another unclassified lyssavirus, Shimoni bat virus, has also recently been identified (Kuzmin et al., 2010). Lyssaviruses naturally infect mammals and almost all have been isolated from bats which appear to be the primary natural reservoir (Delmas et al., 2008). Transmission is usually by transfer of saliva in a bite puncture wound. They are neurotropic rhabdoviruses and most have been associated with rabies encephalitis in humans or animals (Rupprecht et al., 2002).
The genus Vesiculovirus comprises an ecologically diverse but genetically similar group of viruses. Viruses causing vesicular stomatitis in cattle, pigs and horses in the Americas are referred to collectively as serotypes of vesicular stomatitis virus (VSV) and classified as the species Vesicular stomatitis Indiana virus, Vesicular stomatitis New Jersey virus and Vesicular stomatitis Alagoas virus. Other recognised mammalian vesiculovirus species include Cocal virus, Piry virus, Carajas virus and Maraba virus that are endemic in the Americas, Isfahan virus from Asia and Chandipura virus which occurs in both Asia and Africa. Most of these viruses have been isolated from phlebotomine sand flies (Lutzomyia, Phlebotomus and Sergentomyia spp.) or black flies (Simulium spp.) which are considered the primary vectors. However, biting midges (Culicoides spp.), mosquitoes and other insects have also been implicated as potential vectors and transmission can also occur by direct contact between infected vertebrates (Comer and Tesh, 1991, Letchworth et al., 1999, Rodríguez, 2002, Stallknecht et al., 2001). Vesiculoviruses can also infect humans and some have been associated with serious disease (Rao et al., 2004). The genus Vesiculovirus also currently contains the recognised species Spring viraemia of carp virus and several other important pathogens of fish, including pike fry rhabdovirus, trout rhabdovirus, sea trout rhabdovirus, and Siniperca chuatsi rhabdovirus, ulcerative disease rhabdovirus, eel virus American and eel virus European X, also appear to be members of the genus (Dietzgen et al., 2012, Hoffmann et al., 2005). Transmission of fish vesiculoviruses can occur by immersion in infected water and does not appear to involve arthropod vectors (Ahne et al., 2002).
The genus Novirhabdovirus comprises a second group of fish rhabdoviruses that appear to have a very different evolutionary history from the fish vesiculoviruses. Species include Infectious hematopoietic necrosis virus and Viral hemorrhagic septicemia virus which are important pathogens of salmonids, but can also infect a very wide range of other fish species (Walker and Winton, 2010). The species Hirame rhabdovirus and Snakehead rhabdovirus are pathogens of marine fish in temperate and tropical regions of Asia, respectively (Johnson et al., 1999, Kim et al., 2005). As for the fish vesiculoviruses, transmission is directly through mucosal surfaces or the skin by immersion in infected water and does not appear to involve an arthropod vector.
The genus Ephemerovirus comprises arthropod-borne rhabdoviruses that have been isolated from mosquitoes and/or biting midges (Culicoides spp.) and infect primarily ruminants (Walker, 2005). The type species Bovine ephemeral fever virus and the newly recognised species Kotonkan virus each cause a disabling febrile disease in cattle and water buffalo (Kemp et al., 1973, St. George and Standfast, 1988). Other ephemeroviruses, including recognised species Berrimah virus and Adelaide River virus, and likely species Obodhiang virus, Kimberley virus, Malakal virus and Puchong virus, have been isolated only from insects or from healthy cattle and their association with disease is not yet known (Walker et al., in press).
Plant-adapted rhabdoviruses are separated taxonomically into two genera, Cytorhabdovirus and Nucleorhabdovirus, based on their site of virus replication and morphogenesis (Dietzgen et al., 2012). Cytorhabdoviruses replicate in the cytoplasm of infected cells and virions bud in association with endoplasmic reticulum membranes. The type species of the genus is Lettuce necrotic yellows virus and it includes eight additional recognised species. Nucleorhabdoviruses replicate in the nuclei of infected plant cells. Morphogenesis occurs at the inner nuclear membrane and virions accumulate in perinuclear spaces. The type species is Potato yellow dwarf virus and there are nine additional recognised species in this genus. Viruses in both genera infect monocot and dicot plants and are transmitted by leafhoppers, planthoppers or aphids in which they also replicate (Jackson et al., 2005).
Two other genera are currently under consideration for inclusion in the Rhabdoviridae. The proposed genus Sigmavirus comprises rhabdoviruses infecting only flies (Drosophila spp.) with which they have co-evolved. They are transmitted vertically and confer CO2 sensitivity on the host. There are five proposed species including Drosophila melanogaster sigmavirus (commonly called sigma virus of Drosophila) which is the most extensively studied of the sigmaviruses (Longdon et al., 2010, Longdon et al., 2011). Like the genus Ephemerovirus, the proposed genus Tibrovirus comprises antigenically related rhabdoviruses that primarily infect cattle and water buffalo, and are transmitted by Culicoides species midges. However, tibroviruses and ephemeroviruses have different evolutionary histories as is reflected in their phylogenetic relationships and genome organisations. Tibrogargan virus and Coastal Plains virus, each isolated in Australia, are currently proposed as species in the new genus (Gubala et al., 2011). Bivens Arm virus, isolated from Culicoides insignis in the USA, also infects cattle and water buffalo and may be a third species in the proposed genus (Gibbs et al., 1989).
Complete genome sequences are also available for several other unclassified animal rhabdoviruses that are considered in this review. Short sequences in the L and N gene have been used to determine phylogenetic relationships between viruses in the established and proposed genera and many other unclassified animal rhabdoviruses (Bourhy et al., 2005, Kuzmin et al., 2006). One monophyletic cluster identified by this approach includes a geographically and ecologically diverse group of viruses termed the Hart Park group because of the close serological relationship of one of the members, Flanders virus, to Hart Park virus. Both of these viruses have been isolated from mosquitoes and infect birds in North America (Boyd, 1972). The Hart Park phylogenetic group also includes Wongabel virus which was isolated from biting midges (Culicoides spp.) in Australia and appears to infect birds, and Ngaingan virus, also isolated from biting midges in Australia but whose vertebrate hosts include marsupials (Doherty et al., 1973, Gubala et al., 2010). Not included in the Hart Park group but related to each other phylogenetically (but not serologically) are tupaia rhabdovirus isolated from hepatocarcinoma cells of a tree shrew (Tupaia belangeri) imported to the USA from Thailand (Kurz et al., 1986), and Durham virus, which was isolated from the brain of a moribund American coot (Fulica americana) in the USA (Allison et al., 2011). Oak Vale virus, isolated from mosquitoes in Australia and infecting feral pigs, and mandarin fish (Siniperca chuatsi) rhabdovirus (SCRV) are other currently unclassified rhabdoviruses that are considered in this review (Cybinski and Muller, 1990, Zhang et al., 2007).
Rhabdoviruses for which complete genome sequences are available are listed in Table 1 .
Table 1.
Rhabdoviruses for which complete genome sequences are currently available.
| Genus and species | Abbreviation | GenBank accessiona | Genome length (nt) | ORFs |
|---|---|---|---|---|
| Vesiculovirus | ||||
| Vesicular stomatitis Indiana virus | VSINV | NC001560 | 11,161 | 6 |
| Vesicular stomatitis Alagoas virus | VSAV | EU373658 | 11,070 | 5 |
| Chandipura virus | CHPV | GU212858 | 11,120 | 6c |
| Cocal virus | COCV | EU373657 | 11,003 | 6c |
| Isfahan virus | ISFV | AJ810084 | 11,088 | 6c |
| Maraba virus | MARAV | HQ660076 | 11,135 | 6 |
| Spring viraemia of carp virus | SVCV | NC002803 | 11,019 | 5 |
| Pike fry rhabdovirus | PFRV | FJ872827 | 11,097 | 5 |
| Eel virus European X | EVEX | FN557213 | 11,806 | 6 |
| Lyssavirus | ||||
| Rabies virus | RABV | NC001542 | 11,932 | 6c |
| Mokola virus | MOKV | NC006429 | 11,940 | 6c |
| Duvenhage virus | DUVV | EU293119 | 11,976 | 6c |
| Lagos bat virus | LBV | EU293108 | 12,016 | 6c |
| European bat lyssavirus 1 | EBLV1 | NC009527 | 11,966 | 6c |
| European bat lyssavirus 2 | EBLV2 | NC009528 | 11,930 | 5 |
| Australian bat lyssavirus | ABLV | NC003243 | 11,822 | 6c |
| Aravan virus | ARAV | EF614259 | 11,918 | 5 |
| Irkut virus | IRKV | EF614260 | 11,980 | 6c |
| Khujand virus | KHUV | EF614261 | 11,903 | 6c |
| West Caucasian bat virus | WCBV | EF614258 | 12,278 | 6c |
| Shimoni bat virus | SHIBV | GU170201 | 12,045 | 6c |
| Ephemerovirus | ||||
| Bovine ephemeral fever virus | BEFV | NC002526 | 14,900 | 12 |
| Berrimah virus | BRMV | HM461974 | 15,024 | 10 |
| Adelaide River virus | ARV | na | 15,119 | 10 |
| Kotonkon virus | KOTV | HM474855 | 15,867 | 11 |
| Obodhiang virus | OBOV | HM856902 | 14,717 | 10 |
| Kimberley virus | KIMV | na | 15430b | 10 |
| Novirhabdovirus | ||||
| Infectious hematopoietic necrosis virus | IHNV | NC001652 | 11,131 | 7c |
| Viral hemorrhagic septicemia virus | VHSV | NC000855 | 11,158 | 7c |
| Hirame rhabdovirus | HRRV | NC005093 | 11,034 | 6 |
| Snakehead rhabdovirus | SHRV | NC000903 | 11,550 | 7c |
| Cytorhabdovirus | ||||
| Lettuce necrotic yellows virus | LNYV | NC007642 | 12,807 | 6 |
| Lettuce yellow mottle virus | LYMoV | NC011532 | 12,926 | 6 |
| Northern cereal mosaic virus | NCMV | NC002251 | 13,222 | 9 |
| Nucleorhabdovirus | ||||
| Potato yellow dwarf virus | PYDV | GU734660 | 12,875 | 7 |
| Maize fine streak virus | MFSV | NC005974 | 13,782 | 7 |
| Maize mosaic virus | MMV | NC005975 | 12,133 | 6b |
| Sonchus yellow net virus | SYNV | NC001615 | 13,720 | 6 |
| Rice yellow stunt virus | RYSV | NC003746 | 14,042 | 7 |
| Taro vein chlorosis virus | TaVCV | NC006942 | 12,020 | 6 |
| Maize Iranian mosaic virus | MIMV | NC011542 | 12,381 | 6b |
| Sigmavirus (proposed genus and species) | ||||
| Drosophila melanogaster sigma virus | DmSV | GQ375258 | 12,625 | 7c |
| Drosophila obscura sigma virus | DoSV | GQ410979 | 12,676 | 7c |
| Tibrovirus (proposed genus and species) | ||||
| Tibrogargan virus | TIBV | GQ294472 | 13,298 | 8 |
| Coastal Plains virus | CPV | GQ294473 | 13,203 | 9 |
| Unassigned species | ||||
| Flanders virus | FLAV | AH012179 | 12,965 | 9 |
| Ngaingan virus | NGAV | NC013955 | 15,764 | 15 |
| Wongabel virus | WONV | NC011639 | 13,196 | 10 |
| Tupaia virus | TRV | NC007020 | 11,440 | 7 |
| Unclassified rhabdoviruses | ||||
| Durham virus | DURV | FJ952155 | 11092b | 7 |
| Oak Vale virus | OAKV | GQ294474 | 9414b | 8 |
| Siniperca chuatsi rhabdovirus | SCRV | NC008514 | 11,545 | 7c |
| Moussa virus | MOUV | FJ985748 | 11,526 | 7c |
Where multiple isolates of a virus has been sequenced, the NCBI GenBank reference sequence is presented (denoted by the prefix NC).
Incomplete terminal UTR sequences.
Includes predicted ORF in the P-gene.
3. Rhabdovirus genome organisation and transcription strategy
The (–) ssRNA rhabdovirus genome ranges in size from approximately 11 kb to almost 16 kb. The five structural protein genes are arranged in the same linear order (3′-N-P-M-G-L-5′) and may be interspersed with one or more additional genes. At each end of the genome, 3′ leader and 5′ trailer sequences of approximately 50–100 nt contain the promoter sequences that initiate replication of the genome and anti-genome, respectively. The 3′ leader and 5′ trailer sequences of plant-adapted rhabdoviruses are considerably longer at 84–206 nt and 145–389 nt, respectively (Jackson et al., 2005). In transcription mode, the RNA-dependent RNA polymerase moves progressively along the genome to transcribe the short leader RNA (l) and then to transcribe, cap and polyadenylate mRNAs from each gene in decreasing molar abundances (Banerjee and Barik, 1992). Critical to this process are transcription initiation (TI) and transcription termination/polyadenylation (TTP) sequences flanking each gene (Barr et al., 2002). These sequences are relatively well conserved in each species and are similar in all species within a genus. The termination of transcription and polyadenylation of mRNAs are critically dependent on a stretch of seven uridine residues that are usually preceded by a guanosine residue. Following polyadenylation, transcription is re-initiated at the next available TI sequence which follows a non-transcribed intergenic region that may be from one to approximately 50 nt in length (Banerjee and Barik, 1992). Corruption of the [U]7 stretch in TPP sequences of animal rhabdoviruses leads to read-through, resulting in polycistronic mRNAs (Barr et al., 1997). In plant-adapted rhabdoviruses, the incorruptible component of the polyadenylation signal is UUMUUUU(U) (Jackson et al., 2005). As discussed below, rhabdovirus genes may contain multiple long open reading frames (ORFs), either overlapping or sequentially in the transcriptional units, and these must be expressed by internal initiation of translation. All evidence suggests that the fundamental processes of genome replication and transcription are common to all rhabdoviruses infecting vertebrates, insects and plants.
