Abstract
Molecular characterisation of a canine coronavirus (CCoV) isolate (BGF), associated with an outbreak of diarrhoea in puppies, showed 92.7% identity with attenuated Insavc-1 strain. Canine coronavirus BGF revealed a full length non-structural protein 3b (nsp 3b), associated with virulence in other coronaviruses, and a highly divergent region at the amino terminal domain of the membrane protein that may be implicated in avoiding the host immune reaction. This new canine coronavirus strain could help to identify virulence factors in coronavirus.
Keywords: Nidovirales, Coronaviridae, Canine coronavirus, Non-structural protein 3b, Membrane protein
Canine coronavirus (CCoV) is a member of the Coronaviridae family of the Nidovirales order (Enjuanes et al., 2000). Coronaviruses are single stranded positive sense RNA viruses between 28 and 31 kb in length. CCoV is classified as group I by antigenic and genetic relationships (Enjuanes et al., 2000, Sanchez et al., 1990). CCoV has been experimentally demonstrated to produce mild gastroenteritis in dogs (Tennant et al., 1991). Clinically, CCoV has usually been found in conjunction with other organisms like canine adenovirus type 1 (Pratelli et al., 2001) or canine parvovirus type 2 (Carmichael, 1999). Changes in the sequences of structural spike glycoprotein (Ballesteros et al., 1997, Hingley et al., 2002, Phillips et al., 2002, Sanchez et al., 1999) and the non-structural protein 3b (nsp 3b) (Kim et al., 2000, Wesley et al., 1990) have been associated with differences in coronavirus strain virulence.
In 2002, an epizootic outbreak of diarrhoea occurred in our Beagle breeding colony. Dogs affected were between 6 and 10 weeks of age. Clinical signs consisted of diarrhoea, with or without mucus, haematochezia, lethargy, dehydration, vomiting, anorexia, pale mucous membranes and an elevated or depressed rectal temperature. Samples from 106 affected animals were taken. Campylobacter jejuni sub. jejuni was found in 68%, Isospora rivolta in 9%, and Giardia canis in 5% of them. CCoV RNA was analysed by use of RT-PCR (Pratelli et al., 1999) and found to be present in all 106 samples.
Campylobacter jejuni sub. jejuni was also isolated from some asymptomatic animals, as has been found by other studies (Fleming, 1983, Fox et al., 1983, Fox et al., 1984, Fox et al., 1988).
Sera from both normal dogs, and dogs in outbreaks of enteritis in kennels usually reveal a high rate of antibodies to CCoV. Seroprevalence studies in several countries indicate that prevalence rates vary from 0 to 80%; in one large study, 45% seroprevelance rates were reported in normal dogs, in contrast to 61% in diarrheic dogs, generally higher rates were observed in enzootically infected kennels (Pratelli, 2000, Tennant et al., 1993).
Recently, canine coronavirus strains that segregate separately from CCoV-Insacv 1 have been isolated from dogs with gastroenteritis (Naylor et al., 2002, Pratelli et al., 2003b). Here, we report the molecular characterization of 9.2 kb of the CCoV-BGF strain genome encoding the spike (S), envelope (E), membrane (M) and nucleoprotein (N) structural proteins. CCoV-BGF was found to have a 92.7% identity with CCoV Insavc-1 strain (Horsburgh et al., 1992) over the whole 9.2 kb analysed and had a full length non-structural protein 3b of 250 amino acids and a highly divergent region at the amino terminal end of the membrane protein.
These characteristics of a strain found to be strongly associated with a significant disease outbreak should help to identify factors associated with virulence in CCoV.
RNA was extracted from faecal samples using RNeasy (QIAGEN, West Sussex, UK) according to the manufacturer’s instructions.
