Abstract
Forty faecal samples were tested by RT-PCR using coronavirus consensus primers to determine faecal shedding of canine coronavirus (CCoV) and canine respiratory coronavirus (CRCoV) in a dog population housed at a rescue centre.
Seven samples were positive for CCoV while all samples were negative for CRCoV. Sequence analysis of five CCoV strains showed a high similarity with transmissible gastroenteritis virus (TGEV) at the N-terminus of the spike protein. All strains contained an open reading frame for the nonstructural protein 7b, which is not present in TGEV, indicating that the strains were related to the previously described CCoV strain UCD-1. Two samples contained CCoV strains with 5′ spike sequences most similar to type II CCoV while one sample was found to contain type I CCoV.
Primers directed to the N gene allowed specific detection of all CCoV strains analysed in this study.
This investigation shows that CCoV strains containing spike proteins similar to TGEV are present in the UK dog population. PCR primers directed to conserved regions of the CCoV genome are recommended for detection of CCoV in clinical samples due to high genetic variability.
Keywords: Canine coronavirus, Spike protein, Transmissible gastroenteritis virus, Sequence analysis
1. Introduction
The Coronaviridae family is organised in three groups according to antigenic and genetic similarities. In dogs, two distinct coronaviruses have been described; canine coronavirus (CCoV), which belongs to group 1 and canine respiratory coronavirus (CRCoV) belonging to group 2. Coronaviruses possess a single stranded, positive sense genome of 27–31 kb in length which contains a large open reading frame for the major nonstructural proteins at the 5′ end and the structural proteins as well as minor nonstructural proteins in the 3′ third of the genome. The structural proteins include the spike glycoprotein (S), the small envelope protein (E), the membrane protein (M) and the nucleocapsid protein (N) (Weiss and Navas-Martin, 2005).
CRCoV has been detected predominantly in respiratory samples but also occasionally in samples from the gastrointestinal tract (Decaro et al., 2007, Erles et al., 2003, Yachi and Mochizuki, 2006). A high prevalence of CRCoV has been found in kennelled dog populations in which infectious respiratory disease is common.
Canine coronavirus exhibits a predominantly enteric tropism and causes gastroenteritis particularly in young dogs (Pratelli, 2006). However a recently described strain, CCoV CB/05 has been shown to cause a fatal disease which was characterised by systemic spread of the virus (Decaro et al., 2008). CCoV is divided into two genotypes; type I is characterised by genetic similarity to feline coronavirus (FCoV) type I in the spike and membrane genes. One CCoV strain, UCD-1, contains a spike gene with a high sequence identity at the 5′ end to that of the porcine transmissible gastroenteritis virus (TGEV) (Wesley, 1999).
The aim of this study was to investigate faecal shedding of coronaviruses in kennelled dogs and to characterise detected coronavirus strains by sequence analysis.
2. Materials and methods
2.1. Clinical samples
Faecal samples were collected from dogs housed at a rehoming centre. None of the dogs showed clinical signs of gastroenteritis and the general health status of all dogs was good. In total faecal samples from 40 dogs were analysed.
2.2. RNA extraction and RT-PCR
RNA was extracted using the RNeasy Mini Kit (Qiagen, Crawley, UK). 25–50 mg of faeces was suspended in 350 μl Buffer RLT and centrifuged. The extraction was then performed on the supernatant as per the manufacturer's protocol for animal cells.
Reverse transcription was carried out using 7 μl of total RNA, 0.5 μg random hexameres (GE Healthcare) 1 μl of ImPromII reverse transcriptase (Promega) and a MgCl2 concentration of 3 mM in a total reaction volume of 20 μl.
For PCR, 2 μl of cDNA was added to a reaction mix of 1× PCR buffer, 2.5 mM MgCl2, dNTP mix (final concentration of each deoxynucleotide 0.2 mM) and 1.25 U of Taq polymerase (Promega).
PCR for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA was carried out using the primers and conditions as described by Grone et al. (1996).
PCR for the specific detection of CRCoV was carried out using the nested PCR for the spike gene as described elsewhere (Erles et al., 2003).