4. Rhabdovirus accessory genes
The five major structural proteins of VSV and RABV are amongst the most intensely studied of all viral proteins. Although there are reported variations in their ancillary functions, the primary functions and structural characteristics of these proteins are conserved across all rhabdoviruses and have been reviewed in detail on many previous occasions (Albertini et al., 2008, Assenberg et al., 2010, Coll, 1995, Jackson et al., 2005, Jayakar et al., 2004, Redinbaugh and Hogenhout, 2005, Roche et al., 2008, Rupprecht et al., 2002). In contrast, the number and locations of accessory genes varies widely both within and between taxa (Fig. 1, Fig. 2 ). In most cases, levels of amino acid sequence identity between cognate accessory proteins are quite low and, with a few exceptions, their specific functions do not appear to be conserved between taxa. As discussed in detail below, some occur as alternative or overlapping ORFs within the major structural protein genes; many occur in the regions between the major structural protein genes as new genes containing single or multiple ORFs. Over 35 rhabdovirus accessory genes have been identified to date and it is likely that many more will emerge as the number of complete genome sequences continues to grow.
Fig. 1.
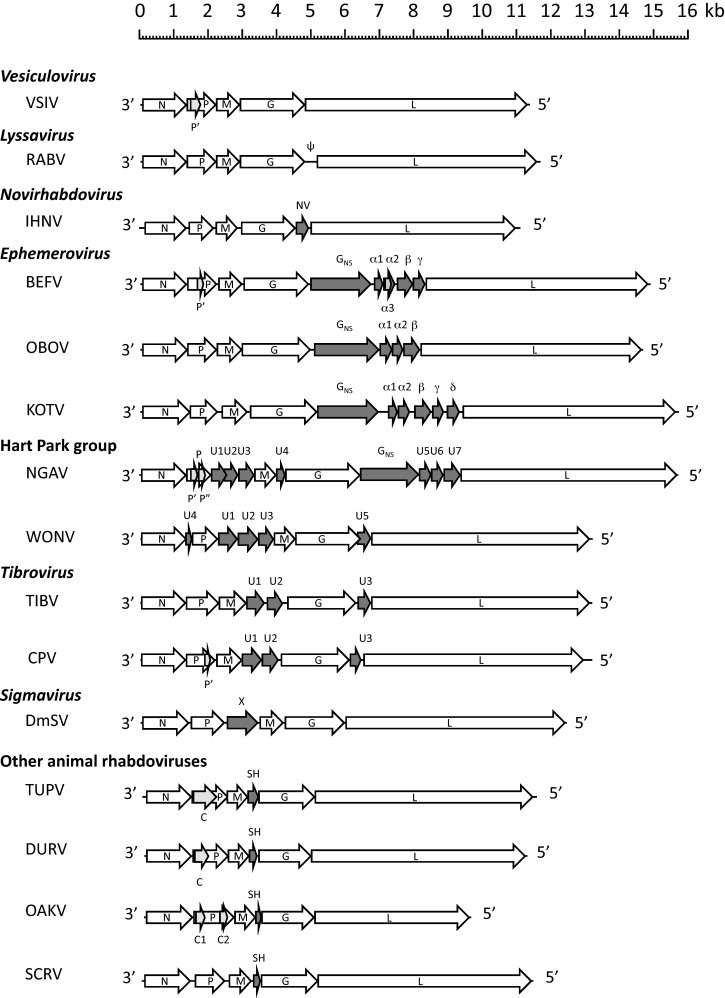
Genome organisation of animal rhabdoviruses (including sigma virus of Drosophila). The positions of ORFs are indicated and those encoding accessory proteins are shaded. Abbreviations of virus names are as listed in Table 1.
Fig. 2.
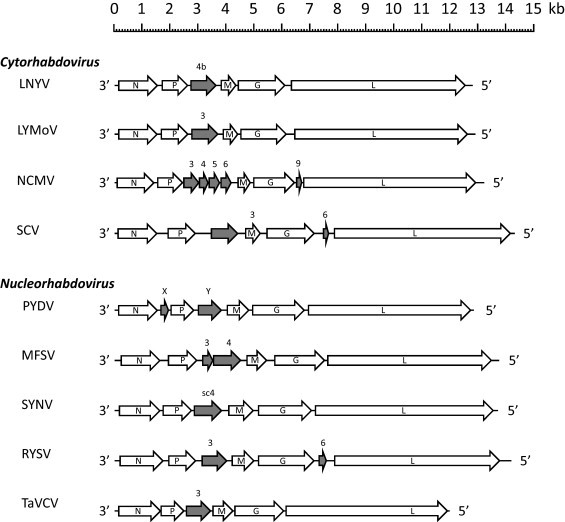
Genome organisation of plant-adapted rhabdoviruses. The positions of ORFs are indicated and those encoding accessory proteins are shaded. Abbreviations of virus names are as listed in Table 1.
4.1. Vesiculovirus P and M genes
Although the vesiculovirus genome is the smallest and arguably the least complex of all rhabdoviruses, containing only the 5 canonical structural protein genes, additional proteins are expressed from both the P and M genes (Fig. 1). The P genes of most vesiculoviruses encode a small highly basic protein (C) in an alternative ORF. In VSV New Jersey and VSV Indiana, two forms of the C protein (C and C′) are expressed in infected cells by translation initiation at alternative start codons (Peluso et al., 1996, Spiropoulou and Nichol, 1993). There is evidence that the C protein is associated with nucleocapsids and increases the activity and fidelity of viral RNA polymerase activity in vitro (Peluso et al., 1996). However, C and C′ are not essential for replication in cell culture and a VSV infectious clone lacking these proteins has been shown to have identical growth characteristics to wild-type virus (Kretzschmar et al., 1996). The predicted sizes of the putative C proteins in viruses examined to date range from 41 amino acids (Piry virus) to 93 amino acids (Cocal virus). Interestingly, although spring viraemia of carp virus has the potential to encode highly basic proteins of 83 and 57 amino acids in an alternative ORF in the P gene (Hoffmann et al., 2002, Teng et al., 2007), there appears to be no alternative ORFs in the pike fry rhabdovirus P gene (Chen et al., 2009), suggesting its function may not be conserved across all fish vesiculoviruses. However, these data should be viewed cautiously as a single point mutation during adaptation to cell culture would be sufficient and inhibit expression of these accessory proteins by codon modification. It has also been reported that a small (7 kDa) protein is expressed from the VSV P gene from the same ORF as the P protein (Herman, 1986). The presence of alternative ORFs in the P gene of vesiculoviruses mirrors the situation in paramyxoviruses in which the complex expression of the C and V family of proteins in overlapping open reading frame or by RNA editing is very well characterised (Nagai and Kato, 2004). The paramyxovirus C and V proteins have various functions associated with infection and pathogenesis including regulation of viral RNA synthesis, virion assembly and budding, inhibition of apoptosis, and in binding to components of the Jak/STAT signalling pathway to antagonize the cellular interferon response (Fontana et al., 2008, Irie et al., 2010, Kurotani et al., 1998).
The VSV M protein is a multi-functional protein that, as well as being a structural component of virions, is involved during infection in inhibition of viral transcription, assembly and budding, inhibition of host gene expression and perturbation of the cell cytoskeleton (Black and Lyles, 1992, Blondel et al., 1990, Coulon et al., 1990). The M protein also blocks the host anti-viral response and promotes apoptosis both indirectly through inhibition of host gene expression and directly by targeting the extrinsic pathway and promoting expression of pro-apoptotic genes (Kopecky et al., 2001, Pearce and Lyles, 2009). The VSV M gene is also expressed in infected cells from two alternative initiation codons in the same frame, producing the M2 and M3 proteins (Jayakar and Whitt, 2002). Little is known of the function of these proteins but they appear to be involved in cell rounding and cytopathology. Although alternative initiation codons are available in the M proteins of other vesiculoviruses, it is not known if they are utilised.
4.2. Lyssavirus ‘pseudogenes’ (ψ) and P genes
The lyssavirus genome also conforms to the basic canonical arrangement of the genes encoding the five major structural proteins but lyssaviruses share the common characteristic of a very long 3′ non-coding region in the G gene (Fig. 1). The region varies in length from approximately 400 to 700 nt in different lyssaviruses and, in one lineage of laboratory-adapted RABV strains, it is truncated by an additional TTP signal 70 nucleotides downstream of the G protein ORF, leading to the suggestion that it may be the remnant of an ancestral gene or pseudogene (ψ) (Conzelmann et al., 1990, Morimoto et al., 1989, Tordo et al., 1986). However, although the long 3′ non-coding region is preserved, the additional TTP is absent from wild-type rabies viruses isolated directly from infected animals and from other lyssaviruses (Ravkov et al., 1995). In West Caucasian bat virus (WCBV), the region extends 697 nucleotides beyond the G protein ORF and, uniquely, contains an ORF of 180 nucleotides but there is no evidence of expression of the second ORF in WCBV-infected cells (Kuzmin et al., 2008). Universal preservation of the ψ region in wild-type lyssaviruses suggests it has functional significance but a RABV infectious clone lacking the region displayed normal growth characteristics in fibroblasts and neuronal cells in culture and no difference in axonal spread or pathogenicity in mice (Ceccaldi et al., 1998, Schnell et al., 1994). Nevertheless, there is some evidence that, in conjunction with the G protein, the RABV ψ region does contribute to neuroinvasiveness from peripheral sites of infection (Faber et al., 2004). Proposed functions of the region include attenuation of L gene expression, stabilisation of the G mRNA and interaction of the mRNA with host cell proteins (Ravkov et al., 1995).
Like the vesiculoviruses, lyssaviruses also commonly encode small, highly basic proteins in alternative ORFs in the P gene (Nadin-Davis et al., 2002). The lyssavirus ORFs usually occur in the same region of the P gene as vesiculovirus C ORFs but they vary in size in different isolates and do not appear to be selectively retained by a locally constrained mutation frequency (Nadin-Davis et al., 2002). It is not known if lyssavirus C proteins are expressed and the variability of their detection in different isolates suggests they may have little functional relevance. However, if their role is to facilitate infection in vivo, sequence data on cell culture-adapted isolates should be regarded cautiously as introduced point mutations in the hypervariable P gene may be selected to interrupt functional C ORFs.
In addition to the full-length P protein, RABV P gene also encodes four smaller products (P2–P5) in the same reading frame as a result of leaky scanning of the ribosome on the P mRNA (Chenik et al., 1995). Due to a nuclear localisation signal (NLS) in the C-terminal domain and a nuclear export signal (NES) in the N-terminal domain, the various forms of the P protein accumulate differentially in the cell such that the P1 (P) and P2 forms are cytoplasmic whereas P3, P4 and P5 are transported to the nucleus (Moseley et al., 2007, Pasdeloup et al., 2005). The P isoforms antagonise cellular interferon responses by several different mechanisms including inhibiting the phosphorylation of cytoplasmic interferon regulatory factor 3 (IRF3), binding to nuclear and cytoplasmic STAT1 and STAT2, and interacting with promyelocytic leukaemia (PML) nuclear bodies (Blondel et al., 2002, Brzozka et al., 2005, Brzozka et al., 2006, Chelbi-Alix et al., 2006). The P3 isoform has also been shown to mediate the sequestration of STAT1 away from the nucleus by a microtubule-associated mechanism, preventing nuclear import and activation of the interferon response (Moseley et al., 2009). The interferon-antagonistic activity of the P protein isoforms are important determinants of the pathogenicity of RABV in mice (Ito et al., 2010, Rieder et al., 2011).
4.3. Novirhabdovirus NV genes
In the region of the genome occupied by the lyssavirus ψ non-coding region (i.e., between the G and L genes), the novirhabdoviruses contain an additional gene of approximately 500 nucleotides (Kurath et al., 1985, Kurath et al., 1997, Kurath and Leong, 1985) (Fig. 1). The non-virion (NV) ORF is flanked by TI and TTP sequences, and encodes a neutral to mildly acidic protein of 110–122 amino acids (13.2–13.6 kDa) (Kurath and Leong, 1985, Schutze et al., 1996). The NV proteins of infectious haematopoietic necrosis virus (IHNV) and viral hemorrhagic septicaemia virus (VHSV) are expressed at low levels in infected cells but do not occur in virions (Basurco and Benmansour, 1995, Schutze et al., 1996). The level of amino acid sequence conservation amongst NV proteins is quite low (e.g., VHSV and Hirame rhabdovirus share only 16.5% identity) but the NV gene is present in all four novirhabdovirus species and there is evidence that they are functionally equivalent (Kurath et al., 1997, Thoulouze et al., 2004). The construction of novirhabdovirus infectious clones lacking the NV gene has indicated that it is not essential for virus replication in cell culture but reports vary on the replication efficiency and pathogenicity of the recombinant viruses (Biacchesi, 2011). Deletion of the NV gene from an IHNV infectious clone resulted in reduced replication efficiency in EPC fish epithelial cells and reduced cumulative mortalities following intraperitoneal injection of rainbow trout (Oncorhynchus mykiss) (Biacchesi et al., 2000, Thoulouze et al., 2004) and similar observations have been reported for an NV-knockout infectious clone of VHSV in yellow perch (Perca flavescens) (Ammayappan et al., 2010). However, snakehead rhabdovirus (SHRV) infectious clones either lacking the NV gene or in which the NV ORF was truncated by point mutations were shown to replicate normally in EPC cells, and retained pathogenicity when injected intraperitoneally into zebra fish (Danio rerio) (Alonso et al., 2004, Johnson et al., 2000). It has been suggested that this apparent difference in the biological response to inhibition of NV expression may reflect the natural host range of the viruses which infect cold water fish species (VHSV and IHNV) or warm water fish species (SHRV) and have corresponding temperature ranges for efficient virus replication (Biacchesi, 2011). Recent evidence suggests that the VHSV NV protein has an anti-apoptotic function early in infection in vitro (Ammayappan and Vakharia, 2011). However, as the M protein of IHNV has pro-apoptotic activity (Chiou et al., 2000) and rhabdoviruses generally are known to promote apoptosis, the biological role of the NV protein remains unresolved.