cDNA of 9 kb length containing the structural proteins S, E, M and N was cloned from a faecal positive sample using pCR 2.1 (Invitrogen, Paisley, UK), according to the manufacturer’s instructions, in three large partially overlapping fragments (Fig. 1A and B ). RT-PCR consisted of an initial reverse transcription using Superscript III (Invitrogen, Paisley, UK), according to the manufacturer’s instructions, followed by a PCR reaction using the Expand Long Template PCR system (Roche, East Sussex, UK) according to the manufacturer’s instructions. Once the consensus sequence was obtained, overlapping RT-PCRs of approximately 600 nucleotides were designed and run using HotStart Taq polymerase (QIAGEN, West Sussex, UK). PCRs were carried out as follows: 50 ng of cDNA template from reverse transcription (Fig 1A) was added to a PCR mix containing 2 mM dNTPs, 10 pmol of each primer, and 2.5 units of HotStartTaq™ DNA polymerase (QIAGEN, West Sussex, UK) in PCR buffer 1x (QIAGEN, West Sussex, UK) to a final concentration of 1.5 mM MgCl2. PCR conditions consisted of an initial activation step of 95 °C for 15 min followed by 35 cycles of denaturation at 94 °C for 15 s, annealing at 55–60 °C, depending on the primer pair, for 45 s and extension at 72 °C for 1 min, and a final extension step at 72 °C for 10 min. PCR products for cloning and sequencing were cleaned using QIAquick (QIAGEN, West Sussex, UK). The most representative viral population was obtained by sequencing three independent RT-PCR reactions from three randomly chosen clinical samples. Direct sequencing from clinical samples was done to avoid in vitro selection of attenuated strains (Sanchez et al., 1999).
Fig. 1.
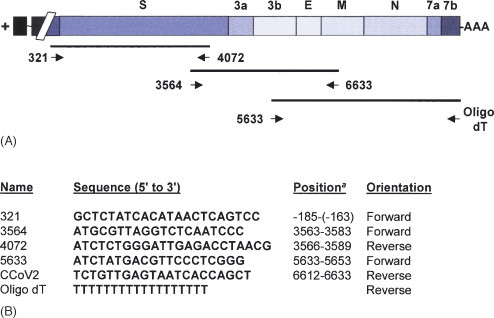
(A) Schematic representation of the cloning process for the last third of CCoV-BGF genome. (B) Oligonucleotides used for the cloning. (a) Nucleotide position starting from the first codon of the spike gene.
Sequencing was done using the Big Dye Version 1.0 Sequence Reaction Kit (Applied Biosystems, Foster City, CA, USA). Samples were sequenced by use of an ABI Prism 3700 Capillary Sequencer (Applied Biosystems, Foster City, CA, USA).
Sequences were edited and analysed using EditSeq and SeqMan of Lasergene programs (DNAStar Konstanz, Germany). PSORT II (Nakai and Horton, 1999), SignalP-HMM (Nielsen et al., 1997) and TMHMM (Sonnhammer et al., 1998) were used to predict subcellular localization sites, signal peptides and transmembrane helices, respectively. A search against the PROSITE database was done to identify the different protein domains (Falquet et al., 2002). A sequence similarity search was performed for the different structural proteins using the Fasta3 program (Pearson and Lipman, 1988) against the SWall-SPTR protein sequence database (EMBL). Phylogenetic analyses were done using Phylip 3.6 through the Phylogeny Interface Environment (PIE) at the Rosalind Franklin Centre for Genomics Research (Fig. 2 ).
Fig. 2.
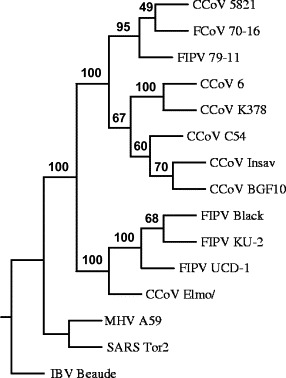
Phylogenetic tree of spike protein of canine and feline coronaviruses. Phylogenetic tree was constructed by the protein sequence parsimony method (protpars) using Phylip 3.6 through the Phylogeny Interface Environment (PIE) at the Rosalind Franklin Centre for Genomics Research. Bootstrap values were calculated on 1000 replicates randomising the input order 41 times. The tree was rooted with MHV A59, SARS Tor2 and IBV Beaudette. FIPV, feline infectious peritonitis virus; FCoV, feline coronavirus; CCoV, canine coronavirus; MHV, mouse hepatitis virus; SARS, severe acute respiratory syndrome; IBV, infectious bronchitis virus.
The spike protein was a polypeptide of 1453 amino acids, two amino acids longer than that of the attenuated Insavc-1 strain. When compared to Insacv-1, the nucleotide sequence contained 325 point mutations and the protein 90 amino acid changes. A putative signal peptide—probably cleaved between Cys-18 and Thr-19 by the signal peptidase—was represented by the first 18 amino acids. A transmembrane domain was found between residues 1393 and 1415. The protein contained 36 potential N-glycosylation sites, 1 potential leucine zipper domain (1346–1367) that is highly conserved among coronaviruses (Luo et al., 1999), and 1 cysteine rich region (1408–1436). The best identity was obtained against CCoV Insavc-1 (93.8%). Minor amino acid differences in the sequence of the spike protein have been shown to change the virulence of even very close isolates (Sanchez et al., 1999). In the phylogenetic analysis, three different canine coronavirus clusters were identified for the spike protein. CCoV-BGF spike protein fitted into the cluster of canine coronaviruses containing no feline sequences (Fig. 2).
