Abstract
The 3′ untranslated region (UTR) of porcine arterivirus genome plays a pivotal role for virus replication, yet the properties of 3′ UTR remain largely undefined. We conducted site-directed mutagenesis to the 3′ UTR of the type II porcine reproductive and respiratory syndrome virus (PRRSV). Serial deletions of the 3′ UTR showed that at least 40 nucleotides immediately following the ORF7 stop codon were dispensable for the viability of PRRSV in cultured cells. We then constructed a chimeric PRRSV cDNA clone using type II PRRSV as the backbone containing the 3′ UTR from the type I PRRSV. The chimeric virus was viable and shared similar properties with the parental virus. Our results provided the first description of the 40nt dispensable region in type I PRRSV 3′ UTR, and further predicted structure demonstrated that the high-order structure of 3′ UTR might play significant roles in its function.
Keywords: Porcine reproductive and respiratory syndrome virus, Replication, Secondary structure, 3′ untranslated region (UTR)
1. Introduction
Positive strand virus genomes serve as both a messenger RNA and templates for genomic RNA synthesis, encapsidation and subgenomic mRNA transcription (Pasternak et al., 2006, Snijder and Meulenberg, 1998). The coordination among different stages of virus infection has been the focus for RNA virology. It is believed that un-translated region (UTR) of the RNA genome plays a central role in balancing these processes, via different sequence motifs, alternative high-order structures, and interacting with other viral or host factors (Chang and Luo, 2006, Garneau et al., 2008, Liu and Leibowitz, 2010, Uzri and Gehrke, 2009, Zust et al., 2008). For instance, complete 3′ UTR deletion of poliovirus genome has deleterious effect on virus replication (Rohll et al., 1995, Todd et al., 1997). In addition, it has been reported that positive-strand RNA virus 3′ UTR can control viral gene translation as an independent enhancer or indirect interaction with the internal ribosome entry site of the 5′ UTR (Koh et al., 2002, Lopez de Quinto et al., 2002). The 3′ UTR structure of porcine reproductive and respiratory syndrome virus (PRRSV) genome remains largely unknown.
Coronaviruses and arteriviruses belong to the order Nidovirales, of which RNA synthesis include genomic RNA replication and subgenomic mRNA (sgmRNA) transcription. It is believed that both genomic RNA and subgenomic RNA synthesis begin with the synthesis of the minus-strand RNA at the 3′ end of the genome (Pasternak et al., 2006, Sawicki et al., 2007), during which 3′ UTR plays an essential role in Nidovirus replication. It was reported that the primary sequence of an essential region, a bulged stem-loop, and a pseudoknot structure of mouse hepatitis virus (MHV) 3′ UTR are crucial for the virus viability, and such multiple, sometimes mutually exclusive elements may serve as a molecular switch to balance the different stages of virus replication (Goebel et al., 2004a, Goebel et al., 2004b, Hsue and Masters, 1997, Liu and Leibowitz, 2010, Williams et al., 1999).
PRRSV is a member of Arteriviridae, together with equine arteritis virus (EAV), mouse lactate dehydrogenase-elevating virus (LDV), and simian haemorrhagic fever virus (SHFV) (den Boon et al., 1991, Pasternak et al., 2006, Snijder and Meulenberg, 1998). PRRSV genome is about 15 kb in length, containing at least nine open reading frames (ORF) flanked by 5′- and 3′-UTR. Based on the overall genetic relationship, PRRSV is further classified as two genotypes, European (type I) and North American (type II), between which 60% genetic identity of the overall genomic sequences exists (Mateu and Diaz, 2008). PRRSV is known for its genetic heterogeneity, up to 21% intra-genotypic diversity for type II PRRSV has been reported (Key et al., 2001, Mateu and Diaz, 2008, Meng, 2000). Interestingly, these two distinct PRRSV genotypes were identified almost simultaneously as the causative agents of blue-ear disease in two different continents (Collins et al., 1992, Plagemann, 2003, Wensvoort et al., 1991), which demonstrat that these two genotype virus share the same key functional component in virus replication or transcription process.
Similar to the coronavirus, arterivirus RNA synthesis is initiated at the genomic 3′ end, where the RNA-dependent RNA polymerase (RdRp) binds and initiates the synthesis of the negative strand genome-length RNA and a set of subgenomic RNAs, formed by a unique discontinuous transcription mechanism (Beerens et al., 2007, Meng et al., 1996, Snijder and Meulenberg, 1998). These negative-sense RNAs then serve as templates for the synthesis of progeny viral RNA genome and a series of 5′ and 3′ co-terminal subgenomic RNAs (Pasternak et al., 2006). The UTR sequence and structural elements are believed to play a vital regulatory role in these RNA synthesis processes. It was reported that the Lelystad virus (LV), the prototypic type I PRRSV can tolerate 7 nt deletion, but 30 nt deletion in the 114 nt length of LV 3′ UTR abolished RNA replication (Verheije et al., 2001, Verheije et al., 2002). With a shorter 3′ UTR of 59 nt, the EAV 3′ genomic terminus forms up to five stem-loop (SL) structures, with the SL5 located in 3′ UTR forming a pseudoknot with the SL4 coding for nucleocapsid (N) protein (Beerens and Snijder, 2007). Interestingly, part of the SL4 analogue, located in PRRSV LV 3′ UTR, is also engaged in a kissing-loop interaction with a 7 nt sequence coding for the LV N protein (Beerens and Snijder, 2007, Verheije et al., 2001, Verheije et al., 2002). These results demonstrated that arterivirus replication is also regulated by structural elements residing in the 3′ end of the genomic RNA. Previous studies find that the 911 nucleotides from the 3′ end of the viral genome contains the minimal cis-acting element required for type II PRRS viral replication (Choi et al., 2006). Beyond that, little is known about regulatory elements for PRRSV type II, which has the longest 3′ UTR (151 nt) among the arteriviruses. In particular, it remains unresolved that if the 3′ UTR of the two genotypes of PRRSV play a similar role in PRRSV replication process, despite of the distinct relationship in primary sequence and the length.
We hypothesized that the extra 40 nt located in the 5′ proximal of the 3′ UTR of type II PRRSV is nonessential for virus replication, which may also be regulated by structural RNA elements at the genomic 3′ end. Based on the full-length infectious cDNA clone of pAPRRS (Yu et al., 2009, Yuan and Wei, 2008), we conducted reverse genetic manipulations to the PRRSV 3′ UTR, and found that 1) the first 40 nt of the 5′ proximal region of type II PRRSV 3′ UTR is dispensable for viral infectivity in cultured cells; and surprisingly, 2) the entire 3′ UTR of the type I PRRSV is fully functional in the backbone of pAPRRS, despite that only 63% pair-wise genetic identity between the two 3′ UTR. Our data provide clues that the high order structural elements residing in 3′ UTR might play a critical role for porcine arterivirus replication.