For the detection of coronaviruses in faecal samples the following PCR protocols were used. For primers CENP 1 and CENP 2 directed to the CCoV nucleocapsid protein gene (Table 1 ) the following cycling conditions were used: 95 °C for 5 min, 35 cycles of 95 °C for 1 min, 53 °C for 40 s and 72 °C for 30 s followed by a final extension at 72 °C for 10 min. The primers were designed to amplify a 280 bp product corresponding to 68–347 nt of the nucleocapsid gene of CCoV. The 5′ end of the spike gene was amplified using a consensus forward primer located in ORF1b (CEPol-1) and reverse primers specific for either TGEV (TGSp-2), CCoV type I (CESp-10) or CCoV type II (CESp-6). The primer sequences are given in Table 1. Primer pair CEPol-1-TGSp-2 amplified a 370 bp product corresponding to 20,168–20,537 nt of the TGEV Purdue genome, which consisted of 198 nt of the 3′ end of ORF1b and the first 172 nt of the spike gene. Primer pair CEPol-1-CESp-10 was designed to amplify a 1108 bp product consisting of 167 nt of the 3′ end of ORF1b and the first 941 nt of the spike gene. Primer pair CEPol-1-CESp-6 amplified a 578 bp product consisting of 197 nt of the 3′ end of ORF1b and the first 381 nt of the spike gene. The cycling conditions were 95 °C for 5 min, 35 cycles of 95 °C for 1 min, 50 °C for 1 min and 72 °C for 1 min followed by a final extension for 10 min at 72 °C. In addition primers EL1F and EL1R directed to the spike gene of type I CCoV (Pratelli et al., 2004) were used, which amplify a 346 bp product corresponding to 3538–3883 nt of the spike gene of CCoV strain Elmo. A nested PCR primer set to the CCoV spike gene (Naylor et al., 2002) amplified a 1086 bp product using the outer primers, corresponding to 81–1166 nt of the spike gene of CCoV strain Insavc. The spike gene consensus primers 55 and 56 for all coronaviruses (Wise et al., 2006) amplified 3549–4188 nt of the spike gene of CCoV Insavc (640 bp product). Primers M5 and M6 amplified the complete sequence of the CCoV membrane protein gene (Pratelli et al., 2003). The consensus primers Conscoro 5 and Conscoro 6 were based on the polymerase gene (corresponding to 13,602–13,852 nt of the genome of TGEV strain Purdue). These primers detect both, CCoV and CRCoV (Erles et al., 2003). The region containing ORFs 7a and 7b was amplified using primers N3SN and R3AS as previously described (Wesley, 1999).
Table 1.
Primer sequences.
| Primer | Sequence 5′–3′ | Position | GenBank accession number |
|---|---|---|---|
| CENP1 | CTC-GTG-GYC-GGA-AGA-ATA-AT | 7270-7289 | D13096 |
| CENP2 | GCA-ACC-CAG-AMR-ACT-CCA-TC | 7549-7530 | D13096 |
| CEPol-1 | TCT-ACA-ATT-ATG-GCT-CTA-TCA-C | 309-330 | D13096 |
| CESP-6 | TTG-CAC-ATC-ATC-TCT-ATA-AGC | 886-866 | D13096 |
| CESP-10 | GAT-TTA-TCT-GAT-ARC-AGR-AAC-CA | 941-919 | AY307020 |
| TGSP-2 | TAA-TCA-CCT-AAM-ACC-ACA-TCT-G | 20537-20516 | NC_002306 |
The primer positions refer to the corresponding GenBank sequence.
2.3. Analytic restriction digest of Conscoro PCR product
Using the Qiaquick PCR purification kit (Qiagen) 30 μl of the PCR product was purified. Restriction digest was performed using 15 μl of purified product and AluI (Promega) in the appropriate buffer for 1 h at 37 °C. The 251 bp product amplified by primers conscoro 5/6 contained a restriction site for AluI at position 124 for CRCoV generating two fragments of 124 and 127 bp length. In PCR products generated from CCoV the restriction site was located at position 43 and digest generated two products of 43 and 208 bp. The products were analysed by gel electrophoresis on 1.5% agarose gels and visualised using ethidium bromide.