The nsp3 protein was a polypeptide of 250 amino acids. When compared to that of the Insacv-1 strain, the nucleotide sequence contained 18 point mutations and the protein 9 amino acid changes. Three transmembrane domains were found between residues 49–66, 73–95 and 99–121.
The envelope protein was a polypeptide of 82 amino acids, 2 amino acids longer than that of the attenuated Insavc-1 strain. When compared to Insacv-1, the nucleotide sequence contained 22 point mutations and the protein 9 amino acid changes. A transmembrane domain was found between residues 20–42. The best identity was obtained against CCoV Insavc-1 (89%).
The membrane protein was 262 amino acids long. When compared to that of the Insacv-1 strain, the nucleotide sequence contained 91 point mutations and the protein 37 amino acid changes. Four transmembrane domains were predicted between residues 5–27, 47–69, 76–98 and 113–135 (Fig. 3A ). The protein contained two potential glycosylation sites and three potential phosphorylation sites. The best identity was obtained against CCoV 259/01 (88.2%).
Fig. 3.
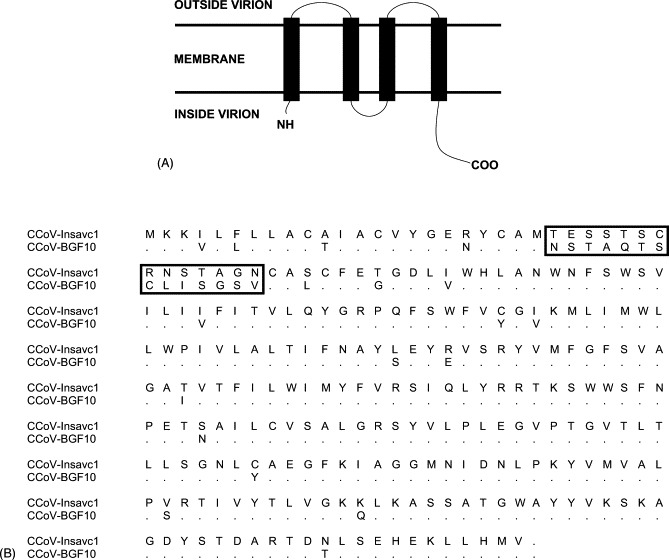
(A) Topology of the CCoV-BGF membrane protein as predicted by TMHMM method. (B) Comparison of the membrane protein sequence from CCoV Insavc-1 and CCoV-BGF10 with the highly variable sequence highlighted.
The nucleoprotein was 382 amino acids long. When compared to that of the Insacv-1 strain, the nucleotide sequence contained 86 point mutations and the protein 23 amino acid changes. The protein contained 3 potential glycosylation sites, 13 potential phosphorylation sites, and a bipartite nuclear targeting sequence (199-KKLGVDTEKQQQRARSK-255). The nucleoprotein of transmissible gastroenteritis virus (TGEV), mouse hepatitis virus (MHV) and infectious bronchitis virus (IBV) have been shown experimentally to localise in the nucleolus (Chen et al., 2002, Wurm et al., 2001). The best identity was obtained against CCoV 1–71 (93.5%).
The CCoV BGF strain was found to be closely related to the attenuated Insavc-1 strain (overall amino acid identity of 92.7%) (Fig. 2). We compared both strains because data for all the structural proteins are only available for those two strains and also because CCoV-Insacv-1 is attenuated and a very well characterized strain (Horsburgh et al., 1992) that has been used as a vaccine. Previously, a strain of CCoV have been classified as “novel third CCoV subgroup” (Naylor et al., 2002) based on sequence data from a small part of the highly variable 5′ terminal spike region. Based on the whole spike sequence phylogeny, we found two different subgroups. The first contains only the CCoV recovered from diarrheic dogs in Italy (Pratelli et al., 2003a), although it is difficult to discard a recombination event with feline infectious peritonitis virus (FIPV) in that case.
The second group contains the remaining CCoVs sequenced to date, including CCoV-BGF (Fig. 2). We found that for the other three structural proteins, the CCoV-BGF is not grouped with the known CCoV sequences. A lack of data from the last third of the genome containing the coronavirus structural proteins of the different CCoV isolates prevented a powerful phylogenetic analysis. Nevertheless, the high homology between Insavc-1 and BGF strains could help to identify CCoV genomic regions involved in virulence.