2. Materials and methods
2.1. Viruses and cells
MARC-145 cells (ATCC No. CRL-12219, Manassas, VA) were grown in MEM (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen, Gaithersburg, MD) and 100 ug/ml penicillin and 100 UI/ml streptomycin. The parental and mutant viruses were cultured as described previously (Yuan and Wei, 2008).
2.2. Site-directed PCR-based mutagenesis
The infectious cDNA clone pAPRRS of type II PPRSV (Yuan and Wei, 2008) was used as a template and parental cDNA clone. pTB5c, a subclone covering the 3′ terminus (nt 11210–15521) of APRRS genome (GenBank accession no. GQ330474), was used as the shuttle plasmid for mutagenesis PCR (Yuan and Wei, 2008). Six oligonucleotides primers (Table 1 ) containing appropriate restriction enzyme sites and the desired mutations were designed and synthesized based on APRRS genomic sequences, and Quick-Change PCR mutagenesis (Stratagene) was carried out on the pTB5c plasmid, as instructed by the supplier. The infectous cDNA clone containing the 40 bp deletion was produced by three way ligation. Two primer pairs SF13013/SRLVAscUR and SFLVAscUR/Qst (Table 1) were used to amplify two PCR fragments, then the PCR fragments were digested with single enzyme site Spe I and Pac I, Pac I and Xho I (immediately downstream of the genomic poly(A)-tail), respectively. The full-length cDNA clone pAPRRS was treated with Spe I and Xho I, and then the gel-purified pAPRRS Spe I/Xho I was used as the vector arm for assembly of the full-length cDNA clones. Three purified fragments were used for the three-way ligation, yielding the full-length mutant clones p3URAD40. To investigate whether the inserted Asc I cutting site affects the virus replication, a splicing-overlap-extension (S.O.E) PCR was used to construct full-length clone p3URD40, in which the polylinker was eliminated. Primer pairs SF13103 and SR15270-40U, SF15411N and Qst (Table 1) were used to amplify two PCR fragments, using pAPRRS as template. Two purified PCR fragments were used as templates for second round PCR, subsequently primer SF13103 and Qst were used to amplify. The final mutant fragment was cloned into pCR-Blunt II-TOPO vector named pK3URD40. The plasmid pK3URD40 and parental pAPRRS were digested with Spe I and Xho I, and then the purified DNA fragments were ligated by T4 DNA ligase (Fermentas, SANGON, Shanghai) to construct the full-length clone p3URD40. Chimeric plasimd pSURLV was constructed by using primer pairs SF13013/LVAscUR, and LVAscUF/Qst, but the second PCR fragment was amplified with type I PRRSV cDNA as template, pLV5c (a kind gift of Dr. Zhuang) was the cDNA clone covering ORF7-3′ UTR of the PRRSV Lelystad virus (GenBank Accession Number: M96262). All mutations were verified by PCR restriction enzyme mapping and nucleotide sequencing.
Table 1.
Oligonucleotides used in this study.
| Primer* | Sequence (5′-3′)† | Purpose |
|---|---|---|
| SF13103 | CTTGactagtGTTTACGCCTGGTTG | Mutation |
| Qst | gagtgacgaggactcgagcgcatgcTTTTTTTTTTTTTT | Mutation |
| SFCDU40 | ttaattaagggcgcgccTGGGCTGGCATTCTTGAG | Deletion |
| SRCDU40 | ttaattaagggcgcgccTCATGCTGAGGGTGATGCTGTG | Deletion |
| SFLVAscUR | ggcgcgccttaattaaaTCATGCTGTGGTGATGCTGTGACG | Swapping |
| SRLVAscUR | tttaattaaggcgcgccTTTGACAGTCAGGTGAATGGCCGC | Swapping |
| SF15411N | CCTCAGCATGAGAATGTGTGGTGAATGGCACTG | Deletion |
| SR15370-40U | CCACACATTCTCATGCTGAGGGTGATGCTGTG | Deletion |
| VR3 | AATTTCGGCCGCATGGTTTTCGCCAATTAAATCTTACCCCCACACGGTCGC | Probe |
| LVR3 | AATTTCGTCACATGGTTCCTGCCTGATTAAGTATGACCCCCCATGTTATCG | Probe |
Primer names are organized in groups. Prefixes: Qst, reverse transcription primer, SF, forward PCR primer; SR, reverse PCR primer; LVR3 and VR3 are used for probe.
Restriction sites introduced by PCR are lowercase.
2.3. RNA transcription and transfection
10 μg of the full-length cDNA clone plasmid pAPRRS and its derivatives (pSURLV, p3URD40, p3URAD40) were linearized by using restriction endonuclease Xho I. In vitro transcripts were generated with the T7 mMESSAGER mMACHINE kit (Ambion Inc, Austin, TX) according to the supplier's instruction. The purity and integrity of the synthetic RNAs were verified by non-denatured 1% agarose gel electrophoresis (AGE) and the quantity and integrity of the synthetic RNA were measured based on electrophoresis results. MARC-145 cell transfections were carried out with 2 μl in vitro transcribed RNA, using the DMRIE-C according to the supplier's protocol (Invitrogen, Carlsbad, CA). Briefly, the transcription RNA mixture were added to each monolayer cells in 6-well-plate (Cornings) for 30–45 min at room temperature, then the mixtures were removed and the monolayers were washed twice with phosphate buffered saline (PBS), followed by replenishing with 2 ml/well of MEM containing 2% FBS (Invitrogen), and incubated at 37 °C with 5% CO2. When the tansfected cells developed 80% of cytopathic effect (CPE), the supernatant was harvested, aliquoted, and stored at −70 °C. The primary transfectant viruses were designated as passage 0 (P0) of the rescued viruses. The P1 virus stocks were obtained by infecting fresh MARC-145 cells with 0.1 of multiplicity of infection (MOI) of P0 viruses. In the same manner, P2-P5 of the rescued viruses were generated and saved for further analysis.