2.4. Cloning and sequencing of PCR products
PCR products were cloned into the pGEM-T-easy vector (Promega). Plasmid DNA (600 ng) was sent to The Sequencing Service (University of Dundee, UK) for sequencing.
2.5. Phylogenetic analysis
DNA sequences were aligned using ClustalX (Thompson et al., 1997). Phylip (Phylogeny Inference Package, version 3.6) was used to perform bootstrap and maximum parsimony analyses (Felsenstein, 2004). A consensus tree was calculated using Consense (Phylip 3.6). The resulting trees were drawn using the Treeview program (Page, 1996).
2.6. GenBank accession numbers
The following accession numbers were assigned: partial spike protein gene 04-0709 (FJ009112), 03-3684 (FJ009113), complete M protein gene 04-0709 (FJ009114), 04-0377 (FJ009115), partial N protein gene 04-0377 (FJ009116), 04-0709 (FJ009117), 03-3684 (FJ009118).
3. Results
3.1. PCR analysis for CRCoV and CCoV
Forty faecal samples were collected from dogs housed at a rehoming kennel. The samples were analysed to determine the presence of canine coronaviruses in faecal samples. The results of all PCR analyses are summarised in Table 2 .
Table 2.
PCR analysis of canine faecal samples. Only samples positive using at least one primer set are shown.
| Primer set | Cons coro 5-6 | Spike CCoV nested | CEPol-CESp-6 | CEPol-CESp-10 | EL1F-EL1R | CEPol-TGSp-2 | Spike 55-56 | M5-6 | CENP 1-2 | N3SN-R3AS |
|---|---|---|---|---|---|---|---|---|---|---|
| Region | ORF1b | S gene | S gene (CCoV type II) | S gene (CCoV type I) | S gene (CCoV type I) | S gene (TGEV) | S gene | M gene | N gene | ORF 7a/b |
| CCoV strain | ||||||||||
| 03-3489 | Pos | Neg | Neg | Neg | Neg | Pos | Pos | Pos | Pos | Pos |
| 03-3575 | Pos | Neg | Neg | Neg | Neg | Pos | Neg | Pos | Pos | Pos |
| 03-3684 | Pos | Pos | Pos | Neg | Neg | Neg | Pos | Neg | Pos | Pos |
| 03-3734 | Pos | Neg | Neg | Neg | Neg | Pos | Pos | Pos | Pos | Pos |
| 03-3814 | Pos | Pos | Pos | Neg | Neg | Pos | Pos | Pos | Pos | Pos |
| 04-0377 | Pos | Neg | Neg | Neg | Pos | Neg | Neg | Pos | Pos | Pos |
| 04-0709 | Pos | Neg | Neg | Neg | Neg | Pos | Pos | Pos | Pos | Pos |
All faecal samples were negative for CRCoV by nested PCR based on the CRCoV spike gene.
Two out of 40 samples were positive for CCoV by PCR using a nested primer set based on the spike gene of CCoV (Naylor et al., 2002).
PCR analysis using consensus coronavirus primers based on the polymerase gene detected seven positive samples. Subsequent restriction digests and sequence analyses determined that these samples contained CCoV. As the CCoV specific nested PCR directed to the spike gene had failed to detect five out of seven samples further sets of primers were tested to find a method for detecting UK strains of CCoV.
Primers M5 and M6 had previously been described to be able to detect type I and type II CCoV (Pratelli et al., 2003). Using these primers six out of seven samples were positive. PCR using a newly designed primer set (CENP1/2) located in the N gene detected all seven positive samples.