A deletion of 61 bp (isolates 5 and 10) to 64 bp (isolate 27) was found in the intergenic S-3a region. Deletions in this region have already been described in virulent TGEV variants (McGoldrick et al., 1999) and may be associated with increased nsp 3b translation levels. The length of the transcription regulatory sequences has been shown to influence the level of transcription and translation in TGEV (Alonso et al., 2002). Interestingly, a nsp 3b protein of 250 amino acids is present, instead of the truncated form of 171 amino acids described for the attenuated Insavc-1 strain (Horsburgh et al., 1992). The truncated nsp 3b has been associated in survey studies with attenuated strains (Horsburgh et al., 1992, Kim et al., 2000, Wesley et al., 1990). The only reported experimental study of virulence of coronavirus group 1 involving the nsp 3b protein is difficult to interpret because of lack of experimental details such as animal health status, and the absence of administration of a known attenuated strain as a control (Festing et al., 2002, Sola et al., 2003). Therefore, we think that an increased amount of the full-length nsp 3b protein could be partially responsible for the increased in virulence that appears to be associated with CCoV-BGF.
The CCoV-BGF membrane protein accumulates most of the changes when compared to the Insacv-1 strain at the 5′ end of the gene. A region of high divergence was found between residues 24 and 37 of the membrane protein (Fig. 3B). This region has previously been shown to be highly variable in other isolates and it has been postulated that it may be involved in escaping the host immune response in CCoV (Pratelli et al., 2002). The importance of the membrane protein in eliciting the humoral host immune response is not clear because monoclonal antibodies against TGEV membrane protein do not completely neutralise virus infection (Sanchez et al., 1990, Risco et al., 1995). We assume that the presence of an additional membrane domain in CCoV-BGF, predicted by TMHMM (Fig. 3A), could be helping the virus to avoid degradation by host proteases (Armstrong et al., 1984) and therefore to evade the T-cell response.
Canine coronavirus is highly seroprevalent in the UK (Tennant et al., 1993). Field samples will identify more accurately virulence factors such as the spike and nsp 3b proteins. In vitro and in vivo studies using the newly generated infectious cDNAs (Almazan et al., 2000, Yount et al., 2000) will help to study not only virulence but also protein variants like the highly variable amino terminal domain of the M protein that could be involved in escaping the host immune response. Generation of a CCoV-BGF infectious cDNA is currently under development in our laboratory.
Acknowledgements
We thank Amita Shortland and Helen L. White for their support with clinical work. Jaymini Patel and Nicola Crocker for assisting sequencing. Also, we thank Ghislaine M. Poirier and Ander Izeta for critical reading.
Footnotes
GeneBank accession number: AY342160.
References
- Almazan F, Gonzalez J.M, Penzes Z, Izeta A, Calvo E, Plana-Duran J, Enjuanes L. Engineering the largest RNA virus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. USA. 2000;97:5516–5521. doi: 10.1073/pnas.97.10.5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso S, Izeta A, Sola I, Enjuanes L. Transcription regulatory sequences and mRNA expression levels in the coronavirus transmissible gastroenteritis virus. J. Virol. 2002;76:1293–1308. doi: 10.1128/JVI.76.3.1293-1308.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong J, Niemann H, Smeekens S, Rottier P, Warren G. Sequence and topology of a model intracellular membrane protein, E1 glycoprotein, from a coronavirus. Nature. 1984;308:751–752. doi: 10.1038/308751a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballesteros M.L, Sanchez C.M, Enjuanes L. Two amino acid changes at the N-terminus of transmissible gastroenteritis coronavirus spike protein result in the loss of enteric tropism. Virology. 1997;227:378–388. doi: 10.1006/viro.1996.8344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmichael L.E. Canine viral vaccines at a turning point—a personal perspective. Adv. Vet. Med. 1999;41:289–307. doi: 10.1016/S0065-3519(99)80022-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Wurm T, Britton P, Brooks G, Hiscox J.A. Interaction of the coronavirus nucleoprotein with nucleolar antigens and the host cell. J. Virol. 2002;76:5233–5250. doi: 10.1128/JVI.76.10.5233-5250.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enjuanes, L., Spaan, W., Snijder, E., Cavanagh, D., 2000. Nidovirales. In: Regenmortel, M.H.V., Fauquet, C.M., Bishop, D.H.L., Carstens, E.B., Estes, M.K., Lemon, S.M., Maniloff, J., Mayo, M.A., McGeoch, D.J., Pringle, C.R., Wickner, R.B. (Eds.), Virus Taxonomy: Seventh Report of the International Committee on Taxonomy of Viruses. Academic Press, New York, pp. 827–834.