2.4. Viral RNA purification, RT-PCR, and nucleotide sequencing
Viral genomic RNA was isolated directly from the supernatant of the P5 virus infected cell cultures using QIAamp Viral RNA Mini Kit (QIAgen, Hilden, Germany), according to the manufacturer's protocol. The first strand cDNA synthesis was carried out at 42 °C for 1 h using avian myeloblastosis virus (AMV) reverse transcriptase (TaKaRa, Dalian, China) with an anchored poly(T) primer Qst (Table 1), using the purified viral RNA as the template. Subsequently, the cDNA was used as template for amplification of mutation site flanking sequencing, ranging from nt15008 (GQ330474) to the end of the 3′UTR of the respective rescued virus. Oligonucleotide SF15008, which located upstream of the 3′ UTR was used as sense primer, and primer Qst was used as antisense primer (Table 1). The PCR consists of 38 cycles, each comprising 95 °C denaturation for 20 s, 30 s annealing at a temperature calculated to be 5 °C below the melting temperature (Tm) of the involved primers, and 1 min of extention at 72 °C. The PCR products were separated on 1% AGE, purified by QIAquick gel-purification kit (QIAgen), and subjected to nucleotide sequencing. Besides, all rescued viruses were verified by full-length sequence. The primers for full-length sequence are available upon request.
2.5. Northern blot analysis
The MARC-145 cells were infected with a 0.01 MOI of the rescued viruses (P1). Total cellular RNAs were isolated at 36 hours post inoculation (hpi) using the RNAWiz reagent per the instruction of the supplier (Ambion, Austin, TX). Nothern blot was carried out as previously described (Yu et al., 2009) with minor modifications. Briefly, 2 μl of total cellular RNAs per lane were loaded and separated by 1% formaldhyde-denatured AGE. The separated RNAs were transferred to the BrightStar-Plus membrane (Ambion, Austin, TX) for 10 h, cross-linked by UV light, pre-hybridized at 42 °C for 2 h, and probed with the genotypically specific 3′ UTR oligodeoxynucleotide probe VR3 and LVR3 (Table 1), respectively. The filters were hybridized overnight, and washed with low-stringency buffers (Ambion, Austin, TX), and were incubated with alkaline phosphatase-conjugated streptavidin, followed by reaction with the chemiluminescent substrate CDP-STAR (Ambion, Austin, TX). The overlaid film was developed by exposure for 6 h in a dark cassette in the dark room.
2.6. Viral plaque assay and growth kinetics of the rescued viruses
For plaque assays, the sub-confluent MARC-145 cells in 6-well-plate were incubated with 200 μl of 10-fold serial dilution of P5 virus, and incubated at room temperature (RT) for 1 h. The inoculum was then discarded, followed by washing the cell monolayer with PBS for three times, and overlaid with a pre-made MEM (Invitrogen) containing 1% low melting agarose and 2% FBS (Invitrogen). The solidified agarose plate was inversely (top down) placed into the incubator, the cell monolayer at 5 dpi was stained with 5% crystal violet in 20% ethanol. To determine the viral one-step growth curve, P5 virus (1 MOI) of the rescued viruses were inoculated in the sub-confluent MARC-145 cells in a six-well-plate, as described before (Yu et al., 2009). Then 200 μl of supernatant of the infected cells was collected and replenished with the same volume of fresh medium at the indicated time points (4, 8, 12, 16, 20 and 24 hpi, respectively), and stored at −70 °C for virus titration. The virus titers for each time point were determined as described previously (Yu et al., 2009).
2.7. Indirect immunofluorescence analysis (IFA)
Subconfluent MARC-145 cell monolayer was infected with a 0.01 MOI by primary passage (P0) of the rescued virus, incubated for 72 hpi. The cell monolayer was washed twice by PBS, then fixed by cold methanol for 10 min at room temperature. The fixed cells were processed by 0.1% BSA at room temperature for 30 min, and then incubated at 37 °C for 2 h with anti-N monoclonal antibody of PRRSV (Kindly provided by Dr. Ying Fang at South Dakota State University) at 1:600 dilution in phosphate-buffered saline (PBS). After five times wash, cells were incubated at 37 °C for 1 h with Alexa Fluor 568 anti-mouse IgG (H + L) (Invitrogen). Finally, cells were washed five times with PBS and visualized under a Olympus inverted fluorescence microscope fitted with a camera.
2.8. In silico analysis of RNA structure
The secondary structure of 3′ UTR was predicted by using the Mfold version 3.2 web server (http://frontend.bioinfo.rpi.edu/applications/mfold/cgi-bin/rna-form1.cgi), under default folding conditions (37 °C, 1 M NaCl, no divalent ions, and no limit on distance between paired bases) (Zuker, 2003), and the secondary structures were edited by the use of RNAviz version 2 (De Rijk et al., 2003).
3. Results
3.1. PRRSV 3′ UTR is genetically diversified yet with inter-typically conserved domains
There is no report on characterization of possible 3′ UTR cis-acting elements of type II PRRSV yet, which is the focus of this study. To do so, we compared the 3′ UTR nucleotide sequences of five representative strains of each type I and type II PRRSV. The genotype I strains include LV (GenBank Accession No. M96262), SD01-08 (DQ489311), EuroPRRS (AY366525), and LV421 (AY588319), and five type II strains including VR2332 (AY150564), JX143 (EF488048), p129 (AF494042), CH1a (AY032626) and APRRS (GQ330474). As shown in Fig. 1 , the type I PRRSV 3′ UTR is 37 nt shorter than that of type II, which located at immediately downstream of the ORF7 stop codon. However, the 3′ UTR sequences are extremely intra-genotypically conserved. In addition, the pair-wise genetic identity between the two genotypes is 63%, and yet inter-typically conserved domains, shaded in black, do exist. We arbitrarily designated these inter-typically conserved domains as CS1 (conserved sequence), CS2, a possible long bulged stem-loop region LSL, and VR represents variable region. The main goal of this study is to investigate the possible effect of these 3′ UTR sequence on PRRSV virus replication. To facilitate reverse genetic manipulation, we inserted a stretch of sequence consisting of Pac I and Asc I recognition sites. Next, utilizing the Asc I site, we deleted the extra 40 nt region of type II PRRSV 3′ UTR, compared with that of type I, and the resultant full-length plasmid was designated as pURAD40 (Fig. 2 ). Utilizing SOE-PCR, we then took out the Pac I and Asc I site in order to eliminate the possible adverse effect of the Pac I and Asc I recognition sequence. Finally, based on pURD40, we replaced the entire 3′ UTR sequence with that of type I, such that the pSURLV is the full-length cDNA clone in the backbone of type II PRRSV containing the type I 3′ UTR. All full-length cDNA clones were verified by restriction mapping and nucleotide sequencing.