Novirhabdoviruses also encode small basic proteins in alternative reading frames in the P gene but their functional relevance is unknown.
4.4. Sigma virus X gene
Sigmaviruses also contain a single additional gene but it is located between the P and M genes (Teninges and Bras-Herreng, 1987) (Fig. 1). The X gene encodes a polypeptide (PP3) of 298–321 amino acids (Landes-Devauchelle et al., 1995). Although amino acid sequence identity between the PP3 proteins of sigmaviruses infecting Drosophila melanogaster and Drosophila obscura is relatively low (∼11%), sequence similarity is relatively high and each is predicted with high probability to have an N-terminal signal peptide. Each also has three predicted N-glycosylation sites near the N-terminus and so the mature X protein appears to be a secreted glycoprotein. Although original reports suggested that PP3 of Drosophila melanogaster sigmavirus may be distantly related to reverse transcriptase of retrotransposons, the relevant amino acid motifs do not appear to be conserved in the Drosophila obscura sigmavirus PP3 protein (Landes-Devauchelle et al., 1995, Longdon et al., 2010). As sigmaviruses have co-evolved with their hosts and are transmitted only vertically in ecological time frames, further studies of the function of PP3 may reveal interesting aspects of virus–insect interactions and innate anti-viral immunity in insects (Longdon et al., 2011, Tsai et al., 2008).
4.5. Accessory genes in plant-adapted rhabdoviruses
The genomes of plant rhabdoviruses are 12–14.5 kb in size and have the same basic organisation as their animal-infecting counterparts, 3′-N-P-M-G-L-5′ (Fig. 2). The unique feature of plant-adapted rhabdoviruses is that they encode a viral movement protein (MP) between P and M genes which facilitates cell-to-cell transport (Jackson et al., 2005). There are no characteristic genus-specific differences in genome size or genome organisation between cytorhabdoviruses and nucleorhabdoviruses. However, nucleorhabdoviruses replicate in the nuclei of infected plant cells and most viral proteins are directed to the nucleus, while cytorhabdoviruses replicate in the cytoplasm (as all other rhabdoviruses) and viral proteins are mainly localised to the cell periphery and endomembranes (Jackson et al., 2005). Individual members of both genera may encode up to four additional accessory proteins of unknown function interspersed between N-P, P-M and G-L genes (Fig. 2). The number and location of these additional genes transcends the two genera. Rice yellow stunt virus (RYSV) and rice transitory yellowing virus (RTYV) are strains of the same virus and the nucleotide sequences of their genomes are 98.5% identical (Hiraguri et al., 2010). All other fully sequenced plant rhabdoviruses are only distantly related and have been classified taxonomically as members of distinct virus species.
Genes located between the P and M genes encode 30–35 kDa proteins that appear to be involved in viral intercellular cell-to-cell movement across plasmodesmata, symplastic connections that traverse the plant cell wall of neighbouring cells (Benitez-Alfonso et al., 2010). ORFs at this position encode the sc4, 4b and Y proteins of sonchus yellow net virus (SYNV), lettuce necrotic yellows virus (LNYV) and potato yellow dwarf virus (PYDV), respectively (Bandyopadhyay et al., 2010, Dietzgen et al., 2006, Scholthof et al., 1994). Other plant-adapted rhabdoviruses also contain accessory genes at this location. Single accessory genes (designated P3) are located at this position in rice yellow stunt virus (RYSV), rice transitory yellowing virus (RTYV), maize mosaic virus (MMV), maize Iranian mosaic virus (MIMV), taro vein chlorosis virus (TaVCV), lettuce yellow mottle virus (LYMoV) and strawberry crinkle virus (SCV) (Heim et al., 2007, Hiraguri et al., 2010, Huang et al., 2005, Massah et al., 2008, Reed et al., 2005, Revill et al., 2005, Schoen et al., 2004). However, maize fine streak virus (MFSV) contains two genes (P3 and P4) and in northern cereal mosaic virus (NCMV) there are four genes (P3, P4, P5 and P6) at this position in the genome (Fig. 2) (Tanno et al., 2000, Tsai et al., 2005).
Secondary structure predictions have shown that the SYNV sc4, LNYV 4b, LYMoV P3, MMV P3, MFSV P4, RYSV P3 and PYDV Y proteins have some structural similarities in common with those of the ‘30K’ superfamily of plant virus movement proteins (Melcher, 2000). BlastP database searches and detailed secondary structure prediction of the LNYV 4b and LYMoV P3 proteins also revealed close similarities with the movement proteins of viruses in the family Flexiviridae which induce movement-associated tubules in infected cells (Dietzgen et al., 2006, Heim et al., 2007). LYMoV P3 has a 58% amino acid sequence identity with LNYV 4b (Heim et al., 2007). The membrane and cell periphery associations of SYNV sc4 also suggest a role in cell-to-cell movement.
Direct experimental evidence of cell-to-cell movement function exists only for RYSV P3 which is the most characterised protein encoded in the P-M region (Huang et al., 2005). P3 has been shown to trans-complement cell-to-cell movement of a movement-defective heterologous plant virus and binds non-specifically ssRNA in vitro, a common property of MPs of numerous plant viruses (Benitez-Alfonso et al., 2010). Moreover, pull-down assays have shown that P3 interacts with the N protein, providing support for involvement of the nucleocapsid core in cell-to-cell movement (Huang et al., 2005).
When expressed as fusion proteins with green fluorescent protein (GFP), PYDV Y and SYNV sc4 have been shown to localize to the cell periphery (Bandyopadhyay et al., 2010, Goodin et al., 2007) whereas MFSV P4 localized to nuclei (Tsai et al., 2005). The putative MPs of SYNV and PYDV self-interact in bimolecular fluorescence complementation (BiFC) assays (Bandyopadhyay et al., 2010, Min et al., 2010). PYDV Y and M proteins interacted in planta and the complex accumulated inside the nucleus, while SYNV sc4 interacted with a soluble truncated form of G protein and complexes were seen on the nuclear envelope and on punctuate loci along the cell periphery. Sc4 has also been shown to interact with microtubule-associated host proteins and cytoplasm-tethered transcription activators have been implicated in the intercellular movement of SYNV (Min et al., 2010). The available information for these four nucleorhabdoviruses indicates potentially different cell-to-cell movement pathways.
The functions of the other accessory genes, including the four unique NCMV ORFs located between P and M genes, are unknown. Three plant rhabdoviruses, NCMV, SCV and RYSV (and the synonymous RTYV) contain a short ORF between the G and L genes (Fig. 2). The NCMV P9, SCV P6, and RYSV P6 genes are predicted to encode proteins of 52, 68, and 93 amino acids, respectively. They share no identifiable sequence similarities but limited similarity can be identified with other rhabdovirus proteins. The RYSV P6 protein shares 24% sequence identity with the N-terminal 110 amino acids of the SYNV L protein and has limited sequence identity with the NV proteins of novirhabdoviruses (up to 25%) and non-coding regions preceding the L gene in Hendra virus (38.3%) and RABV (36.6%). The SCV P6 and NCMV P9 proteins are moderately basic (pI ∼ 8.9) but RYSV P6 is highly acidic (pI 3.49). The large negative charge of this protein is reminiscent of the acidic N-terminal domain of the RYSV L protein, although they lack sequence similarity. Of the three small proteins, only RYSV P6 has been characterised experimentally (Huang et al., 2003). P6 is phosphorylated in vitro on both serine and threonine residues. It has also been shown to be associated with virions and may have a structural role but it is present only in low amounts during infection. The function of proteins encoded in this genome position is yet unknown. However, distant similarities of RYSV P6 with the L proteins of RYSV and SYNV may suggest a close evolutionary relationship with L proteins and potentially an ancillary role in rhabdovirus replication.
PYDV is unique amongst the sequenced plant-adapted rhabdoviruses in that it has an ORF X of unknown function located between the N and P genes. The 11 kDa X protein has a high content of proline, aspartic and glutamic acids, with a predicted pI of 4.5. These properties are similar to those of MFSV P3 (10 kDa, pI 5.4). However, while MFSV P3-GFP fusions accumulate in punctuate loci in the cytoplasm (Tsai et al., 2005), PYDV X is positive in yeast-based nuclear import assay, indicating its likely nuclear localization (Bandyopadhyay et al., 2010).
Like the animal rhabdoviruses, some cytorhabdoviruses and nucleorhabdoviruses present alternative ORFs in the P gene, but it is unknown if these are expressed in infected plants or vector insects (Callaghan and Dietzgen, unpublished).
4.6. Ephemerovirus accessory genes
Ephemerovirus genomes are amongst the largest and most complex of known rhabdoviruses with multiple accessory genes located in a region that extends up to 4.2 kb between the G and L genes (Fig. 1). Perhaps the most interesting of these is a second glycoprotein (GNS) gene that immediately follows the G gene (Walker et al., 1992). The GNS gene occurs in all ephemeroviruses and, until recently, has been regarded as a defining characteristic of the genus. The GNS proteins are 60–90 kDa type 1 transmembrane glycoproteins that are highly glycosylated with N-linked glycans (Walker et al., 1992, Wang and Walker, 1993). They share significant amino sequence identity and structural homology with the G proteins of ephemeroviruses and other rhabdoviruses, and appear to have arisen by recombination or gene duplication (see below). The bovine ephemeral fever virus (BEFV) GNS protein has been detected in infected cells but is not present in virions (Walker et al., 1991). Recombinant GNS expressed from a vaccinia virus vector has been detected in association with amorphous structures at the cell surface but not with budding virus particles (Hertig et al., 1996). Despite the structural similarity, the BEFV GNS protein shares no neutralising epitopes with the G protein (Johal et al., 2008) and polyclonal rabbit antibody to the BEFV GNS protein does not neutralise virus produced in either mammalian or insect cell cultures (Hertig et al., 1996). Furthermore, unlike the G protein, BEFV GNS does not induce spontaneous cell fusion at low pH (Johal et al., 2008), suggesting it functions neither in cell attachment nor fusion in vertebrates or invertebrates. In an attenuated vaccine strain of BEFV (Joubert and Walker, unpublished data) and in a highly cell-culture-adapted strain of Adelaide River virus (ARV), corruptions in the TTP sequence at the end of the G gene allow expression of the GNS protein only as a polycistronic mRNA (Wang and Walker, 1993) but this has not been observed in other wild-type or cell culture adapted ephemeroviruses. The function of ephemerovirus GNS proteins is currently unknown.
Immediately downstream of the GNS gene lies a set of relatively small accessory genes bounded by TI and TTP sequences that vary in number in different ephemeroviruses (McWilliam et al., 1997, Wang et al., 1994). The α gene occurs in all ephemeroviruses and contains two consecutive ORFs (α1 and α2) that are expressed from a single transcript due to the universal absence of intervening TTP and TI sequences. The α1 ORF encodes proteins of 10.5–14.5 kDa, each with a predicted N-terminal ectodomain containing clusters of aromatic residues, a central transmembrane domain and a highly basic C-terminal endodomain (Fig. 3 ). Although sequence identity between the α1 protein is relatively low, the structure is highly conserved and strongly suggests they function as viroporins (Gonzalez and Carrasco, 2003, McWilliam et al., 1997). The α2 ORF encodes proteins that range in size from 10.7 kDa to 14.2 kDa in different ephemeroviruses and have no identifiable motifs that may suggest a particular function. They share identifiable amino acid sequence homology with significantly higher levels of identity (up to 59%) between the more closely related viruses. Conservation of the α2 ORF in all ephemeroviruses and the preservation of sequence identity suggest that it is of functional significance. In most ephemeroviruses the α2 ORF initiation codon is in close proximity to the α1 termination codon suggesting expression may occur by translation re-initiation as has been described for several other RNA viruses (Powell, 2010). In BEFV, ORF α2 also contains a 51 amino-acid ORF (α3) in an alternative reading frame but it does not occur in other ephemeroviruses and its functional significance is unknown (McWilliam et al., 1997).
Fig. 3.

An alignment of the sequences of viroporin-like proteins encoded in the genomes of 10 animal rhabdoviruses. The alignment has been structured to emphasise the predicted N-terminal domains, transmembrane domains (shaded), and highly basic C-terminal domains. Aromatic residues in the N-terminal domain basic amino acids in the C-terminal domain are in bold and underlined. Abbreviations of virus names are as listed in Table 1.
All ephemeroviruses also contain the β gene which is located immediately downstream of the α gene and is expressed as a monocistronic transcript encoding a 16.9–18.4 kDa neutral protein (McWilliam et al., 1997, Wang et al., 1994). The ephemerovirus β proteins share significant amino acid sequence identity and have been detected both in infected cells and in virions. However, in highly adapted laboratory strains of BEFV and Kimberley virus (KIMV) the β proteins are severely truncated by point mutations and are not expressed during infection, indicating that they are not essential for growth in vitro and are not essential components of virions (Walker, Joubert and Blasdell, unpublished data). They have no identifiable homology with any other known proteins but, although their function remains unknown, they may well have a key role in pathogenesis and/or blocking the host immune response.