- Falquet L, Pagni M, Bucher P, Hulo N, Sigrist C.J, Hofmann K, Bairoch A. The PROSITE database, its status in 2002. Nucleic Acids Res. 2002;30:235–238. doi: 10.1093/nar/30.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Festing, M.F., Overend, P., Das, R.G., Borja, M.C., Berdoy, M., 2002. The Design of Animal Experiments: Reducing the Use of Animals in Research through Better Experimental Design. RSM Press, London, 112 pp.
- Fleming M.P. Association of Campylobacter jejuni with enteritis in dogs and cats. Vet. Rec. 1983;113:372–374. doi: 10.1136/vr.113.16.372. [DOI] [PubMed] [Google Scholar]
- Fox J.G, Claps M.C, Taylor N.S, Maxwell K.O, Ackerman J.I, Hoffman S.B. Campylobacter jejuni/coli in commercially reared beagles: prevalence and serotypes. Lab. Anim. Sci. 1988;38:262–265. [PubMed] [Google Scholar]
- Fox J.G, Maxwell K.O, Ackerman J.I. Campylobacter jejuni associated diarrhea in commercially reared beagles. Lab. Anim. Sci. 1984;34:151–155. [PubMed] [Google Scholar]
- Fox J.G, Moore R, Ackerman J.I. Campylobacter jejuni-associated diarrhea in dogs. J. Am. Vet. Med. Assoc. 1983;183:1430–1433. [PubMed] [Google Scholar]
- Hingley S.T, Leparc-Goffart I, Seo S.H, Tsai J.C, Weiss S.R. The virulence of mouse hepatitis virus strain A59 is not dependent on efficient spike protein cleavage and cell-to-cell fusion. J. Neurovirol. 2002;8:400–410. doi: 10.1080/13550280260422703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsburgh B.C, Brierley I, Brown T.D. Analysis of a 9.6 kb sequence from the 3′ end of canine coronavirus genomic RNA. J. Gen. Virol. 1992;73:2849–2862. doi: 10.1099/0022-1317-73-11-2849. [DOI] [PubMed] [Google Scholar]
- Kim L, Hayes J, Lewis P, Parwani A.V, Chang K.O, Saif L.J. Molecular characterization and pathogenesis of transmissible gastroenteritis coronavirus (TGEV) and porcine respiratory coronavirus (PRCV) field isolates co-circulating in a swine herd. Arch. Virol. 2000;145:1133–1147. doi: 10.1007/s007050070114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z, Matthews A.M, Weiss S.R. Amino acid substitutions within the leucine zipper domain of the murine coronavirus spike protein cause defects in oligomerization and the ability to induce cell-to-cell fusion. J. Virol. 1999;73:8152–8159. doi: 10.1128/jvi.73.10.8152-8159.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGoldrick A, Lowings J.P, Paton D.J. Characterisation of a recent virulent transmissible gastroenteritis virus from Britain with a deleted ORF 3a. Arch. Virol. 1999;144:763–770. doi: 10.1007/s007050050541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai K, Horton P. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 1999;24:34–36. doi: 10.1016/s0968-0004(98)01336-x. [DOI] [PubMed] [Google Scholar]
- Naylor M.J, Walia C.S, McOrist S, Lehrbach P.R, Deane E.M, Harrison G.A. Molecular characterization confirms the presence of a divergent strain of canine coronavirus (UWSMN-1) in Australia. J. Clin. Microbiol. 2002;40:3518–3522. doi: 10.1128/JCM.40.9.3518-3522.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- Pearson W.R, Lipman D.J. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips J.J, Chua M.M, Rall G.F, Weiss S.R. Murine coronavirus spike glycoprotein mediates degree of viral spread, inflammation, and virus-induced immunopathology in the central nervous system. Virology. 2002;301:109–120. doi: 10.1006/viro.2002.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratelli A, Elia G, Martella V, Tinelli A, Decaro N, Marsilio F, Buonavoglia D, Tempesta M, Buonavoglia C. M gene evolution of canine coronavirus in naturally infected dogs. Vet. Rec. 2002;151:758–761. [PubMed] [Google Scholar]