Fig. 1.

Comparison of 3′ UTR nucleotide sequence of type I and type II PRRSV. Clustal W program was used to conduct nucleotide sequence alignment of PRRSV 3′ UTR, collected from five representative strains of both genotype I including LV (GenBank Accession# M96262), SD01-08 (DQ489311), EuroPRRS (AY366525), LV421 (AY588319), five type II strains including VR2332 (AY150564), JX143 (EF188048), P129 (AF494042), CH1a (AY032626), and APRRS (GQ330474). Dot (.)at the beginning of the type I panel denote deletion in comparison with the type II 3′ UTR, while black shaded nucleotides are inter-typically conserved domains, arbitrarily designated as CS1, CS2, LSL stands for possible long bulged stem-loop region (see Fig. 7 for detail), and VR represents variable region.
Fig. 2.
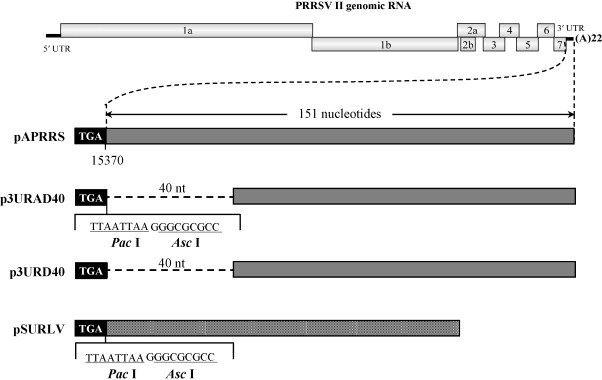
Mutagenesis of PRRSV 3′ UTR. The upper panel shows the PRRSV genomic organization encoding 9 ORFs (denoted by boxed numbers), which is flanked by terminal untranslated regions (UTR) represented by gray bars. The lower panel depicts the 3′UTR mutations, in which restriction enzyme sites were inserted (Pac I and Asc I); Nucleotide deletion was engineered in p3URAD40 and p3URD40, respectively. In pSURLV, the entire 3′UTR of pAPRRS was replaced by that of type I PRRSV marked by the black-striped bar. “TGA” denotes the stop codon of the ORF7.
3.2. The 5′ proximal 40 nt sequence of the 3′ UTR is dispensable for PRRSV viability
To assess the effect variable region of the 3′ UTR on PRRSV replication, p3URAD40, p3URD40, and pAPPRS were linearized by Xho I restriction enzyme that immediately followed the ploy(A)-tail, from which the in vitro transcripts were generated by the use of T7 mRNA mMachine kit (Ambion). About 2 μl of the synthetic RNA was transfected into MARC-145 cell monolayer in duplicated 6-well plates with DMRIE-C (Invitrogen). Surprisingly, all but the mock-transfected cells developed CPE at 3 to 4 dpt. The supernatant was harvested at 5 dpt, when approximately 80% CPE appeared, and the transfectant viral suspensions were designated as P0, named as v3URAD40, v3URD40, and vAPRRS, respectively. Meanwhile, to test that the CPE was PRRSV-specific, the transfected cell monolayer was fixed at 3 dpi, and stained by monoclonal antibody against the PRRSV nucleocapsid protein, followed staining by the Alex fluor-label anti-Mouse antibody. As shown in Fig. 3 , the transfectant virus v3URAD40 and v3URD40 deletion of 40 nt with or without an extra Pac I and Asc I site also developed the same IFA profile, and has no adverse effect on nucleocapsid protein subcellular distribution, demonstrating that the first 40 nt of the 3′ UTR is dispensable for PRRSV viability in cultured cells. Together, these results revealed that the variable region at the 5′ proximal end of the 3′ UTR of the type II PRRSV is flexible and can tolerate deleting.
Fig. 3.
Immunofluoresence assay of the transfected MARC-145 cells by PRRSV 3′UTR mutants. The in vitro RNAs generated from the full-length cDNA template promoted by T7 were synthesized by the T7 mRNA mMachine kit. 2 μl of each synthetic RNA was transfected into MARC-145 cells using DMRIE-C as instructed by the supplier. The cell monolayer was fixed at 72 hpi, and stained by monoclonal antibody against N and Alexa Fluor 568 goat anti-mouse IgG (H + L). The IFA patterns were shown for the parental and mutant viruses, labeled by the rescued viruses vAPRRS, v3URAD40, v3URD40, vSURLV, and the mock control.
3.3. Type I 3′ UTR is fully functional for type II PRRSV replication
Given the fact that inter-genotypic conserved domains exist in 3′ UTR, we next tested if the type I 3′ UTR can be functional in the backbone of the type II full-length cDNA clone. The chimeric clone pSURLV in vitro transcribed RNA was transfected into subconfluent MARC-145 cells in the same manner as described above. The transfected cell monolayer developed CPE by 4 dpt, about 12 hours slower than the parental virus vAPRRS, yet reached about 80% CPE at the end of 5 dpt. The transfection supernatant, designated as vSURLV, was harvested at 5 dpt, and aliquoted as virus stock P0. On the other hand, the duplicate transfected cell dishes were tested by IFA staining as described above. As shown in Fig. 3, vSURLV displayed similar pattern as the parental viruses, vAPRRS and v3URAD40. These results demonstrated for the first time that the 3′ UTR of genotype I is functional for genotype II virus replication in cultured cells, despite the fact that the two types of 3′ UTR are distinctly related, different length and a 37% pair-wise genetic diversity. Based on our results that type II 3′ UTR is fully functional in the backbone of an infectious cDNA clone of European type I (Zhuang et al., unpublished data), and we concluded that the 3′ UTR of PRRSV is interchangeable between different genotypes, which provided foundation for further dissection of cis-acting elements controlling arterivirus replication.