Although absent from the genomes of ARV and Obodhiang virus (OBOV), BEFV, KIMV, kotonkan virus (KOTV) and Berrimah virus (BRMV) each also contain a γ gene, immediately downstream of the β gene (McWilliam et al., 1997). Proteins encoded in the γ ORF are mildly basic and range in size from 11.8 kDa to 13.6 kDa. They share the highest levels of amino acid sequence identity of any of the ephemerovirus accessory proteins but are not related to any other known proteins and contain no identifiable sequence motifs that suggest a function. The γ proteins have been detected in infected cells and in virions. In wild-type viruses, the γ genes are transcribed as monocistronic mRNAs but in highly cell culture adapted strains, expression of the γ protein is severely attenuated by corruption of the TTP sequence at the end of the β gene, allowing expression only as a bicistronic β–γ transcript. Selective suppression of γ gene expression in culture suggests that its function may be important during infection in vivo.
The 15,870 nt KOTV genome is the largest yet discovered in any rhabdovirus due to an additional accessory gene immediately downstream of the γ gene (Walker and Blasdell, unpublished data). The KOTV δ gene is expressed as a monocistronic mRNA encoding a mildly acidic 12.4 kDa protein. The δ protein has been detected in KOTV-infected cells. It is shares significant amino acid sequence identity with the pleckstrin homology (PH) domain of coactivator-associated arginine methyl transferase (CARM1) which is involved in chromatin re-modelling and transcriptional activation of NF-κB dependent gene expression (Covic et al., 2005, Troffer-Charlier et al., 2007). KOTV δ may mimic this domain, inhibiting activation of the anti-viral innate immune response (Joubert and Walker, unpublished data).
Like most other rhabdoviruses, several ephemeroviruses also encode small highly basic proteins in alternative ORFs in the P gene but their functional significance is unclear.
4.7. The complex genomes of the Hart Park group
Unlike the ephemeroviruses, the complex genomes of the Hart Park group of rhabdoviruses contain multiple accessory genes inserted at various locations (Fig. 1). Wongabel virus (WONV) contains five additional ORFs (U1–U5) interspersed at various locations across the 13,196 nt genome (Gubala et al., 2008). The U1, U2 and U3 ORFs are located between the P and M genes and are each bounded by TI and TTP sequences. ORF U1 and ORF U2 encode acidic proteins of similar size and net charge (21.2 kDa/pI 5.2 and 21.9 kDa/pI 4.5, respectively). They have no remarkable characteristics but do share a significant level of amino acid sequence homology (∼16% identity; 60% similarity) and are more closely related to each other than to any other known protein. The WONV U3 gene follows the U2 gene and also encodes a relatively small acidic protein (16.5 kDa/pI 4.6) that is unrelated to any other known proteins but shares sequence homology with the WONV U1 protein (∼22% identity; 57% similarity), including the common sequence [KSxYDFVWPxxxLxxG] in the central region of each protein (Fig. 4A). Preliminary data suggest that the U3 protein binds to the IAP (inhibition of apoptosis) protein apollon and a component of the chromatin remodelling complex and may be involved in promoting apoptosis (Joubert and Walker, unpublished data). WONV ORF U4 is located within the N gene as a second ORF in the 142 nt region that follows the N protein termination codon. It encodes a putative small acidic protein (5.8 kDa/pI 4.0) of 49 amino acids that could only be translated by internal initiation on a bicistronic (N-U4) mRNA. WONV ORF U5 overlaps the G protein ORF and thus also must be translated by internal initiation on a bicistronic (G-U5) mRNA. Translation from the first initiation codon would result in a 14.9 kDa protein with that is similar in size and structure to the ephemerovirus viroporin-like α1 proteins (Fig. 3). Several low molecular weight proteins (range 20–25 kDa) have been detected in infected cells using WONV hyperimmune antiserum but they are larger than the predicted U1–U5 proteins and may well be degradation products of larger viral proteins (Gubala et al., 2008).
Fig. 4.

Clustal X alignments of (A) Wongabel virus (WONV) U1, U2 and U3 proteins and (B) Flanders virus (FLAV) U1 and U2 proteins. A single termination codon interrupting the published sequence of the FLAV U1 protein has been removed to infer the complete amino acid sequence. The FLAV U2 protein is also referred to as the 19K protein. Aligned amino acids are shaded. Fully conserved (*), strongly conserved (:) and weakly conserved (.) amino acids have been assigned by the software.
The Flanders virus (FLAV) genome organisation appears to closely resemble that of WONV. The available sequence data (GenBank AH012179) is dated and appears to be corrupted either by sequencing ambiguities or as a result of cell culture adaptation of the virus. However, through alignment with the WONV genome, it is reasonable to infer that FLAV also contains three accessory genes between the P and M genes and a 13.8 kDa viroporin-like protein encoded in a second ORF in the G gene (Fig. 3). The FLAV U1, U2 and U3 ORFs each appear to encode proteins of 18–19 kDa and, as for the corresponding WONV proteins, they share an unusual level of sequence similarity suggesting a common origin and/or functional association. This is most evident between FLAV U1 and U2 (also referred to as the 19K protein) that share ∼20% amino acid sequence identity and ∼58% similarity (Fig. 4B). There is no second consecutive ORF in the FLAV N gene corresponding to WONV U4 but there is a small alternative ORF within the N gene encoding a polypeptide of 66 amino acids (7.9 kDa). In addition to the N, P, M, G and L proteins, three virus-induced proteins have been reported in FLAV infected cells and in virions (Boyd and Whitaker-Dowling, 1988). However, only one of these proteins (19 kDa) corresponds to the predicted size of any FLAV accessory gene product.
The 15,764 nt Ngaingan virus (NGAV) genome is one of the largest and most complex of known rhabdoviruses (Gubala et al., 2010). As in WONV and FLAV, the NGAV genome contains three ORFs between the P and M genes (U1, U2 and U3) encoding neutral to mildly acidic proteins of 15.1–17.1 kDa that share identifiable sequence similarity. However, unlike WONV and FLAV, ORF U1 and ORF U2 are encoded in overlapping reading frames within the same gene and ORF3 is encoded in a separate gene bounded by TI and TTP sequences. Downstream of the U3 gene, the NGAV genome organisation is very complex and varies significantly from WONV and FLAV. Between the M and G genes the NGAV U4 gene encodes a 9.7 kDa protein with a tyrosine-rich C-terminal domain but otherwise has no remarkable features. Downstream of the G gene, however, is a region that resembles the ephemerovirus genomes including a second glycoprotein gene (GNS), followed by three additional ORFs. The GNS gene encodes a type 1 transmembrane glycoprotein that is most closely related to the ephemerovirus GNS proteins and shares sequence homology and structural characteristics with the NGAV G protein and those of all animal rhabdoviruses (Fig. 5 ). The two accessory genes immediately downstream of the NGAV GNS gene also resemble the ephemerovirus α and β genes in format, size and location. The first contains two overlapping ORFs (U5 and U6) that are similar in size to the ephemerovirus α1 and α2 ORFs. However, the 12.6 kDa U5 protein does not have the characteristic structure of the viroporin-like α1 proteins and, unlike the featureless ephemerovirus α2 proteins, the U6 protein has the structure of a small (10–12 kDa) membrane-anchored class 1 glycoprotein with a signal peptide domain, a transmembrane domain and an N-terminal ectodomain containing a single N-glycosylation site. The NGAV U7 gene contains a single ORF encoding an 18.2 kDa protein that is similar in size to ephemerovirus β proteins but shares no identifiable sequence homology and has no remarkable features.
Fig. 5.
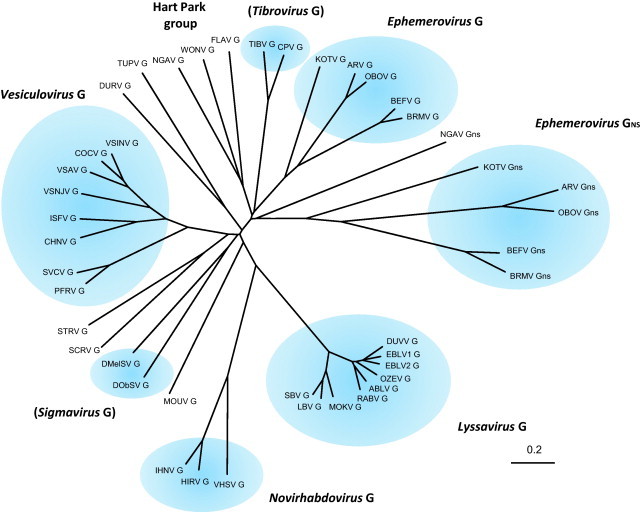
A minimum evolution phylogenetic tree constructed from a Clustal X multiple sequence alignment of animal rhabdovirus G protein and GNS protein sequences. The tree was constructed using the MEGA program. Viruses assigned to established genera and proposed genera are shaded. Abbreviations of virus names are as listed in Table 1.
In addition to these independent accessory genes NGAV encodes two small basic proteins (P′1 and P′2) in alternative reading frames within the P gene and a 10.8 kDa protein in an alternative reading frame in the M gene. If all these potential ORFs in translated, NGAV could potentially express a total of 16 functional proteins.
4.8. Tibrovirus accessory genes
Whole genome sequences are available for two viruses that have recently been proposed as species in a new genus Tibrovirus. The 13.2–13.3 kb Tibrogargan virus (TIBV) and Coastal Plains virus (CPV) genomes share a similar organisation with 3–4 accessory protein genes (Gubala et al., 2011) (Fig. 1). In each virus, ORFs U1 and U2 are located between the M and G genes. Each is bounded by TI and TTP sequences and encodes a protein of 17–20 kDa. The U1 proteins are acidic and the U2 proteins are basic, and there is extensive amino acid sequence homology between the corresponding proteins. An N-terminal signal peptide is predicted with high probability in the CPV U1 protein, suggesting it may be secreted but it is not predicted in the TIBV U1 protein despite the high level of overall sequence homology and so its significance is uncertain. Features of the U2 proteins are unremarkable. The TIBV and CPV genomes also each contain ORF U3 encoded in an independent gene located between the G and L genes. The U3 proteins have the structural characteristics of viroporin-like proteins that occur in ephemeroviruses as the α1 proteins and in WONV and NGAV as the U5 proteins. In each case, the viroporin ORFs are located between the G and L genes, either directly following the G gene or following the GNS gene in those viruses in which it occurs. In TIBV, U4 is an alternative ORF that overlaps the G protein ORF and could potentially encode a very small basic protein (6.6 kDa/pI 9.6). However, U4 does not occur in CPV and its significance, if any, is unclear. As in many other rhabdoviruses, the CPV P gene also contains an alternative ORF encoding a small basic proteins but none is evident in the TIBV P gene.
4.9. The SH genes of other unclassified rhabdoviruses
Several other unclassified rhabdoviruses isolated from insects, mammals, birds and fish share a similar genome organisation (Fig. 1). Tupaia rhabdovirus (TUPV), Durham virus (DURV) and Oak Vale virus (OAKV) each encode small hydrophobic (SH) proteins in ORFs flanked by TI and TTP sequences in the region between the M and G genes (Allison et al., 2011, Springfeld et al., 2005). However, although superficially similar, the putative proteins expressed from these genes have different characteristics. The TUPV ORF encodes a small protein with a predicted signal peptide and a central transmembrane domain that would generate an 8.6 kDa non-glycosylated membrane-anchored protein with short ecto- and endo-domains. In DURV, the SH protein is predicted to have similar membrane topology with predicted signal peptidase cleavage generating a 6.9 kDa non-glycosylated membrane-anchored protein. These proteins resemble the small hydrophobic protein encoded in the NGAV U5 gene (see above). The OAKV SH protein is predicted to have an N-terminal signal peptide but no membrane anchor to generate a 4.2 kDa non-glycosylated secreted product (GenBank GQ294474). The mandarin fish (Siniperca chuatsi) rhabdovirus (SCRV) genome does not contain an independent SH gene but does encode a small hydrophobic protein as an alternative reading frame in the upstream M gene (Tao et al., 2008). The predicted structure is similar to that of the OAKV SH protein with a high probability signal peptidase cleavage site generated a secreted product of only 2.9 kDa. Although it has been reported that the putative SCRV SH protein could not be detected in infected fish cells, this may not be unexpected if it is indeed secreted. The X genes of sigmaviruses also encode a protein that is predicted to be secreted (see above) but it is very much larger than these small peptides. Several of these unclassified rhabdoviruses also encode small basic proteins in alternative ORFs in the P gene.
5. Patterns in the complexity of rhabdovirus genome organisation
Although characterised by remarkable diversity and complexity, a global view of rhabdovirus genomes does reveal some patterns in the nature and location of accessory genes (Fig. 6 ). In various rhabdoviruses, accessory genes occur in each of the junctions between the structural protein genes and as alternative or overlapping ORFs within all structural protein genes except the L gene. Alternative or overlapping ORFs occur least commonly in the N gene (and of course the L gene), possibly reflecting the structural and functional constraints on amino acid sequence variations, and in the G gene that requires sufficient genetic flexibility to evolve under the pressure of neutralising antibody. Alternative ORFs occur most commonly in the P gene in which the preservation of function is far less dependent on conservation of the amino acid sequence, allowing more scope for the evolution and stabilisation of new ORFs in alternative frames (Nadin-Davis et al., 2002, Spiropoulou and Nichol, 1993). The N-P, P-M, M-G, and G-L gene junctions have all been utilised as sites for accessory genes, although in rhabdoviruses examined to date, the P-M and G-L junctions have been utilised more commonly. The primary evolutionary constraint on the occupation of these sites may be the resulting attenuation of transcription in the downstream genes. For example, rare use of the N-P gene junction may be due to the importance of the balance of expression of the N and P proteins in modulating replication and transcription (Banerjee and Barik, 1992).