3.2. Sequence analysis of N gene and M gene PCR products
Sequence analysis of the N gene PCR product showed high sequence identities (97.8% amino acid identity) with porcine coronaviruses for six cDNA samples. Phylogenetic analysis placed these sequences into one cluster and on a branch with porcine coronaviruses (Fig. 1 ). One sample (04-0377) showed a 97.8% amino acid sequence identity with CCoV strains 23/03 and BGF-10. Sequence analysis and phylogenetic analysis of the M gene PCR product confirmed the close relationship of the cDNA from sample 04-0377 with CCoV strain 23/03 (90.1% amino acid identity). The M gene sequences obtained from the other five positive samples were most closely related to CCoV strain CB05 (98.1% amino acid identity) (Fig. 2 ).
Fig. 1.
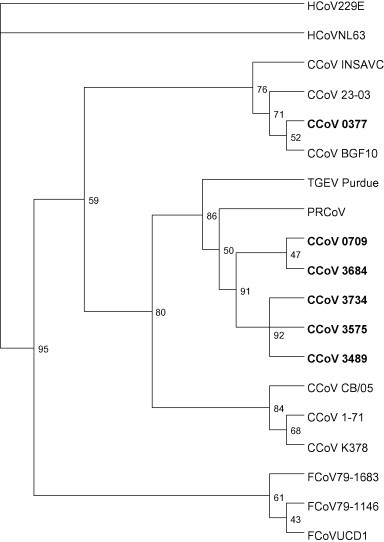
Phylogenetic analysis of the partial nucleotide sequence of the nucleocapsid gene (corresponding to 7290–7529 nt in D13096). GenBank accession numbers were as follows: CCoV 259-01 (AF502583), CCoV 23-03 (AY548235), CCoV BGF-10 (AY342160), CCoV Insavc (D13096), CCoV CB/05 (DQ112226), CCoV GOT-05 (DQ431017), CCoV UPPS1-04 (DQ431018), CCoV HC2 (AY884048), CCoV 1-71 (AY704916), FCoV 79-1146 (AY994055), FCoV 79-1683 (AB086904), FCoV UCD1 (AB086902), HCoV NL-63 (AY567487), HCoV 229E (NC_002645), PRCoV 86/137004 (X60056), TGEV Purdue (AJ271965).
Fig. 2.
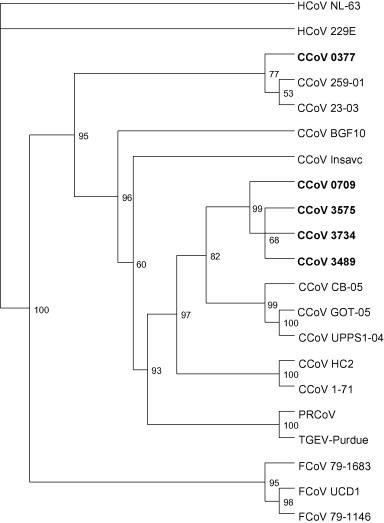
Phylogenetic analysis of the partial nucleotide sequence of the membrane gene (corresponding to 6411–7117 nt in D13096). GenBank accession numbers were as follows: CCoV 23-03 (AY548235), CCoV BGF-10 (AY342160), CCoV Insavc (D13096), CCoV CB/05 (DQ112226), CCoV 1-71 (AB105373), FCoV 79-1146 (AY994055), FCoV 79-1683 (AB086904), FCoV UCD1 (AB086902), HCoV NL-63 (AY567487), HCoV 229E (NC_002645), PRCoV 86/137004 (X60056), TGEV Purdue (AJ271965).
3.3. Sequence analysis of the 5′ end of the spike gene
As the nested PCR for the spike gene had been negative on most samples further primers were designed to obtain sequence information for the 5′ end of the spike gene. The forward primer was designed to be located in a conserved region of ORF1b (primer CEPol-1) while three reverse primers were designed to detect different types of group 1 coronaviruses. The primer sequence for CESp-6 was chosen to amplify the spike gene of viruses similar to CCoV type II and feline coronavirus type II. Primer TGSp-2 was based on the spike gene sequence of TGEV and CCoV strain UCD-1. The primer sequence for CESp-10 was selected to amplify viruses related to type I CCoV and feline coronavirus type I.