3.4. The 3′ UTR mutant PRRSVs are genetically stable in cultured cells
To further characterize the mutant viruses, the rescued viruses were serially inoculated in fresh MARC-145 cells with infection dose of 0.1 MOI. The culture supernatant were harvested and aliquoted as virus stocks of P1 to P5. To investigate the genetic stability, P5 viral suspensions was used for RNA extraction, followed by cDNA synthesis by a anchored-poly(T) primer Qst (Table 1), via reverse transcription. The cDNA was then used for amplification of discrete genomic regions, followed by nucleotide sequencing as described previously (Lv et al., 2008). Fig. 4 A showed the agarose gel electrophoretic pattern of the mutation locus, amplified with primer pair of SF15008 and Qst. The PCR products were gel-purified, and subjected to nucleotide sequencing. The nucleotide sequences were compared by the Clustal W program, as shown in Fig. 4B, the P5 virus retained the engineered mutation without any mutation in the flanking region. Moreover, full-length sequence of rescued virus vSURLV showed some nucleotides sequence alterations in the genome of the rescued viruses. 5 nucleotide differences were identified (Table 2 ) when the DNA sequence of the vSURLV genome was compared to previously published full-length sequences of APRRS (GenBank accession numbers GQ330474). Two of these differences were silent mutations. The other three of the nucleotide mutations resulted in amino acid changes. These were at nucleotides 3033 (G→A), 13854 (P→A) and 13976 (E→Y). These results demonstrated that the 3′ UTR mutants are genetically stable for at least five in vitro passages in cell culture.
Fig. 4.
RT-PCR and nucleotide sequencing of the rescued viruses. For verification if the mutation were retained in the rescued viruses collected from P5 culture supernatant, the target region was amplified as described in the materials and methods. A) The agarose gel electrophoresis of RT-PCR products of v3URAD40 (lane 1), vSURLV (lane 2), and parental virus vAPRRS (lane 4). B) Nucleotide sequence lineup showed that all viruses retained the engineered mutations. The sequence was compared with the parental APRRS (GQ330474), identical sequence displayed as dot (.), while variation was shown the actual nucleotide, natural or artificial deletion denoted as (-). The upstream boxed sequence showed that the vSURLV is homologous with type II APRRS in ORF7, while the downstream box indicated the vSURLV 3′ UTR is identical with type I PRRSV (LV).
Table 2.
Nucleotide differences between the parental APRRS and the vSURLV.
| Nucleotide position within APRRS genomea | Nucleotide in APRRS genome | Nucleotide in vSURLV genome | Change |
|---|---|---|---|
| 3033 | G | C | G → A |
| 13520 | C | T | Silent |
| 13854 | C | G | P → A |
| 13976 | A | T | E → Y |
| 14235 | T | G | Silent |
Nucleotide positions within the APRRS genome are based on GenBank accession numbers GQ330474.
3.5. The mutant PRRSVs displayed the same virological properties with the parental virus
To further characterize the possible effect of 3′ UTR mutation on virological properties, viral plaque assay was conducted. The P5 rescued viruses were inoculated into subconfluent MARC-145 cell monolayer, overlaid with 1% of agarose gel and MEM, and the plaques were stained at 5 dpi with crystal violet. There were no morphology and size differences of the viral plaques between and among v3URAD40, v3URD40, and that of the parental vAPRRS (Fig. 5 A). Interestingly, there is only a little difference between the chimeric vSURLV and vAPRRS, despite the distinct genetic relationship between them. We further assessed the viral growth kinetics. To do so, the P5 viruses were inoculated in fresh MARC-145 cells at a dose of 1 MOI, followed by collecting the infected cultured supernatant at the indicated time points (Fig. 5B). The virus yield of each harvest was determined by measuring plaque numbers. The single step growth curves of v3URAD40, and vURD40 were virtually the same with that of the parental vAPRRS (Fig. 5B), again indicated that the starting region of the 3′ UTR is dispensable for virus infectivity. In the same manner, the single step growth curve of vSURLV was assessed. However, time-point vSURLV titer was one or two log lower than the parental vAPRRS (Fig. 5B). Such difference was particularly obvious at the initial stage of the infection process, demonstrated that the type I 3′ UTR exerted effect on the early stage of virus infection. Further study need to be conducted for elucidating such defects.
Fig. 5.
Virological characterization of the mutant viruses. A) Viral plaque morphology. The rescued viruses at P5 were serially 10-fold diluted and inoculated in fresh MARC-145 cells in 6-well-plates. After one hour adsorption, the infected cell monolayer was overlaid with agarose and stained with crystal violet at 5 dpi. B) One step growth curves of deletion mutant viruses. MARC-145 cells were infected with the P5 mutant virus at an MOI of 1. The cell supernatants were harvested at the indicated time points. Viral titration was conducted by plaque assay on fresh MARC-145 cells, as detailed in the text.
Previous study has showed that 3′ UTR is most likely involved in RNA synthesis, so we next analyzed the subgenomic RNA profiles of the cells infected by the mutant viruses to assess PRRSV UTR mutation effect on virus transcription. Arteriviruses are known to produce a series of 3′ co-terminal sgmRNAs, thus an oligonucleotides probe complementary to the 3′ UTR would display all sets of sgmRNAs. VR3 and LVR3 (Table 1), specific for PRRSV type II and type I, respectively, were synthesized and biotin-labeled for Northern blot analysis. Total RNAs of the infected cells at 36 hpi were separated on duplicate denatured agarose gel, followed by transfer to Nylon membranes, which was then hybridized with biotin-labeled VR3 and LVR3, respectively. Fig. 6 A showed that VR3 probe displayed the same pattern of the sgmRNA (RNA2-RNA7) profiles of total RNAs of the cells infected by the parental vAPRRS, v3URD40, v3URAD40, but not the vSURLV; On the other hand, LVR3 probe only detected sgmRNAs from the cells infected by the chimeric vSURLV, but not these of vAPRRS and derivatives.The northern blot results showed difference in sgmRNA quantity levels between these mutant virus, which might be consequence of RNA membrane transfer efficiency variety. The advanced Northern blotting technique need to be conducted to define the dispensable region effect on virus sgmRNA transcription. This northern blotting indicate this indispensable region has no effect on virus transcription, but it might be affecting different sgmRNA transcription ability.
Fig. 6.
Virus RNA transcription process was identified with Northern Blot. MARC-145 cells in T-75 flasks were infected with the three mutant viruses and the intracellular RNAs were extracted at 36 hpi. The viral genomic RNA and sgmRNAs 2 to 7 were recognized only by genotype specific probe with an oligo complementary to different genotype 3′ UTR respectively, all the sample did two repeat wells, then hybridized with different probe: (A) add type II specific probe VR3 (B) add type I specific probe LVR3.