Fig. 6.
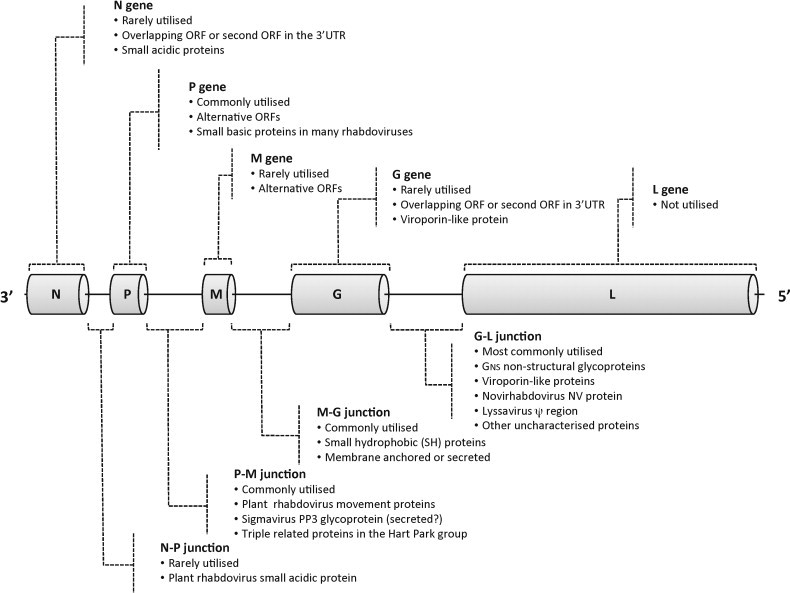
Schematic diagram illustrating the use by rhabdoviruses of the five common structural protein genes (N, P, M, G and L) and the intergenic regions as sites for insertion of ORFs encoding putative accessory proteins genes. The genes are defined as transcriptional units bounded by TI and TTP sequences.
Alternative ORFs in the P gene occur very commonly and usually encode small basic proteins. Although their function has not been examined in any detail, by analogy with paramyxoviruses, they may have multiple roles in replication, virion assembly and in modulating host responses to infection (Fontana et al., 2008, Irie et al., 2010, Kurotani et al., 1998). The P-M gene junction has been utilised as a site of accessory genes in many rhabdoviruses including genes encoding movement proteins in cytorhabdoviruses and nucleorhabdoviruses and the apparently secreted and glycosylated PP3 protein of sigmaviruses. The P-M junction is also the location of the set of three related 15–22 kDa proteins in the Hart Park group viruses that may be involved in the promotion of apoptosis. The M-G gene junction is occupied less commonly but genes encoding small proteins that are either secreted or membrane-anchored do occur here, including the TIBV U1 protein, and the small hydrophobic (SH) proteins of the unclassified viruses TUPV, DURV and OAKV (Allison et al., 2011, Springfeld et al., 2005). Interestingly, in the mandarin fish rhabdovirus (SCRV), and SH protein is encoded just upstream as an alternative ORF in the M gene (Tao et al., 2008).
The G-L gene junction is the most commonly occupied site of accessory genes in animal rhabdoviruses. Genes encoding the non-structural glycoprotein (GNS) invariably occur immediately downstream of the G gene and share a common genetic lineage (Gubala et al., 2010, Walker et al., 1992, Wang and Walker, 1993). Genes encoding viroporin-like proteins also usually occur in this region, immediately following GNS gene in ephemeroviruses (McWilliam et al., 1997, Wang et al., 1994) and following the G gene in FLAV and tibroviruses which lack GNS (Gubala et al., 2011). Interestingly, the WONV viroporin-like protein is encoded immediately upstream of the G-L junction in an ORF that overlaps the end of the G gene. The viroporin-like proteins all have a similar structure with a central hydrophobic domain and a highly basic C-terminal tail, and so may have evolved from a common ancestor (Fig. 3). Many other accessory genes are located in the G-L intergenic region including the novirhabdovirus NV gene, and genes coding proteins of unknown function in ephemeroviruses, NGAV and some plant-adapted rhabdoviruses. The G-L intergenic region is also the site of the lyssavirus ψ region (Tordo et al., 1986). Sequence insertion at this site may be most favoured as it would primarily affect expression of the L protein which is required only in catalytic amounts.
6. The evolution of rhabdovirus genome complexity: gene duplication or recombination?
The ecological diversity of rhabdoviruses suggests they have an ancient evolutionary history, perhaps originating as insect viruses that have adapted to replication in plants or animals (Hogenhout et al., 2003). There is recent evidence that endogenous viral elements (EVEs) derived from rhabdoviruses occur commonly in insect genomes with which they have co-evolved over many millions of years (Katzourakis and Gifford, 2010). Rhabdovirus genomic diversity will also therefore have evolved over geological time frames, possibly involving rare events that may appear highly improbable in a contemporary ecological context. We have argued previously that the evolution of rhabdovirus accessory genes may have occurred by a copy-choice mechanism involving polymerase-jumping (Wang and Walker, 1993). According to this model, upstream relocation of the polymerase during replication would generate a sequence duplication in a process mirroring the downstream relocation that has been proposed for the generation of defective-interfering (DI) genomes (Lazzarini et al., 1981, Perrault, 1981). There seems no prima face reason to suggest that upstream relocation and downstream relocation should not occur with similar frequency but purifying selection would enrich for rapidly replicating DI genomes and quickly remove larger genomes unless the duplication event has provided a specific selective advantage. Although survival of such larger genomes may be exceedingly rare, preservation of the ψ region in lyssaviruses does indicate that the selective advantage of maintaining long stretches of apparently non-functional sequence may sometimes be cryptic (Kuzmin et al., 2008, Ravkov et al., 1995) and sequence similarities in adjacent genes of some rhabdoviruses do suggest that duplication may have contributed to the evolution of accessory genes. Such sequence similarities are evident in the GNS genes in ephemeroviruses and NGAV (Gubala et al., 2010, Walker et al., 1992, Wang and Walker, 1993), the related U1, U2 and U3 genes in the P-M region of Hart Park group viruses (see above), and the P6 gene in rice yellow stunt virus (Huang et al., 2003).
Other possible mechanisms for the acquisition of accessory genes include homologous genetic recombination and lateral gene transfer (LGT). Life-long persistent infections occur commonly in insects and mixed infections with different viruses have been reported (Chen et al., 2004, Mourya et al., 2001, Thavara et al., 2006). Although genetic recombination is believed to occur rarely in (–) ssRNA viruses (Holmes, 2009), possible recombination events have been detected and may have contributed to their long term evolution (Chare et al., 2003, Han et al., 2008, Lukashev, 2005, Qin et al., 2008, Sibold et al., 1999). Unlike gene duplication, in which the new gene must be preserved while evolving a new function, recombination could offer immediate selective advantage in larger more complex genomes by increasing tissue tropism, host range or replication efficiency or more effective blocking of host defensive mechanisms. Homologous genetic recombination must therefore be considered an equally plausible explanation for the generation of genomic structural diversity in rhabdoviruses. LGT also offers a mechanism by which accessory genes could be obtained from the host or an unrelated virus to provide immediate selective advantage (Holmes, 2009). There is evidence that the coronavirus haemagglutinin-esterase glycoprotein was acquired from influenza C virus (Luytjes et al., 1988) and that the envelope glycoprotein of the tick-borne orthomyxovirus Thogoto may have been derived from a baculovirus (Morse et al., 1992). Several other examples of possible LGT involving transfer of host genes to viruses have been reported but these events appear to be very rare and none has yet been reported to our knowledge in rhabdoviruses or other unsegmented (–) ssRNA viruses (Holmes, 2009).
It can also be argued that purifying selection should drive evolution towards smaller and perhaps more efficient genomes rather than large complex genomes. Indeed, the generation of DI particles, which occurs with high frequency during infection in vitro, would appear to provide the mechanism for rapid elimination of redundant sequences (Lazzarini et al., 1981, Perrault, 1981) and phylogenetic analyses do suggest that accessory genes may have been lost during the evolution of some lineages. As shown in Fig. 5, phylogenetic analysis of available rhabdovirus G protein sequences places NGAV with WONV and FLAV to form the Hart Park group and analyses of the L and N protein sequences supports these relationships (Bourhy et al., 2005, Gubala et al., 2010). However, the NGAV GNS protein clusters with the ephemerovirus GNS proteins (Fig. 5), suggesting that it was obtained by a common ancestral virus and subsequently lost from WONV and FLAV. Alternatively, NGAV may have acquired GNS independently in a later event involving recombination with a virus in the ephemerovirus lineage.
All arguments considered, perhaps the most congruent hypothesis is that the evolution of rhabdovirus genomic diversity is a dynamic process in which accessory genes have been gained and lost during the course of continual adaptation and purifying selection.
7. Future research
Despite the importance of rhabdoviruses as pathogens of humans, livestock, fish and plants, it is evident from this review that very little is known of the functions of the proteins encoded in their many accessory genes. In some respects, this is very surprising as five major rhabdovirus structural proteins have been the subject of intensive investigation and have provided some of the most important models for understanding fundamental processes during viral infection. On the basis of available evidence, and by analogy with other viruses that are rich in accessory functions (e.g., HIV, SARS coronavirus), we can anticipate that many of the rhabdovirus accessory proteins may be important determinants of virulence and pathogenesis and function to increase the efficiency of viral replication, block host innate and/or adaptive immune responses, regulate apoptosis or induce cytopathology. Understanding the strategies employed by rhabdoviruses to engage, divert and re-direct cellular processes will not only present opportunities to develop new anti-viral therapies but may also reveal aspects that have broader significance in biology, agriculture and medicine.
One of the most attractive targets for future study is the role of rhabdovirus accessory genes during infection in insects and in transmission to mammals or plants. Despite their importance as vectors of disease, and over 100 years of immunology, the anti-viral defences of insects remain poorly understood (Huszar and Imler, 2008). RNAi has been identified as an important anti-viral mechanism and there is evidence of involvement of the Toll and Imd pathways that function primarily in innate immune responses to bacteria and parasites (Kemp and Imler, 2009). In Drosophila melanogaster, the innate immune response to sigmavirus infection has been shown to involve upregulated peptidoglycan reporter and antimicrobial peptide gene expression (Tsai et al., 2008). However, innate anti-viral sensor and effector mechanisms in insects are largely unexplored and may be critical determinants of their susceptibility to infection and ability to act as efficient vectors. As most rhabdoviruses naturally infect insects, their accessory genes may well be involved in engaging defensive responses and so provide useful tools for exploring invertebrate anti-viral pathways.
There has been a significant increase in the number of fully sequenced rhabdovirus genomes in recent years and the availability of next-generation sequencing platforms, rapid sample preparation methods and new multiplexing technologies will ensure that GenBank depositions continue to grow rapidly. Indeed, it can be anticipated that most or all known rhabdoviruses will be sequenced in the foreseeable future. This will reveal much about the phylogenetic relationships and evolutionary history of rhabdoviruses. It will also present the opportunity to characterise and exploit an increasing array of accessory genes to understand the fundamental processes of transmission, infection and immunity.