Two samples were positive using reverse primer CESp-6. These were samples 03-3684 and 03-3814 that had previously been positive by nested spike gene PCR. Five samples were positive using reverse primer TGSp-2. Sample 03-3814 was positive with both primer sets. None of the samples was positive using reverse primer CESp-10. Samples were subsequently tested using the primer set EL1F-EL1R, which is directed to the spike gene of type I CCoV. Only sample 04-0377 was found to be positive.
Sequence analysis of CEPol-1–CESp-6 PCR products showed a high similarity with CCoV strain 1-71. Products of the CEPol-1-TGSp-2 PCR were closely related to the corresponding regions of TGEV and CCoV strain UCD-1. Fig. 3 shows an alignment of the amino acid sequences obtained by translation of the amplified regions.
Fig. 3.

Sequence alignment of the N-terminal spike region of CCoV strains analysed in the present study with TGEV strain Purdue and the CCoV strains 1-71 and UCD-1.
Further sequence information for the spike gene was obtained using a consensus primer pair located in the 3′ region of the spike gene which was developed for the detection of all coronaviruses (primers 55 and 56) (Wise et al., 2006). Five samples were found to be positive (Table 2).
3.4. Sequence analysis of ORFs 7a and 7b
The region containing open reading frames 7a and 7b was analysed to determine if ORF 7b, which is lacking in TGEV, was present in the samples that had a spike gene related to TGEV. Seven samples were positive using ORF 7 specific primers (Table 2). All positive samples generated a PCR product of over 1000 bp indicating that ORF 7b was present. In addition sequence analysis confirmed the presence of ORF 7b.
Table 3 summarises amino acid identities obtained when comparing CCoV 04-0709, which possesses a “TGEV-like” N-terminal spike protein, to other CCoV strains as well as TGEV strain Purdue and FCoV strain UCD-1. The identities given are based on the analysis of partial sequences obtained by PCR as described above.
Table 3.
Amino acid identities between CCoV strain 04-0709 and other coronaviruses.
| Poly | N-term spike | C-term spike | M | N | 7a | 7b | |
|---|---|---|---|---|---|---|---|
| CCoV 03-3489 | nd | 100% | 99.0% | 99.2% | 98.9% | nd | Nd |
| CCoV 03-3684 | 92.8% | 24% | 99.0% | nd | 97.8% | 100% | 86.9% |
| CCoV 1-71 | 92.8% | 24% | 98.5% | 97.3% | 92.4% | 100% | 90.1% |
| CCoV CB05 | nd | 23% | 98.5% | 98.1% | 93.5% | 100% | 90.6% |
| CCoV 23/03 | nd | 9% | 55.7% | 89% | 84.8% | 95% | 80.8% |
| TGEV Purdue | 92.8% | 83.6% | 97.0% | 95% | 97.8% | 74.3% | - |
| FCoV UCD-1 | nd | 13% | 56.2% | 82.5% | 87.0% | 82% | 59.5% |
4. Discussion
The initial aim of this study was to determine the prevalence of CCoV and CRCoV in faecal samples collected from a kennelled dog population. While all samples were negative for CRCoV by PCR, CCoV was detected in 17.5% of samples tested. The dogs sampled for this study did not show any clinical signs of enteric disease indicating that subclinical infections with CCoV were common in this population. Alternatively the dogs may have recovered from CCoV induced enteritis some time in the past as long term shedding of CCoV for several months has been reported (Pratelli et al., 2001). Dogs housed in kennels are more frequently positive for CCoV as shown by virus isolation and antibody detection (Pratelli et al., 2002, Tennant et al., 1993). A number of PCR assays for CCoV were compared in this study three of which identified seven positive samples (out of 40 tested). The consensus PCR approach offered the advantage of detecting both CCoV and CRCoV but required further processing of the PCR product by restriction digest. Primer pairs CENP1-2 and N3SN-R3AS gave the same results as the consensus primers but were specific for group 1 coronaviruses and are therefore more useful for CCoV detection in clinical samples.