3.6. A highly conserved RNA structure is vital for porcine arterivirus replication
Positive RNA viruses are known to utilize a variety of high-order RNA structure to regulate different stages of virus replication, e.g. RNA synthesis, translation, virion assembly. We conducted in silico analysis by utilizing the Mfold algorism (Zuker, 2003) in order to search for possible structural elements residing in PRRSV 3′ UTR. Fig. 7 B is the predicted overall structure of the parental APRRS 3′ UTR, featured as a long bulged stem-loop (LSL) with several budges. The 40 nt deleted (shaded in grey) in v3URD40 could form branched SL structure with the base of the LSL. Upon deletion of the 40 nt stretch, the v3URD40 displayed the LSL as shown in Fig. 7C. Subsequently, the secondary structure was predicted in the same manner for PRRSV type I LV strain. Although the 3′ UTR of two genotypes are 37% diversified, the secondary structure of type I 3′ UTR (Fig. 7A) is extremely similar to that of the vURD40 3′ UTR (Fig. 7C). These results suggested that the 3′ UTR of both type I and II share the same structure, implying that such structural element is conserved inter-genotypically and may play a critical role for porcine arterivirus replication.
Fig. 7.
The secondary structure of the PRRSV 3′ UTR predicted by Mfold: Location of the extra nucleotides in the type II virus were also shown in predicted secondary structure, the grey marked region is the highly variable region in type II PRRSV, the above region of the lines is conserved secondary structure in two type PRRSV, while the bottom region of the line is variable secondary structure. 1. A). type I, B). type II, C). 3URD40.
4. Discussion
The 3′ UTR of arteriviruses are highly diversified, as the length for PRRSV type II, PRRSV type I, LDV, SHFV, and EAV are 151, 114, 80, 76, and 59 nt, respectively (Snijder and Meulenberg, 1998). It is not unexpected that the extra 40 nt stretch is dispensable for the viability of type II PRRSV, which has the longest 3′ UTR among the arteriviruses. In this study, we demonstrated that all virological properties including plaque morphology, size and viral growth kinetics, as well as the sgmRNA profiles remain unchanged in vURAD40, indicating that this 40 nt stretch located at the 5′ proximal end of the 3′ UTR of the type II PRRSV is indeed nonessential for virus replication in cultured MARC-145 cells. We are attempting to delimit the minimal 3′ UTR cis-acting signals for virus replication, and RNA synthesis processes including these of the minus-strand, genomic and sgmRNAs. The delimitation of maximal nonessential 3′ UTR sequence is under way. The 3′ UTR can influence virus life cycle through different regions, it includes both variable region and conserved region which may affect virus characterization in different aspects. Several studies demonstrated that a hypervariable region (HVR) and some RNA stem-loops of the genomic RNA of the mouse hepatitis virus (MHV) are nonessential for the virus replication in tissue culture, and yet the 3′ UTR HVR played a significant role in viral pathogenesis in vivo (Goebel et al., 2007). Similar observations have been made on flaviviruses including the Dengue virus and bovine viral diarrhea disease virus (BVDV), based on which one can rationally design an attenuated vaccine by deleting the 3′ UTR motif that is nonessential for in vitro culture but critical for in vivo virulence (Hanley et al., 2004, Pankraz et al., 2005). Whether the 40 nt deletion described here has implication on PRRSV in vivo pathogenicity warrants further animal trial.
RNA regulatory elements often function in alternative folds to balance different requirements for discrete stages of virus replication. Our results showed here that the secondary structure of 3′ UTRs between two genotypes are strikingly similar, despite that the relatively low genetic sequence identity. The overall structure of the 3′ UTR of both genotypes are indeed similar as displayed in Fig 7. To further confirm if the long bulged stem-loop is indeed conserved for porcine arterivirus, we predicted the secondary structure of the consensus 3′ UTR sequences, generated from 100 PRRSV type II GenBank deposited 3′ UTR sequences (accession numbers available upon request), which share 96% genetic identity. Not unexpectedly, the LSL structure (Fig. 8 A) remains the same as v3URD40. Using the same parameters, Fig. 8B showed that the overall structure remained the same with that shown in Fig. 7, Fig. 8, implying that the former may play an essential role for PRRSV type II viral replication.
Fig. 8.
Porcine arterivirus contains an inter-typically conserved long bulged stem-loop in 3′ UTR. Mfold program was used for analysing the predicted secondary structures of the 3′ UTR of PRRSV, followed by RNAviz drawing of the structure; (A). the structure for the entire 3′ UTR consensus sequence, obtained from comparison of the available type I PRRSV cited in Fig. 1; (B). The predicted structure from the consensus sequence of the last 111 nt of the type II 3′ UTR, generated from 100 available GenBank deposited sequences (Accession No. available upon request).
Tertiary structure and long-range RNA-RNA interaction may also be factors in regulating virus replication process (Alvarez et al., 2005, Edgil and Harris, 2006, Khromykh et al., 2001). It was reported that high-order structure including pseudoknot are highly conserved among the coronavirus 3′ UTRs (Williams et al., 1999). A similar pseudoknot interaction between the main 3′ UTR loop and the upstream coding sequence for nucleocapsid of EAV was proved to be critical for virus replication (Beerens and Snijder, 2006). Intriguingly, the pseudoknot interacting upstream loop (SL4 counterpart, shaded in Fig. 7C, shaded and circled in Fig. 8B) was found to form another kissing-loop interaction with a 7 nt sequence coding for the N-terminal of the nucleocapsid protein of the type I PRRSV (Verheije et al., 2002). In this study, the predicted structure showed that the long bulged stem-loop could be a critical regulating element for porcine arterivirus replication. Surprisingly, the pseudoknot interacting arms found in EAV formed virtually perfect long stem (shaded base, Fig. 7C, Fig. 8B). Although the exact secondary structure of PRRSV 3′ UTR need to be experimentally confirmed, it is plausible that different viral replication stages can be regulated by the conformational switch between the kissing-loop, pseudoknot, and long stem structure.
The functional replacement of inter-genotypic 3′ UTR also suggests that structural elements may play a vital role for controlling PRRSV replication. For large nidovirus, Hseu and Masters (Hsue and Masters, 1997) found that the entire 3′ UTR of MHV could be replaced by that of bovine coronavirus (BCV), which diverges overall by 31% in nucleotide sequence. Later on, the same lab (Goebel et al., 2004b) reported that the SARS-CoV 3′ UTR can functionally substitute the group II MHV counter, despite that only 38% genetic identity shared by the two 3′ UTRs. In our study, we obtain the similar conclusion as in coronavirus, the heterologous UTR will not affect virus replication and transcription process, however, the chimeric virus vSURLV replication efficiency decreased obviously. Although the chimeric virus can successfully accomplish the life cycle and virus packaging as they share the same molecular switch structure, the subtle difference may affect the virus titer.