References
- Ahne W., Bjorklund H.V., Essbauer S., Fijan N., Kurath G., Winton J.R. Spring viremia of carp (SVC) Diseases of Aquatic Organisms. 2002;52:261–272. doi: 10.3354/dao052261. [DOI] [PubMed] [Google Scholar]
- Albertini A., Schoehn G., Weissenhorn W., Ruigrok R. Structural aspects of rabies virus replication. Cellular and Molecular Life Sciences. 2008;65:282–294. doi: 10.1007/s00018-007-7298-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison A.B., Palacios G., Travassos da Rosa A., Popov V.L., Lu L., Xiao S.Y., DeToy K., Briese T., Lipkin W.I., Keel M.K., Stallknecht D.E., Bishop G.R., Tesh R.B. Characterization of Durham virus, a novel rhabdovirus that encodes both a C and SH protein. Virus Research. 2011;155:112–122. doi: 10.1016/j.virusres.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso M., Kim C.H., Johnson M.C., Pressley M., Leong J.-A. The NV gene of snakehead rhabdovirus (SHRV) is not required for pathogenesis, and a heterologous glycoprotein can be incorporated into the SHRV envelope. Journal of Virology. 2004;78:5875–5882. doi: 10.1128/JVI.78.11.5875-5882.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammayappan A., Kurath G., Thompson T., Vakharia V. A reverse genetics system for the Great Lakes strain of viral hemorrhagic septicemia virus: the NV gene is required for pathogenicity. Marine Biotechnology. 2010 doi: 10.1007/s10126-010-9329-4. [DOI] [PubMed] [Google Scholar]
- Ammayappan A., Vakharia V.N. Nonvirion protein of novirhabdovirus suppresses apoptosis at the early stage of virus infection. Journal of Virology. 2011 doi: 10.1128/JVI.00597-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assenberg R., Delmas O., Morin B., Graham S.C., De Lamballerie X., Laubert C., Coutard B., Grimes J.M., Neyts J., Owens R.J., Brandt B.W., Gorbalenya A., Tucker P., Stuart D.I., Canard B., Bourhy H. Genomics and structure/function studies of Rhabdoviridae proteins involved in replication and transcription. Antiviral Research. 2010;87(2):149–161. doi: 10.1016/j.antiviral.2010.02.322. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay A., Kopperud K., Anderson G., Martin K., Goodin M. An integrated protein localization and interaction map for potato yellow dwarf virus, type species of the genus Nucleorhabdovirus. Virology. 2010;402:61–71. doi: 10.1016/j.virol.2010.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A.K., Barik S. Gene expression of vesicular stomatitis virus genome RNA. Virology. 1992;188:417–428. doi: 10.1016/0042-6822(92)90495-b. [DOI] [PubMed] [Google Scholar]
- Barr J., Whelan S., Wertz G. cis-Acting signals involved in termination of vesicular stomatitis virus mRNA synthesis include the conserved AUAC and the U7 signal for polyadenylation. Journal of Virology. 1997;71:8718–8725. doi: 10.1128/jvi.71.11.8718-8725.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr J.N., Whelan S.P.J., Wertz G.W. Transcriptional control of the RNA-dependent RNA polymerase of vesicular stomatitis virus. Biochimica et Biophysica Acta – Gene Structure and Expression. 2002;1577:337–353. doi: 10.1016/s0167-4781(02)00462-1. [DOI] [PubMed] [Google Scholar]
- Basurco B., Benmansour A. Distant strains of the fish rhabdovirus VHSV maintain a sixth functional cistron which codes for a nonstructural protein of unknown function. Virology. 1995;212:741–745. doi: 10.1006/viro.1995.1534. [DOI] [PubMed] [Google Scholar]
- Benitez-Alfonso Y., Faulkner C., Ritzenthaler C., Maule A.J. Plasmodesmata: gateways to local and systemic virus infection. Molecular Plant–Microbe Interactions. 2010;23:1403–1412. doi: 10.1094/MPMI-05-10-0116. [DOI] [PubMed] [Google Scholar]
- Biacchesi S. The reverse genetics applied to fish RNA viruses. Veterinary Research. 2011;42:e12. doi: 10.1186/1297-9716-42-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biacchesi S., Thoulouze M.I., Bearzotti M., Yu Y.X., Bremont M. Recovery of NV knockout infectious hematopoietic necrosis virus expressing foreign genes. Journal of Virology. 2000;74:11247–11253. doi: 10.1128/jvi.74.23.11247-11253.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black B.L., Lyles D.S. Vesicular stomatitis virus matrix protein inhibits host cell transcription of target genes in vivo. Journal of Virology. 1992;66:4058–4064. doi: 10.1128/jvi.66.7.4058-4064.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondel D., Harmison G.G., Schubert M. Role of matrix protein in cytopathogenesis of vesicular stomatitis virus. Journal of Virology. 1990;64:1716–1725. doi: 10.1128/jvi.64.4.1716-1725.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondel D., Regad T., Poisson N., Pavie B., Harper F., Pandolfi P.P., de Thé H., Chelbi-Alix M.K. Rabies virus P and small P products interact directly with PML and reorganize PML nuclear bodies. Oncogene. 2002;21:7957–7970. doi: 10.1038/sj.onc.1205931. [DOI] [PubMed] [Google Scholar]
- Bourhy H., Cowley J.A., Larrous F., Holmes E.C., Walker P.J. Phylogenetic relationships among rhabdoviruses inferred using the L polymerase gene. Journal of General Virology. 2005;86:2849–2858. doi: 10.1099/vir.0.81128-0. [DOI] [PubMed] [Google Scholar]
- Boyd K.R. Serological comparisons among Hart Park virus and strains of Flanders virus. Infection and Immunity. 1972;5:933–937. doi: 10.1128/iai.5.6.933-937.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd K.R, Whitaker-Dowling P. Flanders virus replication and protein synthesis. Virology. 1988;163:349–358. doi: 10.1016/0042-6822(88)90275-9. [DOI] [PubMed] [Google Scholar]
- Brzozka K., Finke S., Conzelmann K.-K. Identification of the rabies virus alpha/beta interferon antagonist: phosphoprotein P interferes with phosphorylation of interferon regulatory factor 3. Journal of Virology. 2005;79:7673–7681. doi: 10.1128/JVI.79.12.7673-7681.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzozka K., Finke S., Conzelmann K.-K. Inhibition of interferon signaling by rabies virus phosphoprotein P: activation-dependent binding of STAT1 and STAT2. Journal of Virology. 2006;80:2675–2683. doi: 10.1128/JVI.80.6.2675-2683.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccaldi P.E., Fayet J., Conzelmann K.K., Tsiang H. Infection characteristics of rabies virus variants with deletion or insertion in the pseudogene sequence. Journal of Neurovirology. 1998;4:115–119. doi: 10.3109/13550289809113489. [DOI] [PubMed] [Google Scholar]
- Chare E.R., Gould E.A., Holmes E.C. Phylogenetic analysis reveals a low rate of homologous recombination in negative-sense RNA viruses. Journal of General Virology. 2003;84:2691–2703. doi: 10.1099/vir.0.19277-0. [DOI] [PubMed] [Google Scholar]
- Chelbi-Alix M.K., Vidy A., Bougrini J.E., Blondel D. Rabies viral mechanisms to escape the IFN system: the viral protein P interferes with IRF-3, STAT1, and PML nuclear bodies. Journal of Interferon and Cytokine Research. 2006;26:271–280. doi: 10.1089/jir.2006.26.271. [DOI] [PubMed] [Google Scholar]
- Chen H.L, Liu H., Liu Z.X., He J.Q., Gao L.Y., Shi X.J., Jiang Y.L. Characterization of the complete genome sequence of pike fry rhabdovirus. Archives of Virology. 2009;154:1489–1494. doi: 10.1007/s00705-009-0455-0. [DOI] [PubMed] [Google Scholar]
- Chen Y., Zhao Y., Hammond J., Hsu H., Evans J., Feldlaufer M. Multiple virus infections in the honey bee and genome divergence of honey bee viruses. Journal of Invertebrate Pathology. 2004;87:84–93. doi: 10.1016/j.jip.2004.07.005. [DOI] [PubMed] [Google Scholar]
- Chenik M., Chebli K., Blondel D. Translation initiation at alternate in-frame AUG codons in the rabies virus phosphoprotein mRNA is mediated by a ribosomal leaky scanning mechanism. Journal of Virology. 1995;69:707–712. doi: 10.1128/jvi.69.2.707-712.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou P.P., Kim C.H., Ormonde P., Leong J.-A.C. Infectious hematopoietic necrosis virus matrix protein inhibits host-directed gene expression and induces morphological changes of apoptosis in cell cultures. Journal of Virology. 2000;74:7619–7627. doi: 10.1128/jvi.74.16.7619-7627.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll J.M. The glycoprotein G of rhabdoviruses. Acta Virologica. 1995;140:827–851. doi: 10.1007/BF01314961. [DOI] [PubMed] [Google Scholar]
- Comer J.A., Tesh R.B. Phleboromine sand flies as vectors of vesiculoviruses: a review. Parasitologica. 1991;33(Suppl. 1):143–150. [PubMed] [Google Scholar]
- Conzelmann K.K., Cox J.H., Schneider L.G., Thiel H.J. Molecular cloning and complete nucleotide sequence of the attenuated rabies virus SAD B19. Virology. 1990;175:485–499. doi: 10.1016/0042-6822(90)90433-r. [DOI] [PubMed] [Google Scholar]
- Coulon P., Deutsch V., Lafay F., Martinetedelist C., Wyers F., Herman R.C., Flamand A. Genetic evidence for multiple functions of the matrix protein of vesicular stomatitis virus. Journal of General Virology. 1990;71:991–996. doi: 10.1099/0022-1317-71-4-991. [DOI] [PubMed] [Google Scholar]
- Covic M., Hassa P.O., Saccani S., Buerki C., Meier N.I., Lombardi C., Imhof R., Bedford M.T., Natoli G., Hottiger M.O. Arginine methyltransferase CARM1 is a promoter-specific regulator of NF-κB-dependent gene expression. EMBO Journal. 2005;24:85–96. doi: 10.1038/sj.emboj.7600500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cybinski D.H., Muller M.J. Isolation of arboviruses from cattle and insects at two sentinel sites in Queensland Australia, 1979–85. Australian Journal of Zoology. 1990;38:25–32. [Google Scholar]
- Delmas O., Holmes E.C., Talbi C., Larrous F., Dacheux L., Bouchier C., Bouhry H. Genomic diversity and evolution of the lyssaviruses. PLoS One. 2008;3:e2057. doi: 10.1371/journal.pone.0002057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzgen R.G., Callaghan B., Wetzel T., Dale J.L. Completion of the genome sequence of lettuce necrotic yellows virus, type species of the genus Cytorhabdovirus. Virus Research. 2006;118:16–22. doi: 10.1016/j.virusres.2005.10.024. [DOI] [PubMed] [Google Scholar]
- Dietzgen R.G., Calisher C.H., Kurath G., Kuzman I.V., Rodriguez L.L., Stone D.M., Tesh R.B., Tordo N., Walker P.J., Wetzel T., Whitfield A.E. Rhabdoviridae. In: King A.M.Q., Adams M.J., Carstens E.B., Lefkowitz E.J., editors. Virus Taxonomy, Ninth Report of the International Committee on Taxonomy of Viruses. Elsevier; San Diego: 2012. pp. 654–681. [Google Scholar]
- Doherty R.L., Carley J.G., Standfast H.A., Dyce A.L., Kay B.H., Snowdon W.A. Isolation of arboviruses from mosquitoes, biting midges, sandflies and vertebrates collected in Queensland, 1969 and 1970. Transactions of the Royal Society for Tropical Medicine Hygiene. 1973;67:536–543. doi: 10.1016/0035-9203(73)90084-9. [DOI] [PubMed] [Google Scholar]
- Faber M., Pulmanausahakul R., Nagao K., Prosniak M., Rice A.B., Koprowski H., Schnell M.J., Dietzschold B. Identification of viral genomic elements responsible for rabies virus neuroinvasiveness. Proceedings of the National Academy of Sciences United States of America. 2004;101:16328–16332. doi: 10.1073/pnas.0407289101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana J.M., Bankamp B., Rota P.A. Inhibition of interferon induction and signaling by paramyxoviruses. Immunological Reviews. 2008;225:46–67. doi: 10.1111/j.1600-065X.2008.00669.x. [DOI] [PubMed] [Google Scholar]
- Gibbs E.P.J., Calisher C.H., Tesh R.B., Lazuick J.S., Bowen R., Greiner E.C. Bivens Arm virus: a new rhabdovirus isolated from Culicoides insignis in Florida and related to Tibrogargan virus of Australia. Veterinary Microbiology. 1989;19:141–150. doi: 10.1016/0378-1135(89)90079-5. [DOI] [PubMed] [Google Scholar]
- Gonzalez M.E., Carrasco L. Viroporins. FEBS Letters. 2003;552:28–34. doi: 10.1016/s0014-5793(03)00780-4. [DOI] [PubMed] [Google Scholar]
- Goodin M.M., Chakrabarty R., Banerjee R., Yelton S., DeBolt S. Membrane and protein dynamics in live plant nuclei infected with Sonchus yellow net virus, a plant adapted rhabdovirus. Journal of General Virology. 2007;88:1810–1820. doi: 10.1099/vir.0.82698-0. [DOI] [PubMed] [Google Scholar]
- Gubala A., Davis S., Weir R., Melville L., Cowled C., Boyle D. Tibrogargan and Coastal Plains rhabdoviruses: genomic characterisation, evolution of novel genes and seroprevalence in Australian livestock. Journal of General Virology. 2011 doi: 10.1099/vir.0.026120-0. [DOI] [PubMed] [Google Scholar]
- Gubala A., Davis S., Weir R., Melville L., Cowled C., Walker P., Boyle D. Ngaingan virus, a macropod-associated rhabdovirus, contains a second glycoprotein gene and seven novel open reading frames. Virology. 2010;399:98–108. doi: 10.1016/j.virol.2009.12.013. [DOI] [PubMed] [Google Scholar]