Analysis of the 5′ end of the spike gene showed that the majority of the strains analysed in this study were more closely related to TGEV in that region than to CCoV. Previously it had been described that CCoV strain UCD-1, a US field strain isolated in the 1970s, possessed a spike gene similar to that of TGEV (Wesley, 1999). However further studies on the prevalence of this type of CCoV in dogs have not been reported. All strains that were closely related to TGEV at the 5′ end of the spike gene had a complete ORF 7b, which is not present in TGEV, indicating that these were CCoV strains rather than a direct infection of dogs with TGEV. These strains were also more closely related to TGEV in the N gene than to CCoV type I or type II, however they were more closely related to CCoV type II in the M gene. The evolution of CCoV and TGEV appears to be closely linked and it has been proposed that type I CCoV was the ancestor for CCoV type II and that TGEV subsequently evolved from CCoV type II (Lorusso et al., 2008).
All “TGEV-like” CCoV strains in this study were closely related to each other and were likely to be the result of infections occurring during the stay at the kennel following contact between dogs or sharing of common facilities. The prevalence of this type of CCoV in the canine population of the UK and world-wide will therefore have to be determined in future studies. The spike protein carries major antigenic epitopes of coronaviruses. Despite their divergence at the N-terminus of the spike protein cross-neutralisation in vitro between TGEV and CCoV has been reported (Reynolds et al., 1980) and sera raised against CCoV were shown to recognise TGEV spike protein by ELISA (Horzinek et al., 1982). The conserved regions in the spike protein as well as other structural proteins may therefore be sufficient to confer protection in vivo against heterologous challenge.
One dog had a mixed infection with type II CCoV and “TGEV-like” CCoV; such mixed infections have been described to be common for type I and type II CCoV. Overall the prevalence of type II CCoV was low in this kennel; nevertheless this may have been due to the small sample size.
Only one dog was positive for a CCoV strain which grouped with type I CCoVs in phylogenetic analyses and was positive using type I specific spike gene primers. However it was not possible to amplify the 5′ end of the spike gene of this virus despite the use of primers designed to type I CCoV and FCoV strains. It is possible that the spike gene of this virus was distinct from other CCoV strains. Alternatively the amount or quality of the cDNA obtained from this faecal sample may have been insufficient to generate a PCR product when using primers that potentially contained several mismatches due to the variable nature of the 5′ end of the spike gene.
In conclusion the prevalence of CCoV in canine populations may be underestimated if type specific primers only are used for diagnosis. Further studies will be required to determine if the divergent CCoV strains described here are widespread. Nevertheless for diagnostic purposes the use of primers directed to conserved regions is more appropriate. Subsequent typing may then be carried out if required.
Acknowledgement
The authors would like to thank S. Greaves for technical assistance.
References
- Decaro N., Campolo M., Lorusso A., Desario C., Mari V., Colaianni M.L., Elia G., Martella V., Buonavoglia C. Experimental infection of dogs with a novel strain of canine coronavirus causing systemic disease and lymphopenia. Vet. Microbiol. 2008;128(3–4):253–260. doi: 10.1016/j.vetmic.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro N., Desario C., Elia G., Mari V., Lucente M.S., Cordioli P., Colaianni M.L., Martella V., Buonavoglia C. Serological and molecular evidence that canine respiratory coronavirus is circulating in Italy. Vet. Microbiol. 2007;121(3–4):225–230. doi: 10.1016/j.vetmic.2006.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erles K., Toomey C., Brooks H.W., Brownlie J. Detection of a group 2 coronavirus in dogs with canine infectious respiratory disease. Virology. 2003;310(2):216–223. doi: 10.1016/S0042-6822(03)00160-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein, J., 2004. PHYLIP (Phylogeny Inference Package) version 3.6. Department of Genome Sciences, University of Washington, Seattle.