It remains as a puzzle that the simultaneous emergence of the same disease by two distinct PRRSV in Europe and North America, promoting question about the virus origin and evolutionary pathway (Plagemann, 2003). Forsberg (Forsberg, 2005) suggested that the divergent evolution of related viruses on separate continents from a distant common ancestor. Here we demonstrated that the 3′ UTR of both genotype retains the same structure and functional role, despite of the primary sequence diverged significantly along the years of evolution in two different continents.
Three principal conclusions can be drawn from this study, including 1) the 3′ UTR of type II PRRSV can tolerate a deletion of at least 40 nt; 2) a 40nt stretch of sequence is dispensable for virus viability in cultured cells; and 3) a chimeric clone of type II PRRSV bearing the type I 3′UTR is viable. This is the first report that inter-typic 3′ UTR is fully functional in the backbone of the different arterivirus, and first identification of 3′ UTR cis-acting signals for type II PRRSV.
Acknowledgements
This study was supported by the key program of the China Natural Science Foundation (#30530580, #30972204), the National Basic Research Program (#2005CB523202), the China national science and technology special program “Prevention and control of highly pathogenic PRRSV” (2007BAD86B06-3).
References
- Alvarez D.E., Lodeiro M.F., Luduena S.J., Pietrasanta L.I., Gamarnik A.V. Long-range RNA–RNA interactions circularize the dengue virus genome. J. Virol. 2005;79:6631–6643. doi: 10.1128/JVI.79.11.6631-6643.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerens N.B., Selisko S.R., Imbert I., van der Zanden L., Snijder E.J., Canard B. De novo initiation of RNA synthesis by the arterivirus RNA-dependent RNA polymerase. J. Virol. 2007;81:8384–8395. doi: 10.1128/JVI.00564-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerens N., Snijder E.J. RNA signals in the 3′ terminus of the genome of Equine arteritis virus are required for viral RNA synthesis. J. Gen. Virol. 2006;87:1977–1983. doi: 10.1099/vir.0.81750-0. [DOI] [PubMed] [Google Scholar]
- Beerens N., Snijder E.J. An RNA pseudoknot in the 3′ end of the arterivirus genome has a critical role in regulating viral RNA synthesis. J. Virol. 2007;81:9426–9436. doi: 10.1128/JVI.00747-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang K.S., Luo G. The polypyrimidine tract-binding protein (PTB) is required for efficient replication of hepatitis C virus (HCV) RNA. Virus Res. 2006;115:1–8. doi: 10.1016/j.virusres.2005.06.012. [DOI] [PubMed] [Google Scholar]
- Choi Y.J., Yun S.I., Kang S.Y., Lee Y.M. Identification of 5′ and 3′ cis-acting elements of the porcine reproductive and respiratory syndrome virus: acquisition of novel 5′ AU-rich sequences restored replication of a 5′-proximal 7-nucleotide deletion mutant. J. Virol. 2006;80:723–736. doi: 10.1128/JVI.80.2.723-736.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins J.E., Benfield D.A., Christianson W.T., Harris L., Hennings J.C., Shaw D.P., Goyal S.M., McCullough S., Morrison R.B., Joo H.S. Isolation of swine infertility and respiratory syndrome virus (isolate ATCC VR-2332) in North America and experimental reproduction of the disease in gnotobiotic pigs. J. Vet. Diagn. Invest. 1992;4:117–126. doi: 10.1177/104063879200400201. [DOI] [PubMed] [Google Scholar]
- De Rijk P., Wuyts J., Wachter R.D. RnaViz 2: an improved representation of RNA secondary structure. Bioinformatics. 2003;19:299–300. doi: 10.1093/bioinformatics/19.2.299. [DOI] [PubMed] [Google Scholar]
- den Boon J.A., Snijder E.J., Chirnside E.D., de Vries A.A., Horzinek M.C., Spaan W.J. Equine arteritis virus is not a togavirus but belongs to the coronaviruslike superfamily. J. Virol. 1991;65:2910–2920. doi: 10.1128/jvi.65.6.2910-2920.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgil D., Harris E. End-to-end communication in the modulation of translation by mammalian RNA viruses. Virus Res. 2006;119:43–51. doi: 10.1016/j.virusres.2005.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsberg R. Divergence time of porcine reproductive and respiratory syndrome virus subtypes. Mol. Biol. Evol. 2005;22:2131–2134. doi: 10.1093/molbev/msi208. [DOI] [PubMed] [Google Scholar]
- Garneau N.L., Sokoloski K.J., Opyrchal M., Neff C.P., Wilusz C.J., Wilusz J. The 3′ untranslated region of sindbis virus represses deadenylation of viral transcripts in mosquito and Mammalian cells. J. Virol. 2008;82:880–892. doi: 10.1128/JVI.01205-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebel, S.J., B. Hsue, T. F. Dombrowski and Masters, a.P.S., 2004a. Characterization of the RNA components of a putative molecular switch in the 3′ untranslated region of the murine coronavirus genome. J. Virol. 78, 669–82. [DOI] [PMC free article] [PubMed]
- Goebel S.J., Taylor J., Masters P.S. The 3′ cis-acting genomic replication element of the severe acute respiratory syndrome coronavirus can function in the murine coronavirus genome. J. Virol. 2004;78:7846–7851. doi: 10.1128/JVI.78.14.7846-7851.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebel S.J., Miller T.B., Bennett C.J., Bernard K.A., Masters P.S. A hypervariable region within the 3′ cis-acting element of the murine coronavirus genome is nonessential for RNA synthesis but affects pathogenesis. J. Virol. 2007;81:1274–1287. doi: 10.1128/JVI.00803-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanley K.A., Manlucu L.R., Manipon G.G., Hanson C.T., Whitehead S.S., Murphy B.R., Blaney J. J.E. Introduction of mutations into the non-structural genes or 3′ untranslated region of an attenuated dengue virus type 4 vaccine candidate further decreases replication in rhesus monkeys while retaining protective immunity. Vaccine. 2004;22:3440–3448. doi: 10.1016/j.vaccine.2004.02.031. [DOI] [PubMed] [Google Scholar]
- Hsue B., Masters P.S. A bulged stem-loop structure in the 3′ untranslated region of the genome of the coronavirus mouse hepatitis virus is essential for replication. J. Virol. 1997;71:7567–7578. doi: 10.1128/jvi.71.10.7567-7578.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Key K.F., Haqshenas G., Guenette D.K., Swenson S.L., Toth T.E., Meng X.J. Genetic variation and phylogenetic analyses of the ORF5 gene of acute porcine reproductive and respiratory syndrome virus isolates. Vet. Microbiol. 2001;83:249–263. doi: 10.1016/s0378-1135(01)00427-8. [DOI] [PubMed] [Google Scholar]
- Khromykh A.A., Meka H., Guyatt K.J., Westaway E.G. Essential role of cyclization sequences in flavivirus RNA replication. J. Virol. 2001;75:6719–6728. doi: 10.1128/JVI.75.14.6719-6728.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh D.C., Liu D.X., Wong S.M. A six-nucleotide segment within the 3′ untranslated region of hibiscus chlorotic ringspot virus plays an essential role in translational enhancement. J. Virol. 2002;76:1144–1153. doi: 10.1128/JVI.76.3.1144-1153.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, P.H. and Leibowitz, J.L., 2010. RNA Higer-Order Structures Within the Coronavirus 5′ and 3′ Untranslated Regions and Their Roles in Virus Replication. in: S. K. Lal, (1), Molecular Biology of the SARS-Coronavirus. Springer, Berlin Heidelberg, pp. 47-61.