- Gubala A.J., Proll D.F., Barnard R.T., Cowled C.J., Crameri S.G., Hyatt A.D., Boyle D.B. Genomic characterisation of Wongabel virus reveals novel genes within the Rhabdoviridae. Virology. 2008;376:13–23. doi: 10.1016/j.virol.2008.03.004. [DOI] [PubMed] [Google Scholar]
- Han G.-Z., He C.-Q., Ding N.-Z., Ma L.-Y. Identification of a natural multi-recombinant of Newcastle disease virus. Virology. 2008;371:54–60. doi: 10.1016/j.virol.2007.09.038. [DOI] [PubMed] [Google Scholar]
- Heim F., Lot H., Deecolle B., Bassler A., Krczal G., Wetzel T. Complete nucleotide sequence of a putative new rhabdovirus infecting lettuce. Archives of Virology. 2007;153:81–92. doi: 10.1007/s00705-007-1071-5. [DOI] [PubMed] [Google Scholar]
- Herman R.C. Internal initiation of translation on the vesicular stomatitis virus phosphoprotein mRNA yields a second protein. Journal of Virology. 1986;58:797–804. doi: 10.1128/jvi.58.3.797-804.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertig C., Pye A.D., Hyatt A.D., Davis S.S., McWilliam S.M., Heine H.G., Walker P.J., Boyle D.B. Vaccinia virus-expressed bovine ephemeral fever virus G but not G(NS) glycoprotein induces neutralizing antibodies and protects against experimental infection. Journal of General Virology. 1996;77:631–640. doi: 10.1099/0022-1317-77-4-631. [DOI] [PubMed] [Google Scholar]
- Hiraguri A., Hibino H., Hayashi T., Shimizu T., Uehara-Ichiki T., Omura T., Sasaya T. Complete sequence analysis of rice transitory yellowing virus and its comparison to rice yellow stunt virus. Archives of Virology. 2010;155:243–245. doi: 10.1007/s00705-009-0557-8. [DOI] [PubMed] [Google Scholar]
- Hoffmann B., Beer M., Schutze H., Mettenleiter T.C. Fish rhabdoviruses: molecular epidemiology and evolution. Current Topics in Microbiology and Immunology. 2005;292:81–117. doi: 10.1007/3-540-27485-5_5. [DOI] [PubMed] [Google Scholar]
- Hoffmann B., Schutze H., Mettenleiter T.C. Determination of the complete genomic sequence and analysis of the gene products of the virus of spring viremia of carp, a fish rhabdovirus. Virus Research. 2002;84:89–100. doi: 10.1016/s0168-1702(01)00441-5. [DOI] [PubMed] [Google Scholar]
- Hogenhout S.A., Redinbaugh M.G., Ammar E.D. Plant and animal rhabdovirus host range: a bug's view. Trends in Microbiology. 2003;11:264–271. doi: 10.1016/s0966-842x(03)00120-3. [DOI] [PubMed] [Google Scholar]
- Holmes E.C. Oxford University Press; Oxford: 2009. The Evolution and Emergence of RNA Viruses. Oxford Series in Ecology and Evolution. [Google Scholar]
- Huang Y., Geng Y., Ying X., Chen X., Gang R.X. Identification of a movement protein of rice yellow stunt rhabdovirus. Journal of Virology. 2005;79:2108–2114. doi: 10.1128/JVI.79.4.2108-2114.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Zhao H., Luo Z., Chen X., Fang R.-X. Novel structure of the genome of Rice yellow stunt virus: identification of the gene 6-encoded virion protein. Journal of General Virology. 2003;84:2259–2264. doi: 10.1099/vir.0.19195-0. [DOI] [PubMed] [Google Scholar]
- Huszar T., Imler J.L. Drosophila viruses and the study of antiviral host-defense. Advances in Virus Research. 2008;72:227–265. doi: 10.1016/S0065-3527(08)00406-5. [DOI] [PubMed] [Google Scholar]
- Irie T., Nagata N., Igarashi T., Okamoto I., Sakaguchi T. Conserved charged amino acids within Sendai virus C protein play multiple roles in the evasion of innate immune responses. PLoS One. 2010;5:e10719. doi: 10.1371/journal.pone.0010719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito N., Moseley G.W., Blondel D., Shimizu K., Rowe C.L., Ito Y., Masatani T., Nakagawa K., Jans D.A., Sugiyama M. Role of interferon antagonist activity of rabies virus phosphoprotein in viral pathogenicity. Journal of Virology. 2010;84(13):6699–6710. doi: 10.1128/JVI.00011-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson A.O., Dietzgen R.G., Goodin M.M., Bragg J.N., Deng M. Biology of plant rhabdoviruses. Annual Review of Phytopathology. 2005;43:623–660. doi: 10.1146/annurev.phyto.43.011205.141136. [DOI] [PubMed] [Google Scholar]
- Jayakar H.R., Jeetendra E., Whitt M.A. Rhabdovirus assembly and budding. Virus Research. 2004;106:117–132. doi: 10.1016/j.virusres.2004.08.009. [DOI] [PubMed] [Google Scholar]
- Jayakar H.R., Whitt M.A. Identification of two additional translation products from the matrix (M) gene that contribute to vesicular stomatitis virus cytopathology. Journal of Virology. 2002;76:8011–8018. doi: 10.1128/JVI.76.16.8011-8018.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johal J., Gresty K., Kongsuwan K., Walker P.J. Antigenic characterization of bovine ephemeral fever rhabdovirus G and GNS glycoproteins expressed from recombinant baculoviruses. Archives of Virology. 2008;153:1657–1665. doi: 10.1007/s00705-008-0164-0. [DOI] [PubMed] [Google Scholar]
- Johnson M.C., Maxwell J.M., Loh P.C., Leong J.A. Molecular characterization of the glycoproteins from two warm water rhabdoviruses: snakehead rhabdovirus (SHRV) and rhabdovirus of penaeid shrimp (RPS)/spring viremia of carp virus (SVCV) Virus Research. 1999;64:95–106. doi: 10.1016/s0168-1702(99)00071-4. [DOI] [PubMed] [Google Scholar]
- Johnson M.C., Simon B.E., Kim C.H., Leong J.-A.C. Production of recombinant snakehead rhabdovirus: the NV protein. Is not required for viral replication. Journal of Virology. 2000;74:2343–2350. doi: 10.1128/jvi.74.5.2343-2350.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzourakis A., Gifford R.J. Endogenous viral elements in animal genomes. PLoS Genetics. 2010;6:e1001191. doi: 10.1371/journal.pgen.1001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp C., Imler J.L. Antiviral immunity in drosophila. Current Opinion in Immunology. 2009;21:3–9. doi: 10.1016/j.coi.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp G.E., Lee V.H., Moore D.L., Shope R.E., Causey O.R., Murphy F.A. Kotonkan, a new rhabdovirus related to Mokola virus of the rabies serogroup. American Journal of Epidemiology. 1973;98:43–49. doi: 10.1093/oxfordjournals.aje.a121531. [DOI] [PubMed] [Google Scholar]
- Kim D.-H., Oh H.-K., Eou J.-I., Seo H.-J., Kim S.-K., Oh M.-J., Nam S.-W., Choi T.-J. Complete nucleotide sequence of the hirame rhabdovirus, a pathogen of marine fish. Virus Research. 2005;107(1):1–9. doi: 10.1016/j.virusres.2004.06.004. [DOI] [PubMed] [Google Scholar]
- Kopecky S.A., Willingham M.C., Lyles D.S. Matrix protein and another viral component contribute to induction of apoptosis in cells infected with vesicular stomatitis virus. Journal of Virology. 2001;75:12169–12181. doi: 10.1128/JVI.75.24.12169-12181.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzschmar E., Peluso R., Schnell M.J., Whitt M.A., Rose J.K. Normal replication of vesicular stomatitis virus without C proteins. Virology. 1996;216:309–316. doi: 10.1006/viro.1996.0066. [DOI] [PubMed] [Google Scholar]
- Kurath G., Ahern K.G., Pearson G.D., Leong J.C. Molecular cloning of the six mRNA species of infectious hematopoietic necrosis virus, a fish rhabdovirus, and gene order determination by R-loop mapping. Journal of Virology. 1985;53:469–476. doi: 10.1128/jvi.53.2.469-476.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurath G., Higman K.H., Bjorklund H.V. Distribution and variation of NV genes in fish rhabdoviruses. Journal of General Virology. 1997;78:113–117. doi: 10.1099/0022-1317-78-1-113. [DOI] [PubMed] [Google Scholar]
- Kurath G., Leong J.C. Characterization of infectious hematopoietic necrosis virus mRNA species reveals a nonvirion rhabdovirus protein. Journal of Virology. 1985;53:462–468. doi: 10.1128/jvi.53.2.462-468.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurotani A., Kiyotani K., Kato A., Shioda T., Sakai Y., Mizumoto K., Yoshida T., Nagai Y. Sendai virus C proteins are categorically nonessential gene products but silencing their expression severely impairs viral replication and pathogenesis. Genes to Cells. 1998;3:111–124. doi: 10.1046/j.1365-2443.1998.00170.x. [DOI] [PubMed] [Google Scholar]
- Kurz W., Gelderblom H., Flugel R.M., Darai G. Isolation and characterization of a tupaia rhabdovirus. Intervirology. 1986;25:88–96. doi: 10.1159/000149661. [DOI] [PubMed] [Google Scholar]
- Kuzmin I.V., Hughes G.J., Rupprecht C.E. Phylogenetic relationships of seven previously unclassified viruses within the family Rhabdoviridae using partial nucleoprotein gene sequences. Journal of General Virology. 2006;87:2323–2331. doi: 10.1099/vir.0.81879-0. [DOI] [PubMed] [Google Scholar]
- Kuzmin I.V., Mayer A.E., Niezgoda M., Markotter W., Agwanda B., Breiman R.F., Rupprecht C.E. Shimoni bat virus, a new representative of the Lyssavirus genus. Virus Research. 2010;149:197–210. doi: 10.1016/j.virusres.2010.01.018. [DOI] [PubMed] [Google Scholar]
- Kuzmin I.V., Novella I.S., Dietzgen R.G., Padhi A., Rupprecht C.E. The rhabdoviruses: biodiversity, phylogenetics, and evolution. Infection, Genetics and Evolution. 2009;9:541–553. doi: 10.1016/j.meegid.2009.02.005. [DOI] [PubMed] [Google Scholar]
- Kuzmin I.V., Wu X., Tordo N., Rupprecht C.E. Complete genomes of Aravan, Khujand Irkut and West Caucasian bat viruses, with special attention to the polymerase gene and non-coding regions. Virus Research. 2008;136:81–90. doi: 10.1016/j.virusres.2008.04.021. [DOI] [PubMed] [Google Scholar]
- Landes-Devauchelle C., Bras F., Dezelee S., Teninges D. Gene 2 of the sigma rhabdovirus genome encodes the P protein, and gene 3 encodes a protein related to the reverse transcriptase of retroelements. Virology. 1995;213:300–312. doi: 10.1006/viro.1995.0003. [DOI] [PubMed] [Google Scholar]
- Lazzarini R.A., Keene J.D., Schubert M. The origins of defective interfering particles of the negative-strand RNA viruses. Cell. 1981;26:145–154. doi: 10.1016/0092-8674(81)90298-1. [DOI] [PubMed] [Google Scholar]
- Letchworth G.J., Rodriguez L.L., Del Cbarrera J. Vesicular stomatitis. The Veterinary Journal. 1999;157(3):239–260. doi: 10.1053/tvjl.1998.0303. [DOI] [PubMed] [Google Scholar]
- Longdon B., Obbard D.J., Jiggins F.M. Sigma viruses from three species of Drosophila form a major new clade in the rhabdovirus phylogeny. Proceedings of the Royal Society B. 2010;277:35–44. doi: 10.1098/rspb.2009.1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longdon B., Wilfert L., Obbard D.J., Jiggins F.M. Rhabdoviruses in two species of Drosophila: vertical transmission and a recent sweep. Genetics. 2011;188:141–150. doi: 10.1534/genetics.111.127696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukashev A.N. Evidence for recombination in Crimean-Congo hemorrhagic fever virus. Journal of General Virology. 2005;86:2333–2338. doi: 10.1099/vir.0.80974-0. [DOI] [PubMed] [Google Scholar]
- Luytjes W., Bredenbeek P.J., Noten A.F.H., Horzinek M.C., Spaan W.J.M. Sequence of mouse hepatitis virus A59 mRNA 2 Indications for RNA recombination between coronaviruses and influenza C virus. Virology. 1988;166:415–422. doi: 10.1016/0042-6822(88)90512-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massah A., Izadpanah K., Afsharifar A.R. Analysis of nucleotide sequence of Iranian maize mosaic virus confirms its identity as a distinct nucleorhabdovirus. Archives of Virology. 2008;153:1041–1047. doi: 10.1007/s00705-008-0085-y. [DOI] [PubMed] [Google Scholar]
- McWilliam S.M., Kongsuwan K., Cowley J.A., Byrne K.A., Walker P.J. Genome organization and transcription strategy in the complex GNS-L intergenic region of bovine ephemeral fever rhabdovirus. Journal of General Virology. 1997;78:1309–1317. doi: 10.1099/0022-1317-78-6-1309. [DOI] [PubMed] [Google Scholar]
- Melcher U. The ‘30K’ superfamily of viral movement proteins. Journal of General Virology. 2000;81:257–266. doi: 10.1099/0022-1317-81-1-257. [DOI] [PubMed] [Google Scholar]
- Min B.-E., Martin K., Wang R., Tafelmeyer P., Brisges M., Goodin M. A host–factor interaction and localization map for a plant-adapted rhabdovirus implicates cytoplasm-tethered transcription activators in cell-to-cell movement. Molecular Plant–Microbe Interactions. 2010;23:1420–1432. doi: 10.1094/MPMI-04-10-0097. [DOI] [PubMed] [Google Scholar]
- Morimoto K., Ohkubo A., Kawai A. Structure and transcription of the glycoprotein gene of attenuated HEP-Flury strain of rabies virus. Virology. 1989;173:465–477. doi: 10.1016/0042-6822(89)90559-x. [DOI] [PubMed] [Google Scholar]
- Morse M.A., Marriott A.C., Nuttall P.A. The glycoprotein of Thogoto virus (a tick-borne orthomyxo-like virus) is related to the baculovirus glycoprotein GP64. Virology. 1992;186:640–646. doi: 10.1016/0042-6822(92)90030-s. [DOI] [PubMed] [Google Scholar]