- Pratelli A, Martella V, Elia G, Tempesta M, Guarda F, Capucchio M.T, Carmichael L.E, Buonavoglia C. Severe enteric disease in an animal shelter associated with dual infections by canine adenovirus type 1 and canine coronavirus. J. Vet. Med. B Infect. Dis. Vet. Public Health. 2001;48:385–392. doi: 10.1046/j.1439-0450.2001.00466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratelli A, Martella V, Decaro N, Tinelli A, Camero M, Cirone F, Elia G, Cavalli A, Corrente M, Greco G, Buonavoglia D, Gentile M, Tempesta M, Buonavoglia C. Genetic diversity of a canine coronavirus detected in pups with diarrhoea in Italy. J. Virol. Methods. 2003;110:9–17. doi: 10.1016/S0166-0934(03)00081-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratelli A, Martella V, Pistello M, Elia G, Decaro N, Buonavoglia D, Camero M, Tempesta M, Buonavoglia C. Identification of coronaviruses in dogs that segregate separately from the canine coronavirus genotype. J. Virol. Methods. 2003;107:213–222. doi: 10.1016/S0166-0934(02)00246-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratelli A, Tempesta M, Greco G, Martella V, Buonavoglia C. Development of a nested PCR assay for the detection of canine coronavirus. J. Virol. Methods. 1999;80:11–15. doi: 10.1016/S0166-0934(99)00017-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patelli, A., 2000. Canine coronaviral infection. In: Carmichael, L. (Ed.), Recent Advances in Canine Infectious Diseases. Available from URL: http://www.ivis.org/advances/Infect_Dis_Carmichael/pratelli/ivis.pdf.
- Risco C, Anton I.M, Suñe C, Pedregosa A.M, Martin-Alonso J.M, Parra F, Carrascosa J.L, Enjuanes L. Membrane protein molecules of transmissible gastroenteritis coronavirus also expose the carboxy-terminal region on the external surface of the virion. J. Virol. 1995;69:5269–5277. doi: 10.1128/jvi.69.9.5269-5277.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez C.M, Izeta A, Sanchez-Morgado J.M, Alonso S, Sola I, Balasch M, Plana-Duran J, Enjuanes L. Targeted recombination demonstrates that the spike gene of transmissible gastroenteritis coronavirus is a determinant of its enteric tropism and virulence. J. Virol. 1999;73:7607–7618. doi: 10.1128/jvi.73.9.7607-7618.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez C.M, Jimenez G, Laviada M.D, Correa I, Sune C, Bullido M, Gebauer F, Smerdou C, Callebaut P, Escribano J.M, Enjuanes L. Antigenic homology among coronaviruses related to transmissible gastroenteritis virus. Virology. 1990;174:410–417. doi: 10.1016/0042-6822(90)90094-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sola I, Alonso S, Zuniga S, Balasch M, Plana-Duran J, Enjuanes L. Engineering the transmissible gastroenteritis virus genome as an expression vector inducing lactogenic immunity. J. Virol. 2003;77:4357–4369. doi: 10.1128/JVI.77.7.4357-4369.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer E.L, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1998;6:175–182. [PubMed] [Google Scholar]
- Tennant B.J, Gaskell R.M, Jones R.C, Gaskell C.J. Studies on the epizootiology of canine coronavirus. Vet. Rec. 1993;132:7–11. doi: 10.1136/vr.132.1.7. [DOI] [PubMed] [Google Scholar]
- Tennant B.J, Gaskell R.M, Kelly D.F, Carter S.D, Gaskell C.J. Canine coronavirus infection in the dog following oronasal inoculation. Res. Vet. Sci. 1991;51:11–18. doi: 10.1016/0034-5288(91)90023-H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley R.D, Woods R.D, Cheung A.K. Genetic basis for the pathogenesis of transmissible gastroenteritis virus. J. Virol. 1990;64:4761–4766. doi: 10.1128/jvi.64.10.4761-4766.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurm T, Chen H, Hodgson T, Britton P, Brooks G, Hiscox J.A. Localization to the nucleolus is a common feature of coronavirus nucleoproteins, and the protein may disrupt host cell division. J. Virol. 2001;75:9345–9356. doi: 10.1128/JVI.75.19.9345-9356.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yount B, Curtis K.M, Baric R.S. Strategy for systematic assembly of large RNA and DNA genomes: transmissible gastroenteritis virus model. J. Virol. 2000;74:10600–10611. doi: 10.1128/jvi.74.22.10600-10611.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]