- Lopez de Quinto S., Saiz M., de la Morena D., Sobrino F., Martinez-Salas E. IRES-driven translation is stimulated separately by the FMDV 3′-NCR and poly(A) sequences. Nucleic Acids Res. 2002;30:4398–4405. doi: 10.1093/nar/gkf569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv J., Zhang J., Sun Z., Liu W., Yuan S. An infectious cDNA clone of a highly pathogenic porcine reproductive and respiratory syndrome virus variant associated with porcine high fever syndrome. J. Gen. Virol. 2008;89:2075–2079. doi: 10.1099/vir.0.2008/001529-0. [DOI] [PubMed] [Google Scholar]
- Mateu E., Diaz I. The challenge of PRRS immunology. Vet. J. 2008;177:345–355. doi: 10.1016/j.tvjl.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X.J. Heterogeneity of porcine reproductive and respiratory syndrome virus: implications for current vaccine efficacy and future vaccine development. Vet. Microbiol. 2000;74:309–329. doi: 10.1016/S0378-1135(00)00196-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X.J., Paul P.S., Morozov I., Halbur P.G. A nested set of six or seven subgenomic mRNAs is formed in cells infected with different isolates of porcine reproductive and respiratory syndrome virus. J. Gen. Virol. 1996;77:1265–1270. doi: 10.1099/0022-1317-77-6-1265. [DOI] [PubMed] [Google Scholar]
- Pankraz A., Thiel H.J., Becher P. Essential and nonessential elements in the 3′ nontranslated region of Bovine viral diarrhea virus. J. Virol. 2005;79:9119–9127. doi: 10.1128/JVI.79.14.9119-9127.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasternak A.O., Spaan W.J., Snijder E.J. Nidovirus transcription: how to make sense? J. Gen. Virol. 2006;87:1403–1421. doi: 10.1099/vir.0.81611-0. [DOI] [PubMed] [Google Scholar]
- Plagemann P.G. Porcine reproductive and respiratory syndrome virus: origin hypothesis. Emerg. Infect. Dis. 2003;9:903–908. doi: 10.3201/eid0908.030232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohll J.B., Moon D.H., Evans D.J., Almond J.W. The 3′ untranslated region of picornavirus RNA: features required for efficient genome replication. J. Virol. 1995;69:7835–7844. doi: 10.1128/jvi.69.12.7835-7844.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki S.G., Sawicki D.L., Siddell S.G. A contemporary view of coronavirus transcription. J. Virol. 2007;81:20–29. doi: 10.1128/JVI.01358-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Meulenberg J.J. The molecular biology of arteriviruses. J. Gen. Virol. 1998;79:961–979. doi: 10.1099/0022-1317-79-5-961. [DOI] [PubMed] [Google Scholar]
- Todd S., Towner J.S., Brown D.M., Semler B.L. Replication-competent picornaviruses with complete genomic RNA 3′ noncoding region deletions. J. Virol. 1997;71:8868–8874. doi: 10.1128/jvi.71.11.8868-8874.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzri D., Gehrke L. Nucleotide sequences and modifications that determine RIG-I/RNA binding and signaling activities. J. Virol. 2009;83:4174–4184. doi: 10.1128/JVI.02449-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheije M.H., Kroese M.V., Rottier P.J., Meulenberg J.J. Viable porcine arteriviruses with deletions proximal to the 3′ end of the genome. J. Gen. Virol. 2001;82:2607–2614. doi: 10.1099/0022-1317-82-11-2607. [DOI] [PubMed] [Google Scholar]
- Verheije M.H., Olsthoorn R.C., Kroese M.V., Rottier P.J., Meulenberg J.J. Kissing interaction between 3′ noncoding and coding sequences is essential for porcine arterivirus RNA replication. J. Virol. 2002;76:1521–1526. doi: 10.1128/JVI.76.3.1521-1526.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wensvoort G., Terpstra C., Pol J.M., ter Laak E.A., Bloemraad M., de Kluyver E.P., Kragten C., van Buiten L., den Besten A., Wagenaar F. Mystery swine disease in The Netherlands: the isolation of Lelystad virus. Vet. Q. 1991;13:121–130. doi: 10.1080/01652176.1991.9694296. [DOI] [PubMed] [Google Scholar]
- Williams G.D., Chang R.Y., Brian D.A. A phylogenetically conserved hairpin-type 3′ untranslated region pseudoknot functions in coronavirus RNA replication. J. Virol. 1999;73:8349–8355. doi: 10.1128/jvi.73.10.8349-8355.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D., Lv J., Sun Z., Zheng H., Lu J., Yuan S. Reverse genetic manipulation of the overlapping coding regions for structural proteins of the type II porcine reproductive and respiratory syndrome virus. Virology. 2009;383:22–31. doi: 10.1016/j.virol.2008.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S., Wei Z. Construction of infectious cDNA clones of PRRSV: separation of coding regions for nonstructural and structural proteins. Sci. China C Life Sci. 2008;51:271–279. doi: 10.1007/s11427-008-0023-y. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zust R., Miller T.B., Goebel S.J., Thiel V., Masters P.S. Genetic interactions between an essential 3′ cis-acting RNA pseudoknot, replicase gene products, and the extreme 3′ end of the mouse coronavirus genome. J. Virol. 2008;82:1214–1228. doi: 10.1128/JVI.01690-07. [DOI] [PMC free article] [PubMed] [Google Scholar]