- Moseley G.W., Filmer R.P., DeJesus M.A., Jans D.A. Nucleocytoplasmic distribution of rabies virus P protein is regulated by phosphorylation adjacent to C-terminal nuclear import and export signals. Biochemistry. 2007;46:12053–12061. doi: 10.1021/bi700521m. [DOI] [PubMed] [Google Scholar]
- Moseley G.W., Lahaye X., Roth D.M., Oksayan S., Filmer R.P., Rowe C.L., Blondel D., Jans D.A. Dual modes of rabies P-protein association with microtubules: a novel strategy to suppress the antiviral response. Journal of Cell Science. 2009;122:3652–3662. doi: 10.1242/jcs.045542. [DOI] [PubMed] [Google Scholar]
- Mourya D.T., Thakare J.R., Gokhale M.D., Powers A.M., Hundekar S.L., Jayakumar P.C., Bondre V.P., Shouche Y.S., Padbirdi V.S. Isolation of chikungunya virus from Aedes aegypti mosquitoes collected in the town of Yawat, Pune District, Maharashtra State, India. Acta Virologica. 2001;45:305–309. [PubMed] [Google Scholar]
- Nadin-Davis S.A., Abdel-Malik M., Armstrong J., Wandeler A.I. Lyssavirus P gene characterisation provides insights into the phylogeny of the genus and identifies structural similarities and diversity within the encoded phosphoprotein. Virology. 2002;298:286–305. doi: 10.1006/viro.2002.1492. [DOI] [PubMed] [Google Scholar]
- Nagai Y., Kato A. Accessory genes of the paramyxoviridae, a large family of nonsegmented negative-strand RNA viruses, as a focus of active investigation by reverse genetics. Current Topics in Microbiology and Immunology. 2004;283:197–248. doi: 10.1007/978-3-662-06099-5_6. [DOI] [PubMed] [Google Scholar]
- Pasdeloup D., Poisson N., Raux H., Gaudin Y., Ruigrok R.W., Blondel D. Nucleocytoplasmic shuttling of the rabies virus P protein requires a nuclear localization signal and a CRM1-dependent nuclear export signal. Virology. 2005;334:284–293. doi: 10.1016/j.virol.2005.02.005. [DOI] [PubMed] [Google Scholar]
- Pearce A.F., Lyles D.S. Vesicular stomatitis virus induces apoptosis primarily through Bak rather than Bax by inactivating Mcl-1 and Bcl-XL. Journal of Virology. 2009;83:9102–9112. doi: 10.1128/JVI.00436-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peluso R.W., Richardson J.C., Talon J., Lock M. Identification of a set of proteins (C′ and C) encoded by the bicistronic P gene of the Indiana serotype of vesicular stomatitis virus and analysis of their effect on transcription by the viral RNA polymerase. Virology. 1996;218:335–342. doi: 10.1006/viro.1996.0202. [DOI] [PubMed] [Google Scholar]
- Perrault J. Origin and replication of defective interfering particles. Current Topics in Microbiology and Immunology. 1981;93:151–207. doi: 10.1007/978-3-642-68123-3_7. [DOI] [PubMed] [Google Scholar]
- Powell M.L. Translational termination-reinitiation in RNA viruses. Biochemical Society Transactions. 2010;38:1558–1564. doi: 10.1042/BST0381558. [DOI] [PubMed] [Google Scholar]
- Qin Z., Sun L., Ma B., Cui Z., Zhu Y., Kitamura Y., Liu W. F gene recombination between genotype II and VII Newcastle disease virus. Virus Research. 2008;131:299–303. doi: 10.1016/j.virusres.2007.10.001. [DOI] [PubMed] [Google Scholar]
- Rao B.L., Basu A., Wairagkar N.S., Gore M.M., Arankalle V.A., Thakare J.P., Jadi R.S., Rao K.A., Mishra A.C. A large outbreak of acute encephalitis with high fatality rate in children in Andhra Pradesh, India, in 2003, associated with Chandipura virus. The Lancet. 2004;364:869–874. doi: 10.1016/S0140-6736(04)16982-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravkov E.V., Smith J.S., Nichol S.T. Rabies virus glycoprotein gene contains a long 3′ noncoding region which lacks pseudogene properties. Virology. 1995;206:718–723. doi: 10.1016/s0042-6822(95)80095-6. [DOI] [PubMed] [Google Scholar]
- Redinbaugh M.G., Hogenhout S.A. Plant rhabdoviruses. Current Topics in Microbiology and Immunology. 2005;292:143–163. doi: 10.1007/3-540-27485-5_7. [DOI] [PubMed] [Google Scholar]
- Reed S.E., Tsai C.-W., Willie K.J., Redinbaugh M.G., Hogenhout S.A. Shotgun sequencing of the negative-sense RNA genome of the rhabdovirus maize mosaic virus. Journal of Virological Methods. 2005;129:91–96. doi: 10.1016/j.jviromet.2005.05.013. [DOI] [PubMed] [Google Scholar]
- Revill P., Trinh X., Dale J., Harding R. Taro vein chlorosis virus: characterization and variability of a new nucleorhabdovirus. Journal of General Virology. 2005;86:491–499. doi: 10.1099/vir.0.80591-0. [DOI] [PubMed] [Google Scholar]
- Rieder M., Brzozka K., Pfaller C.K., Cox J.H., Stitz L., Conzelmann K.-K. Genetic dissection of interferon-antagonistic functions of rabies virus phosphoprot inhibition of interferon regulatory factor 3 activation. Is important for pathogenicity. Journal of Virology. 2011;85:842–852. doi: 10.1128/JVI.01427-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche S., Albertini A., Lepault J., Bressanelli S., Gaudin Y. Structures of vesicular stomatitis virus glycoprotein: membrane fusion revisited. Cellular and Molecular Life Sciences. 2008;65:1716–1728. doi: 10.1007/s00018-008-7534-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez L.L. Emergence and re-emergence of vesicular stomatitis in the United States. Virus Research. 2002;85:211–219. doi: 10.1016/s0168-1702(02)00026-6. [DOI] [PubMed] [Google Scholar]
- Rupprecht C.E., Hanlon C.A., Hemachudha T. Rabies re-examined. Lancet Infectious Diseases. 2002;2:327–343. doi: 10.1016/s1473-3099(02)00287-6. [DOI] [PubMed] [Google Scholar]
- Schnell M.J., Mebatsion T., Conzelmann K.K. Infectious rabies viruses from cloned cDNA. EMBO Journal. 1994;13:4195–4203. doi: 10.1002/j.1460-2075.1994.tb06739.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoen C.D., Limpens W., Moller I., Groeneveld L., Klerks M.M., Lindner J.L. The complete gnomic sequence of strawberry crinkle virus, a member of the Rhabdoviridae. Acta Horticologica. 2004;656:45–50. [Google Scholar]
- Scholthof K.G., Hillman B.I., Modrell B., Heaton L.A., Jackson A.O. Characterization and detection of sc4: a sixth gene encoded by sonchus yellow net virus. Virology. 1994;204:279–288. doi: 10.1006/viro.1994.1532. [DOI] [PubMed] [Google Scholar]
- Schutze H., Enzmann P.J., Mundt E., Mettenleiter T.C. Identification of the non-virion (NV) protein of fish rhabdoviruses viral haemorrhagic septicaemia virus and infectious haematopoietic necrosis virus. Journal of General Virology. 1996;77(Pt 6):1259–1263. doi: 10.1099/0022-1317-77-6-1259. [DOI] [PubMed] [Google Scholar]
- Sibold C., Meisel H., Kruger D.H., Labuda M., Lysy J., Kozuch O., Pejcoch M., Vaheri A., Plyusnin A. Recombination in Tula hantavirus evolution: analysis of genetic lineages from Slovakia. Journal of Virology. 1999;73:667–675. doi: 10.1128/jvi.73.1.667-675.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiropoulou C.F., Nichol S.T. A small highly basic protein is encoded in overlapping frame within the P gene of vesicular stomatitis virus. Journal of Virology. 1993;67:3103–3110. doi: 10.1128/jvi.67.6.3103-3110.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springfeld C., Darai G., Cattaneo R. Characterization of the Tupaia rhabdovirus genome reveals a long open reading frame overlapping with P and a novel gene encoding a small hydrophobic protein. Journal of Virology. 2005;79:6781–6790. doi: 10.1128/JVI.79.11.6781-6790.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St. George T.D., Standfast H.A. Bovine ephemeral fever. In: Monath T.P., editor. vol. 2. CRC Press; Boca Raton: 1988. pp. 71–86. (The Arboviruses: Epidemiology and Ecology). [Google Scholar]
- Stallknecht D.E., Perzak D.E., Bauer L.D., Murphy M.D., Howerth E.W. Contact transmission of vesicular stomatitis virus New Jersey in pigs. American Journal of Veterinary Research. 2001;62:516–520. doi: 10.2460/ajvr.2001.62.516. [DOI] [PubMed] [Google Scholar]
- Tanno F., Nakatsu A., Toryama S., Kojima M. Complete nucleotide sequence of northern cereal mosaic virus and its genome organization. Archives of Virology. 2000;145:1373–1384. doi: 10.1007/s007050070096. [DOI] [PubMed] [Google Scholar]
- Tao J.J., Zhou G.Z., Gui J.F., Zhang Q.Y. Genomic sequence of mandarin fish rhabdovirus with an unusual small non-transcriptional ORF. Virus Research. 2008;132:86–96. doi: 10.1016/j.virusres.2007.10.018. [DOI] [PubMed] [Google Scholar]
- Teng Y., Liu H., Lv J.Q., Fan W.H., Zhang Q.Y., Qin Q.W. Characterization of complete genome sequence of the spring viremia of carp virus isolated from common carp (Cyprinus carpio) in China. Archives of Virology. 2007;152:1457–1465. doi: 10.1007/s00705-007-0971-8. [DOI] [PubMed] [Google Scholar]
- Teninges D., Bras-Herreng F. Rhabdovirus sigma, the hereditary CO2 sensitivity agent of drosophila: nucleotide sequence of a cDNA clone encoding the glycoprotein. Journal of General Virology. 1987;68:2625–2638. doi: 10.1099/0022-1317-68-10-2625. [DOI] [PubMed] [Google Scholar]
- Thavara U., Siriyasatien P., Tawatsin A., Asavadachanukorn P., Anantapreecha S., Wongwanich R., Mulla M.S. Double infection of heteroserotypes of dengue viruses in field populations of Aedes aegypti and Aedes albopictus (Diptera: Culicidae) and serological features of dengue viruses found in patients in southern Thailand. Southeast Asian Journal of Tropical Medical Public Health. 2006;37:468–476. [PubMed] [Google Scholar]
- Thoulouze M.-I., Bouguyon E., Carpentier C., Bremont M. Essential role of the NV protein of novirhabdovirus for pathogenicity in rainbow trout. Journal of Virology. 2004;78:4098–4107. doi: 10.1128/JVI.78.8.4098-4107.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tordo N., Poch O., Ermine A., Keith G., Rougeon F. Walking along the rabies genome: is the large G–L intergenic region a remnant gene? Proceedings of the National Academy of Sciences United States America. 1986;83:3914–3918. doi: 10.1073/pnas.83.11.3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troffer-Charlier N., Cura V., Hassenboehler P., Moras D., Cavarelli J. Functional insights from structures of coactivator-associated arginine methyltransferase 1 domains. EMBO Journal. 2007;26:4391–4401. doi: 10.1038/sj.emboj.7601855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C.W., McGraw E.A., Ammar E.-D., Dietzgen R.G., Hogenhout S.A. Drosophila melanogaster mounts a unique Immune response to the rhabdovirus sigma virus. Applied and Environmental Microbiology. 2008;74:3251–3256. doi: 10.1128/AEM.02248-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C.W, Redinbaugh M.G., Willie K.J., Reed S., Goodin M., Hogenhout S.A. Complete genome sequence and in planta subcellular localization of maize fine streak virus proteins. Journal of Virology. 2005;79:5304–5314. doi: 10.1128/JVI.79.9.5304-5314.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker P.J. Bovine ephemeral fever in Australia and the world. Current Topics in Microbiology and Immunology. 2005;292:57–80. doi: 10.1007/3-540-27485-5_4. [DOI] [PubMed] [Google Scholar]
- Walker, P.J., Blasdell, K., Joubert, D.A. Ephemeroviruses: arthropod-borne rhabdoviruses of ruminants, with large and complex genomes. In: Dietzgen, R.G., Kuzmin, I.V. (Eds.), Rhabdoviruses: Molecular Taxonomy, Evolution, Genomics, Ecology, Cytopathology and Control. Horizon Scientific Press, London, in press.
- Walker P.J., Byrne K.A., Cybinski D.H., Doolan D.L., Wang Y.H. Proteins of bovine ephemeral fever virus. Journal of General Virology. 1991;72:67–74. doi: 10.1099/0022-1317-72-1-67. [DOI] [PubMed] [Google Scholar]
- Walker P.J., Byrne K.A., Riding G.A., Cowley J.A., Wang Y., McWilliam S. The genome of bovine ephemeral fever rhabdovirus contains two related glycoprotein genes. Virology. 1992;191:49–61. doi: 10.1016/0042-6822(92)90165-l. [DOI] [PubMed] [Google Scholar]
- Walker P.J., Winton J.R. Emerging viral diseases of fish and shrimp. Veterinary Research. 2010;41:e51. doi: 10.1051/vetres/2010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., McWilliam S.M., Cowley J.A., Walker P.J. Complex genome organization in the GNS-L intergenic region of Adelaide River rhabdovirus. Virology. 1994;203:63–72. doi: 10.1006/viro.1994.1455. [DOI] [PubMed] [Google Scholar]
- Wang Y., Walker P.J. Adelaide river rhabdovirus expresses consecutive glycoprotein genes as polycistronic mRNAs: new evidence of gene duplication as an evolutionary process. Virology. 1993;195:719–731. doi: 10.1006/viro.1993.1423. [DOI] [PubMed] [Google Scholar]
- Zhang Q.Y., Tao J.J., Gui J.F. Isolation and characterization of a rhabdovirus from co-infection of two viruses in mandarin fish. Aquaculture. 2007;262:1–9. [Google Scholar]