- Grone A., Weckmann M.T., Capen C.C., Rosol T.J. Canine glyceraldehyde-3-phosphate dehydrogenase complementary DNA: polymerase chain reaction amplification, cloning, partial sequence analysis, and use as loading control in ribonuclease protection assays. Am. J. Vet. Res. 1996;57(3):254–257. [PubMed] [Google Scholar]
- Horzinek M.C., Lutz H., Pedersen N.C. Antigenic relationships among homologous structural polypeptides of porcine, feline, and canine coronaviruses. Infect. Immun. 1982;37(3):1148–1155. doi: 10.1128/iai.37.3.1148-1155.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorusso A., Decaro N., Schellen P., Rottier P.J.M., Buonavoglia C., Haijema B.-J., de Groot R.J. Gain, preservation and loss of a group 1a coronavirus accessory glycoprotein. J. Virol. 2008;82(20):10312–10317. doi: 10.1128/JVI.01031-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naylor M.J., Walia C.S., McOrist S., Lehrbach P.R., Deane E.M., Harrison G.A. Molecular characterization confirms the presence of a divergent strain of canine coronavirus (UWSMN-1) in Australia. J. Clin. Microbiol. 2002;40(9):3518–3522. doi: 10.1128/JCM.40.9.3518-3522.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page R.D. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 1996;12(4):357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Pratelli A. Genetic evolution of canine coronavirus and recent advances in prophylaxis. Vet. Res. 2006;37(2):191–200. doi: 10.1051/vetres:2005053. [DOI] [PubMed] [Google Scholar]
- Pratelli A., Decaro N., Tinelli A., Martella V., Elia G., Tempesta M., Cirone F., Buonavoglia C. Two genotypes of canine coronavirus simultaneously detected in the fecal samples of dogs with diarrhoea. J. Clin. Microbiol. 2004;42(4):1797–1799. doi: 10.1128/JCM.42.4.1797-1799.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratelli A., Elia G., Martella V., Palmieri A., Cirone F., Tinelli A., Corrente M., Buonavoglia C. Prevalence of canine coronavirus antibodies by an enzyme-linked immunosorbent assay in dogs in the south of Italy. J. Virol. Methods. 2002;102(1–2):67–71. doi: 10.1016/S0166-0934(01)00450-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratelli A., Martella V., Elia G., Decaro N., Aliberti A., Buonavoglia D., Tempesta M., Buonavoglia C. Variation of the sequence in the gene encoding for transmembrane protein M of canine coronavirus (CCV) Mol. Cell. Probes. 2001;15(4):229–233. doi: 10.1006/mcpr.2001.0364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratelli A., Martella V., Pistello M., Elia G., Decaro N., Buonavoglia D., Camero M., Tempesta M., Buonavoglia C. Identification of coronaviruses in dogs that segregate separately from the canine coronavirus genotype. J. Virol. Methods. 2003;107(2):213–222. doi: 10.1016/S0166-0934(02)00246-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds D., Garwes D., Lucey S. Differentiation of canine coronavirus and porcine transmissible gastroenteritis virus by neutralization with canine, porcine and feline sera. Vet. Microbiol. 1980;5(4):203–290. [Google Scholar]
- Tennant B.J., Gaskell R.M., Jones R.C., Gaskell C.J. Studies on the epizootiology of canine coronavirus. Vet. Rec. 1993;132(1):7–11. doi: 10.1136/vr.132.1.7. [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25(24):4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss S.R., Navas-Martin S. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol. Mol. Biol. Rev. 2005;69(4):635–664. doi: 10.1128/MMBR.69.4.635-664.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley R.D. The S gene of canine coronavirus, strain UCD-1, is more closely related to the S gene of transmissible gastroenteritis virus than to that of feline infectious peritonitis virus. Virus Res. 1999;61(2):145–152. doi: 10.1016/S0168-1702(99)00032-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise A.G., Kiupel M., Maes R.K. Molecular characterization of a novel coronavirus associated with epizootic catarrhal enteritis (ECE) in ferrets. Virology. 2006;349(1):164–174. doi: 10.1016/j.virol.2006.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yachi A., Mochizuki M. Survey of dogs in Japan for group 2 canine coronavirus infection. J. Clin. Microbiol. 2006;44(7):2615–2618. doi: 10.1128/JCM.02397-05. [DOI] [PMC free article] [PubMed] [Google Scholar]


